ABSTRACT
Sulbactam is one of four β-lactamase inhibitors in current clinical use to counteract drug resistance caused by degradation of β-lactam antibiotics by these bacterial enzymes. As a β-lactam itself, sulbactam is susceptible to degradation by β-lactamases. I investigated the Michaelis-Menten kinetics of sulbactam hydrolysis by 14 β-lactamases, representing clinically widespread groups within all four Ambler classes, i.e., CTX-M-15, KPC-2, SHV-5, and TEM-1 for class A; IMP-1, NDM-1, and VIM-1 for class B; Acinetobacter baumannii ADC-7, Pseudomonas aeruginosa AmpC, and Enterobacter cloacae P99 for class C; and OXA-10, OXA-23, OXA-24, and OXA-48 for class D. All of the β-lactamases were able to hydrolyze sulbactam, although they varied widely in their kinetic constants for the reaction, even within each class. I also investigated the inactivation kinetics of the inhibition of these enzymes by sulbactam. The class A β-lactamases varied widely in their susceptibility to inhibition, the class C and D enzymes were very weakly inhibited, and the class B enzymes were essentially or completely unaffected. In addition, we measured the sulbactam turnover number, the sulbactam/enzyme molar ratio required for complete inhibition of each enzyme. Class C enzymes had the lowest turnover numbers, class A enzymes varied widely, and class D enzymes had very high turnover numbers. These results are valuable for understanding which β-lactamases ought to be well inhibited by sulbactam. Moreover, since sulbactam has intrinsic antibacterial activity against Acinetobacter species pathogens, these results contribute to understanding β-lactamase-mediated sulbactam resistance in Acinetobacter, especially due to the action of the widespread class D enzymes.
KEYWORDS: sulbactam, β-lactamase, kinetics, turnover number
INTRODUCTION
The β-lactam antibacterial drugs, including penicillins, cephalosporins, carbapenems, and monobactams, are widely used to treat bacterial infections. Unfortunately, this widespread use has led to the spread of resistance mediated by serine β-lactamase and metallo-β-lactamase enzymes that degrade the β-lactams. To counter resistance due to serine β-lactamases, β-lactams can be combined with β-lactamase inhibitors (1–4). Three of the four β-lactamase inhibitors in current clinical use, namely, sulbactam, tazobactam, and clavulanic acid (but not the diazabicyclooctanone avibactam), are themselves β-lactams and therefore are potentially subject to degradation by β-lactamases. Many clinical bacterial isolates contain multiple β-lactamase genes (5). Since the ability of β-lactamase inhibitors to inactivate β-lactamases is variable, these combinations of multiple enzymes may be capable of degrading both the β-lactam antibacterial drug and the β-lactamase inhibitor. Therefore, it is important to understand the susceptibilities of β-lactamase inhibitors to inactivation by β-lactamases.
As a β-lactamase inhibitor, sulbactam (Fig. 1) has been combined in clinical practice with ampicillin (6), cefoperazone (7), and ceftriaxone (8). In addition to its use as a β-lactamase inhibitor, sulbactam has intrinsic antibacterial activity against Acinetobacter species and a few other pathogens (9, 10). Since many clinical Acinetobacter strains express one or more β-lactamases, including class D enzymes that are not potently inhibited by sulbactam (3, 11), resistance to sulbactam is common (12).
FIG 1.
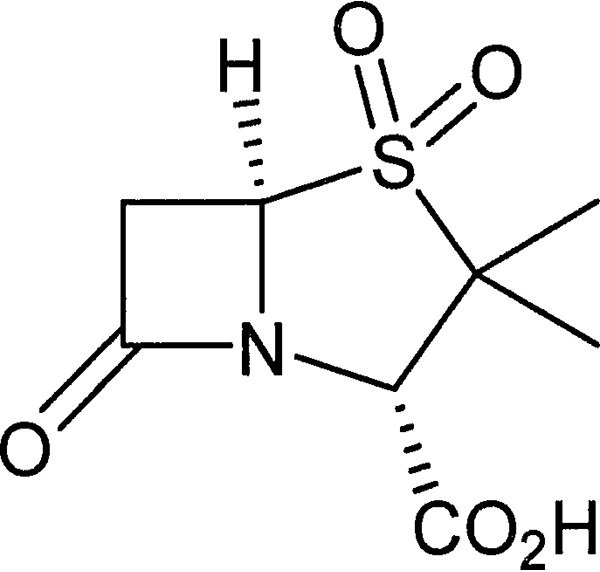
Structure of sulbactam.
This report supplies measurements of the steady-state kinetics of sulbactam hydrolysis by 14 purified β-lactamase enzymes, representing clinically important families within all four Ambler classes into which β-lactamases are grouped (13, 14), as well as the kinetics of inhibition of these enzymes by sulbactam. The class A serine β-lactamases were CTX-M-15, KPC-2, SHV-5, and TEM-1. The class B metallo-β-lactamases were IMP-1, NDM-1, and VIM-1. The class C serine β-lactamases were Acinetobacter baumannii ADC-7, Pseudomonas aeruginosa AmpC, and Enterobacter cloacae P99. The class D serine β-lactamases were OXA-10, OXA-23, OXA-24, and OXA-48.
RESULTS
Sulbactam hydrolysis by β-lactamases.
For the class A β-lactamases tested, there was a wide range of kinetic constants for sulbactam hydrolysis (Table 1). The Kms ranged from ≤2 μM for SHV-5 and TEM-1 to >6.25 mM for CTX-M-15, and the kcats ranged from 0.07 s−1 for SHV-5 to 14 s−1 for CTX-M-15. The kcat/Km values ranged from 470 M−1 s−1 for CTX-M-15 to ∼1,000,000 M−1 s−1 for TEM-1.
TABLE 1.
Steady-state kinetic constants for hydrolysis of sulbactam by β-lactamases, determined by initial rate and progress curve analyses
| Enzymea |
Km (mM) |
kcat (s−1) |
kcat/Km (M−1 s−1) |
|||
|---|---|---|---|---|---|---|
| Initial rate | Progress curve | Initial rate | Progress curve | Initial rate | Progress curve | |
| Class A | ||||||
| CTX-M-15 | >6.25 | 30 | ND | 14 | ND | 470 |
| KPC-2 | 1.4 | 1.1 | 7.4 | 14 | 5,400 | 13,000 |
| SHV-5 | <0.0025 | NT | 0.072 | NT | >29,000 | NT |
| TEM-1 | ∼0.002 | NT | 1.8 | NT | ∼1,000,000 | NT |
| Class B | ||||||
| IMP-1 | 10 | 12 | 170 | 160 | 17,000 | 13,000 |
| NDM-1 | 3 | ND | 430 | ND | 140,000 | ND |
| VIM-1 | 0.96 | 0.89 | 13 | 13 | 13,000 | 15,000 |
| Class C | ||||||
| ADC-7 | 0.26 | 1.1 | 0.22 | 0.55 | 830 | 500 |
| AmpC | 0.42 | 0.82 | 0.34 | 0.52 | 800 | 630 |
| P99 | 0.55 | 0.76 | 0.52 | 0.55 | 950 | 720 |
| Class D | ||||||
| OXA-10 | 1.5 | 2.0 | 2.0 | 2.2 | 1,300 | 1,100 |
| OXA-23 | 1.7 | 0.97 | 16 | 14 | 9,400 | 14,000 |
| OXA-24 | 9.7 | 8.4 | 55 | 42 | 5,700 | 5,000 |
| OXA-48 | 3.9 | 2.5 | 43 | 48 | 14,000 | 19,000 |
AmpC, Pseudomonas aeruginosa class C chromosomal AmpC; P99, Enterobacter cloacae class C chromosomal β-lactamase; ADC-7, Acinetobacter baumannii class C chromosomal β-lactamase; NT, not tested; ND, could not be determined.
In contrast to the highly varied class A β-lactamases, the class B, C, and D β-lactamases had more similar kinetic constants within each class, with only a 10-fold range of kcat/Km values. The class C enzymes had lower Kms than the class B and D enzymes but also had lower kcats, resulting in lower kcat/Kms than for the class B and D enzymes. TEM-1 had the highest kcat/Km, by far, of the 14 enzymes tested.
Two methods were employed here to measure the kinetics of sulbactam hydrolysis by β-lactamases, i.e., traditional initial rate measurements and progress curve analysis by numerical integration. The two methods gave similar results (Table 1). Individual kinetic constants for CTX-M-15 could not be measured using initial rates because the Michaelis plot was linear, due to the very high Km, but the constants could be measured by progress curve analysis (see Fig. S2 in the supplemental material).
Interesting features of the reaction of sulbactam with β-lactamases were revealed by considering the entire reaction progress curves rather than just the initial rates. Whereas the simple 2-step steady-state kinetic mechanism (E + S ↔ ES → E + P) was sufficient for numerical integration in some cases (CTX-M-15, IMP-1, and VIM-1), other enzymes required more complex models that included formation of a covalent complex or inhibited state, dissociation of the covalent complex, and substrate inhibition due to binding of sulbactam to that complex or inhibited state (see Materials and Methods and Fig. S3 to S5). The possibility that kinetic mechanisms other than the selected ones could perform equally well in the progress curve analysis cannot be excluded, however.
A particularly interesting observation from the progress curve analysis is the crossing of the progress curves at different sulbactam concentrations that was seen with class C β-lactamases, an example of which is shown in Fig. S5 for E. cloacae P99. This was interpreted as being due to substrate inhibition by sulbactam binding to an inhibited state of the enzyme that accumulates gradually. No significant substrate inhibition was observed in the Michaelis plots, based on initial rates (Fig. S1). The equilibrium dissociation constants for this interaction with A. baumannii ADC-7, P. aeruginosa AmpC, and E. cloacae P99 were 84 nM, 340 nM, and 180 nM, respectively.
Inhibition of β-lactamases by sulbactam.
The values of the second-order rate constant (kinact/Ki) for inhibition of the serine β-lactamases varied widely, from 0.4 M−1 s−1 for OXA-24 to 129,000 M−1 s−1 for SHV-5 (Table 2). For CTX-M-15 and OXA-23, the inhibition appeared not to be time dependent on the time scale of the measurements; therefore, a value for the equilibrium inhibition constant Ki (the equilibrium dissociation constant of the inhibitor) was determined. It is likely that the inhibition of these enzymes was, in fact, time dependent but the value of the off-rate constant koff was so high that the time dependence was not observable. CTX-M-15 was potently inhibited by sulbactam, with a Ki of 0.018 μM. In contrast, OXA-23 was weakly inhibited, with a Ki of 130 μM.
TABLE 2.
Inhibition constants for inhibition of β-lactamases by sulbactama
| Enzymeb | kinact/Ki (M−1 s−1) | koff (s−1) | Ki (μM) |
|---|---|---|---|
| Class A | |||
| CTX-M-15 | 0.018 | ||
| KPC-2 | 26 | 0.0007 | |
| SHV-5 | 129,000 | 0.002 | |
| TEM-1 | 5,300 | 0.001 | |
| Class B | |||
| VIM-1 | 2,400 | ||
| Class C | |||
| ADC-7 | 44 | 0 | |
| AmpC | 32 | 0.00003 | |
| P99 | 19 | 0 | |
| Class D | |||
| OXA-10 | 22 | 0 | |
| OXA-23 | 130 | ||
| OXA-24 | 0.4 | 0.0008 | |
| OXA-48 | 4 | 0.0002 |
Examples of the global fit of the data to the kinetic model are shown in Fig. S3 for SHV-5, CTX-M-15, and P99.
AmpC, Pseudomonas aeruginosa class C chromosomal AmpC; P99, Enterobacter cloacae class C chromosomal β-lactamase; ADC-7, Acinetobacter baumannii class C chromosomal β-lactamase.
Estimates of koff, the rate constant for dissociation/hydrolysis of sulbactam from the enzyme, were obtained from the same global progress curve-fitting used to obtain the kinact/Ki measurements. In some cases (A. baumannii ADC-7, E. cloacae P99, and OXA-10), koff was too low to measure by this method and therefore was given as zero. Measured values ranged from 3 × 10−5 s−1 for P. aeruginosa AmpC to 0.002 s−1 for SHV-5. Examples of nitrocefin hydrolysis progress curves for β-lactamases with weakly time-dependent inhibition (SHV-5), apparent time-independent inhibition (CTX-M-15), and strongly time-dependent inhibition (E. cloacae P99) by sulbactam are shown in Fig. S6.
The metallo-β-lactamases IMP-1 and NDM-1 were not significantly inhibited by sulbactam concentrations up to at least 5 mM. VIM-1, in contrast, was inhibited by millimolar concentrations of sulbactam, and the Ki was calculated to be 2.4 mM. Inhibition was not time dependent.
Sulbactam turnover numbers for inhibition of β-lactamases.
The turnover number may be described as the average number of molecules of an inhibitor per enzyme molecule required to achieve 100% inhibition, given sufficient time. Since sulbactam is both a substrate (Table 1) and an inhibitor (Table 2) of serine β-lactamases, turnover numbers greater than 1 are expected, as reflected in the literature (see Discussion).
The lowest sulbactam turnover numbers measured here were for the class C enzymes, with ratios ranging from 9 to 33 (Table 3 and Fig. S7). Turnover numbers for the class A enzymes varied widely, from 70 for CTX-M-15 to 14,000 for TEM-1. High turnover numbers were measured for all four of the class D enzymes tested. The turnover number for OXA-24 was unmeasurably high, with no inhibition observed under the conditions of the experiment.
TABLE 3.
Turnover numbers for sulbactam with β-lactamases, measured after 1 h or 5 h of incubationa
| Enzymeb | Turnover number |
|
|---|---|---|
| 1–2 h | 5–6 h | |
| Class A | ||
| CTX-M-15 | 70 | 280 |
| KPC-2 | 2,300 | 10,000 |
| SHV-5 | 440 | 660 |
| TEM-1 | 14,000 | 9,400 |
| Class C | ||
| ADC-7 | 9 | 9 |
| AmpC | 16 | 18 |
| P99 | 30 | 33 |
| Class D | ||
| OXA-10 | 6,400 | 2,900 |
| OXA-23 | 18,000 | 23,000 |
| OXA-24 | ≫34,000 | ≫34,000 |
| OXA-48 | 43,000 | 42,000 |
Experimental data are shown in Fig. S3.
AmpC, Pseudomonas aeruginosa class C chromosomal AmpC; P99, Enterobacter cloacae class C chromosomal β-lactamase; ADC-7, Acinetobacter baumannii class C chromosomal β-lactamase.
In most cases, the turnover numbers measured after a 1- to 2-hour preincubation of sulbactam with the β-lactamase were approximately the same as those measured after a 5- to 6-hour preincubation. For CTX-M-15, KPC-2, SHV-5, and OXA-23, however, the turnover numbers were higher after the longer preincubation. This indicates that complete inhibition of these enzymes was not achieved within the time frame of the shorter preincubation. This result is consistent with the relatively high sulbactam koff values of these enzymes, which prevents the enzymes from being completely inhibited by the sulbactam concentrations used, allowing a substantial amount of the sulbactam to be degraded during the course of the long preincubation. In contrast, the turnover number for OXA-10 decreased with incubation time. This indicates that the shorter incubation time was insufficient to achieve full inhibition of this enzyme. Owing to the negligible off-rate constant of the sulbactam-OXA-10 complex, however, more complete inhibition was achieved after the longer incubation. Turnover numbers were not measured for the metallo-β-lactamases, because IMP-1 and NDM-1 were not inhibited in the range of concentrations tested and the inhibition of NDM-1 was purely competitive and not time dependent.
DISCUSSION
Sulbactam hydrolysis by β-lactamases.
The Km of 1.4 mM measured for KPC-2 was higher than the value of 135 μM reported previously by Papp-Wallace et al. (15). The reason for this is not known but could be related to differences in the buffer conditions used for testing (10 mM phosphate-buffered saline [PBS] [pH 7.4] in the study by Papp-Wallace et al. [15] versus 0.1 M sodium phosphate, 10 mM NaHCO3, 0.005% Triton X-100 in this study). The closely related enzyme KPC-3 (differing from KPC-2 by a single amino acid residue) was reported by Alba et al. (16) to have a sulbactam Km of 30 μM, a kcat of 4 s−1, and a kcat/Km of 1 × 105 M−1 s−1.
Results reported here for TEM-1 are consistent with previously published measurements. Brenner and Knowles (17) reported a sulbactam Km of 0.8 μM and a kcat of 2 s−1 for RTEM (TEM-1), such that the kcat/Km was 2.5 × 106 M−1 s−1. Imtiaz et al. (18) and Meroueh (19) also reported a sulbactam kcat of 2 s−1 for TEM-1, and Delaire et al. (20) reported a kcat of 0.84 s−1.
SHV-5 differs from SHV-1 at only two adjacent residues but is considered to be hypersusceptible to mechanism-based inhibition as a result (21). Consistent with this conception, Thomson et al. (22) reported a sulbactam kcat of 730 s−1 for SHV-1, compared with the measurement reported here of 0.072 s−1 for SHV-5. Since the two residues in question, i.e., Gly-234 and Glu-235 in SHV-1 versus Ser-234 and Lys-235 in SHV-5 (when numbering from the initiator Met; residues 238 and 239 when numbering the active site nucleophile as Ser-70 or residues 238 and 240 when using the canonical Ambler class A β-lactamase alignment, which omits residue 239) are adjacent to the active site, it is not surprising that the substitution results in an enormous change in kcat, rendering SHV-5 far slower than SHV-1 at sulbactam hydrolysis.
Measurements reported here of the kinetic constants for hydrolysis of sulbactam by CTX-M-15 appear to be the first reported for a member of the clinically important CTX-M family of β-lactamases. Interestingly, CTX-M-15 had by far the lowest catalytic efficiency for sulbactam hydrolysis among the four class A enzymes tested, due to its comparatively high Km.
The reported kinetic constants for sulbactam hydrolysis by metallo-β-lactamases range widely. The kinetic constants measured here for IMP-1, NDM-1, and VIM-1 are within this range. Marcoccia et al. (23) reported Km, kcat, and kcat/Km values for NDM-1 of 1.4 mM, 50 s−1, and 36,000 M−1 s−1, respectively. Franceschini et al. (24) reported Km, kcat, and kcat/Km values for VIM-1 of 194 μM, 10 s−1, and 52,000 M−1 s−1, respectively. The BlaB metallo-β-lactamase of Chryseobacterium meningosepticum had a Km of 1.4 mM, a kcat of 470 s−1, and a kcat/Km of 3.4 × 105 M−1 s−1 (25). The IND-5 metallo-β-lactamase of Chryseobacterium indologenes had a sulbactam Km of 1.3 mM, a kcat of 3.3 s−1, and a kcat/Km of 2,500 M−1 s−1 (26). Metallo-β-lactamases from Aeromonas hydrophila, P. aeruginosa, and Bacteroides fragilis had kcat/Kms of 10, 13,700, and 5,900 M−1 s−1, respectively (27). The A. hydrophila A2 metallo-β-lactamase, Pseudomonas maltophilia L-1 metallo-β-lactamase, and Bacillus cereus metallo-β-lactamase II were reported (28) to have sulbactam Kms of 37 μM, 76 μM, and 5.2 mM, respectively, kcats of 0.12 s−1, 210 s−1, and 10 s−1, respectively, and kcat/Kms of 3,240 M−1 s−1, 2.8 × 106 M−1 s−1, and 1,900 M−1 s−1, respectively. The kinetic constants shown here for the hydrolysis of sulbactam by class C and class D β-lactamases appear to be the first such measurements reported.
Inhibition of β-lactamases by sulbactam.
I measured the second-order rate constants (kinact/Ki) for sulbactam inhibition of each β-lactamase or the Ki, as appropriate (Table 2). These results are consistent with the generally held view that sulbactam has utility as an inhibitor of class A enzymes only (3). Potent inhibition was seen with CTX-M-15, SHV-5, and TEM-1. In comparison, Faheem et al. (29) reported a kinact/Ki value of 29,000 M−1 s−1 for CTX-M-15 and a Ki of 62 nM, similar to our Ki result of 18 nM. Imtiaz et al. (18) and Meroueh et al. (19) reported a kinact/Ki of 125 M−1 s−1 for TEM-1. In contrast, Labia et al. (30) and Bret et al. (31) reported kinact/Ki values of 1,300 and 2,200 M−1 s−1, respectively. Therefore, the kinact/Ki measurement of 5,300 M−1 s−1 for TEM-1 reported here is higher than the earlier reports.
Published reports include other examples of potent inhibition of class A β-lactamases. Mariotte-Boyer et al. (32) reported a kinact/Ki of 1,325 M−1 s−1 for the class A NMC-A carbapenemase from E. cloacae. Thomson et al. (22) reported a kinact/Ki of 6,500 M−1 s−1 for SHV-1. Data reported by Yamaguchi et al. (33) and Sawai and Tsukamoto (34) were used to calculate kinact/Ki values of 1,100 M−1 s−1 and 2,300 M−1 s−1, respectively, for type 1b (TEM-2-type) penicillinase, and Labia et al. (30) similarly reported 1,000 M−1 s−1. Therrien et al. (35) reported a kinact/Ki of 3,300 M−1 s−1 for the class A PSE-4 (also called CARB-1) β-lactamase.
Sulbactam had much lower potency against the class A β-lactamase KPC-2, however. This enzyme differs from the other class A enzymes in having potent carbapenemase activity. Similarly low inhibitory activities of sulbactam were observed with the class B, C, and D enzymes. In comparison, a kinact/Ki value of 16 M−1 s−1 was calculated for Citrobacter freundii class C GN346 cephalosporinase from the data reported by Yamaguchi et al. (33). In contrast, higher kinact/Ki values of 220 M−1 s−1 and 2,300 M−1 s−1 were calculated for Proteus morgana cephalosporinase and Proteus vulgaris cephalosporinase, respectively, based on the data reported by Sawai and Tsukamoto (34). Measurements of kinetic constants for the inhibition of class B and D β-lactamases by sulbactam do not appear to have been published previously.
Sulbactam turnover numbers for inhibition of β-lactamases.
The sulbactam turnover number of TEM-1 measured here agreed closely with the value of 10,000 reported by Imtiaz et al. (18), Meroueh et al. (19), and Therrien et al. (35). Delaire et al. (20) and Labia et al. (30), however, reported much lower values of 415 and 525, respectively. Our value of 2,300 for the sulbactam turnover number with KPC-2, after a 1- to 2-h preincubation, was similar to the values of 1,000 and 1,500, following a 15-min incubation, reported by Papp-Wallace et al. (15, 36).
Some other reported β-lactamase turnover numbers with sulbactam include 2,280 (30) and 5,200 (33) for the class A enzyme TEM-2, 1,225 for the class A NMC-A carbapenemase from E. cloacae (32), 13,000 for SHV-1 (22), 13,000 for the class A SGM-1 β-lactamase (37), 40,000 for the class A OHIO-1 β-lactamase (38), 131 for the class A PSE-4 (CARB-1) β-lactamase (35), and 80 for the Citrobacter freundii class C GN346 cephalosporinase (33). Except for the finding for PSE-1, these published values are consistent with our observations of high turnover numbers for class A enzymes and much lower turnover numbers for class C enzymes. No sulbactam turnover number measurements for class D β-lactamases appear to have been published previously.
Sulbactam is considered to be useful primarily as a class A β-lactamase inhibitor (3). Indeed, potent inhibition of the class A enzymes CTX-M-15, SHV-5, and TEM-1 was observed (Table 2), although KPC-2 was much more weakly inhibited. However, relatively high catalytic efficiencies for hydrolysis of sulbactam by SHV-5 and TEM-1 (Table 1), as well as a very high turnover number for TEM-1, were also seen. From these measurements, the utility of sulbactam against class A β-lactamase-expressing clinical strains ought to be limited to a subset of class A enzymes. Since sulbactam is used clinically in combination with ampicillin, cefoperazone, and ceftriaxone, the sensitivity of these β-lactams to hydrolysis by the β-lactamases in clinical strains must also be considered.
Based on the observations reported here, it might be expected that sulbactam would be effective as an inhibitor of class C enzymes, if A. baumannii ADC-7, P. aeruginosa AmpC, and E. cloacae P99 are representative. The catalytic efficiencies of these enzymes for sulbactam hydrolysis were relatively low, which should result in relatively slow degradation of sulbactam, and the relatively low turnover numbers should contribute to effective inhibition. However, the inhibitory efficiencies (kinact/Ki) were also relatively low, leading to low rates of inactivation of the enzymes by sulbactam. A sufficiently high level of class C β-lactamase expression, low outer membrane permeability to the β-lactam partner, and/or a sufficient degree of active efflux may override the capacity of sulbactam to inhibit β-lactamase activity. Moreover, clinical strains often express multiple β-lactamase enzymes, which could overcome the effectiveness of sulbactam by combining an enzyme that degrades it effectively with an enzyme that degrades its β-lactam partner effectively.
The class B metallo-β-lactamases studied here showed high catalytic efficiencies for sulbactam hydrolysis and weak or no inhibition by sulbactam. Thus, sulbactam lacks utility against metallo-β-lactamases, and the expression of class B enzymes can be expected to reduce or to eliminate the utility of sulbactam as either a β-lactamase inhibitor or an antibacterial drug.
One use of the measurements described is to ascertain which β-lactamases pose the greatest threat to the use of sulbactam as an antibiotic (as opposed to a β-lactamase inhibitor) for the treatment of A. baumannii infections. Sulbactam has antibacterial activity against A. baumannii due to its inhibition of PBP3 (10, 39), but this activity is compromised in current clinical strains due to the expression of multiple β-lactamases, including class D enzymes (40). This study provides some of the first measurements of the kinetic constants for sulbactam hydrolysis by class D β-lactamases and for inhibition of those enzymes by sulbactam. The relatively high catalytic efficiencies of class D enzymes such as OXA-23 and OXA-24 (which are often found in A. baumannii clinical strains) with sulbactam as a substrate, the relatively low efficiencies of inactivation, and the very high turnover numbers combine to make sulbactam a good substrate for hydrolysis by these enzymes and a poor inhibitor. Thus, the expression of these enzymes by bacteria at sufficient levels should effectively degrade sulbactam, lowering its antibacterial potency. An effective class D β-lactamase inhibitor, especially one that is a poor substrate for these enzymes, is needed to counter this problem. ETX2514, a broad-spectrum diazabicyclooctenone β-lactamase inhibitor (11), likely satisfies this requirement, since ETX2514 inhibits many class D β-lactamases in addition to class A and C enzymes and is not significantly degraded by them. Indeed, the addition of 4 mg/liter ETX2514 significantly restored the activity of sulbactam against a global collection of 1,131 clinical isolates of A. baumannii collected in 2014, reducing its MIC90 from >32 mg/liter to 4 mg/liter (41). The combination of sulbactam and ETX2514 is currently in clinical development for the treatment of A. baumannii infections.
MATERIALS AND METHODS
Materials and experimental conditions.
β-Lactamases were purified as described in reference 11 and references therein. ADC-7 (42) was a gift from the laboratory of Robert A. Bonomo of Case Western Reserve University. The free acid of sulbactam was from U.S. Pharmacopeia (Rockville, MD). The sodium salt of sulbactam was from Dr. Friedrich Eberth Arzneimittel GmbH (Ursensollen, Germany). The latter was used when high concentrations of sulbactam were required, in order to avoid acidification of the buffer with the acid form of sulbactam. All serine β-lactamase experiments were performed at ambient temperature in a buffer composed of 0.1 M sodium phosphate (pH 7.0), 10 mM sodium bicarbonate, and 0.005% Triton X-100. All metallo-β-lactamase experiments were performed at ambient temperature in buffer composed of 50 mM HEPES-NaOH (pH 7.0), 1 μM ZnSO4, and 0.005% Triton X-100.
Sulbactam hydrolysis by β-lactamases.
Hydrolysis of sulbactam was monitored as an increase in absorbance at 235 nm (17). Assays were performed in a volume of 100 μl in clear 96-well acrylic plates (Corning Life Sciences, Tewksbury, MA). Absorbance was measured at 3- to 6-s intervals for 10 min, with a SpectraMax Plus plate reader (Molecular Devices, Sunnyvale, CA). Progress curves from triplicate wells were averaged. Slopes were measured from the linear initial phase of each progress curve. Two-fold serial dilutions of sulbactam were used, varying the range of concentrations as needed. The enzyme concentration was also varied as needed (see Fig. S1 in the supplemental material). Enzyme concentrations were relatively high, to permit substantial product formation prior to the onset of inhibition by sulbactam. Km and Vmax values for each enzyme were obtained by fitting a Michaelis plot of initial rate versus sulbactam concentration to the Michaelis-Menten equation. Vmax was converted to kcat by using the enzyme concentration and a measured extinction coefficient difference between hydrolyzed and intact sulbactam, for the path length of the 100-μl assay format, of 416 M−1. For TEM-1, the Km was estimated by extrapolation and was consistent with published measurements (17, 18, 20). A Michaelis plot could not be obtained for SHV-5 due the low sulbactam Km. Vmax was calculated for SHV-5 based on the rate of hydrolysis of 50 μM sulbactam by 500 nM SHV-5.
In addition to the traditional, initial rate-based analysis of the reaction kinetics, progress curve analysis was employed, when feasible, using Global Kinetic Explorer software (KinTek) to perform numerical integration. The set of progress curves at a range of sulbactam concentrations for 10-min reactions with each enzyme were fit globally to either a simple model (CTX-M-15 [Fig. S2] and class B enzymes), depicted by E + S ↔ ES (with rate constants for forward and reverse reactions of k+1 and k−1, respectively) and ES ↔ E + P (with rate constants for forward and reverse reactions of k+2 and k−2, respectively), with Km = (k−1 + k+2)/k+1 and k+2 = kcat, or a more complex model incorporating one or more of the following, in order of increasing complexity: formation of an inhibited state or covalent inhibition of the enzyme by sulbactam (class D enzymes [see Fig. S3 for OXA-23]) plus resolution of the covalent complex by hydrolysis (KPC-2 [Fig. S4]) plus substrate inhibition due to sulbactam binding to the inhibited state or covalent complex (AmpC and P99 [Fig. S5] and ADC-7), depicted by E + S ↔ ES, ES ↔ E + P, ES ↔ EI (formation of covalent complex or inhibited state), EI ↔ E + P (koff for covalent complex), and EI + S ↔ EIS (substrate inhibition). There was close agreement between the two methods regarding the values of Km and kcat (Table 1), and the progress curve method allowed estimation of kinetic constants for CTX-M-15 (Fig. S2), which could not be measured by the initial rate method because of the very high Km. In the above description, E, S, P, and I represent the enzyme, the substrate (sulbactam), the product of sulbactam hydrolysis, and inhibitor, respectively.
Inhibition of β-lactamases by sulbactam.
The second-order rate constants for time-dependent inhibition (kinact/Ki) of serine β-lactamase-catalyzed nitrocefin hydrolysis, or in some cases the equilibrium inhibition constant (Ki) instead, for sulbactam were measured as described in reference 11, with 100 μM nitrocefin as the substrate. For ADC-7, the nitrocefin Km was 400 μM under the same conditions as used for the other enzymes, and the enzyme concentration used was 16 pM. The maximal sulbactam concentration tested was 5 mM.
For the metallo-β-lactamases IMP-1, NDM-1, and VIM-1, the nitrocefin Kms were 6.0, 3.2, and 12.4 μM, respectively (data not shown). The substrate was 100 μM nitrocefin. The enzyme concentrations used were 300 pM IMP-1, 1.8 nM NDM-1, and 200 pM VIM-1. The maximal sulbactam concentrations tested were 5 mM for IMP-1 and NDM-1 and 66.7 mM for VIM-1. The Ki for inhibition of VIM-1 by sulbactam was calculated by using the formula for a competitive inhibitor, Ki = IC50/(1 + [S]/Km), where IC50 is the 50% inhibitory concentration and [S] is the nitrocefin concentration.
Sulbactam turnover numbers for inhibition of serine β-lactamases.
Each enzyme was incubated at either 3 μM (CTX-M-15, SHV-5, P. aeruginosa AmpC, E. cloacae P99, and A. baumannii ADC-7) or 0.3 μM (KPC-2, TEM-1, and class D enzymes) with a set of 2-fold serial dilutions of sulbactam, with the highest sulbactam concentration being either 1 or 10 mM, respectively. After 1 to 2 h or 5 to 6 h at ambient temperature, the enzyme-sulbactam mixtures were diluted 1:333,000 from 3 μM or 1:33,000 from 0.3 μM into reaction mixtures with 100 μM nitrocefin. The triplicate 45-μl reaction mixtures in clear polystyrene 384-well plates were monitored at 490 nm for 10 min with a SpectraMax Plus plate reader. Control wells in which enzyme was replaced with buffer were included for background subtraction. Data for the triplicate wells were averaged, the background was subtracted, the initial rate of absorbance increase was measured, and the percent inhibition was calculated for each sulbactam/enzyme ratio. The extrapolated point of intersection with the horizontal axis of the percent inhibition versus sulbactam/enzyme ratio curve was taken to be the turnover number.
Supplementary Material
ACKNOWLEDGMENTS
I thank Alita Miller, Thomas Durand-Réville, and Ruben Tommasi for critical reading of the manuscript and Robert A. Bonomo and colleagues at Case Western Reserve University for providing purified ADC-7 protein.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01612-17.
REFERENCES
- 1.Bush K, Bradford PA. 2016. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K. 2015. A resurgence of β-lactamase inhibitor combinations effective against resistant Gram-negative pathogens. Int J Antimicrob Agents 46:483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin W, Panunzio M, Biondi S. 2014. β-Lactam antibiotic renaissance. Antibiotics 3:193–215. doi: 10.3390/antibiotics3020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanheira M, Farrell SE, Deshpande LM, Mendes RE, Jones RN. 2013. Prevalence of β-lactamase-encoding genes among Enterobacteriaceae bacteremia isolates collected in 26 U.S. hospitals: report from the SENTRY antimicrobial surveillance program (2010). Antimicrob Agents Chemother 57:3012–3020. doi: 10.1128/AAC.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betrosian AP, Douzinas EE. 2009. Ampicillin-sulbactam: an update on the use of parenteral and oral forms in bacterial infections. Expert Opin Drug Metab Toxicol 5:1099–1112. doi: 10.1517/17425250903145251. [DOI] [PubMed] [Google Scholar]
- 7.Pfaller MA, Flamm RK, Duncan LR, Mendes RE, Jones RN, Sader HS. 2017. Antimicrobial activity of tigecycline and cefoperazone/sulbactam tested against 18,386 Gram-negative organisms from Europe and the Asia-Pacific region (2013–2014). Diagn Microbiol Infect Dis 88:177–183. doi: 10.1016/j.diagmicrobio.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Xin X, Jian L, Xia X, Jia B, Huang W, Li C, Wang C, Zhou L, Sun X, Tang X, Huang Y, Zhu Y, Zhang W. 2013. A multicentre clinical study on the injection of ceftriaxone/sulbactam compared with cefoperazone/sulbactam in the treatment of respiratory and urinary tract infections. Ann Clin Microbiol Antimicrob 12:38. doi: 10.1186/1476-0711-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. In vitro activities of the β-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam alone or in combination with β-lactams against epidemiologically characterized multidrug-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother 48:1586–1592. doi: 10.1128/AAC.48.5.1586-1592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Liu Q, Chen Z, Li C. 2017. Efficacy of sulbactam for the treatment of Acinetobacter baumannii complex infection: a systematic review and meta-analysis. J Infect Chemother 23:278–285. doi: 10.1016/j.jiac.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Durand-Réville TF, Guler S, Comita-Prevoir J, Chen B, Bifulco N, Huynh H, Lahiri S, Shapiro AB, McLeod SM, Carter NM, Moussa SH, Velez-Vega C, Olivier NB, McLaughlin R, Gao N, Thresher J, Palmer T, Andrews B, Giacobbe RA, Newman JV, Ehmann DE, deJonge B, O'Donnell J, Mueller JP, Tommasi RA, Miller AA. 2017. ETX2514 is a broad spectrum β-lactamase inhibitor for the treatment of drug resistant Gram-negative infections, including those caused by Acinetobacter baumannii. Nat Microbiol 2:17104. doi: 10.1038/nmicrobiol.2017.104. [DOI] [PubMed] [Google Scholar]
- 12.Jones RN, Flonta M, Gurler N, Cepparulo M, Mendes RE, Castanheira M. 2014. Resistance surveillance program report for selected European nations (2011). Diagn Microbiol Infect Dis 78:429–436. doi: 10.1016/j.diagmicrobio.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Hall BG, Barlow M. 2005. Revised Ambler classification of β-lactamases. J Antimicrob Chemother 55:1050–1051. doi: 10.1093/jac/dki130. [DOI] [PubMed] [Google Scholar]
- 14.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. 2010. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob Agents Chemother 54:890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 49:4760–4762. doi: 10.1128/AAC.49.11.4760-4762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner DG, Knowles JR. 1981. Penicillanic acid sulfone: an unexpected isotope effect in the interaction of 6α- and 6β-monodeuterio and of 6,6-dideuterio derivatives with RTEM β-lactamase from Escherichia coli. Biochemistry 20:3680–3687. doi: 10.1021/bi00516a003. [DOI] [PubMed] [Google Scholar]
- 18.Imtiaz U, Billings EM, Knox JR, Mobashery S. 1994. A structure-based analysis of the inhibition of class A β-lactamases by sulbactam. Biochemistry 33:5728–5738. doi: 10.1021/bi00185a009. [DOI] [PubMed] [Google Scholar]
- 19.Meroueh SO, Roblin P, Golemi D, Maveyraud L, Vakulenko SB, Zhang Y, Samama J-P, Mobashery S. 2002. Molecular dynamics at the root of expansion of function in the M69L inhibitor-resistant TEM β-lactamase from Escherichia coli. J Am Chem Soc 124:9422–9430. doi: 10.1021/ja026547q. [DOI] [PubMed] [Google Scholar]
- 20.Delaire M, Labia R, Samama J-P, Masson J-M. 1992. Site-directed mutagenesis at the active site of Escherichia coli TEM-1 β-lactamase. J Biol Chem 267:20600–20606. [PubMed] [Google Scholar]
- 21.Kalp M, Bethel CR, Bonomo RA, Carey PR. 2009. Why the extended-spectrum β-lactamases SHV-2 and SHV-5 are “hypersusceptible” to mechanism-based inhibitors. Biochemistry 48:9912–9920. doi: 10.1021/bi9012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JM, Distler AM, Bonomo RA. 2007. Overcoming resistance to β-lactamase inhibitors: comparing sulbactam to novel inhibitors against clavulanate resistant SHV enzymes with substitutions at Ambler position 244. Biochemistry 46:11361–11368. doi: 10.1021/bi700792a. [DOI] [PubMed] [Google Scholar]
- 23.Marcoccia F, Bottoni C, Sabatini A, Colapietro M, Mercuri PS, Galleni M, Kerff F, Matagne A, Celenza G, Amicosante G, Perilli M. 2016. Kinetic study of laboratory mutants of NDM-1 metallo-β-lactamase and the importance of an isoleucine at position 35. Antimicrob Agents Chemother 60:2366–2372. doi: 10.1128/AAC.00531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschini N, Caravelli B, Docquier J-D, Galleni M, Frère J-M, Amicosante G, Rossolini GM. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob Agents Chemother 44:3003–3007. doi: 10.1128/AAC.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vessillier S, Docquier J-D, Rival S, Frere J-M, Galleni M, Amicosante G, Rossolini GM, Franceschini N. 2002. Overproduction and biochemical characterization of the Chryseobacterium meningosepticum BlaB metallo-β-lactamase. Antimicrob Agents Chemother 46:1921–1927. doi: 10.1128/AAC.46.6.1921-1927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perilli M, Caporale B, Celenza G, Pellegrini C, Docquier JD, Mezzatesta M, Rossolini GM, Stefani S, Amicosante G. 2007. Identification and characterization of a new metallo-β-lactamase, IND-5, from a clinical isolate of Chryseobacterium indologenes. Antimicrob Agents Chemother 51:2988–2990. doi: 10.1128/AAC.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prosperi-Meys C, Llabres G, de Seny D, Soto RP, Valladares MH, Laraki N, Frere J-M, Galleni M. 1999. Interaction between class B β-lactamases and suicide substrates of active-site serine β-lactamases. FEBS Lett 443:109–111. doi: 10.1016/S0014-5793(98)01689-5. [DOI] [PubMed] [Google Scholar]
- 28.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere JM. 1993. An overview of the kinetic parameters of class B β-lactamases. Biochem J 291:151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faheem M, Rehman MT, Danishuddin M, Khan AU. 2013. Biochemical characterization of CTX-M-15 from Enterobacter cloacae and designing a novel non-β-lactam-β-lactamase inhibitor. PLoS One 8:e56926. doi: 10.1371/journal.pone.0056926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labia R, Lelievre V, Peduzzi J. 1980. Inhibition kinetics of three R-factor-mediated β-lactamases by a new β-lactam sulfone (CP45899). Biochim Biophys Acta 611:351–357. doi: 10.1016/0005-2744(80)90071-6. [DOI] [PubMed] [Google Scholar]
- 31.Bret L, Chaibi EB, Chanal-Claris C, Sirot D, Labia R, Sirot J. 1997. Inhibitor-resistant TEM (IRT) β-lactamases with different substitutions at position 244. Antimicrob Agents Chemother 41:2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariotte-Boyer S, Nicolas-Chanoine MH, Labia R. 1996. A kinetic study of NMC-A β-lactamase, an Ambler class A carbapenemase also hydrolyzing cephamycins. FEMS Microbiol Lett 143:29–33. doi: 10.1111/j.1574-6968.1996.tb08457.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi A, Hirata T, Sawai T. 1983. Kinetic studies on inactivation of Citrobacter freundii cephalosporinase by sulbactam. Antimicrob Agents Chemother 24:23–30. doi: 10.1128/AAC.24.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawai T, Tsukamoto K. 1982. Cefoxitin, N-formimidoyl thienamycin, clavulanic acid, and penicillanic acid sulfone as suicide inhibitors for different types of β-lactamases produced by Gram-negative bacteria. J Antibiot (Tokyo) 35:1594–1602. doi: 10.7164/antibiotics.35.1594. [DOI] [PubMed] [Google Scholar]
- 35.Therrien C, Kotra LP, Sanschagrin F, Mobashery S, Levesque RC. 2000. Evaluation of inhibition of the carbenicillin-hydrolyzing β-lactamase PSE-4 by the clinically used mechanism-based inhibitors. FEBS Lett 470:285–292. doi: 10.1016/S0014-5793(00)01342-9. [DOI] [PubMed] [Google Scholar]
- 36.Papp-Wallace KM, Taracila M, Wallace CJ, Hujer KM, Bethel CR, Hornick JM, Bonomo RA. 2010. Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci 19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamoureaux TL, Vakulenko V, Toth M, Frase H, Vakulenko SB. 2013. A novel extended-spectrum β-lactamase, SGM-1, from an environmental isolate of Sphingobium sp. Antimicrob Agents Chemother 57:3783–3788. doi: 10.1128/AAC.00808-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin S, Thomas M, Mark S, Anderson V, Bonomo RA. 1999. OHIO-1 β-lactamase mutants: the Arg244Ser mutant and resistance to β-lactams and β-lactamase inhibitors. Biochim Biophys Acta 1432:125–136. doi: 10.1016/S0167-4838(99)00025-4. [DOI] [PubMed] [Google Scholar]
- 39.Penwell WF, Shapiro AB, Giacobbe RA, Gu R-F, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BLM, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. doi: 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans BA, Amyes SGB. 2014. OXA-β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller A, Hackel M, Bouchillon S, DeJonge B, Tommasi R, Mueller J. 2016. Global surveillance of the activity of sulbactam combined with the novel β-lactamase inhibitor ETX2514 against clinical isolates of Acinetobacter baumannii from 2014. Open Forum Infect Dis 3(Suppl 1):2243. doi: 10.1093/ofid/ofw172.1791. [DOI] [Google Scholar]
- 42.Hujer KM, Hamza NS, Hujer AM, Perez F, Helfand MS, Bethel CR, Thomson JM, Anderson VE, Barlow M, Rice LB, Tenover FC, Bonomo RA. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 β-lactamase: defining a unique family of class C enzymes. Antimicrob Agents Chemother 49:2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


