ABSTRACT
Interactions between simeprevir (hepatitis C virus [HCV] NS3/4A protease inhibitor) and ledipasvir (HCV NS5A replication complex inhibitor) were investigated in treatment-naive HCV genotype 1-infected patients without cirrhosis, treated with simeprevir-sofosbuvir-ledipasvir in a two-panel, phase 2, open-label study. Patients had stable background treatment with sofosbuvir (400 mg once daily [QD]). In panel 1 (n = 20), the effect of ledipasvir (90 mg QD) on simeprevir (150 mg QD) was studied. Patients received simeprevir and sofosbuvir from days 1 to 14; steady-state pharmacokinetics (PK) of simeprevir was assessed (day 14). On day 15, ledipasvir was added and steady-state PK of simeprevir in the combination was evaluated (day 28). In panel 2 (n = 20), the effect of simeprevir on ledipasvir was investigated. From days 1 to 14, patients received ledipasvir and sofosbuvir and steady-state PK of ledipasvir was assessed (day 14). On day 15, simeprevir was added and a full PK profile was obtained (day 28). The least-squares mean maximum plasma concentration and area under the concentration-time curve (90% confidence interval) increased 2.3-fold (2.0- to 2.8-fold) and 3.1-fold (2.4- to 3.8-fold) for simeprevir, respectively (panel 1), and 1.6-fold (1.4- to 1.9-fold) and 1.7-fold (1.6- to 2.0-fold) for ledipasvir, respectively (panel 2), in the presence versus the absence of the other drug. All patients achieved sustained virologic responses 12 weeks after treatment end. Adverse events, mainly grade 1/2, occurred in 80% of patients; the most common was photosensitivity (45%). Due to the magnitude of interaction and the limited amount of safety data available, the use of this treatment combination is not recommended. (This study has been registered at ClinicalTrials.gov under registration no. NCT02421211.)
KEYWORDS: drug-drug interactions, hepatitis C virus, hepatitis C virus genotype 1, ledipasvir, pharmacokinetics, simeprevir, sofosbuvir, treatment-naive
INTRODUCTION
Hepatitis C virus (HCV) infection is a leading cause of liver disease, with an estimated 71 million individuals infected worldwide (1). Chronic HCV infection is a slow, progressive disease characterized by persistent hepatic inflammation, which can develop into cirrhosis, hepatic decompensation, and hepatocellular carcinoma (2–4). Newer treatment options for HCV do not include interferon (IFN) or ribavirin, and they generally are comprised of combinations of two or more direct-acting antivirals (DAAs).
Simeprevir in combination with sofosbuvir has demonstrated antiviral efficacy for HCV genotype 1 and genotype 4 infections when administered for 12 weeks. Simeprevir is an oral, once-daily (QD) HCV NS3/4A protease inhibitor and sofosbuvir is an oral QD HCV nucleotide analogue NS5B polymerase inhibitor. Another 2-DAA combination regimen comprising ledipasvir (an HCV NS5A replication complex inhibitor) and sofosbuvir is approved for the treatment of chronic HCV genotype 1, 4, 5, and 6 infections. Treatment duration with this regimen is 12 weeks, but it can be shortened to 8 weeks in treatment-naive patients without cirrhosis and with HCV RNA levels of <6,000,000 IU/ml (5).
Adding a third anti-HCV agent (for example, simeprevir) with a mechanism of action different from that of a 2-DAA regimen (such as ledipasvir-sofosbuvir) may lead to increased efficacy and allow for shorter treatment duration (6). However, before such combinations can be administered in large patient trials, it is important to understand the potential for drug-drug interactions (DDI) within the combination.
Simeprevir is a substrate and inhibitor of P-glycoprotein (P-gp), breast cancer resistance protein (BCRP), and organic anion-transporting polypeptide B1 (OATP1B1). Simeprevir is transported by OATP1B1/3 into the liver, where it undergoes metabolism by cytochrome CYP3A. Simeprevir is a mild inhibitor of intestinal (not hepatic) CYP3A4 and a mild inhibitor of CYP1A2 (7). Clinically significant interactions have been observed with CYP3A inducers and inhibitors such as ritonavir and efavirenz and with cyclosporine, which is an inhibitor of multiple transporters, such as OATP and P-gp. The coadministration of simeprevir with these drugs is not recommended (7).
Sofosbuvir is a substrate of P-gp and BCRP but not a substrate of OATP1B1/3. It is a prodrug that is converted to an active metabolite, which is subsequently converted to GS-331007, the predominant circulating moiety of sofosbuvir and which represents >90% of sofosbuvir-related entities and is not a substrate of P-gp or OATP1B1/3. Sofosbuvir is not metabolized by CYP enzymes, nor is it an inducer or inhibitor of these enzymes (8). A clinically significant decrease in exposure was observed for sofosbuvir with potent P-gp inducers such as rifampin, and coadministration is not recommended.
Ledipasvir is a substrate and inhibitor of both P-gp and OATP1B1/3 and an inhibitor of BCRP. It is neither a substrate nor an inhibitor or inducer of CYP enzymes (8, 9). In a clinical DDI study, ledipasvir increased digoxin (a P-gp substrate) area under the plasma concentration-time curve (AUC) by 34%, and therefore, the combination should be used with caution. Ledipasvir-sofosbuvir also increased exposure of rosuvastatin (an OATP1B1/3 and BCRP substrate) in clinical trials 7-fold, and coadministration is contraindicated (5, 7, 8).
The interaction between sofosbuvir and simeprevir in patients with HCV has been investigated previously (10). Sofosbuvir has no effect on the plasma concentration of simeprevir; however, there was an increase in sofosbuvir concentrations {a 3.16-fold (90% confidence interval [CI]: 2.25 to 4.44) increase in AUC at steady state (AUCτ) and a 1.91-fold (90% CI: 1.26 to 2.90) increase in the maximum plasma concentration (Cmax)} but not in that of the major metabolite GS-331007. No dose adjustment is required for either agent when coadministered (10).
The interaction between sofosbuvir and ledipasvir was studied with healthy volunteers. Sofosbuvir AUC was increased 2.3-fold by ledipasvir; however, the pharmacokinetics (PK) of GS-331007 was not affected (9). The interaction is likely to be mediated via inhibition of P-gp and BCRP by ledipasvir.
A DDI study with healthy volunteers showed that coadministration of simeprevir at 150 mg QD with ledipasvir at 30 mg QD, which is lower than the recommended dose, increased the Cmax and AUCτ of simeprevir 2.61-fold (90% CI: 2.39- to 2.86-fold) and 2.69-fold (90% CI: 2.44- to 2.96-fold) and those of ledipasvir 1.81-fold (90% CI: 1.69- to 2.94-fold) and 1.92-fold (90% CI: 1.77- to 2.07-fold), respectively (5). However, simeprevir has much higher (2- to 3-fold) exposure in patients with HCV than in healthy volunteers (7). As simeprevir is a perpetrator of the interaction with ledipasvir, the higher exposure in patients may be expected to result in a greater magnitude of interaction with ledipasvir. Similarly, administration of ledipasvir at the higher, clinically recommended dose of 90 mg QD may potentially result in a greater magnitude of interaction with simeprevir (5).
Therefore, this phase 2 HPC2017 study (ClinicalTrials registration no. NCT02421211) aimed to investigate the PK interactions between simeprevir and ledipasvir (administered at the recommended dose) in treatment-naive, HCV genotype 1-infected patients without cirrhosis. As the efficacy of the two-drug combination of simeprevir and ledipasvir is unknown, patients were treated with simeprevir and ledipasvir on a stable background of sofosbuvir. The fixed-dose combination of sofosbuvir at 400 mg QD and ledipasvir at 90 mg QD is an approved treatment regimen for this patient population. In order to minimize the duration of the study, only patients with HCV RNA levels of <6,000,000 IU/ml at screening were eligible for this study since a treatment duration of 8 weeks of ledipasvir with sofosbuvir is approved for these patients, and the addition of simeprevir to this regimen would not be expected to negatively impact efficacy.
RESULTS
Patients.
In total, 50 patients were screened and 40 received at least one dose of study treatment; these patients comprised the intent-to-treat (ITT) population (20 in each panel). All 40 (100%) randomized patients completed both the study treatment period and the follow-up period (see Fig. S1 in the supplemental material). In total, 10 patients failed screening due to the following reasons: HCV RNA levels of >6,000,000 IU/ml (5/10), HCV RNA levels of >6,000,000 IU/ml and body mass index (BMI) out of range (1/10), BMI that was too high (1/10), no suitable veins (1/10), no suitable veins/drug abuse/no stable medication intake (1/10), or withdrawal of consent before dosing (no reason given; 1/10). There were no major protocol deviations in this study.
Baseline patient demographic and disease characteristics are presented in Table 1. The majority of patients were female (55%) and white (98%) and had HCV genotype 1b infection (80%).
TABLE 1.
Patient demographics and baseline disease characteristics (ITT population)a
| Parameter | Value for patients receiving SMV at 150 mg QD + LDV-SOF at 90/400 mg QD |
||
|---|---|---|---|
| Panel 1 (n = 20) | Panel 2 (n = 20) | Total (n = 40) | |
| Age (yrs), median (range) | 50.5 (25–70) | 51.0 (26–62) | 51.0 (25–70) |
| Male, no. (%) | 8 (40) | 10 (50) | 18 (45) |
| BMI (kg/m2), median (range) | 24.4 (19.0–30.7) | 25.0 (20.2–33.2) | 25.1 (19.0–33.2) |
| ≥30, no. (%) | 2 (10) | 2 (10) | 4 (10) |
| Race, no. (%) | |||
| White/black | 19 (95)/1 (5) | 20 (100)/0 | 39 (98)/1 (3) |
| IL28B genotype, no. (%) | |||
| CC | 9 (45) | 5 (25) | 14 (35) |
| CT | 5 (25) | 12 (60) | 17 (43) |
| TT | 6 (30) | 3 (15) | 9 (23) |
| HCV genotype/subtype, no. (%) | |||
| 1a | 5 (25) | 3 (15) | 8 (20) |
| 1b | 15 (75) | 17 (85) | 32 (80) |
| Baseline HCV RNAb (log10 IU/ml; median [range]) | 6.2 (4.6–6.9) | 5.9 (5.0–6.9) | 6.0 (4.6–6.9) |
BMI, body mass index; HCV, hepatitis C virus; ITT, intent-to-treat; LDV, ledipasvir; QD, once daily; SMV, simeprevir; SOF, sofosbuvir.
All patients had HCV RNA levels of <6,000,000 IU/ml at screening. HCV RNA levels shown here are at baseline (day 1).
Simeprevir NS3 resistance-associated substitutions (RASs) were observed at baseline in 3/40 patients (8%): Q80K and R155K each in 1 genotype 1a-infected patient and D168E in 1 genotype 1b-infected patient. Ledipasvir NS5A RASs were observed at baseline in 8/40 patients (either alone or in combination with another baseline polymorphism), all infected with genotype 1b: L31I in 2 patients, L31M in 3 patients, and Y93H in 3 patients. NS5B RASs were observed in 7/40 patients (18%): C316N in 2 genotype 1b-infected patients, C316N plus L159F in 4 genotype 1b-infected patients, and C316N plus N142S in 1 genotype 1b-infected patient. The baseline presence of L159F and/or C316N has been associated with sofosbuvir failure posttransplant in HCV genotype 1b-infected patients (11). The sofosbuvir NS5B RAS S282T was not observed at baseline.
Pharmacokinetics.
PK parameters for ledipasvir and simeprevir in both panels 1 and 2 (in the presence of sofosbuvir) are presented in Tables 2 and 3 and in Tables S2 and S3 in the supplemental material. The PK parameters for ledipasvir increased when simeprevir was coadministered in panel 2 (minimum plasma concentration [Cmin] increased from 283 to 485 ng/ml, Cmax increased from 507 to 832 ng/ml, and AUCτ increased from 8,944 to 15,562 ng · h/ml [Table 2]). In panel 1, the PK parameters for simeprevir also increased upon coadministration with ledipasvir (Cmin increased from 936 to 4,394 ng/ml, Cmax increased from 4,193 to 9,951 ng/ml, and AUCτ increased from 54,735 to 166,157 ng · h/ml [Table 3]).
TABLE 2.
PK parameters for ledipasvir in panel 2a
| Parameter | Value for patients receiving the following: |
|
|---|---|---|
| LDV-SOF, 90/400 mg QD (reference, day 14) | LDV-SOF, 90/400 mg QD, + SMV,150 mg QD (test, day 28) | |
| No. | 20 | 18b |
| Ctrough, ng/ml | 332 (56.1) | 554 (61.6) |
| Cmin, ng/ml | 283 (55.7) | 485 (55.1) |
| Cmax, ng/ml | 507 (48.5) | 832 (50.1) |
| Tmax, h | 4.07 (1.00–8.00) | 6.00 (3.93–10.00) |
| AUCτ, ng · h/ml | 8,944 (50.0) | 15,562 (50.3) |
| Css,avg, ng/ml | 372 (50.3) | 648 (50.1) |
All values are geometric means (coefficients of variation, expressed as percents) except in the case of Tmax, for which values are medians (ranges). AUCτ, area under the curve at steady state; Cmax, maximum plasma concentration; Cmin, minimum plasma concentration; Css,avg, average steady-state plasma concentration; Ctrough, trough plasma concentration; LDV, ledipasvir; PK, pharmacokinetic; QD, once daily; SMV, simeprevir; SOF, sofosbuvir; Tmax, time to Cmax.
Due to day 28 PK profile not being taken for 2 patients; n = 17 for Cmax, Tmax, AUCτ, and Css,avg due to missing PK samples for an additional patient around expected Cmax time.
TABLE 3.
PK parameters for simeprevir in panel 1a
| Parameter | Value for patients receiving the following: |
|
|---|---|---|
| SMV, 150 mg, + SOF, 400 mg QD (reference, day 14) | SMV, 150 mg QD, + LDV-SOF, 90/400 mg QD (test, day 28) | |
| No. | 20b | 20c |
| Ctrough, ng/ml | 1,548 (138.5) | 5,250 (76.4) |
| Cmin, ng/ml | 936 (156.7) | 4,394 (62.4) |
| Cmax, ng/ml | 4,193 (94.0) | 9,951 (56.8) |
| Tmax, h | 6.00 (4.00–12.00) | 6.00 (0.47–10.00) |
| AUCτ, ng · h/ml | 54,735 (115.3) | 166,157 (65.3) |
| Css,avg, ng/ml | 2,284 (115.2) | 6,914 (65.4) |
All values are geometric means (coefficient of variations, expressed as percents) except in the case of Tmax, for which values are medians (ranges).
n = 19 for Ctrough due to a missing PK sample at trough.
n = 19 for Cmax, Tmax, AUCτ, and Css,avg due to a missing PK sample around the time of expected Cmax.
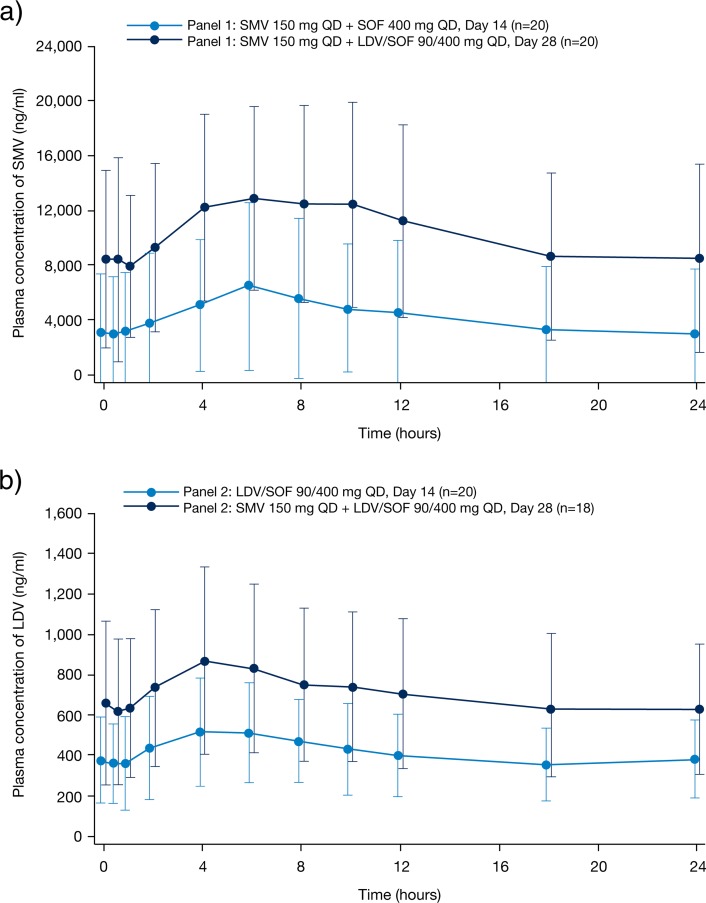
Simeprevir least-squares (LS) mean ratios of the PK parameters Cmin, Cmax, and AUCτ increased 4.7-fold (90% CI: 3.4- to 6.5-fold), 2.3-fold (90% CI: 2.0- to 2.8-fold), and 3.1-fold (90% CI: 2.4- to 3.8-fold), respectively, when the drug was administered with ledipasvir versus without ledipasvir (panel 1) (Table 4; Fig. 1a). There was considerable interpatient variability, with coefficients of variation of 68%, 46%, and 58% for the individual ratios of Cmin, Cmax, and AUCτ, respectively (Table S4). The maximum observed ratios for the increase in an individual patient's exposure were up to 16-fold, 5.3-fold, and 9.0-fold for Cmin, Cmax, and AUCτ, respectively.
TABLE 4.
Statistical PK results for simeprevir and ledipasvir at day 28 versus day 14
| Drug and parameter | LS mean ratio (day 28 vs day 14) (n = 20) | 90% CI |
|---|---|---|
| Simeprevir | ||
| Cmin, ng/ml | 4.69 | 3.40–6.47 |
| Cmax, ng/ml | 2.34a | 1.95–2.81 |
| AUCτ, ng · h/ml | 3.05a | 2.43–3.84 |
| Ledipasvir | ||
| Cmin, ng/ml | 1.74b | 1.55–1.97 |
| Cmax, ng/ml | 1.64c | 1.45–1.86 |
| AUCτ, ng · h/ml | 1.75c | 1.56–1.96 |
n = 19 at day 28 due to a missing PK sample around time of Cmax.
n = 18 at day 28 due to missing PK profiles from 2 patients.
n = 17 at day 28 due to missing PK profiles from 2 patients and missing samples around Cmax for 1 patient.
FIG 1.
Mean (standard deviation [SD]) plasma concentration-time curves of simeprevir (a) and ledipasvir (b) at day 14 and day 28 (ITT population). ITT, intent to treat; LDV, ledipasvir; QD, once daily: SMV, simeprevir; SOF, sofosbuvir.
The plasma concentrations of ledipasvir, as demonstrated by LS mean ratios of Cmin, Cmax, and AUCτ, were 1.7-fold (90% CI: 1.5- to 2.0-fold), 1.6-fold (90% CI: 1.4- to 1.9-fold), and 1.7-fold (90% CI: 1.6- to 2.0-fold) higher, respectively, when administered with simeprevir versus without simeprevir (panel 2) (Table 4; Fig. 1b). The interpatient variability for the ratio of PK parameters was lower for ledipasvir than for simeprevir, with coefficients of variation of 30%, 29%, and 27% for the individual ratios of the PK parameters Cmin, Cmax, and AUCτ, respectively (Table S4). The maximum observed ratios for the increase in an individual patient's exposure to ledipasvir in the presence versus absence of simeprevir were up to 3.4-fold, 2.8-fold, and 2.8-fold for Cmin, Cmax, and AUCτ, respectively.
Efficacy.
All 40 patients (100%) achieved sustained virologic response 12 weeks after treatment end (SVR12) with simeprevir plus ledipasvir and sofosbuvir regardless of IL28B genotype and the presence of baseline RASs.
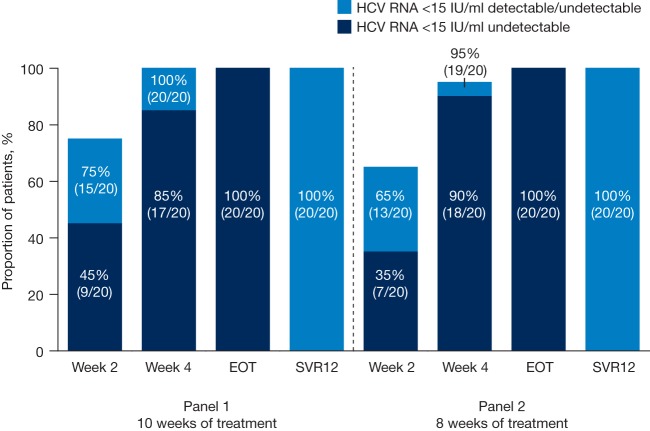
Overall, 39/40 patients (98%) had plasma HCV RNA levels of <15 IU/ml (detectable or undetectable) at week 4 of treatment. From week 6 onwards, all patients had plasma HCV RNA levels of <15 IU/ml (detectable or undetectable). Additional on-treatment response data are presented in Fig. 2. The patient not achieving an HCV RNA level of <15 IU/ml (detectable or undetectable) at week 4 of treatment had HCV genotype 1b infection, an IL28B CC genotype, and the ledipasvir RAS L31M in NS5A at baseline.
FIG 2.
On-treatment and posttreatment virologic responses in HPC2017 (ITT population). EOT, end of treatment; HCV, hepatitis C virus; ITT, intent-to-treat; SVR12, sustained virologic response 12 weeks after EOT.
Safety.
In total, adverse events (AEs) occurred in 32/40 patients (80%). Of these, 30/40 patients (75%) experienced a grade 1 or 2 AE, and 2/40 (5%) experienced a grade 3 AE (Table 5). The grade 3 AEs, recorded for 1 patient with a tibia fracture and 1 patient with a fibula fracture, were also reported as serious AEs (SAEs) and were both considered unrelated to the study drugs. In total, 26/40 (65%), 16/40 (40%), and 8/40 (20%) patients experienced AEs considered at least possibly related to simeprevir, ledipasvir-sofosbuvir, and sofosbuvir, respectively.
TABLE 5.
Summary of adverse events (ITT population)
| AE | No. (%) experiencing AE with SMV, 150 mg QD, + LDV-SOF, 90/400 mg QD |
||
|---|---|---|---|
| Panel 1 (n = 20) | Panel 2 (n = 20) | Total (n = 40) | |
| Any AE | 17 (85) | 15 (75) | 32 (80) |
| Grade 1 AE | 9 (45) | 14 (70) | 23 (58) |
| Grade 2 AE | 6 (30) | 1 (5) | 7 (18) |
| Grade 3 AE | 2 (10) | 0 | 2 (5) |
| Grade 4 AE | 0 | 0 | 0 |
| Serious AE | 2 (10) | 0 | 2 (5) |
| AE leading to permanent discontinuation | 0 | 0 | 0 |
| Most common AEsa | |||
| Photosensitivity reaction | 11 (55) | 7 (35) | 18 (45) |
| Pruritus | 3 (15) | 3 (15) | 6 (15) |
| Headache | 1 (5) | 3 (15) | 4 (10) |
| Asthenia | 2 (10) | 1 (5) | 3 (8) |
| Fatigue | 2 (10) | 1 (5) | 3 (8) |
| Upper abdominal pain | 2 (10) | 1 (5) | 3 (8) |
| Hot flush | 2 (10) | 1 (5) | 3 (8) |
In ≥7.5% of patients overall. AE, adverse event.
No AE-related treatment discontinuations were observed. The most frequent AEs reported during treatment for 3 or more patients (≥7.5%) were photosensitivity reaction (18/40 [45%]), pruritus (6/40 [15%]), headache (4/40 [10%]), upper abdominal pain (3/40 [8%]), asthenia (3/40 [8%]), fatigue (3/40 [8%]), and hot flush (3/40 [8%]) (Table 5). All of these AEs were either grade 1 or 2.
Notably, all photosensitivity reactions were grade 1, and many were reported as sunburn. Of the 18 patients (45%) with ≥1 photosensitivity event(s), 7 (18%) did not always apply sun protection to all parts of the body. All photosensitivity reactions were considered related (possibly, probably, or likely) to simeprevir, and in 5/40 patients (12.5%) photosensitivity reactions were considered related to ledipasvir-sofosbuvir. The frequency of photosensitivity events may have been related to the increased plasma concentrations of simeprevir and ledipasvir, although the range of exposures among the patients with AEs varied widely.
Grade 3 treatment-emergent laboratory abnormalities were observed in panel 1 in 2/20 (10%) patients. Of these, 1 patient (5%) had an asymptomatic pancreatic amylase elevation (associated with a grade 1 lipase elevation), and 1 patient (5%) had hyperbilirubinemia. No grade 3 treatment-emergent laboratory abnormalities were observed in panel 2.
DISCUSSION
The goal of the current study was to investigate the PK interactions between simeprevir and ledipasvir in treatment-naive, HCV genotype 1-infected patients without cirrhosis in order to obtain a better understanding of the drug interactions and safety and efficacy of the 3-DAA regimen comprising simeprevir, ledipasvir, and sofosbuvir. In this study, PK interactions were observed, with increases in the exposure of both ledipasvir and simeprevir. As the sofosbuvir background remained constant throughout the study, and the interactions between sofosbuvir-simeprevir and sofosbuvir-ledipasvir are already well known, the PK of sofosbuvir was not assessed.
Simeprevir exhibits nonlinear PK, and CYP3A4 and OATP1B1/3 have been shown to be key determinants in the nonlinear PK of simeprevir in healthy Caucasians (12). In clinical studies with chronic HCV-infected patients, a 2- to 3-fold-higher mean simeprevir plasma exposure was observed after dosing at 150 mg QD compared to exposures in healthy Caucasians (13, 14). In addition to physiological and biochemical differences in HCV patients compared to healthy volunteers, activities of enzymes and transporters may differ, resulting in PK differences in plasma and in the liver for simeprevir. Interestingly, Morcos et al. (15) recently published data showing that the increase in midazolam exposure in HCV-infected patients in the presence of ritonavir is on average 11-fold. This is 2.2-fold less pronounced than the published interaction of ritonavir on midazolam in healthy patients, thus providing evidence for a reduction in CYP3A activity in the HCV population (16, 17).
Despite the higher plasma simeprevir concentrations in this study in HCV-infected patients than in healthy volunteers, the change in plasma ledipasvir concentrations in this study was comparable with that reported previously when simeprevir was coadministered with ledipasvir at 30 mg in healthy volunteers (Cmax, 1.6-fold versus 1.8-fold increase, and AUCτ, 1.7-fold versus 1.9-fold increase, respectively) (5, 8). Ledipasvir is an inhibitor of P-gp (8), OATP1B1/3 (5), and BCRP (8), and simeprevir is a substrate and inhibitor of P-gp, BCRP, and OATP1B1 (7). The increase in ledipasvir exposure is likely caused by inhibition of P-gp and OATP1B1/3 by simeprevir. The similar magnitudes of interaction in HCV-infected patients despite higher perpetrator (simeprevir) exposure may be explained by a combination of factors: first, by the lower expression of OATP transporters in patients with hepatic impairment versus healthy volunteers (18), and second, by the fact that the interaction via P-gp inhibition takes place in the gastrointestinal tract, where it is the luminal concentration of drug that would drive the interaction, rather than the systemic circulating drug concentration (19). The luminal concentrations are expected to be the same in HCV-infected patients and healthy volunteers. Interestingly, the interaction between ledipasvir and atazanavir led to a similar increase in the plasma ledipasvir concentration upon coadministration, which did not impact dosing recommendations (5).
The increase in the plasma simeprevir concentrations among the HCV-infected patients in this study were similar to those observed when simeprevir was coadministered with ledipasvir at 30 mg in healthy volunteers (Cmax, 2.3-fold versus 2.6-fold increase, and AUCτ, 3.1-fold versus 2.7-fold increase, respectively) (5). The increase in simeprevir exposure is likely caused by inhibition of P-gp, BCRP, and OATP1B1/3 by ledipasvir. BCRP has been shown to be a significant interaction pathway for ledipasvir-sofosbuvir: concurrent administration with rosuvastatin (a BCRP substrate) increased rosuvastatin exposure 7-fold, and there have been reports of rhabdomylosis when ledipasvir was used in combination with atorvastatin, an OATP substrate (8, 20, 21). The lack of further interaction at the high dose of ledipasvir may be explained by saturation of the transporters already at the lower dose of ledipasvir. Thus, there was no effect of chronic HCV infection or of the ledipasvir dose on the drug interactions of the combination.
Increases in plasma simeprevir concentrations of magnitudes similar to those observed with ledipasvir in this study were also observed in East Asian patients receiving simeprevir at 150 mg in combination with (pegylated) IFN and ribavirin compared with Caucasian patients (2.1-fold-higher exposure in East Asian versus Caucasian patients) (22). This difference appeared to be mainly driven by the smaller liver volume and body surface area and the slightly lower CYP3A4 abundance in East Asian versus Caucasian patients (23). The smaller liver volume resulted in a lower simeprevir distribution volume and a lower liver-mediated clearance. Notably, the increased simeprevir concentrations in East Asian patients were not associated with adverse safety findings, and therefore, no adverse safety findings are anticipated from the similarly increased simeprevir concentrations in this study, in the presence of ledipasvir (22). However, in patients with combinations of different characteristics, such as patients of East Asian origin who would use ledipasvir, the increases in simeprevir exposure may be higher than in the current study and the clinical implications when given in combination with ledipasvir are not known.
The safety profile of the 3-DAA combination was comparable to the individual safety profiles for simeprevir and for the ledipasvir-sofosbuvir combination. All AEs were grade 1/2, with the exception of the two grade 3 SAEs that were unrelated to the study drugs. However, photosensitivity was reported more frequently in this study (by 45% of patients) than in other IFN-free studies of simeprevir (by ≤5% of patients), despite similar guidance in the study protocol regarding sun-protective measures (10, 24, 25). All photosensitivity reactions were grade 1 and were considered at least possibly related to simeprevir; many were reported as sunburn, and this higher frequency of events may also be attributed to the treatment phase having taken place in Belgium during the summer months. Notably, the range of simeprevir and ledipasvir exposures among all patients with photosensitivity reaction AEs varied widely, and although advised, 7/40 patients (18%) who reported photosensitivity reactions did not always apply sun protection to all parts of the body.
High SVR12 rates, 100%, were reported in this study for HCV genotype 1-infected, treatment-naive, noncirrhotic patients with HCV RNA levels of <6,000,000 IU/ml at screening after 10 and 8 weeks of treatment in panel 1 and panel 2, respectively. Notably, all patients achieved SVR12 in this study regardless of the presence of simeprevir, ledipasvir, and sofosbuvir RAVs at baseline. The maximal efficacy reported in this study after 10 or 8 weeks of treatment builds upon the SVR12 rates reported for HCV genotype 1-infected patients in the OPTIMIST-1 study, in which treatment-naive and -experienced patients received 12 weeks of treatment with simeprevir plus sofosbuvir (97%) (24). In addition, results from the current study are in agreement with SVR12 rates from the phase 3 ION-3 study in which treatment-naive, HCV genotype 1-infected patients without cirrhosis received ledipasvir-sofosbuvir for 8 (94%) or 12 (95%) weeks (26).
In the phase 2a study, AL-335-604, a 3-DAA regimen comprising of simeprevir in combination with two experimental HCV drugs, AL-335 and odalasvir, taken for 6 or 8 weeks resulted in an SVR12 rate of 100%. This was an improvement on the SVR12 rate of 84% achieved by patients receiving 8 weeks of treatment with a 2-DAA regimen comprised of AL-335 and odalasvir in the same study. These results also support the addition of a third anti-HCV agent with a different mechanism of action to a 2-DAA regimen, due to increased efficacy which enables shorter treatment durations (6).
A limitation of the current study was the inclusion of only treatment-naive, predominantly Caucasian patients without cirrhosis. In the current study, the terminal elimination half-lives could not be calculated because the dosing interval was too short to allow reliable calculation in comparison to the half-lives of the drugs. The half-life of simeprevir is 41 h in patients with HCV (14), and the half-life of ledipasvir is 47 h (9).
In conclusion, major DDI were observed between simeprevir and ledipasvir, with 3.1-fold-higher simeprevir exposure (AUC) and 1.7-fold-higher ledipasvir exposure (AUC) in the combination. Despite these increases, the combination was generally well tolerated, although a higher incidence of mild photosensitivity was observed. The treatment regimen of simeprevir plus ledipasvir-sofosbuvir as used in this study was highly efficacious in treatment-naive, HCV genotype 1-infected patients, as all patients achieved SVR12. However, in view of the magnitude of interaction, and the relatively limited amount of safety data available, the use of this treatment combination is not recommended.
MATERIALS AND METHODS
Patients and study design.
HPC2017 was a phase 2, open-label, randomized study conducted at five hospital sites in Belgium and initiated on 27 May 2015. The study was approved by the relevant Institutional Review Board or Independent Ethics Committee at each study center and met the principles of the Declaration of Helsinki and guidelines of good clinical practice. All patients provided written informed consent to participate.
Eligible adults (18 to 70 years of age) had documented chronic HCV genotype 1 infection (diagnosed >6 months prior to screening), with plasma HCV RNA levels of >10,000 IU/ml and <6,000,000 IU/ml at screening and BMIs of between 18.0 and 35.0 kg/m2. Eligible patients did not have cirrhosis, as determined by a FibroTest/FibroSure (Laboratory Corporation of America, Raritan, NJ) score of ≤0.75 and an aspartate aminotransferase (AST)/platelet ratio of ≤2 or a FibroScan (Echosens, Paris, France) result of ≤14.6 kPa. Patients were required to be naive to any treatment with an approved or investigational drug, including DAAs, IFN-based treatments, and vaccines. Women of childbearing potential were required to have a negative serum pregnancy test at screening and were required to take contraceptive measures (including at least one barrier method, along with either a second barrier method, hormone-based contraceptive, or an intrauterine device) up to 4 weeks after the end of treatment. No clinically significant drug interactions have been observed or are expected when contraceptives are used with simeprevir, ledipasvir, or sofosbuvir (5, 7, 11).
Exclusion criteria included infection with HCV genotypes other than genotype 1, infection with HIV type 1 or 2, any evidence of liver disease of non-HCV etiology, history of malignancy within 5 years before screening, or significant comorbidity. Patients were excluded if they had any known allergies, hypersensitivity, or intolerance to any of the study drugs. Patients with significant laboratory abnormalities such as total serum bilirubin of >1.5× the upper limit of normal, albumin at <30 g/liter, platelet count at <90 × 103/μl, hemoglobin at <11 g/dl (male patients) or <10 g/dl (female patients), absolute neutrophil count at <1.50 × 103/μl or <1.00 × 103/μl (black patients), alanine aminotransferase (ALT) and/or AST at >10× the upper limit of normal, estimated glomerular filtration rate at <30 ml/min/1.73 m2, or end-stage renal disease requiring hemodialysis were also excluded from the study.
Concomitant use of immunomodulators such as cyclosporine, systemic corticosteroids, amiodarone, moderate or strong CYP3A or P-gp inducers (such as rifampin, ritonavir, and grapefruit juice), and moderate or strong CYP3A inhibitors (such as ketoconazole and erythromycin) and the use of rosuvastatin were prohibited.
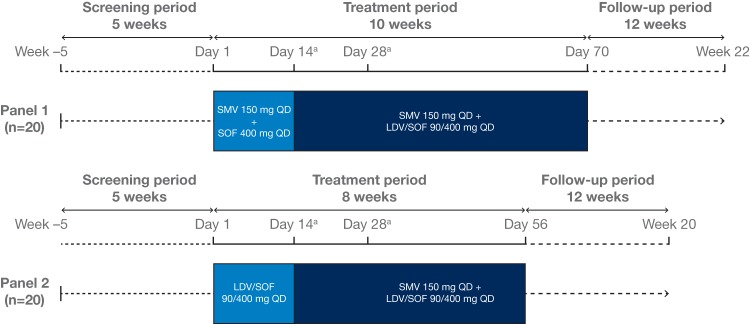
Patients were randomly assigned to 1 of 2 panels (Fig. 3). The formulations and doses used in this study are those approved for each drug: a 150-mg QD simeprevir capsule, a 400-mg QD sofosbuvir tablet, or a fixed-dose combination tablet of 400 mg of sofosbuvir with 90 mg of ledipasvir administered QD (Harvoni). Patients assigned to panel 1 started with a combination of simeprevir and sofosbuvir from day 1 to day 14. After 2 weeks, sofosbuvir was replaced by treatment with ledipasvir-sofosbuvir for a further 8 weeks. Patients assigned to panel 2 started with a combination of ledipasvir-sofosbuvir. After 2 weeks, ledipasvir-sofosbuvir was combined with simeprevir for a further 6 weeks.
FIG 3.
Study design. aPK sampling occurred on day 14 and day 28. LDV, ledipasvir; PK, pharmacokinetic; QD, once daily; SMV, simeprevir; SOF, sofosbuvir.
All study drugs had to be taken orally, once daily in the morning with food (i.e., during or within 15 min of meal consumption) at approximately the same time every day. On days of rich serial PK sampling (day 14 and day 28), patients should have fasted for at least 10 h prior to dosing, and the study drugs were to be taken on-site within 15 min after completion of a standardized breakfast consisting of (or the equivalent of) 4 slices of bread, 2 slices of ham and/or cheese, butter, jelly, and 2 cups (up to 480 ml) of decaffeinated coffee or decaffeinated tea with milk and/or sugar, if desired (containing approximately 21 g of fat, 67 g of carbohydrates, 19 g of proteins, and 533 cal). The standardized breakfast should have been eaten completely within 30 min.
Patients were instructed to continue intake of all study drugs received in order to complete the indicated treatment duration, and they were provided a medication diary to record the dates and times of intake of all study drugs. On days of rich serial PK blood sampling, the patients were observed overnight, and the following times were recorded: the actual dates and times of study drug intake on the day of the visit and the previous and next day, the start and end times of the accompanying meal, and the dates and times of PK blood sampling. It was also documented whether patients took their drugs in a fed state on the day prior to the visit.
Patients received treatment during the summer months and, in line with label recommendations, were advised to use sun-protective measures (such as wearing a hat, sunglasses, protective clothing, or sunscreen), limit exposure to natural sunlight, and avoid artificial sunlight during treatment with simeprevir (13).
Sample size calculation.
Using an estimated intrapatient variability (root mean squared error) of 0.72 on the log (lne) scale for the exposure PK parameters of simeprevir, a sample size of 18 patients who had completed treatment per panel was considered sufficient for the point estimate of the ratio of the geometric mean of simeprevir PK parameters after combined administration of simeprevir with ledipasvir and sofosbuvir, versus simeprevir administered only with sofosbuvir, to fall within 75% and 133% of the true value with 90% confidence (panel 1). This CI is wider than the interval commonly used in studies (80 to 125%) because the relatively high variability of simeprevir would require too large a sample size, which would be difficult to achieve in a patient trial. In addition, both simeprevir and ledipasvir have a large therapeutic index, and such changes in plasma concentrations are not anticipated to be associated with an impact on efficacy or safety. The same intrapatient variability was assumed for ledipasvir-sofosbuvir and yielded the same precision in 90% CI for the ratio of ledipasvir with simeprevir and sofosbuvir versus ledipasvir and sofosbuvir administered alone (panel 2).
The expected SVR12 rates ranged from 90 to 95%; the corresponding 95%, 2-sided exact CIs for each panel (20 patients per panel) or potential pooling of panels are presented in Table S1.
In addition, with a total sample size of 40 patients, the probability of observing an AE with an incidence of 1% is 33%, and the probabilities of observing AEs with incidences of 0.1%, 0.5%, and 0.8% are 4%, 18%, and 27%, respectively.
Procedures.
Blood samples for determination of HCV RNA levels were collected at screening, baseline (day 1), days 7, 15, 28, 42, 56, and 70 (panel 1 only), and weeks 4 and 12 of follow-up.
The baseline and day 15 samples were taken predose. HCV RNA was measured using the Roche COBAS AmpliPrep/COBAS TaqMan HCV Quantitative Test v2.0 (Roche Diagnostics, Pleasanton, CA), with a lower limit of quantification of 15 IU/ml.
Population sequencing of the HCV NS3/4A, NS5A, and NS5B gene regions was performed pretreatment in all patients, to identify preexisting sequence polymorphisms, and postbaseline in patients not achieving SVR, to characterize emerging viral variants. A sample for IL28B genotyping was collected at baseline.
Blood samples for PK sampling were taken predose and at 0.5, 1, 2, 4, 6, 8, 10, 12, 18, and 24 h after study drug intake on days 14 and 28. Full PK profiles were determined over 24 h for simeprevir (panel 1) or ledipasvir (panel 2) on day 14 and for both simeprevir and ledipasvir on day 28. On days 14 and 28, PK samples for sofosbuvir were collected in both panels and stored for analysis in case of a need to explain unexpected outcomes in the trial. The samples were not analyzed.
Safety and tolerability were evaluated throughout the study until the last study-related visit in a consistent manner and in accordance with the World Health Organization Toxicity Grading Scale for Determining the Severity of Adverse Events. AEs were monitored throughout the study until posttreatment follow-up week 4. From week 4 of follow-up onwards, reporting was limited to SAEs. Blood samples for serum chemistry, hematology (including coagulation), and urinalysis were collected at screening, at days 1, 7, 15, 28, 42, 56, and 70 (panel 1 only), and at weeks 4 and 12 of follow-up. Graded treatment-emergent laboratory abnormalities for amylase, neutrophils and precursors, ALT, AST, and bilirubin were recorded. Physical examination and vital-sign evaluation were conducted at screening, days 1, 15, 28, 42, 56, and 70 (panel 1 only), and weeks 4 and 12 of follow-up. Electrocardiograms were performed at screening and at further time points at the investigators' discretion.
Outcomes.
The primary endpoints of this study were the steady-state PK parameters (Cmin, Cmax, and AUCτ) of simeprevir before (day 14) and after (day 28) the addition of ledipasvir to the treatment regimen (panel 1) and of ledipasvir before (day 14) and after (day 28) the addition of simeprevir (panel 2).
The secondary endpoints included the safety and tolerability of simeprevir plus ledipasvir-sofosbuvir, on-treatment and posttreatment virologic responses, and changes from baseline in the HCV NS3/4A, NS5A, and NS5B sequences in patients not achieving SVR.
Bioanalysis.
The plasma concentrations of simeprevir were measured using validated liquid chromatography methods with tandem mass spectrometric detection (LC-MS/MS; range: 2.00 to 2,000 ng/ml), as previously described (27). A 100-μl aliquot was fortified with TMC435-d3 internal standard working solution. Sample preparation was done via protein precipitation. The supernatant was analyzed via LC-MS/MS detection using positive turbo-ion spray. Ledipasvir samples were measured at a commercial laboratory (PPD, Middleton, WI) using LC-MS/MS (range: 0.500 to 200 ng/ml). A 100-μl matrix aliquot was fortified with [13C2, 2H6]ledipasvir internal standard working solution. Analytes were isolated through liquid/liquid extraction. The eluate was evaporated under a nitrogen stream and the remaining residue was reconstituted. The final extract was analyzed via LC-MS/MS detection using positive ion electrospray. None of the plasma concentrations of simeprevir or ledipasvir were below the lower limit of quantification.
Pharmacokinetic analysis.
PK parameters were measured using the validated computer program Phoenix WinNonlin (version 6.2.1; Tripos L.P., St. Louis, MO). Noncompartmental analysis model 200 (extravascular input, plasma data) was applied for the PK analysis. Furthermore, Microsoft Excel (version 2007; Microsoft, Redmond, WA) and SAS (version 9.3; SAS Institute Inc., Cary, NC) were used. PK parameters included Cmax, predose plasma concentration (C0), average steady-state plasma concentration (Css,avg), time to Cmax (Tmax), and AUCτ. AUCs were calculated by linear-linear trapezoidal summation.
Statistical analysis.
All analyses were performed on patients from the ITT population (all patients who received at least one dose of study drug). The PK analysis population (where these samples were evaluable) was used for the PK parameters as indicated in the PK tables.
For the primary endpoint, descriptive statistics were calculated for the plasma concentrations of simeprevir and ledipasvir at each sampling time point and for the derived PK parameters (Cmin, Cmax, and AUCτ). Plasma concentration-time graphs were also produced.
Statistical analyses were performed for simeprevir in panel 1 and ledipasvir in panel 2 using day 28 as the test and day 14 as the reference. The primary PK parameters were Cmin, Cmax, and AUCτ on the log scale. All observations for test and reference, paired and unpaired, were included in the statistical analysis. The LS means of the log-transformed primary parameters of simeprevir and ledipasvir of the test and reference were estimated with a linear mixed-effects model, controlling for treatment as a fixed effect and subject as a random effect. A 90% CI was constructed around the difference between the LS means of the test and reference. Both the difference between the LS means and the 90% CIs were retransformed to the original scale and are reported.
Secondary efficacy and safety endpoints were analyzed descriptively. AEs were coded according to Medical Dictionary for Regulatory Activities (MedDRA) preferred terms (https://www.meddra.org/how-to-use/basics/hierarchy).
Data are presented for both the primary and final analyses. The primary PK analysis was conducted when PK sampling results at both day 14 and day 28 were available for all patients. The final analysis was performed to assess key secondary endpoints when all patients had completed 12 weeks of follow-up after the end of treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank all patients and their families for participating in this study, and we thank all study investigators. Medical writing assistance was provided by Kimberley Haines of Complete Medical Communications, funded by Janssen.
This study was funded by Janssen. Janssen was responsible for the study design and the collection and interpretation of data. Medical writing support was funded by Janssen.
S.B., Y.H., F.N., H.V.V., and C.M. were involved in the acquisition and interpretation of study data and in the critical revision of the manuscript for important intellectual content. M.B., L.V., V.V.E., V.H., P.V.R., J.V.D.L., and S.O.-M. were involved in the study concept and design and in the acquisition, analysis, and interpretation of study data. They were also involved in the critical revision of the manuscript for important intellectual content. D.L. provided statistical analysis of the study data and was involved in the critical revision of the manuscript for important intellectual content.
Stefan Bourgeois has acted as a consultant for AbbVie, Bristol-Myers Squibb, Gilead Sciences, Janssen, and MSD and has given sponsored lectures for Gilead Sciences and Janssen. Yves Horsmans has acted as a consultant for AbbVie, Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline, Janssen, MSD, and Novartis. Frederik Nevens has received grant support from AbbVie, Astelas, Ferring, Janssen-Cilag, Novartis, and Roche and has acted as a consultant for AbbVie, Bristol-Myers Squibb, CAF, Durect, Gore, Intercept, Janssen-Cilag, MSD, Novartis, Ono Pharma, and Promethera Biosciences. Hans van Vlierberghe has received grant support from AbbVie, Astellas, Ferring, Gilead Sciences, Johnson & Johnson, and Novartis, has acted as a consultant for AbbVie, Bristol-Myers Squibb, Gilead Sciences, and Johnson & Johnson, and has given sponsored lectures for Gilead Sciences. Christophe Moreno has received grant support from AbbVie, Astellas, Gilead, Janssen, MSD, and Roche and has acted as a consultant for AbbVie, Bristol-Myers Squibb, Gilead, Janssen, and MSD. Maria Beumont, Leen Vijgen, Donghan Luo, and Sivi Ouwerkerk-Mahadevan are employees of Janssen and are Johnson & Johnson stockholders. Veerle van Eygen, Vera Hillewaert, Pieter Van Remoortere, and Jolanda van de Logt are employees of Janssen.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01217-17.
REFERENCES
- 1.The Polaris Observatory HCV Collaborators. 2017. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2:161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 3.Westbrook RH, Dusheiko G. 2014. Natural history of hepatitis C. J Hepatol 61:S58–S68. doi: 10.1016/j.jhep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. 2017. Hepatitis C—fact sheet 164. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs164/en/. [Google Scholar]
- 5.Gilead. 2017. Highlights of prescribing information, Harvoni. http://www.gilead.com/~/media/Files/pdfs/medicines/liver-disease/harvoni/harvoni_pi.pdf.
- 6.Gane E, Stedman C, McClure M, Apelian D, Westland C, Vuong J, Patel M, Kakuda T, Chanda S, Blatt L, Beigelman L, Smith D, Fry J. 2017. Short duration treatment with AL-335 and odalasvir, with or without simeprevir, in treatment naïve patients with hepatitis C infection with or without cirrhosis. J Hepatol 66(1):S82. doi: 10.1016/S0168-8278(17)30429-4. [DOI] [Google Scholar]
- 7.Ouwerkerk-Mahadevan S, Snoeys J, Peeters M, Beumont-Mauviel M, Simion A. 2016. Drug-drug interactions with the NS3/4A protease inhibitor simeprevir. Clin Pharmacokinet 55:197–208. doi: 10.1007/s40262-015-0314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talavera Pons S, Boyer A, Lamblin G, Chennell P, Chatenet FT, Nicolas C, Sautou V, Abergel A. 2017. Managing drug-drug interactions with new direct-acting antiviral agents in chronic hepatitis C. Br J Clin Pharmacol 83:269–293. doi: 10.1111/bcp.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.German P, Mathias A, Brainard D, Kearney BP. 2016. Clinical pharmacokinetics and pharmacodynamics of ledipasvir/sofosbuvir, a fixed-dose combination tablet for the treatment of hepatitis C. Clin Pharmacokinet 55:1337–1351. doi: 10.1007/s40262-016-0397-0. [DOI] [PubMed] [Google Scholar]
- 10.Lawitz E, Sulkowski MS, Ghalib R, Rodriguez-Torres M, Younossi ZM, Corregidor A, DeJesus E, Pearlman B, Rabinovitz M, Gitlin N, Lim JK, Pockros PJ, Scott JD, Fevery B, Lambrecht T, Ouwerkerk-Mahadevan S, Callewaert K, Symonds WT, Picchio G, Lindsay KL, Beumont M, Jacobson IM. 2014. Simeprevir plus sofosbuvir, with or without ribavirin, to treat chronic infection with hepatitis C virus genotype 1 in non-responders to pegylated interferon and ribavirin and treatment-naive patients: the COSMOS randomised study. Lancet 384:1756–1765. doi: 10.1016/S0140-6736(14)61036-9. [DOI] [PubMed] [Google Scholar]
- 11.Gilead. 2017. SOVALDI highlights of prescribing information. http://www.gilead.com/~/media/Files/pdfs/medicines/liver-disease/sovaldi/sovaldi_pi.pdf.
- 12.Snoeys J, Beumont M, Monshouwer M, Ouwerkerk-Mahadevan S. 2016. A mechanistic understanding of the non-linear pharmacokinetics and inter-subject variability of simeprevir: a PBPK-guided drug development approach. Clin Pharmacol Ther 99:224–234. doi: 10.1002/cpt.206. [DOI] [PubMed] [Google Scholar]
- 13.Janssen Research & Development. 2017. Olysio™ (simeprevir) US Prescribing Information. Janssen Therapeutics, Titusville, NJ: http://www.olysio.com/shared/product/olysio/prescribing-information.pdf. [Google Scholar]
- 14.Reesink HW, Fanning GC, Farha KA, Weegink C, Van Vliet A, Van't Klooster G, Lenz O, Aharchi F, Mariën K, Van Remoortere P, de Kock H, Broeckaert F, Meyvisch P, Van Beirendonck E, Simmen K, Verloes R. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 15.Morcos PN, Chang L, Kulkarni R, Giraudon M, Shulman N, Brennan BJ, Smith PF, Tran JQ. 2013. A randomised study of the effect of danoprevir/ritonavir or ritonavir on substrates of cytochrome P450 (CYP) 3A and 2C9 in chronic hepatitis C patients using a drug cocktail. Eur J Clin Pharmacol 69:1939–1949. doi: 10.1007/s00228-013-1556-y. [DOI] [PubMed] [Google Scholar]
- 16.Mathias AA, German P, Murray BP, Wei L, Jain A, West S, Warren D, Hui J, Kearney BP. 2010. Pharmacokinetics and pharmacodynamics of GS-9350: a novel pharmacokinetic enhancer without anti-HIV activity. Clin Pharmacol Ther 87:322–329. doi: 10.1038/clpt.2009.228. [DOI] [PubMed] [Google Scholar]
- 17.Greenblatt DJ, Peters DE, Oleson LE, Harmatz JS, MacNab MW, Berkowitz N, Zinny MA, Court MH. 2009. Inhibition of oral midazolam clearance by boosting doses of ritonavir, and by 4,4-dimethyl-benziso-(2H)-selenazine (ALT-2074), an experimental catalytic mimic of glutathione oxidase. Br J Clin Pharmacol 68:920–927. doi: 10.1111/j.1365-2125.2009.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smolders EJ, de Kanter CT, van Hoek B, Arends JE, Drenth JP, Burger DM. 2016. Pharmacokinetics, efficacy, and safety of hepatitis C virus drugs in patients with liver and/or renal impairment. Drug Saf 39:589–611. doi: 10.1007/s40264-016-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodges LM, Markova SM, Chinn LW, Gow JM, Kroetz DL, Klein TE, Altman RB. 2011. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharmacogenet Genomics 21:152–161. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghanta V, Strauss M, Collazo-Maldonado R. 2016. Life threatening rhabdomyolysis from the combination of ledipasvir/sofosbuvir and high dose statins. Am J Kidney Dis 67:A48. doi: 10.1053/j.ajkd.2016.03.124. [DOI] [Google Scholar]
- 21.Hill L. 2015. Hepatitis C virus direct-acting antiviral drug interactions and use in renal and hepatic impairment. Top Antivir Med 23:92–96. [PMC free article] [PubMed] [Google Scholar]
- 22.Wei L, Han T, Yang D, Heo J, Shang J, Cheng J, Chen X, Xie Q, Kim JH, Kalmeijer R, Ouwerkerk-Mahadevan S, Hoeben E, Lenz O, Verbinnen T, Sinha R, Li M, Scott J, Peeters M, Witek J. 2016. Simeprevir plus peginterferon/ribavirin for HCV genotype 1-infected treatment-naive patients in China and South Korea. J Gastroenterol Hepatol 31:912–920. doi: 10.1111/jgh.13288. [DOI] [PubMed] [Google Scholar]
- 23.Snoeys J, Beumont M, Monshouwer M, Ouwerkerk-Mahadevan S. 2017. Elucidating the plasma and liver pharmacokinetics of simeprevir in special populations using physiologically based pharmacokinetic modelling. Clin Pharmacokinet 56:781–792. doi: 10.1007/s40262-016-0476-2. [DOI] [PubMed] [Google Scholar]
- 24.Kwo P, Gitlin N, Nahass R, Bernstein D, Etzkorn K, Rotjer S, Schiff E, Davis M, Ruane P, Younes Z, Kalmeijer R, Sinha R, Peeters M, Lenz O, Fevery B, De La Rosa G, Scott J, Witek J. 2016. Simeprevir plus sofosbuvir (12 and 8 weeks) in hepatitis C virus genotype 1-infected patients without cirrhosis: OPTIMIST-1, a phase 3, randomized study. Hepatology 64:370–380. doi: 10.1002/hep.28467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawitz E, Matusow G, DeJesus E, Yoshida EM, Felizarta F, Ghalib R, Godofsky E, Herring RW, Poleynard G, Sheikh AM, Tobias H, Kugelmas M, Kalmeijer R, Peeters M, Lenz O, Fevery B, De La Rosa G, Scott J, Sinha R, Witek J. 2016. Simeprevir plus sofosbuvir in patients with chronic hepatitis C virus genotype 1 infection and cirrhosis: a phase 3 study (OPTIMIST-2). Hepatology 64:360–369. doi: 10.1002/hep.28422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M, Rustgi V, Chojkier M, Herring R, Di Bisceglie AM, Pockros PJ, Subramanian GM, An D, Svarovskaia E, Hyland RH, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Pound D, Fried MW. 2014. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 370:1879–1888. doi: 10.1056/NEJMoa1402355. [DOI] [PubMed] [Google Scholar]
- 27.Vanwelkenhuysen I, de Vries R, Timmerman P, Verhaeghe T. 2014. Determination of simeprevir: a novel, hepatitis C protease inhibitor in human plasma by high-performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 958:43–47. doi: 10.1016/j.jchromb.2014.02.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.