Abstract
N-methyl-d-aspartate (NMDA) receptors are glutamate- and glycine-gated channels that flux Na+ and Ca2+ into postsynaptic neurons during synaptic transmission. The resulting intracellular Ca2+ transient is essential to physiological and pathological processes related to synaptic development, plasticity, and apoptosis. It also engages calmodulin (CaM) to reduce subsequent NMDA receptor activity in a process known as Ca2+-dependent inactivation (CDI). Here, we used whole-cell electrophysiology to measure CDI and computational modeling to dissect the sequence of events that underlies it. With these approaches, we estimate that CaM senses NMDA receptor Ca2+ influx at ∼9 nm from the channel pore. Further, when we controlled the frequency of Ca2+ influx through individual channels, we found that a kinetic model where apoCaM associates with channels before their activation best predicts the measured CDI. These results provide, to our knowledge, novel functional evidence for CaM preassociation to NMDA receptors in living cells. This particular mechanism for autoinhibitory feedback reveals strategies and challenges for Ca2+ regulation in neurons during physiological synaptic activity and disease.
Introduction
Glutamate exerts fast excitatory actions in the mammalian central nervous system by gating cation-selective tetrameric ion channels. Among these, N-methyl-d-aspartate (NMDA) receptors composed of GluN1 and GluN2 subunits have characteristically high conductance, high Ca2+-permeability, and slow kinetics, such that their activation produces a significant local rise in intracellular Ca2+. Specifically, NMDA receptor-mediated Ca2+ transients initiate signaling pathways essential for synaptic development and plasticity and can cause the pathologic apoptosis that underlies excitotoxicity.
Given its critical role in fundamental neurologic processes, NMDA receptor activity is under tight developmental and regional control, and is sensitive to multiple extra- and intracellular cues. Specifically, Ca2+ levels affect NMDA receptor currents through several distinct mechanisms. External Ca2+ ions bind directly to residues in the outer vestibule of the obligatory GluN1 subunit to reduce channel conductance and gating (1, 2, 3), whereas internal Ca2+ ions act indirectly through slow and fast mechanisms. Slow, minute-scale processes initiate Ca2+-dependent changes in receptor phosphorylation by kinases (e.g., protein kinase A, protein kinase C, and CaMKII) and/or phosphatases (e.g., calcineurin) to affect channel gating and/or permeation (4, 5, 6, 7), whereas a fast, millisecond-scale process, known as Ca2+-dependent inactivation (CDI), which requires calmodulin (CaM) binding to intracellular portions of the GluN1 subunit (8, 9), reduces the NMDA receptor current by increasing its macroscopic desensitization (10, 11).
Numerous other ion channels display CaM-mediated CDI, suggesting that this regulatory mechanism is highly conserved across several protein families and life forms. CaM is ubiquitous to living cells, is highly expressed in neural tissue, and is enriched in dendritic spines (12). CaM control of NMDA receptor activity, primarily by mediating CDI, is a major mechanism for tuning the excitatory postsynaptic current during development and across brain regions (13, 14, 15, 16). However, the process remains insufficiently understood.
NMDA receptors have several CaM binding sites, each with different CaM binding properties. The intracellular sequence immediately adjacent to the M4 transmembrane helix of GluN1, known as C0, is required for CDI (10, 17). In addition to strongly binding calcified CaM, several in vitro studies observed that the C0 peptide can bind CaM in the absence of Ca2+ (18, 19, 20). Therefore, CaM may reside on C0, and this proximity to the channel pore positions it optimally for rapid activation by incoming Ca2+ (21). However, results remain inconsistent (8, 22) and whether CDI of NMDA receptors occurs by Ca2+ binding to freely diffusing or resident apoCaM is unknown. Yet, each mechanism has distinct implications for how intracellular Ca2+ transients tune synaptic responses and how disease-associated CaM mutations influence excitatory glutamatergic transmission.
Here we modeled possible mechanisms of CaM-mediated NMDA receptor CDI and tested these with electrophysiological measurements in several rigorously controlled conditions. Results provide the first evidence in living cells that apoCaM preassociates with NMDA receptors and show that this resident apoCaM is sensitive to elevated [Ca2+] within nanodomains of the channel pore.
Methods
Molecular biology
Rat GluN1-2a (U08262.1), GluN2A (M91561.1), and GFP cDNAs were cloned in pcDNA3.1 (+) and were transiently transfected at a 1:1:1 ratio into HEK 293 cells (ATTC CRL-1573, passages 26–32) using the Ca2+ phosphate precipitation method (23). Transfected cells were maintained in DMEM (10% FBS) supplemented with 10 mM Mg2+ and were used for experiments 24 h posttransfection. All CaM constructs were gifts from Drs. Inoue and Ben Johny (24). We introduced all mutations using the QuikChange method (Stratagene, Amsterdam, Netherlands). We verified each construct by sequencing before and after subcloning, and after plasmid amplification (QIAGEN, Valencia, CA).
Western blotting
We transfected HEK293 cells with the indicated constructs; and harvested and lysed them 24 h posttransfection, with 1% SDS lysis buffer, and removed debris and contaminating DNA/RNA by centrifugation 15 min at 13 RPM. Equal amounts of proteins in the supernatant were loaded onto 5% SDS polyacrylamide gel to achieve separation between endogenous CaM (17 kD), GFP (37 kD), YFP-CaM (44 kD), and GluN1-2a (100 kD). Proteins were electrotransferred onto PDF membranes as described in Cummings et al. (25) and probed with rabbit anti-CaM antibody (ab45689, lot: GR123440-4; 1:2500; Abcam, http://www.abcam.com/); mouse anti-GFP (MAB1083, lot: NG1720568; 1:5000; Millipore, Billerica, MA), and rabbit anti-GluN1 pan (MAB1586, lot: 2739489, 1:1000; Millipore). Hybridizations were done overnight at 4° for primary antibodies, and 1 h at room temperature for secondary antibodies (rabbit or mouse anti-HRP, 1:10,000).
Fluorometry
The relative expression of YFP-CaM constructs was quantified as the ratio of fluorescence intensity from a single cell expressing YFP to background autofluorescence from untransfected cells on the same day of recording. All measurements were from cells after electrophysiological recording. Fluorescence was measured with a photomultiplier tube (Hamamatsu Photonics, Hamamatsu City, Japan) and recorded in the software pClamp10.3 (Molecular Devices, Sunnyvale, CA). Data were analyzed in MATLAB 2011a (The MathWorks, Natick, MA).
Electrophysiology
Macroscopic currents were recorded with the whole-cell patch clamp-technique, using borosilicate pipettes (4–5 MΩ) filled with (intracellular) 135 mM CsCl, 35 mM CsOH, 4 mM MgATP, 0.3 mM Na2GTP, and either BAPTA or EGTA at the specified concentration, buffered to pH 7.4 (CsOH). For Ca2+ dialysis experiments, recording pipettes contained (intracellular) 135 mM CsCl, 35 mM CsOH, 4 mM MgATP, 0.3 mM Na2GTP, 10 mM HEDTA, pH 7.4 with CsOH. Free Ca2+ calculations were done with the software MAXC (www.maxchelator.stanford.edu) with CaCl2. Extracellular solutions contained 150 mM NaCl, 2.5 mM KCl, 10 mM HEPBS, 0.1 mM EDTA, and 0.1 mM glycine; CaCl2 was added to produce the indicated free Ca2+ concentrations according to the software MAXC; they were applied using a pressurized perfusion system with a digitally controlled solenoid valve. Series resistance was monitored to ascertain seal quality. In all experiments, cells were bathed in PBS unless otherwise indicated.
Unitary currents were recorded with the cell-attached patch-clamp technique from membrane patches containing a single receptor (26). Borosilicate pipettes (15–25 MΩ) contained (extracellular) 150 mM NaCl, 2.5 mM KCl, 10 mM HEPBS, 0.1 mM EDTA, 0.1 mM Glycine, 1 mM Glutamate, pH 8 with NaOH, and with CaCl2 buffered according to the software MAXC. Ca2+-only solutions contained 75 mM CaCl2, 2.5 mM KCl, 10 mM HEPBS, 0.1 mM EDTA, 0.1 mM Glycine, 1 mM Glutamate, pH 8 with Ca(OH)2. Stationary single-channel data were acquired, processed, and analyzed in the software QuB (www.qub.buffalo.edu). Currents were idealized using the SKM algorithm (27). Single-channel voltage-ramps were recorded and processed in the software pClamp. Analysis of voltage-ramp data was done using custom scripts in MATLAB 2011a (The MathWorks). During voltage ramps, cells were bathed in 142 mM KCl, 5 mM NaCl, 10 mM HEPES, 5 mM EGTA, 1.7 mM MgCl2, pH = 7.4 with KOH to offset the resting membrane potential of HEK293 cells. Applied voltage was initially stepped from 0 to +100 mV and ramped to −60 mV over 4 s, then stepped back to 0 mV. All sweeps were averaged to yield a time-averaged 〈i〉-amplitude as a function of recording potential (E). To correct for leak current and capacitive transient current during the step pulses, sweeps that contained no channel activity (typically 5–13 sweeps/patch) were averaged to produce a baseline current and this was subtracted from the original 〈i〉-E waveform. The unitary Ca2+ current was determined as described in Jahr and Stevens (28) first by fitting 〈i〉-E data with
| (1) |
where Erev is the reversal potential, γin and γout are the inward and outward conductance, respectively, and p reflects the sharpness of the transition between inward and outward current. The extrapolated γin calculated for several external Ca2+ served to evaluate the steepness of the conductance/[Ca2+] dependency (Eq. 7),
| (2) |
where γmin is the limiting inward conductance in the presence of high extracellular Ca2+ (75 mM), 0.5 is the activity coefficient of Ca2+ in physiological solution, and h is a coefficient that sets the steepness with which the limiting conductance is reached. In our cells, we found h = 9.5 pS/mM for GluN1-2a/GluN2A. From these data, the fractional Ca2+ current (fCa) was calculated directly as
| (3) |
where γmax is the limiting inward conductance in the absence of extracellular Ca2+. Table S1 summarizes these results.
Statistics
Statistical tests were performed with custom scripts in R and MATLAB 2011a. The nonparametric Mann-Whitney U-test was used for comparison because CDI data tended to follow nonnormal distributions as determined from the Lillifore’s and Anderson-Darling tests. Fittings of CDI models to CDIEQ-Po data and Jahr models to single-channel voltage-ramp data were performed with custom software with the built-in statistics toolbox fittype command using the least-squares method in MATLAB 2011a.
Results
Define new metric to evaluate specifically CDI of NMDA receptors
Presently the literature documents several metrics to quantify the extent of NMDA receptor CDI but the reported values are difficult to reconcile across experimental conditions, most likely due to uncontrolled confounding factors. Aside from effects on response kinetics, external Ca2+ controls NMDA receptor conductance (3); and aside from Ca2+, the kinetics and extent of NMDA receptor macroscopic desensitization vary with external glycine concentrations (29, 30), mutations (31), and posttranslational modifications (6, 32, 33). To exclude contributions from these separate Ca2+-dependent and Ca2+-independent forms of NMDA receptor current modulation, we developed, to our knowledge, a new method to specifically monitor changes in CDI.
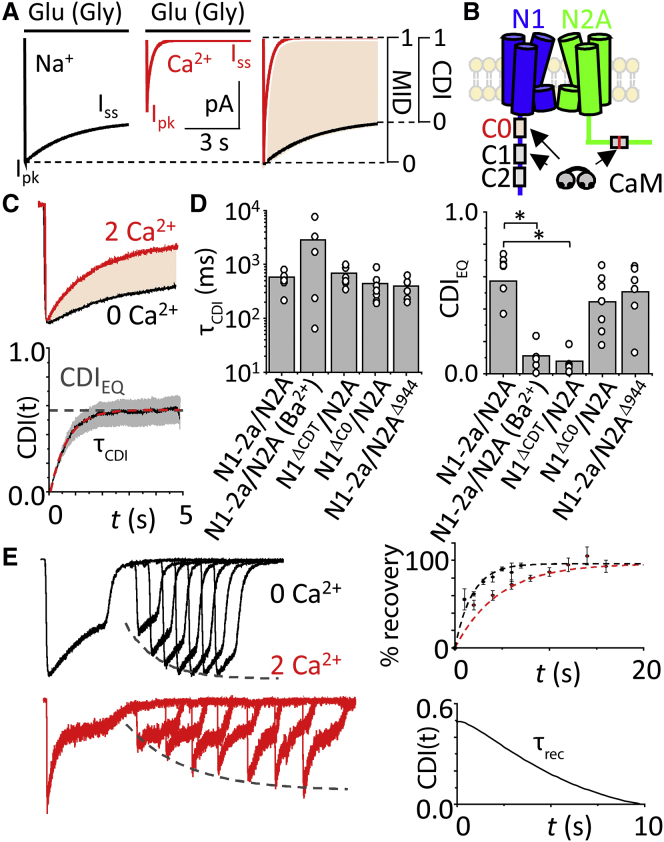
First, we aimed to separate CDI from the intrinsic desensitization of macroscopic NMDA receptor currents (macroscopic intrinsic desensitization, MID). MID can be observed and measured in Ca2+-free solutions as the steady-state fractional residual current (1 − Iss/Ipk), and can theoretically take values from zero (nondesensitizing) to unity (fully desensitizing) (Fig. 1 A). In external Ca2+, currents are smaller and desensitize deeper and faster. We define CDI specifically as the increased desensitization of the macroscopic response observed in Ca2+, relative to Ca2+-free conditions (Eq. 4); it can also take values from zero (noninactivating) to unity (fully inactivating) (Fig. 1 A):
| (4) |
Similar methodology helped define the CDI of voltage-dependent Ca2+ channels (34, 35, 36). To validate the use of this metric for NMDA receptors, we quantified CDI in experimentally recorded currents from wild-type NMDA receptors and from mutants known to lack CDI. Previous reports showed that NMDA receptor CDI requires CaM binding to the GluN1 C-terminal domain C0 segment (10). However, CaM also binds to the alternatively spliced C1 cassette (8) (Fig. 1 B). To eliminate possible confounding effects of Ca2+ mediated by the C1 cassette, we used the naturally occurring splice variant GluN1-2a (N1-2a), which lacks C1 (10). We expressed wild-type NMDA receptors (N1-2a/N2A, WT) and wild-type YFP-tagged calmodulin (YFP-CaMWT) in HEK293 cells and recorded whole cell currents successively without (0 mM) and with (2 mM) Ca2+. We elicited currents with pulses of supersaturating glutamate (1 mM, EC50, 1–3 μM) (37) sufficiently long such that both MID and CDI reached steady-state values (5 s), in background glycine (0.1 mM, Gly EC50 <1 μM) (38, 39) (Fig. 1 C), to prevent glycine-dependent desensitization (40). We found that 2 mM Ca2+ decreased the steady-state current by 58.8 ± 8.2%. This compares well with values reported with this same metric in neurons (41%) (11) and recombinant (N1-1a/N2A) receptors (46.1%) (41). Next, we used Eq. 4 to calculate CDI as defined here, at all time-points, I(t), in paired recordings (5 sweeps/cell). We found that CDI equilibrated to a midrange value, CDIEQ = 0.55 ± 0.09 (N = 7), close to previously reported magnitudes, and estimate for the first time, to our knowledge, its relatively fast onset, τCDI = 0.56 s (Fig. 1 C), which falls well within our chosen observation time (5 s).
Figure 1.
Measuring pure CDI of NMDA receptors. (A) Shown here are simulated NMDA receptor currents using a declining exponential function. In the absence of external Ca2+, (black) currents display (MID, Ipk/Iss) with values between zero and unity; in 2 mM external Ca2+, (red) currents have lower peaks and increased desensitization; superimposed traces normalized to Ipk illustrate the additional desensitization due specifically to Ca2+-dependent inactivation (shaded area). (B) Given here is a schematic of GluN1 (N1, blue) with alternatively spliced cassettes C0, C1, and C2, and GluN2A (N2A, green), with W1017 highlighted (red line), illustrating reported CaM-binding sites. (C) (Top) Paired whole-cell currents from HEK293 cells coexpressing N1-2a and N2A subunits with YFP-CaMWT were recorded in the absence of divalent cations (black) and with Ca2+ (red), are shown normalized to peak and superimposed. (Bottom) CDI (black) is quantified with Eq. 4 along the recording coordinate and averaged across cells, with standard error (gray shaded). CDI-time curves were fit with an exponential of form CDI(t) = CDIEQ + A × e−t/τCDI. (D) Shown here is the CDI time constant (left) and steady-state levels for the conditions indicated (∗p < 0.05, Mann-Whitney U-test). (E) (Left) Exemplar whole-cell recordings of N1-2a and N2A subunits with YFP-CaMWT were recorded using a recovery from MID protocol in the absence (black) and presence (red) of external Ca2+. (Dashed curve) Shown here is an exponential function fitted to the data. (Top right) Fraction of recovery from MID defined as the percent of peak current at the test pulse relative to the conditioning pulse. Data represent mean ± SE of N = 5 cells. (Dashed curve) Shown here is a fitted exponential function. (Bottom right) Shown here is recovery from the CDI curve, derived from fitted exponential curves to recovery of MID data above, using Eq. 4. To see this figure in color, go online.
As expected, receptors lacking the GluN1 C-terminal domain (N1ΔCTD) had no significant CDI (CDIEQ = 0.08 ± 0.07, N = 6, P = 0.0005 relative to WT) (Fig. 1 D). Therefore, Eq. 4 correctly reports CDI regardless of intrinsic differences in the conductance or gating kinetics of the NMDA receptor preparation used (42). Moreover, we found that adding back the C0 cassette to truncated receptors (N1ΔC0) rescued CDI to wild-type values (τCDI = 0.44 ± 0.10 s, CDIEQ = 0.44 ± 0.06, N = 7, P = 0.2 relative to WT), further validating the robustness of our CDI measure.
In addition to the two CaM binding-sites in GluN1 subunits, CaM also binds to the GluN2A subunit (43). To learn whether this site (W1017) contributes to CDI, we removed it by truncating the GluN2A subunit at position 944 (N2AΔ944) and assayed the mutated receptors (N1-2a/N2AΔ944). This mutant had WT-like CDI (τCDI = 0.40 ± 0.07 s, CDIEQ = 0.40 ± 0.08, N = 6, P = 0.6 relative to WT) consistent with previous reports that the C0 portion of GluN1 subunits is necessary and sufficient for NMDA receptor CDI (Fig. 1 D) (9, 10). Last, we verified the Ca2+ dependence of CDI by substituting external Ca2+ with Ba2+ (9, 41). As expected, our measurements also indicated the absence of CDI in these conditions (CDIEQ = 0.07; N = 5, P = 0.0002) (Fig. 1 D).
Next, we determined the kinetics of recovery from CDI using a classic two-pulse protocol consisting of a conditioning glutamate pulse (5 s) followed after increasingly longer resting intervals by a test glutamate pulse (2 s) (44), with glycine present continuously. We performed this protocol in Ca2+-free and 2 mM Ca2+ external solutions (Fig. 1 E, left) and fitted with exponential functions. Values of the corresponding curves were used to determine the recovery time from CDI using Eq. 4, τrec = 9.3 s (Fig. 1 E, right). This value is substantially shorter than values reported in previous studies, which did not isolate CDI from MID (45).
Together, these results validate the use of Eq. 4 as a robust, rigorous, and specific metric for NMDA receptor CDI across experimental conditions and preparations. They also establish for the first time, to our knowledge, that CDI equilibrates and recovers faster than previously reported. In subsequent analyses, we use the equilibrium CDI value (CDIEQ) as a consistent index of CDI magnitude.
Define a cellular system to specifically isolate CDI
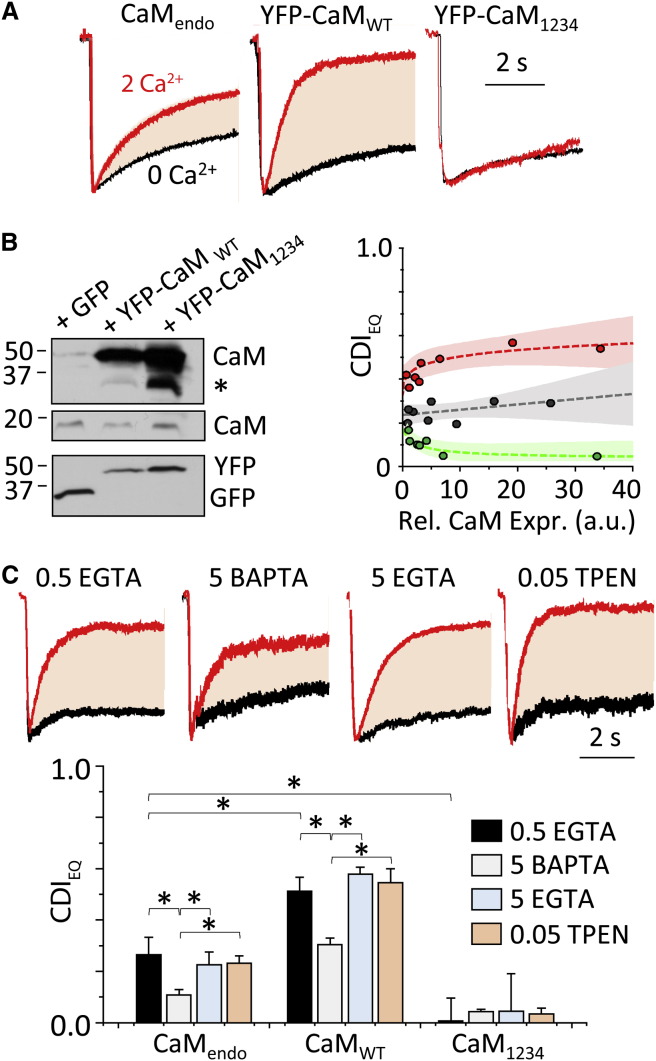
HEK293 cells express low endogenous levels of CaM and CaM-modulated enzymes (46). In the preceding experiments, we overexpressed recombinant CaM to preclude possible variability in the observed CDI due to uncontrolled levels of endogenous CaM. Thus, we asked whether experimental manipulations in CaM expression levels affect NMDA receptor CDI in our system (Fig. 2 A). In cells transfected with only NMDA receptor subunits (N1-2a, N2A, and GFP) the measured CDI (0.26 ± 0.07) was approximately half that observed in cells that overexpressed recombinant CaMWT (Fig. 1 C). This suggests that the levels of endogenous CaM (CaMendo) are insufficient to produce full inactivation, and that the extent of NMDA receptor CDI may have been underestimated in previous studies (45). To test this possibility, we overexpressed recombinant CaM1234, a CaM mutant with Asp to Ala substitutions at all four Ca2+-binding sites rendering it Ca2+ insensitive (47). We reasoned that if the increase in CDI produced by CaM overexpression reflects limitations on CDI imposed by subsaturating levels of endogenous CaM, overexpressing CAM1234 would reduce CDI even further by a dilution effect. Consistent with this scenario, we found that overexpressing CaM1234, abolished CDI (0.01 ± 0.09; P = 0.004 relative to CaMendo). Taken together, these results indicate that in HEK cells, endogenous levels of CaM produce about one-half of the CDI observed with CaM overexpression and, therefore, manipulating cellular CaM levels can effectively modulate NMDA receptor CDI.
Figure 2.
CaM levels modulate CDI magnitude. (A) Traces illustrate paired whole-cell recordings obtained from HEK293 cells expressing NMDA receptors and CaM as indicated, normalized to peak amplitude and superimposed. (B) (Left) Given here are Western blots of proteins isolated from HEK cells transfected with GFP and YFP-CaM, WT or mutant (indicated at top), and probed with antibodies specific for CaM, GFP, and YFP (indicated at right); (∗) nonspecific bands from CaM primary antibody are indicated. (Right) CDI dependency on cell-to-cell variation in relative CaM expression (measured as YFP fluorescence) is shown for CaMWT (red) CaMendo (GFP, gray), and CaM1234 (green). Shaded regions reflect the upper and lower 95% confidence intervals of the fits (bold lines). (C) (Top) Representative whole-cell currents were recorded under indicated intracellular buffering conditions. (Bottom) Shown here are summary results from measurements as in (A) with the indicated metal chelators included in the recording pipette (mean ± SE; ∗p < 0.05, Mann-Whitney U-test). To see this figure in color, go online.
This conclusion is supported by Western blots, which show that in HEK293 cells transfected with either wild-type or mutant CaM, we detected substantially more intense staining for the higher molecular weight CaM (YFP-CaM) relative to untagged CaM (endogenous) (Fig. 2 B, top). The blots also showed a lack of low-molecular-weight species in the YFP-stained gel, confirming the integrity of the YFP-CaM protein through the experiment. The measured CDI was independent of cell-to-cell variability in the level of overexpressed CaM, as evaluated by fluorescence intensity, thus controlling for possible nonspecific effects of overexpressed proteins (Fig. 2 B, bottom). Importantly, intracellular EGTA, a slow Ca2+ buffer, or TPEN (a nonspecific divalent chelator) did not affect CDI in cells expressing either CaM construct, whereas intracellular BAPTA, a fast Ca2+ buffer, substantially reduced CDI in all cases, as previously reported (Fig. 2 C). Intracellular chelators themselves did not exhibit any effect on MID (Fig. 2 C, top).
Our observation that even strong intracellular Ca2+ buffering (with BAPTA) preserves a significant fraction of CDI is consistent with previous results and supports the hypothesis that the Ca2+ sensor (i.e., the CaM pool responsible for CDI), whether freely diffusing or associated with the channel, resides proximally to the fluxing pore (11). To discern between the preassociation and the Ca2+-triggered association models, we set up to estimate the distance between the Ca2+-source (channel pore) and the Ca2+ sensor (CaM) by first calibrating the magnitude of CDI to intracellular levels of Ca2+ and then ascertaining the distance at which fluxed Ca2+ reaches the concentration necessary to produce the experimentally observed CDI.
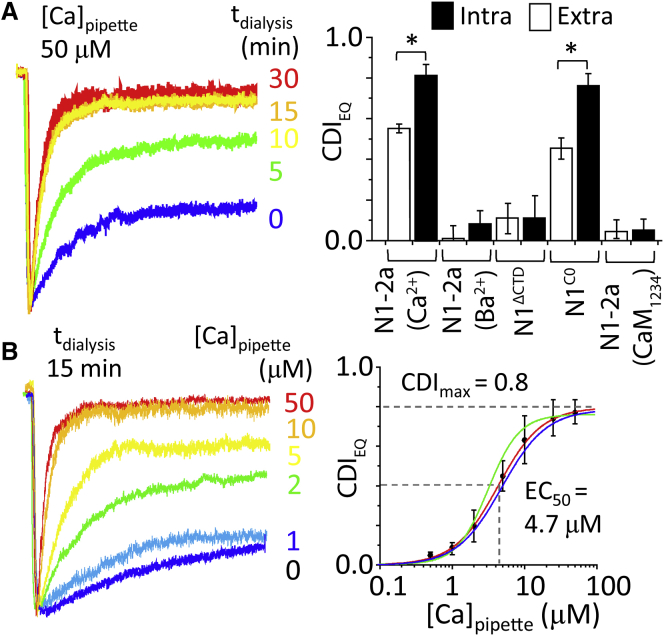
Calibrate the magnitude of CDI to intracellular [Ca2+] levels
We manipulated intracellular [Ca2+] by filling the recording electrode with buffered solutions of controlled free [Ca2+] (HEDTA, KD = 4 μM). In HEK293 cells expressing N1-2a/N2A receptors and CaMWT, we recorded whole-cell currents in response to glutamate applications (1 mM for 5 s) with background glycine (0.1 mM). Relative to traces recorded in 0 μM intracellular Ca2+, which remained stable from sweep to sweep, with 50 μM Ca2+, we observed a slow time-dependent decrease in steady-state current levels (Fig. 3 A), as expected for slow dialysis upon break-in. We calculated CDI with Eq. 4 for each sweep by relating its waveform to that recorded at break-in (0 min), ahead of substantial [Ca2+] influx. Fitting a monoexponential function to the time-CDIEQ data, we found that CDI equilibrated within ∼8 min of break-in (data not shown). In subsequent experiments, we used 15 min as a consistent time point to measure CDI for a given intracellular [Ca2+]. We verified that the reduction in NMDA receptor current observed upon Ca2+ dialysis was Ca2+-dependent, CaM-dependent, and C0-dependent as required for CDI (Fig. 3 A). With this approach we varied the intracellular free [Ca2+] between 0.5 and 50 μM (Fig. 3 B) and fitted the Hill equation to the Ca2+ concentration-dependent increase in CDI to estimate a half-maximal dose EC50 = 4.7 ± 0.3 μM for free [Ca2+], a maximal effect CDImax = 0.80, and a cooperativity factor n = 1.8 (N = 4–7 for each Ca2+ dose).
Figure 3.
Determining CDI sensitivity to [Ca2+]. (A) (Left) Given here are whole-cell NMDA receptor currents recorded with 50 μM free Ca2+ in pipette, at indicated times after break-in. (Right) Shown here is a summary of CDI values at 15 min postbreak-in, with indicated constructs and permeating divalent cation (Ca2+ or Ba2+; Intra, intracellular; Extra, extracellular). (B) (Left) Currents were recorded from NMDA receptors with indicated free [Ca2+] in the pipette recorded at 15 min postbreak-in. (Right) Given here are intracellular Ca2+ dose-response data (points) and fitted Hill function for WT receptors (red); pretreated with calcineurin inhibitor (green, EC50 = 3.9 μM; CDImax = 0.76); or N1ARPAAR (blue, EC50 = 5.2 μM; CDImax = 0.78; blue), ∗P < 0.05, Mann-Whitney U-test. To see this figure in color, go online.
These results show that even in maximally effective intracellular free [Ca2+], a considerable residual current persists, and indicates that fully inactivated NMDA receptors continue to gate, with a lower open probability (Po). This is consistent with previous single-channel recordings of inactivated channels (48, 49). In our hands, the measured CDI sensitivity to internal free [Ca2+] was independent of the Ca2+-dependent phosphatase calcineurin (6, 50), and was not caused by binding of effluxed Ca2+ ions at the external DRPEER motif (1, 2). First, treatment with an inhibitor of calcineurin (6 μM FK-506, 30 min) had no effect on the measured [Ca2+] sensitivity of CDI (EC50 = 4.9 ± 1.2 μM; Fig. 3 B, blue). Second, a mutant NMDA receptor that lacks the external Ca2+-binding site (N1ARPAAR) had WT-like sensitivity (EC50 = 2.8 ± 0.6 μM; Fig. 3 B, green). Thus, the dialysis assay measured precisely and specifically the CDI produced by controlled concentrations of free intracellular [Ca2+]. Together these experiments demonstrate that CDI magnitude is a robust reporter of the free [Ca2+] that engages CaM to initiate CDI. At what distance from the mouth of the NMDA receptor channel can we expect to find this concentration?
Estimate the distance between the Ca2+source and CaM
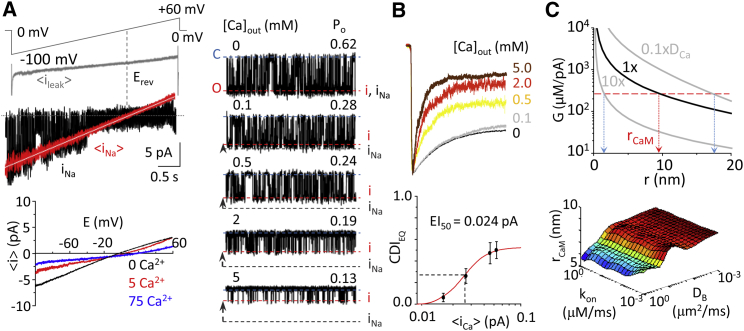
We simulated spatiotemporal Ca2+ diffusion profiles from a point source based on Fick’s law in hemispherically symmetrical space as described in the literature (35, 51, 52). This model predicts that channel openings produce steep spatiotemporal [Ca2+] gradients near the mouth of the channel, which can be approximated for a given radial distance and a known time-averaged unitary Ca2+ flux.
We first determined the unitary fractional Ca2+ current (iCa) of N1-2a/N2A channels at each external Ca2+ tested from conductance changes measured by single-channel voltage ramps (Fig. 4 A; Table S1). Our values were similar to those obtained from native rat hippocampal receptors using this method (28) and with optical measurements (53, 54). We scaled the unitary iCa by the equilibrium Po measured from stationary single-channel records for each extracellular Ca2+ concentration (Fig. 4 B; Table S1) to estimate an average unitary Ca2+ flux at steady-state (〈iCa〉). Because CDI measured from steady-state macroscopic currents reflects the ensemble inactivation produced by the steady-state time-averaged [Ca2+], the CDI versus [Ca2+] relationship can be transformed to a CDI versus 〈iCa〉 function. We varied extracellular Ca2+ between 0 and 5 mM (in constant 150 mM Na+) and found that half-maximal CDI occurred for EI50 = 0.02 pA Ca2+ current. Hereafter, we restricted our analysis to these conditions to avoid reduction in unitary current due to Ca2+-block (55). To relate these data into CDI sensitivity to intracellular [Ca2+], we determined the [Ca2+]/flux ratio, G = [Ca2+]/〈iCa〉, as EC50/EI50, and for N1-2a/N2A channels, we found this to be G = 235 μM/pA.
Figure 4.
Dependence of CDI on unitary Ca2+ influx. (A) (Left, top) Shown here is a schematic of voltage-ramp protocol and average leak current (〈ileak〉) detected during cell-attached voltage ramps with no channel activity (gray). Given here are single-channel Na+-only currents (iNa) from single sweep (black) and averaged over 70–100 sweeps (red). The averaged current (〈i〉) was recorded at multiple external [Ca2+] (0–75 mM). (Bottom) The 〈i〉 was fit with the Jahr and Stevens model (white line) to determine conductance (γin), reversal potential (Erev), and fractional Ca2+ current (fCa) (28). (Right) Given here are representative stationary unitary currents in cell-attached patches at the indicated external [Ca2+]. Unitary current amplitude values (i) C and O indicate closed and open current levels. (B) (Top) Whole-cell currents were recorded with the indicated external [Ca2+] (5 mM BAPTA internal). (Bottom) Shown here is CDI measured with increasing external [Ca2+] plotted against the equivalent unitary Ca2+ current corrected for channel opening, 〈iCa〉 (see Table S1). EI50 = 0.024 pA was extracted from the Hill equation fitted to data. (C) (Top) Simulated steady-state Ca2+ spatial profile during channel opening is corrected for channel Po to reflect the time-averaged Ca2+ signal driving CDIEQ in macroscopic recordings plotted for various Ca2+ diffusion coefficients. Experimentally derived [Ca2+]/flux ratio (EC50/EI50, red horizontal dashed line) predicts rCaM = 9 nm. When Ca2+ diffusion coefficient (DCa) is varied across several orders of magnitude, rCaM likely resides between 2 and 16 nm (vertical pink dashed lines). (C) (Bottom) Systematic error in the model was evaluated as BAPTA kinetic (kon) and diffusion (DB) parameters were varied across several orders of magnitude; the estimated distance of rCaM remains approximately stable for a given value of DCa. To see this figure in color, go online.
With this experimental value, we estimated the distance at which CaM resides (rCaM), using Ca2+ diffusion simulations (56, 57) and, using Eq. 5, found this to be 9 nm:
| (5) |
Given that Eq. 5 assumes no buffer saturation, which may not hold in close proximity to the pore, we also used an alternative approach. We simulated Ca2+ diffusion from a point source using a discretized version of Fick’s laws assuming hemispherical symmetry in the presence of diffusible buffer (see Table S2 and Supporting Material) (35, 51, 52). Results of the Ca2+ diffusion simulation show that the temporal Ca2+ profile rises within a submillisecond timescale to a steady-state value during channel opening and decays to background Ca2+ within a submillisecond timescale of channel closing. Thus, relative to the slow millisecond timescale gating kinetics of the channel, the temporal Ca2+ profile in the presence of buffer during channel opening can be approximated well as a stepwise pulse from 0 μM to a steady-state value during channel opening. Therefore, for simplicity we used the steady-state spatial free [Ca2+] profile when determining rCaM. The free Ca2+ profile from the simulation is transformed to reflect the single-channel G factor by normalizing the concentration to the measured unitary Ca2+ current, iCa. Finally, this unitary open channel G factor (Go) is transformed to reflect channel stochasticity in the steady-state macroscopic response according to the previously derived relation (52):
| (6) |
where G is the steady-state macroscopic gain, n is the Hill coefficient determined from intracellular dialysis that we found to be 1.8 (Fig. 3 B), Po is the single-channel steady-state open probability, and Go is the free intracellular [Ca2+] generated by channel opening normalized by the unitary Ca2+ current, iCa. Assuming an initial Ca2+ diffusion coefficient (DCa) of 0.4 μm2/ms (35, 51), we calculated that G corresponded to rCaM = 9 nm (Fig. 4 C, top), consistent with our first approach. However, DCa values are approximate; we varied this parameter, and found that for the range between 10 × DCa and 0.1 × DCa, rCaM varied between 3 and 16 nm. Further, systematically varying other buffer diffusion (DB) and buffer binding kinetics (kon) parameters also had little effect on the estimate of rCaM for a given value of DCa (Fig. 4 C, bottom). Therefore, in all cases, CaM likely exists well within the Ca2+ nanodomain (<50 nm) generated by the channel, and is likely temporally coupled to the receptor’s activity.
Probing CaM/receptor interaction models
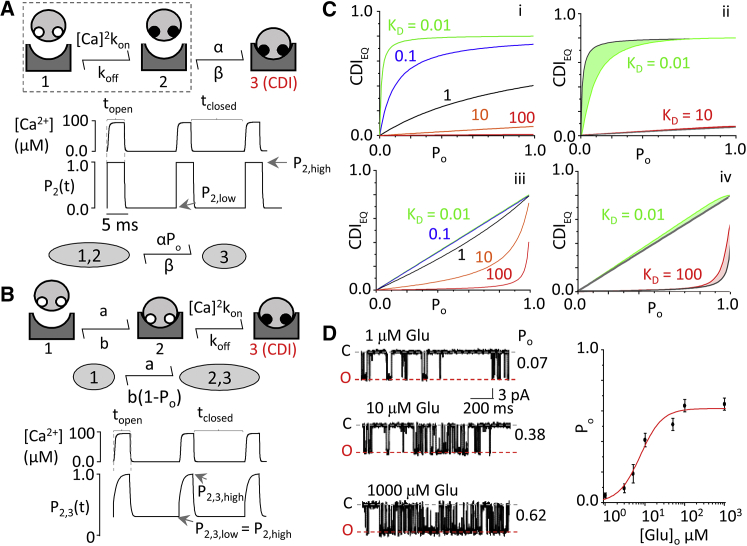
Our observations that CaM1234 can ablate CDI in HEK293 cells and that the receptive CaM resides within nanometers distance from the pore are consistent with a model where apoCaM resides on the C0 segment of the N1. However, given the low level of CaM endogenous to HEK293 cells, it is conceivable that overexpression of CaM1234 excludes CaMendo from the nanodomain volume, thus reducing its access to the channel. To more rigorously test whether apoCaM preassociates with the channel, we considered the behavior of two conceptual models of CaM/channel interaction and their response to a train of Ca2+ spikes produced by channel gating (Fig. 5, A and B). Using reported in vitro values of C0 affinity for apoCaM (19) and holoCaM (8), we found that each model predicted unique time-dependent behaviors. Next, we considered these behaviors as we varied the frequency of the Ca2+ trains, by systematically varying the channel Po (Fig. 5 C, i and iii). To simplify the three-state conceptual models into two-state models, we restricted our analysis to only the fast Ca2+-binding lobe of CaM (N-lobe) (Fig. 5, A and B; see Supporting Material). For each model, we derived the equation that predicts the steady-state magnitude of CDI (CDIEQ) as a function of steady-state Ca2+ influx as determined from channel open probability (Po). For holoCaM association, we have model 1:
| (7) |
where α is the holoCaM binding rate, β is the holoCaM unbinding rate, and CDImax corrects for the low Po of inactivated channels. We measured CDImax directly with our Ca2+ dialysis assay, and found it to be 0.8 (Fig. 3). For (apoCaM preassociation), we have model 2:
| (8) |
where a is the apoCaM binding rate, and b is the apoCaM unbinding rate. Importantly, even when varying apoCaM and holoCaM affinities for C0, which are known only under in vitro conditions (10, 20), the predicted CDIEQ-Po relations remained distinct from each other (Fig. 5 C). Therefore, these relationships impose no assumptions regarding the values of CaM affinity for the channel, and allow us to keep them as free parameters. In addition, we took two important precautions (see Supporting Material). First, we measured CDI in HEK293 cells loaded with 5 mM BAPTA to restrict Ca2+ elevations to local Ca2+ nanodomains, which synchronize with channel gating and, therefore, with Po. Second, we coexpressed channels with CaM34, to restrict Ca2+ binding to the N-lobe. In our cells, mutants with ablated Ca2+-binding at either the N- or C-lobe were fully able to elicit CDI (data not shown). Biochemically, N- and C-lobe mutants are equally capable of displacing actinin from GluN1 C0, which is a likely molecular determinant of CDI (18).
Figure 5.
CDI models reveal unique, testable behaviors. (A) (Top) Model 1 represents NMDA receptor association with holoCaM with subsequent Ca2+. apoCaM (open white circles) binds Ca2+ (solid circles) before binding to the channel. (Middle) Time-dependent behavior of subsystem (dashed box) is given, representing Ca2+ binding to CaM using only fast, N-lobe kinetics in response to a train of Ca2+ influx from a gating channel. State 2 (CaM bound with Ca2+) of the subsystem is pulsatile between P2,high and P2,low occupancy probabilities, in tight synchrony with gating. (Bottom) Shown is simplification of a three-state model given rapid equilibration of its subsystem. (B) (Top) Model 2 represents NMDA receptor association with apoCaM with subsequent Ca binding. (Middle) Given is a simplified model 2 assuming N-lobe kinetics. (Bottom) Shown is behavior of condensed state 2, 3 in response to a train of Ca2+ in-flux. The pulsatile system oscillates between P2,3,high and P2,3,low. P2,3,low is set by the fraction of channels preassociated with apoCaM at rest, P2,high. (C) (i) Given here is evaluation of Eq. 7’s corresponding model 1 across all values of Po. As CaM affinity for the channel is increased, the system exhibits saturation kinetics. As affinity decreases, the system becomes linear. (ii) Deviations of the simplified model in Eq. 7 from numerical integration to the full three-state model did not affect the overall shape or predicted behavior. (iii) Given here is evaluation of Eq. 8’s corresponding model 2 across all values of Po. As CaM affinity for the channel is increased, the system exhibits a linear trend. As affinity decreases, the system uniquely develops an upward curvature. (iv) Deviations of the simplified model in Eq. 8 from numerical integration in the full three-state model were minimal and did not affect the overall shape or predicted behavior. (D) (Left) Exampled here is single-channel activity elicited by indicated glutamate concentration. (Right) Given here is a calibration curve for glutamate concentration and Po determined from cell-attached single-channel recordings. To see this figure in color, go online.
Resident apoCaM mediates CDI
To control the steady-state Po, we varied the glutamate concentration used to elicit current. Using cell-attached single-channel recordings, we first calibrated the glutamate concentration to the channel equilibrium Po. The resulting [Glu]-Po relation allowed the [Glu]-CDIEQ relation measured from whole-cell recordings to be transformed to a CDIEQ-Po relation (Fig. 5 D). Under these conditions, the measured CDIEQ-Po relationship is roughly linear, as predicted by model 2 (Fig. 6 A). Fitting Eqs. 7 and 8 to these data estimated the physiological apoCaM KD = b/a = 0.19 μM (R2 = 0.929; sum of squares errors = 0.021). We noted that model 1 could also fit the data when holoCaM koff ≫ kon, with a predicted holoCaM KD = β/α = 0.49 μM (R2 = 0.934; sum of squares errors = 0.019). However, this is unlikely to be true given substantial evidence that holoCaM has much higher affinity for C0 than apoCaM (10, 20, 58), weakening the case for model 1.
Figure 6.
CaM preassociates with NMDA receptors. (A) Both model 1 and model 2 describe well the dependency of CDI on Po (varied by changing glutamate concentrations). (B) (Top) C0 sequence indicates CaM binding region. Residues are color-coded by property relevant to CaM binding: positive charge (blue), negative charge (red), hydrophobic (gray). (Bottom) Whole-cell currents recorded from N1-2aW858A/N2A receptors have altered CDI at all glutamate concentrations tested. (C) (Top) Given here are unitary currents of high-Po mutant N1-2aA652Y, W858A/N2A. (Bottom) Given here are whole-cell currents from the mutants indicated. (D) Shown here is a CDIEQ-Po profile with pooled data from N1-2aA652Y/N2A (black circles) and high-Po double mutant N1-2aA652Y, W858A/N2A (red circle) fit with model 1 (holoCaM association, gray) and model 2 (apoCaM association, blue) (N = 4). To see this figure in color, go online.
We further probed the models by altering CaM affinity for the channel. The main feature distinguishing the models is the upward curvature revealed by model 2 at low affinity values. A previous study pointed to several hydrophobic residues on GluN1 C0 (850–862) as critical for CaM binding (Fig. 6 B, top) (59); specifically, a Trp residue is a critical anchoring site for CaM to many target proteins (60, 61). We introduced a point mutation at W858 on C0 (9, 20), which clearly perturbed CDI (Fig. 6 B, bottom). However, because WT channels have a maximum Po = 0.62, our ability to resolve the upward curvature was limited. We improved the resolution of the CDIEQ-Po relation by incorporating data obtained with the high-Po mutant (31), GluN1A652Y (Po = 0.81) (Fig. 6 C, top). GluN1A652Y itself did not prevent CDI (Fig. 6 C, bottom left), and the double mutation GluN1A652Y, W858 also reduced CDI (Fig. 6 C, bottom right). With these data, we observe clear upward curvature, which is only predicted by model 2 (Figs. 5 C and 6 D). Fitting the model to this pooled dataset, and assuming GluN1A652Y does not perturb CaM/channel binding, predicted an apoCaM KD = b/a = 18.17 μM, nearly two orders-of-magnitude larger than predicted for wild-type, and consistent with a critical role for C0 W858 in binding CaM.
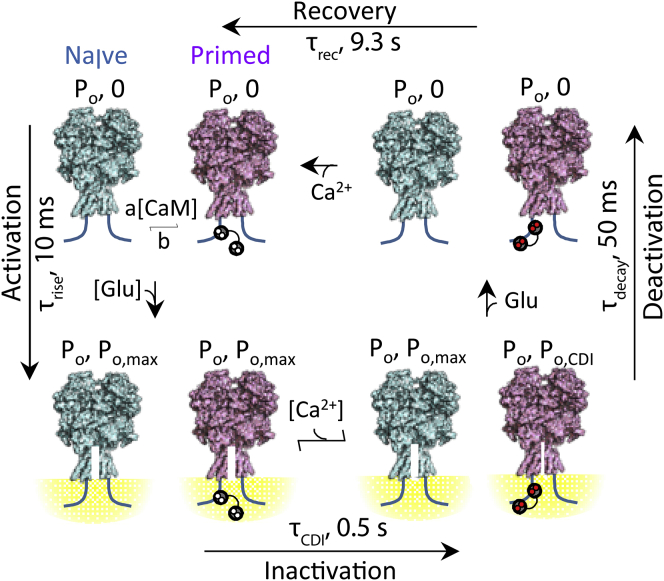
Based on our collective data and consistent with previous results we propose a kinetic mechanism for activity-dependent CDI of NMDA receptors with the following distinguishing features (Fig. 7). At rest, NMDA receptors interconvert between primed (apoCaM bound) and CaM-free forms. Upon agonist binding, both forms activate and initially gate with similar high Po to initiate Ca2+ influx. The fluxed Ca2+ reaches ∼140 μM at ∼9 nm distance from the pore, engages apoCaM, and CaCaM-bound channels switch into low Po (inactivated) to initiate CDI within 0.5 s. Upon glutamate removal, channels deactivate, the Ca2+ gradient dissipates, and closed channels recover from inactivation into the resting state, within 9.2 s. This model incorporates numerous means of CDI regulation, including reciprocal affinities of apoCaM and NMDA receptors, apoCaM affinity for Ca2+, CaM concentration, and NMDA receptor Ca2+-permeability and gating kinetics. Therefore, the model predicts that changes in these parameters, whether physiological or pathological, will influence the rate and extent of NMDA receptor CDI.
Figure 7.
Proposed sequence of molecular events underlying CDI. The model posits that at all times, NMDA receptors exist in a dynamic equilibrium of CaM-free (teal) and CaM-bound (magenta) receptors, which depends on endogenous levels of CaM, and the mutual affinities of the two partners. Upon binding agonist (Glu), receptors transition into highly active (Po, max) Ca2+-permeable conformations, leading to current influx (τrise) and the formation of intracellular Ca2+ nanodomains (yellow). Intracellular Ca2+ binds CaM, according to the local level of Ca2+ in the vicinity of CaM, and only CaM-bound channels transition into inactivated conformations (τCDI), which have very low activity (Po,CDI), leading to a population of active and inactivated channels. Upon glutamate removal, both active and inactivated receptors lose glutamate and deactivate with unique time constants, and the current decays with complex kinetics (τdecay). As intracellular [Ca2+] dissipates, Ca2+ will dissociate from CaM, returning inactivated channels to their resting state (τrec). To see this figure in color, go online.
Discussion
We used, to our knowledge, a new rigorously controlled quantification of NMDA receptor CDI, which is independent from the channel’s intrinsic kinetics and conductance. This allowed us to compare CDI across mutants and experimental conditions and to estimate, by two methods, that the pool of CaM responsible for CDI responds to NMDA receptor Ca2+-flux within ∼9 nm of the channel pore. In addition, we present evidence that CaM associates with NMDA receptors before receptor activation and measure relatively short inactivation (0.5 s) and recovery (9.2 s) times. To our knowledge, this is the first electrophysiological evidence for apoCaM preassociation with NMDA receptors in living cells. These results and conclusions are consistent with recent experimental data indicating that brief synaptic events initiate NMDA receptor CDI (13). Thus, in addition to a serving as a protective feedback mechanism at extrasynaptic sites, CDI likely serves multiple physiological roles, including modulating Ca2+ influx during synaptic transmission.
A standardized metric for Ca2+-dependent inactivation
A lack of consistency in the mathematical definition of NMDA receptor CDI across existing studies has likely contributed to the high variability of CDI values reported in the literature and hampered the rigorous investigation of this process. Some studies quantified CDI as the percentage of macroscopic desensitization, which assumes that no desensitization occurs in the absence of Ca2+ (9, 22, 41, 62); this approach most likely overestimated CDI. To address this limitation, subsequent studies aimed to eliminate the contribution of glycine-dependent desensitization by focusing on currents elicited with varied agonist concentrations (11). However, agonist concentration also influences channel Po and thus the influx of Ca2+ that triggers CDI (Fig. 6); this approach most likely underestimated CDI. In addition, variability in the preparations used whether due to differences in the NMDA receptor isoforms expressed, or in the levels of endogenous CaM, may have contributed to the broad range of reported CDI values. Here, we adapted a metric initially developed for voltage-gated channels (34, 35, 36), which corrects for intrinsic Ca2+-independent desensitization (Eq. 4). With this approach, we show that in some cases endogenous levels of CaM may be insufficient to support full CDI of NMDA receptors (Fig. 2), and that fully inactivated channels gate with substantial open probabilities (Fig. 3).
ApoCaM preassociation is a conserved mechanism of ion channel regulation
CaM is an ancient, highly conserved Ca2+ sensor, which through direct interaction with a variety of target proteins, imparts Ca2+ sensitivity onto fundamental cellular processes. This general mechanism assumed initially that the interaction between CaM and its target proteins is Ca2+-dependent. However, ensuing evidence demonstrated that CaM may interact with targets independently of Ca2+ (63). The multiplicity of Ca2+ permeable channel families that associate with apoCaM, including Cav (64), Nav (5, 65), TMEM16A/16B (66), RyR (67), SK channels (68), and IP3R (69), suggests that preassociation with apoCaM is an evolutionarily conserved mode of ion channel regulation. In support of this hypothesis, mutations that upset the apoCaM interaction with Ca2+ or with the channel correlate with pernicious diseases (70, 71, 72, 73). Our functional evidence for apoCaM preassociation with NMDA receptors adds NMDA receptors to the list of channels that use this regulatory mechanism. Our in situ estimated KD for the NMDA receptor/apoCaM interaction (0.19 μM) is lower than that observed in vitro (19, 20). This may reflect key differences in the undisturbed cellular milieu, which may affect CaM affinity for its targets, such as posttranslational modifications (74). Further, additional factors may control apoCaM association with its targets; for example, within dendritic spines, high levels of neurogranin increase Ca2+ dissociation from CaCaM, and thus increase the fraction of apoCaM within this microenvironment (12, 75). Such a local enrichment of the mobile pool of apoCaM may increase its association with NMDA receptors to affect the range of CDI values.
Divergent mechanisms for local/global Ca2+ signaling
From a physiological standpoint, CDI of Ca2+-permeable channels may represent an autoinhibitory mechanism likely safeguarding against excess Ca2+ influx and its cytotoxic effects. However, aside from being sensitive to this local Ca2+ influx, CDI may reflect the response to other, proximal or more distant, Ca2+ sources (global Ca2+). Sensitivity to local and/or global Ca2+ is ascertained experimentally by examining the effects of intracellular application of fast (BAPTA) and slow (EGTA) Ca2+ buffers (57). This approach established that Cav channels can respond differentially to local and global Ca2+ transients (76) due to distinct affinities of the two CaM lobes for Ca2+, combined with distinct affinities of CaM-binding sites in Cav channels for apoCaM and CaCaM (35).
Our data together with previous studies strongly suggest that in NMDA receptors, the apoCaM and CaCaM sites overlap (10), thus predicting low or nonexistent lobe-specific spatial selectivity to the Ca2+ source. Therefore, both local and global Ca2+ may initiate NMDA receptor CDI. This hypothesis is consistent with evidence suggesting that CDI of NMDA receptors may occur in response to Ca2+ sources other than the target receptors (11, 77). However, if, as proposed in our model, during NMDA receptor activation all primed channels inactivate, additional Ca2+ elevations from other sources (global Ca2+) would play only a minor role during periods when NMDA receptors are active. This mode of primary sensitivity to local Ca2+ may dominate, especially in dendritic spines where spine geometry and endogenous buffering restrict Ca2+ diffusion away from the source.
Insights into the molecular mechanism of CDI
Given that CDI has been predominantly studied using macroscopic currents, which manifest on a slow timescale (>1 s), it was previously deemed unlikely that CDI of NMDA receptors would play a role in modulating fast excitatory synaptic transmission (11). However, in this study we revealed that the rates of inactivation (0.5 s) and recovery (9.3 s) are faster than reported earlier (Fig. 1, C and E). Even based on these new faster values, complete inactivation is unlikely to occur during a single synaptic event (<100 ms; Fig. 7). However, the fraction of primed channels and their gating properties at the time of stimulation will influence the extent to which inactivation develops and set the stage for a broad dynamic CDI range. This activity-dependent autoinhibitory feedback allows the cell to tune individual channels within a designated range of activity. Consistent with this, applying purified CaM on the intracellular leaflet of outside-out membrane patches accelerates the deactivation kinetics of macroscopic currents in response to a brief glutamate pulse, in agreement with the hypothesis that CDI can manifest rapidly (48), including during a single synaptic event (13, 78). Given that CDI of NMDA receptors will also depend on numerous other intracellular factors such as α-actinin and CaMKII (9, 18, 58), derivation of kinetic models for CDI in physiological conditions remains an area of future research.
Physiological roles of NMDA receptor CDI
At central synapses, upon binding glutamate, NMDA receptors transition into highly active (Po, max) Ca2+-permeable conformations (Fig. 7) (79) and their activity sets the decay kinetics of the synaptic response. This decay is critical in setting the pre/postsynaptic spike interval necessary for determining the magnitude and direction of plasticity. To recapitulate the experimental observations, computational models of spike-timing-dependent plasticity required the inclusion of a direct CaM/NMDA receptor inhibitory interaction (15). Specifically, when the postsynaptic spike preceded the presynaptic spike, the inhibitory CaM/NMDA receptor interaction was necessary to decrease the NMDA receptor mediated Ca2+ influx, to predict long-term depression. Although brief synaptic events can initiate NMDA receptor CDI, complete inactivation requires longer periods of activity. Such prolonged activations may occur during periods of high frequency stimulation that engage not only synaptic receptors, but also nonsynaptic receptors through glutamate spillover (80). In this context, factors that affect CDI may serve to control the amplitude and time course of the synaptic current as well as influx at nonsynaptic locations.
In summary, based on the results reported here we put forth a dynamic model of interactions among Ca2+, CaM, and NMDA receptors that describes quantitatively the phenomenon presently referred in the literature as “Ca2+-dependent inactivation”. This model explains how multiple factors/mechanisms can control NMDA receptor CDI by changing the fraction of active/inactivated receptors. However, several specific questions remain regarding NMDA receptor regulation by CDI, including the molecular mechanism of CDI sensitivity to global Ca2+, the kinetic mechanism of CDI, and delineation of the cellular factors regulating CDI. Precise characterization of these events promises to reveal new tools to modulate NMDA receptor activity and may have translational value in addressing Ca2+-related disorders and calmodulinopathies.
Author Contributions
G.J.I. and G.K.P. conceived the work, designed experiments, and drafted and revised the paper. G.J.I. performed data acquisition and analysis.
Acknowledgments
We thank Drs. Takanari Inoue and Manu Ben-Johny (Johns Hopkins) for providing plasmids expressing recombinant CaM; Dr. Michael Tadross (Duke University) for providing Ca2+ diffusion MATLAB code; and Dr. Weinan Sun (Vollum Institute) for his helpful advice with MATLAB coding. We thank Dr. Sophie Belin and Ms. Eileen Kasparek for molecular biology support.
Funding was provided by National Institute of Health (NIH) grants NS098385 and NS097016 to G.K.P.
Editor: Vasanthi Jayaraman.
Footnotes
Supporting Materials and Methods and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30685-9.
Contributor Information
Gary J. Iacobucci, Email: garyiaco@buffalo.edu.
Gabriela K. Popescu, Email: popescu@buffalo.edu.
Supporting Material
References
- 1.Tu Y.C., Yang Y.C., Kuo C.C. Modulation of NMDA channel gating by Ca2+ and Cd2+ binding to the external pore mouth. Sci. Rep. 2016;6:37029. doi: 10.1038/srep37029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe J., Beck C., Wollmuth L.P. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J. Neurosci. 2002;22:10209–10216. doi: 10.1523/JNEUROSCI.22-23-10209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maki B.A., Popescu G.K. Extracellular Ca2+ ions reduce NMDA receptor conductance and gating. J. Gen. Physiol. 2014;144:379–392. doi: 10.1085/jgp.201411244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skeberdis V.A., Chevaleyre V., Zukin R.S. Protein kinase A regulates calcium permeability of NMDA receptors. Nat. Neurosci. 2006;9:501–510. doi: 10.1038/nn1664. [DOI] [PubMed] [Google Scholar]
- 5.Lan J.Y., Skeberdis V.A., Zukin R.S. Protein kinase C modulates NMDA receptor trafficking and gating. Nat. Neurosci. 2001;4:382–390. doi: 10.1038/86028. [DOI] [PubMed] [Google Scholar]
- 6.Tong G., Shepherd D., Jahr C.E. Synaptic desensitization of NMDA receptors by calcineurin. Science. 1995;267:1510–1512. doi: 10.1126/science.7878472. [DOI] [PubMed] [Google Scholar]
- 7.Murphy J.A., Stein I.S., Zukin R.S. Phosphorylation of Ser1166 on GluN2B by PKA is critical to synaptic NMDA receptor function and Ca2+ signaling in spines. J. Neurosci. 2014;34:869–879. doi: 10.1523/JNEUROSCI.4538-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehlers M.D., Zhang S., Huganir R.L. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 1996;84:745–755. doi: 10.1016/s0092-8674(00)81052-1. [DOI] [PubMed] [Google Scholar]
- 9.Krupp J.J., Vissel B., Westbrook G.L. Interactions of calmodulin and α-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J. Neurosci. 1999;19:1165–1178. doi: 10.1523/JNEUROSCI.19-04-01165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S., Ehlers M.D., Huganir R.L. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron. 1998;21:443–453. doi: 10.1016/s0896-6273(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 11.Legendre P., Rosenmund C., Westbrook G.L. Inactivation of NMDA channels in cultured hippocampal neurons by intracellular calcium. J. Neurosci. 1993;13:674–684. doi: 10.1523/JNEUROSCI.13-02-00674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen A., Gerges N.Z. Neurogranin regulates CaM dynamics at dendritic spines. Sci. Rep. 2015;5:11135. doi: 10.1038/srep11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valiullina F., Zakharova Y., Rozov A. The relative contribution of NMDARs to excitatory postsynaptic currents is controlled by Ca2+-induced activation. Front. Cell. Neurosci. 2016;10:12. doi: 10.3389/fncel.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umemiya M., Chen N., Murphy T.H. A calcium-dependent feedback mechanism participates in shaping single NMDA miniature EPSCs. J. Neurosci. 2001;21:1–9. doi: 10.1523/JNEUROSCI.21-01-00001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urakubo H., Honda M., Kuroda S. Requirement of an allosteric kinetics of NMDA receptors for spike timing-dependent plasticity. J. Neurosci. 2008;28:3310–3323. doi: 10.1523/JNEUROSCI.0303-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raman I.M., Tong G., Jahr C.E. Beta-adrenergic regulation of synaptic NMDA receptors by cAMP-dependent protein kinase. Neuron. 1996;16:415–421. doi: 10.1016/s0896-6273(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 17.Rafiki A., Gozlan H., Medina I. The calcium-dependent transient inactivation of recombinant NMDA receptor-channel does not involve the high affinity calmodulin binding site of the NR1 subunit. Neurosci. Lett. 1997;223:137–139. doi: 10.1016/s0304-3940(97)13413-9. [DOI] [PubMed] [Google Scholar]
- 18.Merrill M.A., Malik Z., Hell J.W. Displacement of α-actinin from the NMDA receptor NR1 C0 domain by Ca2+/calmodulin promotes CaMKII binding. Biochemistry. 2007;46:8485–8497. doi: 10.1021/bi0623025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akyol Z., Bartos J.A., Hell J.W. Apo-calmodulin binds with its C-terminal domain to the N-methyl-D-aspartate receptor NR1 C0 region. J. Biol. Chem. 2004;279:2166–2175. doi: 10.1074/jbc.M302542200. [DOI] [PubMed] [Google Scholar]
- 20.Ataman Z.A., Gakhar L., Shea M.A. The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains. Structure. 2007;15:1603–1617. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardingham G.E., Arnold F.J., Bading H. A calcium microdomain near NMDA receptors: on switch for ERK-dependent synapse-to-nucleus communication. Nat. Neurosci. 2001;4:565–566. doi: 10.1038/88380. [DOI] [PubMed] [Google Scholar]
- 22.Wang C., Wang H.G., Pitt G.S. Ca2+/CaM controls Ca2+-dependent inactivation of NMDA receptors by dimerizing the NR1 C termini. J. Neurosci. 2008;28:1865–1870. doi: 10.1523/JNEUROSCI.5417-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson M.G., Alseikhan B.A., Yue D.T. Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 25.Cummings K.A., Belin S., Popescu G.K. Residues in the GluN1 C-terminal domain control kinetics and pharmacology of GluN1/GluN3A N-methyl-d-aspartate receptors. Neuropharmacology. 2017;119:40–47. doi: 10.1016/j.neuropharm.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maki B.A., Cummings K.A., Popescu G.K. One-channel cell-attached patch-clamp recording. J. Vis. Exp. 2014;88 doi: 10.3791/51629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys. J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahr C.E., Stevens C.F. Calcium permeability of the N-methyl-D-aspartate receptor channel in hippocampal neurons in culture. Proc. Natl. Acad. Sci. USA. 1993;90:11573–11577. doi: 10.1073/pnas.90.24.11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer M.L., Vyklicky L., Jr., Clements J. Regulation of NMDA receptor desensitization in mouse hippocampal neurons by glycine. Nature. 1989;338:425–427. doi: 10.1038/338425a0. [DOI] [PubMed] [Google Scholar]
- 30.Cummings K.A., Popescu G.K. Glycine-dependent activation of NMDA receptors. J. Gen. Physiol. 2015;145:513–527. doi: 10.1085/jgp.201411302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murthy S.E., Shogan T., Popescu G.K. Probing the activation sequence of NMDA receptors with lurcher mutations. J. Gen. Physiol. 2012;140:267–277. doi: 10.1085/jgp.201210786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krupp J.J., Vissel B., Westbrook G.L. N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron. 1998;20:317–327. doi: 10.1016/s0896-6273(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 33.Aman T.K., Maki B.A., Popescu G.K. Separate intramolecular targets for protein kinase A control N-methyl-D-aspartate receptor gating and Ca2+ permeability. J. Biol. Chem. 2014;289:18805–18817. doi: 10.1074/jbc.M113.537282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Findeisen F., Minor D.L., Jr. Disruption of the IS6-AID linker affects voltage-gated calcium channel inactivation and facilitation. J. Gen. Physiol. 2009;133:327–343. doi: 10.1085/jgp.200810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tadross M.R., Dick I.E., Yue D.T. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell. 2008;133:1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett C.F., Tsien R.W. The Timothy syndrome mutation differentially affects voltage- and calcium-dependent inactivation of CaV1.2 L-type calcium channels. Proc. Natl. Acad. Sci. USA. 2008;105:2157–2162. doi: 10.1073/pnas.0710501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahum-Levy R., Lipinski D., Benveniste M. Desensitization of NMDA receptor channels is modulated by glutamate agonists. Biophys. J. 2001;80:2152–2166. doi: 10.1016/S0006-3495(01)76188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleckner N.W., Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 1988;241:835–837. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- 39.Chen P.E., Geballe M.T., Wyllie D.J. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J. Physiol. 2008;586:227–245. doi: 10.1113/jphysiol.2007.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benveniste M., Clements J., Mayer M.L. A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J. Physiol. 1990;428:333–357. doi: 10.1113/jphysiol.1990.sp018215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krupp J.J., Vissel B., Westbrook G.L. Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol. Pharmacol. 1996;50:1680–1688. [PubMed] [Google Scholar]
- 42.Maki B.A., Aman T.K., Popescu G.K. C-terminal domains of N-methyl-D-aspartic acid receptor modulate unitary channel conductance and gating. J. Biol. Chem. 2012;287:36071–36080. doi: 10.1074/jbc.M112.390013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajaj G., Hau A.M., Ishmael J.E. Identification of an atypical calcium-dependent calmodulin binding site on the C-terminal domain of GluN2A. Biochem. Biophys. Res. Commun. 2014;444:588–594. doi: 10.1016/j.bbrc.2014.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Howe J.R., Popescu G.K. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat. Neurosci. 2008;11:1373–1375. doi: 10.1038/nn.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina I., Filippova N., Bregestovski P. Calcium-dependent inactivation of heteromeric NMDA receptor-channels expressed in human embryonic kidney cells. J. Physiol. 1995;482:567–573. doi: 10.1113/jphysiol.1995.sp020540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persechini A., Cronk B. The relationship between the free concentrations of Ca2+ and Ca2+-calmodulin in intact cells. J. Biol. Chem. 1999;274:6827–6830. doi: 10.1074/jbc.274.11.6827. [DOI] [PubMed] [Google Scholar]
- 47.Peterson B.Z., DeMaria C.D., Yue D.T. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 48.Rycroft B.K., Gibb A.J. Direct effects of calmodulin on NMDA receptor single-channel gating in rat hippocampal granule cells. J. Neurosci. 2002;22:8860–8868. doi: 10.1523/JNEUROSCI.22-20-08860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rycroft B.K., Gibb A.J. Regulation of single NMDA receptor channel activity by α-actinin and calmodulin in rat hippocampal granule cells. J. Physiol. 2004;557:795–808. doi: 10.1113/jphysiol.2003.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rycroft B.K., Gibb A.J. Inhibitory interactions of calcineurin (phosphatase 2B) and calmodulin on rat hippocampal NMDA receptors. Neuropharmacology. 2004;47:505–514. doi: 10.1016/j.neuropharm.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Kits K.S., de Vlieger T.A., Mansvelder H.D. Diffusion barriers limit the effect of mobile calcium buffers on exocytosis of large dense cored vesicles. Biophys. J. 1999;76:1693–1705. doi: 10.1016/S0006-3495(99)77328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tadross M.R., Tsien R.W., Yue D.T. Ca2+ channel nanodomains boost local Ca2+ amplitude. Proc. Natl. Acad. Sci. USA. 2013;110:15794–15799. doi: 10.1073/pnas.1313898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneggenburger R., Zhou Z., Neher E. Fractional contribution of calcium to the cation current through glutamate receptor channels. Neuron. 1993;11:133–143. doi: 10.1016/0896-6273(93)90277-x. [DOI] [PubMed] [Google Scholar]
- 54.Burnashev N., Zhou Z., Sakmann B. Fractional calcium currents through recombinant GluR channels of the NMDA, AMPA and kainate receptor subtypes. J. Physiol. 1995;485:403–418. doi: 10.1113/jphysiol.1995.sp020738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wollmuth L.P., Sakmann B. Different mechanisms of Ca2+ transport in NMDA and Ca2+-permeable AMPA glutamate receptor channels. J. Gen. Physiol. 1998;112:623–636. doi: 10.1085/jgp.112.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naraghi M., Neher E. Linearized buffered Ca2+ diffusion in microdomains and its implications for calculation of [Ca2+] at the mouth of a calcium channel. J. Neurosci. 1997;17:6961–6973. doi: 10.1523/JNEUROSCI.17-18-06961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neher E. Vesicle pools and Ca2+ microdomains: new tools for understanding their roles in neurotransmitter release. Neuron. 1998;20:389–399. doi: 10.1016/s0896-6273(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 58.Wyszynski M., Lin J., Sheng M. Competitive binding of α-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 59.Leonard A.S., Bayer K.U., Hell J.W. Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and α-actinin. J. Biol. Chem. 2002;277:48441–48448. doi: 10.1074/jbc.M205164200. [DOI] [PubMed] [Google Scholar]
- 60.Mruk K., Farley B.M., Kobertz W.R. Calmodulation meta-analysis: predicting calmodulin binding via canonical motif clustering. J. Gen. Physiol. 2014;144:105–114. doi: 10.1085/jgp.201311140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tidow H., Nissen P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013;280:5551–5565. doi: 10.1111/febs.12296. [DOI] [PubMed] [Google Scholar]
- 62.Vissel B., Krupp J.J., Westbrook G.L. Intracellular domains of NR2 alter calcium-dependent inactivation of N-methyl-D-aspartate receptors. Mol. Pharmacol. 2002;61:595–605. doi: 10.1124/mol.61.3.595. [DOI] [PubMed] [Google Scholar]
- 63.Jurado L.A., Chockalingam P.S., Jarrett H.W. Apocalmodulin. Physiol. Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- 64.Erickson M.G., Liang H., Yue D.T. FRET two-hybrid mapping reveals function and location of L-type Ca2+ channel CaM preassociation. Neuron. 2003;39:97–107. doi: 10.1016/s0896-6273(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 65.Feldkamp M.D., Yu L., Shea M.A. Structural and energetic determinants of apo calmodulin binding to the IQ motif of the NaV1.2 voltage-dependent sodium channel. Structure. 2011;19:733–747. doi: 10.1016/j.str.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang T., Hendrickson W.A., Colecraft H.M. Preassociated apocalmodulin mediates Ca2+-dependent sensitization of activation and inactivation of TMEM16A/16B Ca2+-gated Cl− channels. Proc. Natl. Acad. Sci. USA. 2014;111:18213–18218. doi: 10.1073/pnas.1420984111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore C.P., Rodney G., Hamilton S.L. Apocalmodulin and Ca2+ calmodulin bind to the same region on the skeletal muscle Ca2+ release channel. Biochemistry. 1999;38:8532–8537. doi: 10.1021/bi9907431. [DOI] [PubMed] [Google Scholar]
- 68.Schumacher M.A., Crum M., Miller M.C. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12:849–860. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 69.Patel S., Morris S.A., Taylor C.W. Ca2+-independent inhibition of inositol trisphosphate receptors by calmodulin: redistribution of calmodulin as a possible means of regulating Ca2+ mobilization. Proc. Natl. Acad. Sci. USA. 1997;94:11627–11632. doi: 10.1073/pnas.94.21.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Limpitikul W.B., Dick I.E., Yue D.T. Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca2+ currents and promote proarrhythmic behavior in ventricular myocytes. J. Mol. Cell. Cardiol. 2014;74:115–124. doi: 10.1016/j.yjmcc.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan H.L., Kupershmidt S., Balser J.R. A calcium sensor in the sodium channel modulates cardiac excitability. Nature. 2002;415:442–447. doi: 10.1038/415442a. [DOI] [PubMed] [Google Scholar]
- 72.Weiss L.A., Escayg A., Meisler M.H. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol. Psychiatry. 2003;8:186–194. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- 73.Crotti L., Johnson C.N., George A.L., Jr. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation. 2013;127:1009–1017. doi: 10.1161/CIRCULATIONAHA.112.001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benaim G., Villalobo A. Phosphorylation of calmodulin. Functional implications. Eur. J. Biochem. 2002;269:3619–3631. doi: 10.1046/j.1432-1033.2002.03038.x. [DOI] [PubMed] [Google Scholar]
- 75.Hoffman L., Chandrasekar A., Waxham M.N. Neurogranin alters the structure and calcium binding properties of calmodulin. J. Biol. Chem. 2014;289:14644–14655. doi: 10.1074/jbc.M114.560656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang H., DeMaria C.D., Yue D.T. Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron. 2003;39:951–960. doi: 10.1016/s0896-6273(03)00560-9. [DOI] [PubMed] [Google Scholar]
- 77.Kyrozis A., Goldstein P.A., MacDermott A.B. Calcium entry through a subpopulation of AMPA receptors desensitized neighbouring NMDA receptors in rat dorsal horn neurons. J. Physiol. 1995;485:373–381. doi: 10.1113/jphysiol.1995.sp020736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Medina I., Leinekugel X., Ben-Ari Y. Calcium-dependent inactivation of the monosynaptic NMDA EPSCs in rat hippocampal neurons in culture. Eur. J. Neurosci. 1999;11:2422–2430. doi: 10.1046/j.1460-9568.1999.00664.x. [DOI] [PubMed] [Google Scholar]
- 79.Iacobucci G.J., Popescu G.K. NMDA receptors: linking physiological output to biophysical operation. Nat. Rev. Neurosci. 2017;18:236–249. doi: 10.1038/nrn.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chalifoux J.R., Carter A.G. Glutamate spillover promotes the generation of NMDA spikes. J. Neurosci. 2011;31:16435–16446. doi: 10.1523/JNEUROSCI.2777-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.