ABSTRACT
We previously optimized imipenem and tobramycin combination regimens against a double-resistant clinical Pseudomonas aeruginosa isolate by using in vitro infection models, mechanism-based pharmacokinetic/pharmacodynamic modeling (MBM), and Monte Carlo simulations. The current study aimed to evaluate these regimens in a neutropenic murine thigh infection model and to characterize the time course of bacterial killing and regrowth via MBM. We studied monotherapies and combinations of imipenem with tobramycin in vivo against the double-resistant clinical P. aeruginosa isolate by using humanized dosing schemes. Viable count profiles of total and resistant populations were quantified over 24 h. Tobramycin monotherapy (7 mg/kg every 24 h [q24h] as a 0.5-h infusion) was ineffective. Imipenem monotherapies (continuous infusion of 4 or 5 g/day with a 1-g loading dose) yielded 2.47 or 2.57 log10 CFU/thigh killing at 6 h. At 24 h, imipenem at 4 g/day led to regrowth up to the initial inoculum (4.79 ± 0.26 log10 CFU/thigh), whereas imipenem at 5 g/day displayed 1.75 log10 killing versus the initial inoculum. The combinations (i.e., imipenem at 4 or 5 g/day plus tobramycin) provided a clear benefit, with bacterial killing of ≥2.51 or ≥1.50 log10 CFU/thigh compared to the respective most active monotherapy at 24 h. No colonies were detected on 3×MIC agar plates for combinations, whereas increased resistance (at 3×MIC) emerged for monotherapies (except imipenem at 5 g/day). MBM suggested that tobramycin considerably enhanced the imipenem target site concentration up to 2.6-fold. The combination regimens, rationally optimized via a translational modeling approach, demonstrated substantially enhanced bacterial killing and suppression of regrowth in vivo against a double-resistant isolate and are therefore promising for future clinical evaluation.
KEYWORDS: imipenem, tobramycin, neutropenic thigh infection model, mathematical modeling, population pharmacokinetics and pharmacodynamics
INTRODUCTION
The global spread of multidrug-resistant Pseudomonas aeruginosa isolates, along with the drastic decline in new antibiotics, is causing a serious threat to human health (1–4). Over the past decade, carbapenem resistance has been increasing dramatically and resistant isolates have been found in hospitals worldwide (5, 6). Resistance to aminoglycosides is present in virtually all parts of the world and thus limiting the therapeutic potential of aminoglycoside monotherapies for pseudomonal infections (7). The high prevalence of P. aeruginosa isolates that are resistant to carbapenems and aminoglycosides poses significant challenges for health care professionals, particularly when selecting antibiotic therapies against severe infections (7, 8). This situation is particularly worrying, given the shortage of new antibiotics and the rapid emergence of resistance to all available antibiotics in monotherapies in serious infections (9, 10). Several studies have suggested substantial benefits of β-lactam and aminoglycoside combinations over monotherapies in patients with serious infections (11–14). Therefore, such combinations are considered a tangible option to combat this clinical challenge.
We have previously rationally optimized imipenem and tobramycin combination dosage regimens against a carbapenem- and aminoglycoside-resistant clinical P. aeruginosa isolate via in vitro infection models, mechanism-based modeling, and Monte Carlo simulations (15). The simulations were performed by using the mechanism-based model developed to characterize the time course of bacterial killing and resistance and human population pharmacokinetic models for imipenem and tobramycin (15–17). Our optimized combination regimens (i.e., tobramycin at 7 mg/kg every 24 h [q24h] as a 0.5-h infusion in combination with continuous infusion of imipenem at 4 or 5 g/day with a 1-g loading dose) were predicted to achieve ≥2 log10 CFU/ml of bacterial killing without regrowth in 80.9 or 90.3%, respectively, of simulated patients at 24 and 48 h (15).
Studies on combinations of a β-lactam plus an aminoglycoside in murine infection models (18–20) have demonstrated beneficial effects against susceptible isolates. To the best of our knowledge, there have been no reports of studies that have optimized combination regimens via modeling and subsequently prospectively evaluated them in vivo against a carbapenem- and aminoglycoside-resistant clinical P. aeruginosa isolate.
Therefore, we primarily aimed to prospectively evaluate the impact of the proposed optimized imipenem-plus-tobramycin dosage regimens from earlier in vitro studies (13) in a neutropenic murine thigh infection model with a double-resistant P. aeruginosa isolate using humanized dosing. Furthermore, we aimed to quantify the extent and time course of bacterial load reduction and regrowth in vivo via a mechanism-based model for the humanized regimens of imipenem and tobramycin.
(Part of this work has been presented as a poster at the AAPS Annual Meeting and Exposition, 13 to 17 November 2016, Denver, CO, USA. This work was also subject to oral presentations at the Australian Society of Clinical and Experimental Pharmacologists and Toxicologists [ASCEPT], 28 to 30 November 2016, Melbourne, Australia, and the Annual Meeting of the Population Approach Group of Australia and New Zealand [PAGANZ], 6 to 8 February 2017, Adelaide, Australia.)
RESULTS
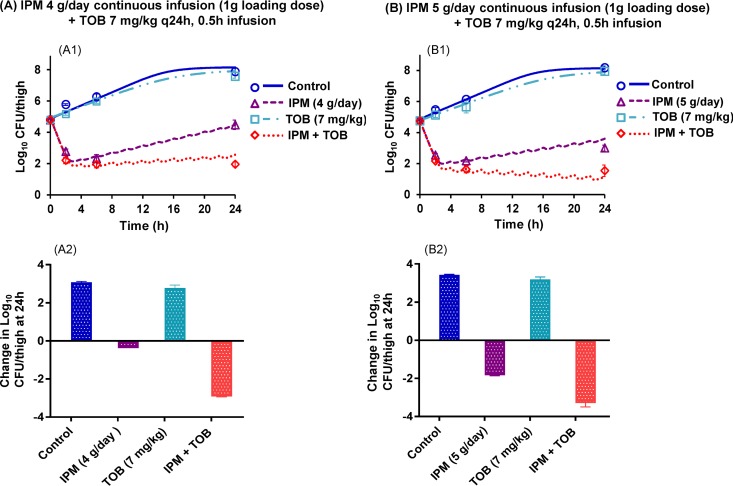
While the bacterial density was ∼105 CFU/thigh at initiation of antibiotic treatment, mice in the control and tobramycin monotherapy groups had growth of 3.1 to 3.4 log10 CFU/thigh at 24 h (Fig. 1). The choice of the initial inoculum was driven by ethical considerations and a desire to maximize the yield of information from the modeling by ensuring that the survival of animals across all groups was not compromised. Tobramycin monotherapy was completely ineffective; however, imipenem monotherapies provided ∼2.5 log10 of bacterial load reduction at 6 h (Fig. 1A1 and B1). Imipenem-plus-tobramycin combinations (i.e., imipenem at 4 or 5 g/day as a continuous infusion plus tobramycin at 7 mg/kg q24h as a 0.5-h infusion) provided a clear benefit, with ≥2.51 log10 and ≥1.50 log10 CFU/thigh of bacterial load reduction at 24 h compared to the respective most active monotherapy (imipenem) (Fig. 1A and B). The data presented as the change in log10 CFU/thigh at 24 h relative to the bacterial load at 0 h demonstrated that the combinations of imipenem at 4 and 5 g/day plus tobramycin were the most effective treatment regimens. (Fig. 1A2 and B2).
FIG 1.
Observed (markers) and population-fitted (lines) viable counts for optimized imipenem (IPM)-plus-tobramycin (TOB) dosage regimens in a mouse thigh infection model with FADDI-PA088 (A1 and B1). The changes in log10 CFU/thigh at 24 h relative to the start of treatment are also presented for all regimens (A2 and B2). Each symbol and bar represents the mean ± the standard deviation of four thighs from two mice.
FIG 2.
The 24-h viable counts of total (on drug-free plates) and resistant (on imipenem [IPM]- and tobramycin [TOB]-containing [3×MIC] agar plates) subpopulations for each treatment regimen. Each bar represents the mean ± the standard deviation of four thighs from two mice. Panel D contains two bars for tobramycin, reflecting experiments 1 and 2.
Viable counts of resistant bacteria quantified on antibiotic-containing agar plates (3×MIC) and total populations of each treatment on drug-free plates at 24 h are shown in Fig. 2. All monotherapies (except the humanized imipenem 5-g/day regimen) resulted in resistant subpopulations (Fig. 2B to D), whereas no colonies (i.e., resistant subpopulations) were detected with the combination regimens (Fig. 2E and F).
MBM.
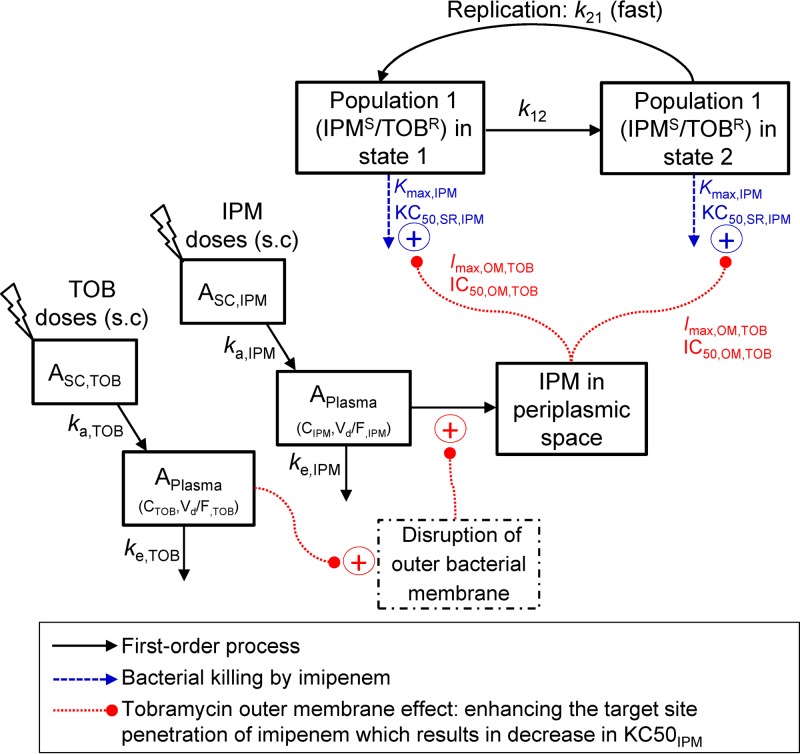
Our mechanism-based pharmacokinetic/pharmacodynamic (PK/PD) modeling (MBM) contained two preexisting bacterial populations with different susceptibilities to imipenem and tobramycin (Fig. 3). This MBM simultaneously described the effects of imipenem plus tobramycin and yielded unbiased and precise curve fits for all monotherapies and combinations (Fig. 1). The coefficient of correlation for the observed versus population-fitted log10 viable counts was ≥0.98.
FIG 3.
The structure of the mechanism-based model of bacterial growth and killing by imipenem (IPM) and tobramycin (TOB) in monotherapies and optimized combination regimens. The first population, i.e., IPMs/TOBr, was susceptible to imipenem and resistant to tobramycin. The second population, i.e., IPMi/TOBr (imipenem intermediate and tobramycin resistant), is not shown. A life cycle growth model (63, 64) was utilized to describe the underlying biology of bacterial replication via two states for each of the two populations. The maximum killing rate constants (Kmax) and the associated antibiotic concentrations (KC50s) causing 50% of Kmax are explained in Table 1. The permeabilizing effect of tobramycin on the bacterial outer membrane (i.e., tobramycin enhancing the target site penetration of imipenem) was applied to both populations. The parameters describing the outer membrane effect (Imax,OM,TOB and IC50,OM,TOB) are explained in Table 1.
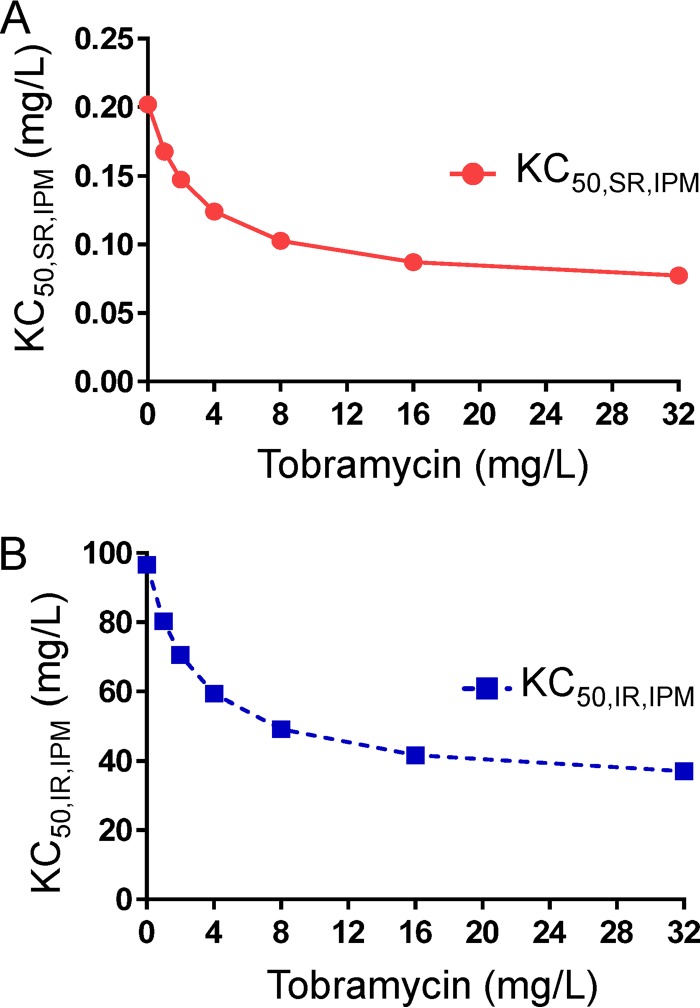
Subpopulation synergy was not sufficient to characterize the time course of bacterial load reduction and regrowth with combination dosage regimens. Mechanistic synergy due to tobramycin enhancing the target site concentration of imipenem (i.e., via the permeabilizing effect of tobramycin on the bacterial outer membrane) was essential to describe the time course of viable count profiles (Fig. 3). The inclusion of mechanistic synergy for both populations significantly enhanced the model's performance. Mechanistic synergy was expressed as a decrease in the imipenem concentration causing 50% killing (KC50,IPM) of both bacterial populations with increasing tobramycin concentrations. The estimated KC50,IPM values for both populations were plotted for a range of tobramycin concentrations, up to the maximal clinically achievable plasma unbound drug concentration following tobramycin administration at 7 mg/kg (q24h, 0.5-h infusion) in humans (Fig. 4). The KC50,IPM for both populations decreased by 2.6-fold in the presence of 32 mg/liter tobramycin compared to the KC50,IPM in the absence of tobramycin (Fig. 4). A decrease in KC50,IPM in the MBM is equivalent to an increase in the target site imipenem concentration. The model-estimated tobramycin concentration required for half-maximal permeabilization of the outer membrane (IC50,OM,TOB) was 2.99 mg/liter (Table 1). All parameters were required to provide unbiased and precise curve fits.
FIG 4.
The outer membrane effect of tobramycin (i.e., tobramycin enhancing the target site penetration of imipenem) resulted in a decrease in the imipenem concentration required for half-maximal bacterial killing (KC50,IPM) of both bacterial populations. The decrease in the KC50,IPM for both populations (SR in panel A and IR in panel B) was estimated up to the maximal clinically achievable plasma unbound tobramycin concentrations following a dose of 7 mg/kg q24h administered as a 0.5-h infusion. Parameters are explained in Table 1.
TABLE 1.
Population mean parameter estimates for the imipenem-plus-tobramycin combination model against FADDI-PA088
| Parameter (unit) | Abbreviation | Population mean value (SE [%]) |
|---|---|---|
| Initial inoculum (log10 CFU/thigh) | LogCFU0 | 4.93 (2.08)a |
| 4.78 (2.01)b | ||
| Max population size (log10 CFU/thigh) | LogCFUmax | 8.00 (1.34) |
| Replication rate constant (h−1) | k21 | 50 (fixed) |
| Mean generation time (min) | ||
| IPMs/TOBr | k12,SR−1 | 142 (9.21) |
| IPMi/TOBr | k12,IR−1 | 142 (9.21) |
| Log10 mutation frequency of IPM | LogMUT,IPM | −3.09 (4.94) |
| Max killing rate constant of imipenem (h−1) | Kmax,IPM | 3.38 (8.65) |
| Imipenem concn causing 50% of Kmax,IPM (mg/liter) | ||
| IPMs/TOBr | KC50,SR,IPM | 0.202 (35.4) |
| IPMi/TOBr | KC50,IR,IPM | 96.6 (16.1) |
| Permeabilization of outer membrane by tobramycin | ||
| Max fractional decrease of KC50,IPM by tobramycin via outer membrane disruption | Imax,OM,TOB | 0.647 (0.44–0.816)c |
| Tobramycin concn causing 50% of Imax,OM,TOB (mg/liter) | IC50,OM,TOB | 2.99 (38.3) |
| Hill coefficient for imipenem | HillIPM | 2.05 (16.1) |
| SD of residual error on log10 scale | SDCFU | 0.114 (24.9) |
Experiment 1, imipenem at 4 g/day with a 1-g loading dose plus tobramycin at 7 mg/kg.
Experiment 2, imipenem at 5 g/day with a 1-g loading dose plus tobramycin at 7 mg/kg.
95% confidence interval.
DISCUSSION
This is the first study that presents a prospective evaluation of PK/PD model-based optimized imipenem-plus-tobramycin combination dosage regimens in a neutropenic murine thigh infection model against a carbapenem- and aminoglycoside-resistant clinical P. aeruginosa isolate. Furthermore, the extent and time course of bacterial load reduction and regrowth were characterized by an MBM for humanized regimens of imipenem and tobramycin.
In this study, we found that tobramycin monotherapy was ineffective, while imipenem monotherapy (4 or 5 g/day) significantly decreased bacterial counts during murine thigh infections compared with those observed in untreated control mice. Monotherapy results were in agreement with the time for which the unbound (free) imipenem concentration remained above the MIC (fT>MIC; i.e., 38 and 43%) for imipenem regimens (21) and the area under the unbound tobramycin concentration-time curve over 24 h divided by the MIC (fAUC/MIC; i.e., 3.1) (22). Combinations demonstrated substantially enhanced killing compared to the monotherapies. No colonies were detected on 3×MIC agar plates for combination regimens as opposed to monotherapies, except for imipenem 5-g/day monotherapy. Therefore, considered globally, our optimized combination dosage regimens were clearly beneficial in regard to enhancement of bacterial killing and suppression of resistance at 24 h in the murine thigh infection model compared to monotherapies.
We used a robust approach to develop humanized regimens of imipenem and tobramycin in mice based on the human and murine PK of both antibiotics (23, 24). While we did not measure plasma drug concentrations, our humanized doses and dosing frequencies in mice over 24 h were within the range of total doses and frequencies reported in murine studies (18, 25, 26). Furthermore, our total daily doses in mice, expressed in mg/kg, were 10.1- to 10.9-fold higher than the corresponding human doses; this is similar to the factor for mouse-to-human animal scaling for dose equivalence (27, 28).
Our results considerably add to the findings from relatively few published studies on β-lactam-plus-aminoglycoside combinations in murine models of infection with P. aeruginosa. In the past, studies with various in vivo infection models showed that β-lactam-plus-aminoglycoside combinations were synergistic against P. aeruginosa (18–20, 29–34). Studies evaluated the combinations of imipenem plus tobramycin in a murine septicemia model (19) and meropenem plus tobramycin in a murine pneumonia model (18) against double-susceptible P. aeruginosa isolates (imipenem MIC, 0.5 mg/liter; meropenem MIC, 0.5 to 1 mg/liter; tobramycin MIC, 0.5 to 1 mg/liter). Ticarcillin plus tobramycin was synergistic against P. aeruginosa, prevented regrowth in intraperitoneal infections in mice, and was found to be more efficacious than carbenicillin-plus-tobramycin combinations (20). None of these studies prospectively evaluated rationally optimized combination dosage regimens in a murine thigh infection model against a P. aeruginosa isolate that was carbapenem and aminoglycoside resistant. Furthermore, the extent and time course of bacterial killing and regrowth have not been characterized on the basis of an MBM for the humanized regimens of imipenem and tobramycin.
Our MBM indicated that tobramycin enhanced the target site concentration of imipenem, expressed as an up to 2.6-fold decrease in KC50,IPM (Fig. 4). An approximately 2.99-mg/liter concentration of tobramycin was required to half-maximally permeabilize the outer membrane (IC50,OM,TOB) of this isolate and thus achieve the half-maximal extent of mechanistic synergy (Table 1). The mechanistic synergy term in the MBM was informed by studies that demonstrated a disruption of the outer membrane of P. aeruginosa by albumin-conjugated aminoglycosides (35, 36). The outer membrane of P. aeruginosa presents a formidable barrier (37, 38), and its disruption is likely to enhance the target site penetration of imipenem (39, 40). Overall, an MBM incorporating both mechanistic synergy and subpopulation synergy excellently described the time course of bacterial load reduction and regrowth. This MBM had a structure similar to that previously described (15) and thus translated well from in vitro static concentration time-kill (SCTK) experiments to in vivo data. The synergy mechanism proposed by our mechanism-based PK/PD model (i.e., tobramycin permeabilizing the outer bacterial membrane) was confirmed by electron micrographs of ultrastructural damage, loss of cytosolic green fluorescent protein (GFP) from a GFP-expressing strain, and nitrocefin uptake results from our recently published studies (41).
It has been reported that effective and early antibiotic therapy significantly increases survival in an immunocompetent murine infection model of septic shock (42, 43). The target endpoint (i.e., ≥2 log10 killing at 24 and 48 h) described in our Monte Carlo simulation (15) was based on a study by Drusano et al. (44). In the current study, in the neutropenic murine thigh infection model, our rationally optimized combination regimens provided >2.5 log10 killing at 24 h relative to the start of treatment. These regimens, which were optimized on the basis of SCTK, MBM, and Monte Carlo simulations, therefore translated well to the murine model. Thus, our results suggest that the optimized imipenem-plus-tobramycin regimens may provide a beneficial effect in reducing the bacterial load to an extent where the immune system in immunocompetent patients can achieve bacterial clearance and are worthy of future clinical evaluation. Future studies may be directed at evaluating the optimized combination regimen against other isolates.
Imipenem at 4 g/day was predicted to be sufficient for a high percentage of patients, as evidenced by very high success rates in Monte Carlo simulations (15, 40); however, imipenem at 5 g/day may be required to achieve effective and early effects in serious infections, patients with high renal function, or both. Therapeutic drug monitoring may be a valuable strategy to optimize imipenem exposure and guide the therapy of critically ill patients (45–47).
Optimization of dosage regimens using in vitro infection models plus MBM with subsequent in vivo evaluation of the optimized regimens is highly beneficial to minimize the number of animals and in vivo studies required. The current in vivo study, together with our previously published in vitro study, supports the robustness of this translational approach. However, only very few studies have previously utilized in vitro to in vivo translation via modeling; these include studies on a meropenem-and-levofloxacin combination (48) and monotherapy with colistin (49) and meropenem (50).
In summary, the rationally optimized imipenem-plus-tobramycin combination dosage regimens demonstrated substantially enhanced killing in vivo against a double-resistant clinical P. aeruginosa isolate. Mechanism-based PK/PD modeling suggested that the permeabilizing effect of tobramycin contributed to this considerably increased activity. Thus, these imipenem-plus-tobramycin combination regimens, which were optimized via a robust translational modeling approach, are expected to be highly promising for future clinical studies.
MATERIALS AND METHODS
Antibiotics.
Stock solutions of imipenem (Merck Sharp & Dohme Pty. Ltd., NSW, Australia) and tobramycin (AK Scientific, Inc., Union City, CA) were prepared in sterile, distilled water and filter sterilized with a Millex-GV 0.22-μm polyvinylidene difluoride syringe filter (Merck Millipore Ltd., Cork, Ireland).
Bacterial isolate and susceptibility testing.
The imipenem- and tobramycin-resistant P. aeruginosa isolate (FADDI-PA088; MICimipenem, 16 mg/liter; MICtobramycin, 32 mg/liter) was obtained from the collection at Monash University. All susceptibility testing was performed in cation-adjusted Mueller-Hinton II broth (BBL, BD, Sparks, MD). Viable counting was conducted on cation-adjusted Mueller-Hinton II agar (CAMHA; Medium Preparation Unit, The University of Melbourne). European Committee on Antimicrobial Susceptibility Testing breakpoints were used to define carbapenem and aminoglycoside resistance (51).
Humanized dosing scheme.
Our previously optimized regimens for the simulated PK in critically ill patients were continuous infusion of imipenem at 4 or 5 g/day with a 1-g loading dose plus tobramycin at 7 mg/kg q24h as a 0.5-h infusion (15). We performed simulations to identify the imipenem and tobramycin regimens in mice that approximate the plasma unbound drug exposures observed in humans. These simulations used the murine pharmacokinetics of imipenem and tobramycin (23, 24). Humanized regimens of imipenem at 60 and 77 mg/kg given to the mice subcutaneously (s.c.) every 2 h (i.e., total daily doses of 720 and 924 mg/kg) were selected to represent the imipenem 4- and 5-g/day continuous infusions with a 1-g loading dose in humans. The percentage of the dosing interval during which the fT>MIC was 38 and 43% for the imipenem regimens of 60 and 77 mg/kg every 2 h. The humanized regimen of tobramycin (i.e., a total daily dose of 73 mg/kg) was fractionated in decreasing doses injected s.c. every 4 h as 33.3% at 0 h; 16.65% each at 4, 8, and 12 h; and 8.37% at 16 and 20 h to represent the human dose of tobramycin at 7 mg/kg q24h given as a 0.5-h infusion. The fAUC/MIC for the humanized regimen of tobramycin was 3.1 for this isolate. Each of the above imipenem and tobramycin regimens was evaluated alone and in combinations.
Murine thigh infection model.
All animal experiments were approved by the Monash Institute of Pharmaceutical Sciences animal ethics committee (approval number MIPS.2016.01). The animals were maintained in accordance with the criteria of the Australian code of practice for the care and use of animals for scientific purposes. Seven-week-old male, 25- to 30-g Swiss mice were obtained from Monash Animal Services (Clayton, Victoria, Australia). Mice were rendered neutropenic by intraperitoneal administration of 150 mg/kg of cyclophosphamide (Baxter Healthcare Pty. Ltd., New South Wales, Australia) 4 days before infection plus 100 mg/kg 1 day before infection (52, 53). Thigh infections were established by injecting 50 μl of a freshly prepared bacterial suspension at a density of 2 × 106 CFU/ml of FADDI-PA088 (i.e., ∼105 CFU) into each posterior thigh muscle of the mice under isoflurane anesthesia (52, 53). The bacterial inoculum was confirmed by quantitative cultures. Several clinical studies reported similar bacterial densities in blood, soft tissue, wound, and urinary tract infections in patients (54–59). Previously published animal studies have also used similar inocula to simulate bacterial infections in patients (60–62). Two hours after bacterial inoculation (t = 0 h) and before initiation of therapy, two mice were euthanized to determine the baseline quantitative cultures of thigh homogenates. Other animals were treated with humanized dosage regimens of imipenem and tobramycin as described above in monotherapy and in combinations. The s.c. injection site was rotated over the neck and flank for animal welfare. For each dosage regimen and sampling time point, two mice (i.e., four thighs) were studied. Following euthanization, both entire posterior thigh muscles from each mouse were aseptically collected at 2, 6, and 24 h after initiation of the treatments and individually homogenized as described previously (52). An untreated control group (n = 2 mice) was included at each time point. Viable counts were determined by manual plating of 100 μl of an undiluted or appropriately diluted thigh homogenate in saline onto CAMHA plates (15, 40). Aliquots of the undiluted or diluted samples at 24 h were plated on agar plates supplemented with imipenem or tobramycin at 3×MIC to quantify resistant subpopulations. Overall, two separate murine thigh infection model experiments were conducted, experiment 1 mimicking imipenem at 4 g/day with a 1-g loading dose plus tobramycin at 7 mg/kg and experiment 2 mimicking imipenem at 5 g/day with a 1-g loading dose plus tobramycin at 7 mg/kg, both using FADDI-PA088.
Mechanism-based PK/PD modeling of imipenem and tobramycin combination regimens. (i) Pharmacokinetic model.
The plasma drug concentration-time profiles of imipenem (23) and tobramycin (24) following s.c. administration to mice were described by one-compartment models. Differential equations 1 (ASC, amount of antibiotic in the s.c. compartment) and 2 (APlasma, amount of antibiotic in the central compartment) describe the pharmacokinetics of imipenem and tobramycin. The plasma antibiotic concentrations (CIPM and CTOB) were calculated via equation 3.
| (1) |
| (2) |
| (3) |
The absorption rate constant (ka), elimination rate constant (ke), apparent volume of distribution (V/F), and unbound fraction in plasma (fu) of imipenem (23) and tobramycin (24) were incorporated into the pharmacodynamic model to drive the time course of bacterial killing in the murine thigh infection model.
(ii) Pharmacodynamic model.
Models with two and three preexisting bacterial subpopulations with different susceptibilities to imipenem and tobramycin were considered. The number of preexisting subpopulations was selected on the basis of model diagnostic plots, biological plausibility of parameter estimates, and the objective function. Two preexisting subpopulations were essential to describe the time course of bacterial killing and resistance. A life cycle growth model was utilized to define the underlying biology of bacterial replication via a vegetative state (i.e., state 1) and a replicating state (i.e., state 2), as described previously (63–65). The first population (IPMs/TOBr) was susceptible to imipenem and resistant to tobramycin, while the second population (IPMi/TOBr) was imipenem intermediate and aminoglycoside resistant (Fig. 3). Each population was described by the two states (i.e., compartments) of the life cycle growth model. Thus, the full model contained four compartments. The total concentration of all viable bacteria (CFUall) is described by the following equation:
| (4) |
The CFUNNx denotes the concentration of viable bacteria for population NN in state x. The differential equation for state 1 of the first population (CFUSR1) incorporates bacterial killing by imipenem (initial conditions are described below) as follows:
| (5) |
The plateau factor (PLAT) is defined as 1 − [CFUall/(CFUall + CFUmax)], with CFUmax being the maximum population size. The factor 2 represents doubling of bacteria during replication (63). The maximum killing rate constant for imipenem (Kmax,IPM), the imipenem concentration causing 50% of Kmax for the first population (KC50,SR,IPM), and the Hill coefficient for imipenem (HillIPM) affected both states (i.e., states 1 and 2) of the population. The effect of tobramycin on the outer membrane (OM_effect; i.e., the synergy term) is described in equation 7 below. The differential equation for state 2 of the first population (CFUSR2) is as follows:
| (6) |
Similar differential equations were used for states 1 and 2 of the second population (i.e., IPMi/TOBr), but with different parameters for KC50,IPM and k12 (15, 40, 66).
Subpopulation synergy.
We considered subpopulation synergy (i.e., imipenem killing bacteria resistant to tobramycin and vice versa) in the model to describe the time course of bacterial killing and prevention of regrowth as previously reported (15, 40, 66).
Mechanistic synergy.
The presence of mechanistic synergy (i.e., one antibiotic enhancing killing by the other antibiotic of one or multiple bacterial populations) was evaluated as described previously (15, 40, 66). Mechanistic synergy was incorporated by assuming that the permeabilizing effect of tobramycin on the bacterial outer membrane (35, 36, 39) enhanced the target site penetration of imipenem. This permeabilizing effect of tobramycin was implemented in the model via the term OM_effect (15) (parameters are explained in Table 1) as follows:
| (7) |
Initial conditions.
We estimated the total initial inoculum (LogCFU0) and the log10 mutation frequencies for the second population (IR) (Fig. 3). Initial conditions were implemented as described previously (40, 63).
Observation model.
The log10 viable counts were fitted by using an additive residual error model. For observations below 100 CFU/thigh (equivalent to fewer than 10 colonies per plate), a previously developed residual error model was utilized to fit the number of colonies per plate (67).
Estimation.
Simultaneous estimation of all PD parameters was performed by using the importance sampling algorithm (pmethod=4) in the parallelized S-ADAPT software (version 1.57) (68) facilitated by the SADAPT-TRAN tool (69, 70). The between-curves variability of the PD parameters was fixed to a final, small coefficient of variation (67). Competing models were evaluated by the objective function (−1×log-likelihood in S-ADAPT), the biological plausibility of the parameter estimates, standard diagnostic plots, and visual predictive checks (71–74).
ACKNOWLEDGMENTS
This work was partly supported by Australian National Health and Medical Research Council (NHMRC) Project Grants (APP1045105 to J.B.B., R.L.N., and C.B.L.; APP1101553 to C.B.L., J.B.B., and R.L.N.; and APP1062040 to C.B.L. and J.B.B.). R.Y. is thankful to the Australian Government and Monash Graduate Education for providing a Monash Graduate Scholarship and Research Training Program Fees Offset Scholarship. C.B.L. is the recipient of an NHMRC Career Development Fellowship (APP1062509).
We have no conflict of interest to declare.
REFERENCES
- 1.Boucher HW, Talbot GH, Benjamin DK Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D. 2013. 10 × '20 progress–development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Infectious Diseases Society of America (IDSA), Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. 2011. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis 52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luepke KH, Mohr JF III. 2017. The antibiotic pipeline: reviving research and development and speeding drugs to market. Expert Rev Anti Infect Ther 15:425–433. doi: 10.1080/14787210.2017.1308251. [DOI] [PubMed] [Google Scholar]
- 4.Martens E, Demain AL. 2017. The antibiotic resistance crisis, with a focus on the United States. J Antibiot (Tokyo) 70:520–526. doi: 10.1038/ja.2017.30. [DOI] [PubMed] [Google Scholar]
- 5.Lee CS, Doi Y. 2014. Therapy of infections due to carbapenem-resistant Gram-negative pathogens. Infect Chemother 46:149-164. doi: 10.3947/ic.2014.46.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zavascki AP, Bulitta JB, Landersdorfer CB. 2013. Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 11:1333–1353. doi: 10.1586/14787210.2013.845523. [DOI] [PubMed] [Google Scholar]
- 7.Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to Gram-negative bacteria. N Engl J Med 362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milatovic D, Braveny I. 1987. Development of resistance during antibiotic therapy. Eur J Clin Microbiol 6:234–244. doi: 10.1007/BF02017607. [DOI] [PubMed] [Google Scholar]
- 10.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 11.Bodey GP, Elting LS, Rodriguez S. 1991. Bacteremia caused by Enterobacter: 15 years of experience in a cancer hospital. Rev Infect Dis 13:550–558. doi: 10.1093/clinids/13.4.550. [DOI] [PubMed] [Google Scholar]
- 12.De Jongh CA, Joshi JH, Newman KA, Moody MR, Wharton R, Standiford HC, Schimpff SC. 1986. Antibiotic synergism and response in Gram-negative bacteremia in granulocytopenic cancer patients. Am J Med 80:96–100. doi: 10.1016/0002-9343(86)90121-X. [DOI] [PubMed] [Google Scholar]
- 13.Legrand M, Max A, Peigne V, Mariotte E, Canet E, Debrumetz A, Lemiale V, Seguin A, Darmon M, Schlemmer B, Azoulay E. 2012. Survival in neutropenic patients with severe sepsis or septic shock. Crit Care Med 40:43–49. doi: 10.1097/CCM.0b013e31822b50c2. [DOI] [PubMed] [Google Scholar]
- 14.Vardakas KZ, Tansarli GS, Bliziotis IA, Falagas ME. 2013. β-Lactam plus aminoglycoside or fluoroquinolone combination versus β-lactam monotherapy for Pseudomonas aeruginosa infections: a meta-analysis. Int J Antimicrob Agents 41:301–310. doi: 10.1016/j.ijantimicag.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Yadav R, Bulitta JB, Nation RL, Landersdorfer CB. 2017. Optimization of synergistic combination regimens against carbapenem- and aminoglycoside-resistant clinical Pseudomonas aeruginosa isolates via mechanism-based pharmacokinetic/pharmacodynamic modeling. Antimicrob Agents Chemother 61:e01011–16. doi: 10.1128/AAC.01011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conil JM, Georges B, Ruiz S, Rival T, Seguin T, Cougot P, Fourcade O, Pharmd GH, Saivin S. 2011. Tobramycin disposition in ICU patients receiving a once daily regimen: population approach and dosage simulations. Br J Clin Pharmacol 71:61–71. doi: 10.1111/j.1365-2125.2010.03793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakka SG, Glauner AK, Bulitta JB, Kinzig-Schippers M, Pfister W, Drusano GL, Sorgel F. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob Agents Chemother 51:3304–3310. doi: 10.1128/AAC.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie A, Liu W, Fikes S, Brown D, Drusano GL. 2013. Impact of meropenem in combination with tobramycin in a murine model of Pseudomonas aeruginosa pneumonia. Antimicrob Agents Chemother 57:2788–2792. doi: 10.1128/AAC.02624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulrich E, Trautmann M, Krause B, Bauernfeind A, Hahn H. 1989. Comparative efficacy of ciprofloxacin, azlocillin, imipenem/cilastatin and tobramycin in a model of experimental septicemia due to Pseudomonas aeruginosa in neutropenic mice. Infection 17:311–315. doi: 10.1007/BF01650716. [DOI] [PubMed] [Google Scholar]
- 20.Comber KR, Basker MJ, Osborne CD, Sutherland R. 1977. Synergy between ticarcillin and tobramycin against Pseudomonas aeruginosa and Enterobacteriaceae in vitro and in vivo. Antimicrob Agents Chemother 11:956–964. doi: 10.1128/AAC.11.6.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavridou E, Melchers RJ, van Mil AC, Mangin E, Motyl MR, Mouton JW. 2015. Pharmacodynamics of imipenem in combination with β-lactamase inhibitor MK7655 in a murine thigh model. Antimicrob Agents Chemother 59:790–795. doi: 10.1128/AAC.03706-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelman B, Gudmundsson S, Leggett J, Turnidge J, Ebert S, Craig WA. 1988. Correlation of antimicrobial pharmacokinetic parameters with therapeutic efficacy in an animal model. J Infect Dis 158:831–847. doi: 10.1093/infdis/158.4.831. [DOI] [PubMed] [Google Scholar]
- 23.Katsube T, Yamano Y, Yano Y. 2008. Pharmacokinetic-pharmacodynamic modeling and simulation for in vivo bactericidal effect in murine infection model. J Pharm Sci 97:1606–1614. doi: 10.1002/jps.21062. [DOI] [PubMed] [Google Scholar]
- 24.Moffie BG, Hoogeterp JJ, Lim T, Douwes-Idema AE, Mattie H. 1993. Effectiveness of netilmicin and tobramycin against Pseudomonas aeruginosa in vitro and in an experimental tissue infection in mice. J Antimicrob Chemother 31:403–411. doi: 10.1093/jac/31.3.403. [DOI] [PubMed] [Google Scholar]
- 25.Daikos GL, Panagiotakopoulou A, Tzelepi E, Loli A, Tzouvelekis LS, Miriagou V. 2007. Activity of imipenem against VIM-1 metallo-beta-lactamase-producing Klebsiella pneumoniae in the murine thigh infection model. Clin Microbiol Infect 13:202–205. doi: 10.1111/j.1469-0691.2006.01590.x. [DOI] [PubMed] [Google Scholar]
- 26.Fantin B, Leggett J, Ebert S, Craig WA. 1991. Correlation between in vitro and in vivo activity of antimicrobial agents against Gram-negative bacilli in a murine infection model. Antimicrob Agents Chemother 35:1413–1422. doi: 10.1128/AAC.35.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma V, McNeill JH. 2009. To scale or not to scale: the principles of dose extrapolation. Br J Pharmacol 157:907–921. doi: 10.1111/j.1476-5381.2009.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.USFDA. 2005. Guidance for industry: estimating the maximum safe starting dose in initial clinical trials for therapeutics in adult healthy volunteers. U.S. Food and Drug Administration, Rockville, MD, USA: https://www.fda.gov/downloads/drugs/guidances/ucm078932.pdf. [Google Scholar]
- 29.Johnson DE, Calia FM, Snyder MJ, Warren JW, Schimpff SC. 1983. Imipenem therapy of Pseudomonas aeruginosa bacteraemia in neutropenic rats. J Antimicrob Chemother 12(Suppl D):89–96. [DOI] [PubMed] [Google Scholar]
- 30.Johnson DE, Thompson B, Calia FM. 1985. Comparative activities of piperacillin, ceftazidime, and amikacin, alone and in all possible combinations, against experimental Pseudomonas aeruginosa infections in neutropenic rats. Antimicrob Agents Chemother 28:735–739. doi: 10.1128/AAC.28.6.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumish RM, Norden CW. 1976. Therapy of neutropenic rats infected with Pseudomonas aeruginosa. J Infect Dis 133:538–547. doi: 10.1093/infdis/133.5.538. [DOI] [PubMed] [Google Scholar]
- 32.Niida M, Sakakibara S, Kawabata T, Maebashi K, Takata T, Hikida M. 2004. Combined effects of arbekacin with biapenem against in vitro and in vivo model of a mixture of MRSA and Pseudomonas aeruginosa. Jpn J Antibiot 57:288–293. (In Japanese.) [PubMed] [Google Scholar]
- 33.Scott RE, Robson HG. 1976. Synergistic activity of carbenicillin and gentamicin in experimental Pseudomonas bacteremia in neutropenic rats. Antimicrob Agents Chemother 10:646–651. doi: 10.1128/AAC.10.4.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fantin B, Carbon C. 1992. In vivo antibiotic synergism: contribution of animal models. Antimicrob Agents Chemother 36:907–912. doi: 10.1128/AAC.36.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. 1993. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol 175:5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadurugamuwa JL, Lam JS, Beveridge TJ. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob Agents Chemother 37:715–721. doi: 10.1128/AAC.37.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obara M, Nakae T. 1991. Mechanisms of resistance to beta-lactam antibiotics in Acinetobacter calcoaceticus. J Antimicrob Chemother 28:791–800. doi: 10.1093/jac/28.6.791. [DOI] [PubMed] [Google Scholar]
- 38.Sato K, Nakae T. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J Antimicrob Chemother 28:35–45. [DOI] [PubMed] [Google Scholar]
- 39.Bulitta JB, Ly NS, Landersdorfer CB, Wanigaratne NA, Velkov T, Yadav R, Oliver A, Martin L, Shin BS, Forrest A, Tsuji BT. 2015. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 59:2315–2327. doi: 10.1128/AAC.04099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav R, Landersdorfer CB, Nation RL, Boyce JD, Bulitta JB. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59:2286–2298. doi: 10.1128/AAC.04379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yadav R, Bulitta JB, Schneider EK, Shin BS, Velkov T, Nation RL, Landersdorfer CB. 11 September 2017. Aminoglycoside concentrations required for synergy with carbapenems against Pseudomonas aeruginosa determined via mechanistic studies and modeling. Antimicrob Agents Chemother. doi: 10.1128/aac.00722-17. [DOI] [PMC free article] [PubMed]
- 42.Kumar A, Haery C, Paladugu B, Kumar A, Symeoneides S, Taiberg L, Osman J, Trenholme G, Opal SM, Goldfarb R, Parrillo JE. 2006. The duration of hypotension before the initiation of antibiotic treatment is a critical determinant of survival in a murine model of Escherichia coli septic shock: association with serum lactate and inflammatory cytokine levels. J Infect Dis 193:251–258. doi: 10.1086/498909. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez-Grande G, Kumar A. 2015. Optimizing antimicrobial therapy of sepsis and septic shock: focus on antibiotic combination therapy. Semin Respir Crit Care Med 36:154–166. doi: 10.1055/s-0034-1398742. [DOI] [PubMed] [Google Scholar]
- 44.Drusano GL, Liu W, Fikes S, Cirz R, Robbins N, Kurhanewicz S, Rodriquez J, Brown D, Baluya D, Louie A. 2014. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210:1319–1324. doi: 10.1093/infdis/jiu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel BM, Paratz J, See NC, Muller MJ, Rudd M, Paterson D, Briscoe SE, Ungerer J, McWhinney BC, Lipman J, Roberts JA. 2012. Therapeutic drug monitoring of beta-lactam antibiotics in burns patients—a one-year prospective study. Ther Drug Monit 34:160–164. doi: 10.1097/FTD.0b013e31824981a6. [DOI] [PubMed] [Google Scholar]
- 46.Pea F, Cojutti P, Sbrojavacca R, Cadeo B, Cristini F, Bulfoni A, Furlanut M. 2011. TDM-guided therapy with daptomycin and meropenem in a morbidly obese, critically ill patient. Ann Pharmacother 45:e37. doi: 10.1345/aph.1P745. [DOI] [PubMed] [Google Scholar]
- 47.Roberts JA, Lipman J. 2009. Pharmacokinetic issues for antibiotics in the critically ill patient. Crit Care Med 37:840–851. doi: 10.1097/CCM.0b013e3181961bff. [DOI] [PubMed] [Google Scholar]
- 48.Louie A, Liu W, VanGuilder M, Neely MN, Schumitzky A, Jelliffe R, Fikes S, Kurhanewicz S, Robbins N, Brown D, Baluya D, Drusano GL. 2015. Combination treatment with meropenem plus levofloxacin is synergistic against Pseudomonas aeruginosa infection in a murine model of pneumonia. J Infect Dis 211:1326–1333. doi: 10.1093/infdis/jiu603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan DD, Friberg LE, Nielsen EI. 2016. A pharmacokinetic-pharmacodynamic (PKPD) model based on in vitro time-kill data predicts the in vivo PK/PD index of colistin. J Antimicrob Chemother 71:1881–1884. doi: 10.1093/jac/dkw057. [DOI] [PubMed] [Google Scholar]
- 50.Kristoffersson AN, David-Pierson P, Parrott NJ, Kuhlmann O, Lave T, Friberg LE, Nielsen EI. 2016. Simulation-based evaluation of PK/PD indices for meropenem across patient groups and experimental designs. Pharm Res 33:1115–1125. doi: 10.1007/s11095-016-1856-x. [DOI] [PubMed] [Google Scholar]
- 51.European Committee on Antimicrobial Susceptibility Testing. 2015. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0. European Committee on Antimicrobial Susceptibility Testing, Basel, Switzerland: http://www.eucast.org/clinical_breakpoints/. [Google Scholar]
- 52.Dudhani RV, Turnidge JD, Nation RL, Li J. 2010. fAUC/MIC is the most predictive pharmacokinetic/pharmacodynamic index of colistin against Acinetobacter baumannii in murine thigh and lung infection models. J Antimicrob Chemother 65:1984–1990. doi: 10.1093/jac/dkq226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudhani RV, Turnidge JD, Coulthard K, Milne RW, Rayner CR, Li J, Nation RL. 2010. Elucidation of the pharmacokinetic/pharmacodynamic determinant of colistin activity against Pseudomonas aeruginosa in murine thigh and lung infection models. Antimicrob Agents Chemother 54:1117–1124. doi: 10.1128/AAC.01114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowler PG, Duerden BI, Armstrong DG. 2001. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naber KG, Llorens L, Kaniga K, Kotey P, Hedrich D, Redman R. 2009. Intravenous doripenem at 500 milligrams versus levofloxacin at 250 milligrams, with an option to switch to oral therapy, for treatment of complicated lower urinary tract infection and pyelonephritis. Antimicrob Agents Chemother 53:3782–3792. doi: 10.1128/AAC.00837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauermann R, Delle-Karth G, Marsik C, Steiner I, Zeitlinger M, Mayer-Helm BX, Georgopoulos A, Muller M, Joukhadar C. 2005. Pharmacokinetics and pharmacodynamics of cefpirome in subcutaneous adipose tissue of septic patients. Antimicrob Agents Chemother 49:650–655. doi: 10.1128/AAC.49.2.650-655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeitlinger MA, Dehghanyar P, Mayer BX, Schenk BS, Neckel U, Heinz G, Georgopoulos A, Muller M, Joukhadar C. 2003. Relevance of soft-tissue penetration by levofloxacin for target site bacterial killing in patients with sepsis. Antimicrob Agents Chemother 47:3548–3553. doi: 10.1128/AAC.47.11.3548-3553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ko HK, Yu WK, Lien TC, Wang JH, Slutsky AS, Zhang H, Kou YR. 2013. Intensive care unit-acquired bacteremia in mechanically ventilated patients: clinical features and outcomes. PLoS One 8:e83298. doi: 10.1371/journal.pone.0083298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luna CM, Videla A, Mattera J, Vay C, Famiglietti A, Vujacich P, Niederman MS. 1999. Blood cultures have limited value in predicting severity of illness and as a diagnostic tool in ventilator-associated pneumonia. Chest 116:1075–1084. doi: 10.1378/chest.116.4.1075. [DOI] [PubMed] [Google Scholar]
- 60.Jin L, Li J, Nation RL, Nicolazzo JA. 2012. Effect of systemic infection induced by Pseudomonas aeruginosa on the brain uptake of colistin in mice. Antimicrob Agents Chemother 56:5240–5246. doi: 10.1128/AAC.00713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffith DC, Corcoran E, Lofland D, Lee A, Cho D, Lomovskaya O, Dudley MN. 2006. Pharmacodynamics of levofloxacin against Pseudomonas aeruginosa with reduced susceptibility due to different efflux pumps: do elevated MICs always predict reduced in vivo efficacy? Antimicrob Agents Chemother 50:1628–1632. doi: 10.1128/AAC.50.5.1628-1632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reyes N, Aggen JB, Kostrub CF. 2011. In vivo efficacy of the novel aminoglycoside ACHN-490 in murine infection models. Antimicrob Agents Chemother 55:1728–1733. doi: 10.1128/AAC.00862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. 2009. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:46–56. doi: 10.1128/AAC.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maidhof H, Johannsen L, Labischinski H, Giesbrecht P. 1989. Onset of penicillin-induced bacteriolysis in staphylococci is cell cycle dependent. J Bacteriol 171:2252–2257. doi: 10.1128/jb.171.4.2252-2257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuji BT, Bulitta JB, Brown T, Forrest A, Kelchlin PA, Holden PN, Peloquin CA, Skerlos L, Hanna D. 2012. Pharmacodynamics of early, high-dose linezolid against vancomycin-resistant enterococci with elevated MICs and pre-existing genetic mutations. J Antimicrob Chemother 67:2182–2190. doi: 10.1093/jac/dks201. [DOI] [PubMed] [Google Scholar]
- 66.Landersdorfer CB, Ly NS, Xu H, Tsuji BT, Bulitta JB. 2013. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 57:2343–2351. doi: 10.1128/AAC.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D'Hondt RE, Jusko WJ, Forrest A, Tsuji BT. 2010. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 54:2051–2062. doi: 10.1128/AAC.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulitta JB, Duffull SB, Kinzig-Schippers M, Holzgrabe U, Stephan U, Drusano GL, Sorgel F. 2007. Systematic comparison of the population pharmacokinetics and pharmacodynamics of piperacillin in cystic fibrosis patients and healthy volunteers. Antimicrob Agents Chemother 51:2497–2507. doi: 10.1128/AAC.01477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bulitta JB, Zhao P, Arnold RD, Kessler DR, Daifuku R, Pratt J, Luciano G, Hanauske AR, Gelderblom H, Awada A, Jusko WJ. 2009. Mechanistic population pharmacokinetics of total and unbound paclitaxel for a new nanodroplet formulation versus Taxol in cancer patients. Cancer Chemother Pharmacol 63:1049–1063. doi: 10.1007/s00280-008-0827-2. [DOI] [PubMed] [Google Scholar]
- 73.Tsuji BT, Okusanya OO, Bulitta JB, Forrest A, Bhavnani SM, Fernandez PB, Ambrose PG. 2011. Application of pharmacokinetic-pharmacodynamic modeling and the justification of a novel fusidic acid dosing regimen: raising Lazarus from the dead. Clin Infect Dis 52(Suppl 7):S513–519. doi: 10.1093/cid/cir166. [DOI] [PubMed] [Google Scholar]
- 74.Landersdorfer CB, Kirkpatrick CM, Kinzig-Schippers M, Bulitta JB, Holzgrabe U, Drusano GL, Sorgel F. 2007. Population pharmacokinetics at two dose levels and pharmacodynamic profiling of flucloxacillin. Antimicrob Agents Chemother 51:3290–3297. doi: 10.1128/AAC.01410-06. [DOI] [PMC free article] [PubMed] [Google Scholar]