ABSTRACT
The novel 63,558-bp plasmid pSA-01, which harbors nine antibiotic resistance genes, including cfr, erm(C), tet(L), erm(T), aadD, fosD, fexB, aacA-aphD, and erm(B), was characterized in Staphylococcus arlettae strain SA-01, isolated from a chicken farm in China. The colocation of cfr and fosD genes was detected for the first time in an S. arlettae plasmid. The detection of two IS431-mediated circular forms containing resistance genes in SA-01 suggested that IS431 may facilitate dissemination of antibiotic resistance genes.
KEYWORDS: Staphylococcus arlettae, cfr, fosD, IS431, circular form
TEXT
Staphylococcus arlettae is a member of the coagulase-negative staphylococci (CoNS) that can serve as a reservoir for various resistance genes, including cfr, and may facilitate the dissemination of resistance genes between different staphylococcal species or even between staphylococci and other bacterial genera (1, 2). The multiresistance gene cfr, which mediates resistance to five antimicrobial classes, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (3), was first described in 2000 (4) and has been identified in a number of staphylococcal species (1, 2, 4–8). Plasmids seem to play an important role in the intra- and intergenus transfer of this gene (9). Thus far, many cfr-carrying plasmids have been described, which differed in structure, size, and presence of additional resistance genes (7). Most of them carry additional resistance genes coding for phenicol resistance (fexA, fexB) (5, 6, 10), macrolide-lincosamide-streptogramin B (MLSB) resistance [erm(B), erm(C), erm(33)] (6, 11, 12) or gentamicin-kanamycin-tobramycin resistance (aacA-aphD) (6). Lately, fosfomycin has gained attention, as it has remained active against both Gram-positive and Gram-negative multidrug-resistant bacteria (13). To date, some fosfomycin resistance genes have been described in various bacteria (13–15). The fosD gene, which mediated fosfomycin resistance, was reported previously in Staphylococcus aureus (16) and Staphylococcus rostri (1). In this study, we reported a novel multiresistance plasmid cocarrying cfr and fosD in S. arlettae.
S. arlettae isolate SA-01, identified by the BD Phoenix-100 diagnostic systems (Sparks, MD), was collected from a fecal sample from a commercial chicken farm in China in July 2015. Antimicrobial susceptibility testing, performed according to the protocols of the Clinical and Laboratory Standards Institute (CLSI) (17–19), indicated that it exhibited high MIC values for florfenicol (>256 mg/liter) and fosfomycin (>128 mg/liter) and showed a linezolid MIC value of 16 mg/liter (Table 1). PCR analysis confirmed that it carried the florfenicol resistance genes cfr, fexA, and fexB, using previously described primers (5, 20) and the fosfomycin resistance gene fosD, using primers F1 (5′-AACTCTAACTTGTGTCCGTCAG-3′) and F2 (5′-GTGGCTTATGGGTTGCGTTA-3′).
TABLE 1.
MICs for S. arlettae SA-01, S. aureus RN4220, and the S. aureus RN4220 transformants carrying plasmid pSA-01
| Bacterial isolate | MICs (mg/liter) ofa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| FFC | CHL | ERY | KAN | FOF | TET | CIP | LZD | |
| SA-01 | >256 | >64 | >128 | 128 | 128 | 128 | >64 | 16 |
| S. aureus RN4220 | 4 | 4 | 0.25 | 0.25 | ≤1 | ≤0.5 | 0.5 | 2 |
| Transformant RN4220+pSA-01 | >256 | >64 | >128 | 128 | 128 | 32 | 0.5 | 16 |
FFC, florfenicol; CHL, chloramphenicol; ERY, erythromycin; KAN, kanamycin; FOF, fosfomycin; TET, tetracycline; CIP, ciprofloxacin; LZD, linezolid.
Conjugation by filter mating (21) and electrotransformation using purified plasmid DNA were performed with S. arlettae SA-01 as donor and S. aureus RN4220 (22) as recipient. Florfenicol (10 mg/liter) or fosfomycin (32 mg/liter) was used as a selection marker. Conjugation assays were failed, but electrotransformation of the plasmid DNA from SA-01 to RN4220 was successful. Compared with RN4220, the transformant (designated RN4220+pSA-01) exhibited drastically increased MICs for florfenicol (>256 mg/liter), erythromycin (>128 mg/liter), kanamycin (128 mg/liter), and fosfomycin (128 mg/liter) (Table 1). PCR results revealed that cfr, fexB, and fosD were detected in the transformant RN4220+pSA-01.
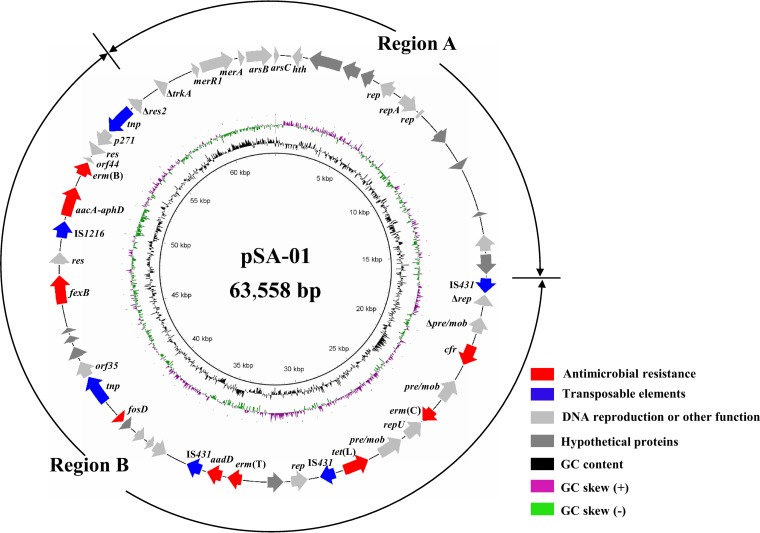
Whole-genome sequencing for transformant RN4220+pSA-01 was performed on the Illumina MiSeq (Majorbio, Shanghai, China) using a 400-bp paired-end TruSeq library with a 2 × 300 run. The paired-end reads were assembled de novo using SOAP v2.04 and GapCloser v1.12. The gaps between different contigs were closed by PCR and sequencing. Sequence analysis was conducted using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A 63,558-bp plasmid (designated pSA-01) with an average GC content of 31.9% was obtained (Fig. 1). The cfr, fexB, and fosD genes were colocated in pSA-01, which also harbored erm(C), erm(T), erm(B), tet(L), aadD, and aacA-aphD (Fig. 1). Cooccurrence of these genes may lead to the persistence and coselection of cfr under selective pressure imposed by the use of aminoglycosides, MLSB compounds, tetracycline, or fosfomycin. Although cfr has been reported to coexist with erm(B) (6, 11), erm(C) (6), fexB (12), or aacA-aphD (6) and so on, to our knowledge, this is the first report for colocation of cfr and fosD in a plasmid from S. arlettae.
FIG 1.
Genetic map of pSA-01. Positions and directions of predicted open reading frames are indicated by colored arrows according to their predicted functions. Truncated coding sequences (CDS) are indicated with a Greek delta symbol.
Based on its genetic content, pSA-01 was divided into two regions, A and B (Fig. 1). Region A was 22,933 bp in size and consisted of the backbone of pSA-01. Three replication genes, including repA and its flanked genes, showed over 97% identity to corresponding regions of plasmid pStO2014-01 from Staphylococcus condimenti (GenBank accession no. CP018777) and pC2014-3 from Staphylococcus equorum (accession no. CP013717), respectively. These genes are essential for plasmid replication. The products of merR1 and merA showed 94% and 90% identity to proteins MerR1 and MerA of the mer operon in Bacillus sp. YR31 (accession no. LC015493), respectively. The genes arsB and arsC showed 85% and 86% identity to the gene coding for the arsenic transporter of Sporosarcina psychrophila (accession no. CP014616) and the gene coding for arsenate reductase of Staphylococcus equorum (accession no. CP013714), respectively. These genes are associated with heavy-metal (mercury and arsenic) resistance (23, 24). Additionally, the remaining 11 genes located in the backbone, including hypothetical protein genes, appear to have been derived from various sources, as their deduced amino acid identities ranged from 70% to 99% to corresponding proteins of Staphylococcus spp., Bacillus spp., and Sporosarcina spp.
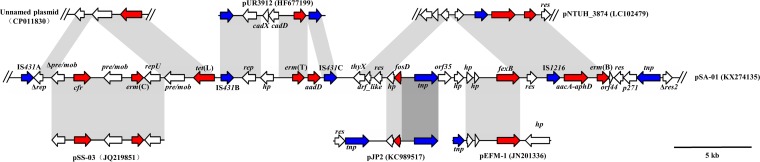
The 40,625-bp region B (nucleotides [nt] 16,317 to 56,941) harbored nine resistance genes, which were carried by segments originating from various sources (Fig. 2). The cfr–erm(C)-carrying segment, which showed 99.5% identity to plasmid pSS-03 (accession no. JQ219851) (6), was found to be inserted into a 4,413-bp fragment showing 99.8% identity to an Enterococcus faecium plasmid (accession no. CP011830) (25). This segment, spanning from Δrep to tet(L), was flanked by two IS431 copies (Fig. 2). Moreover, a 7,240-bp segment comprising erm(T) and aadD was also bracketed by IS431 and showed 99% identity to the chromosomal integrated plasmid pUR3912 (accession no. HF677199) (26, 27) with 80% query coverage. IS431 seemed to mediate the integration of this segment, since pUR3912 could integrate into the chromosomal DNA via IS431 (26). An 8,647-bp fragment containing fosD and fexB seemed to insert into a 7,400-bp aacA–aphD–erm(B)-carrying fragment, which showed 99.8% identity to the corresponding region of pNTUH_3874 (accession no. LC102479) (28). Within the 8,647-bp fragment, the fosD-carrying fragment (1,002 bp) exhibited over 99.9% identity to the corresponding region of pJP2 (accession no. KC989517) (1), but downstream of fosD a 2,320-bp segment containing a transposase gene showed just 91% identity to pJP2. The 3,653-bp fexB-containing segment showed 99.9% identity to plasmid pEFM-1 from Enterococcus faecium (accession no. JN201336) (20). The common DNA segments in these different plasmids suggested recombination between plasmids of different pathogens; pSA-01 was a complex and hybrid plasmid.
FIG 2.
Schematic presentation of region B in plasmid pSA-01 in comparison with other plasmids. Regions of >99% nucleotide sequence identity are marked in light gray, while dark gray represents region of 91.2% nucleotide sequence identity. Arrows indicate the positions and orientations of the genes.
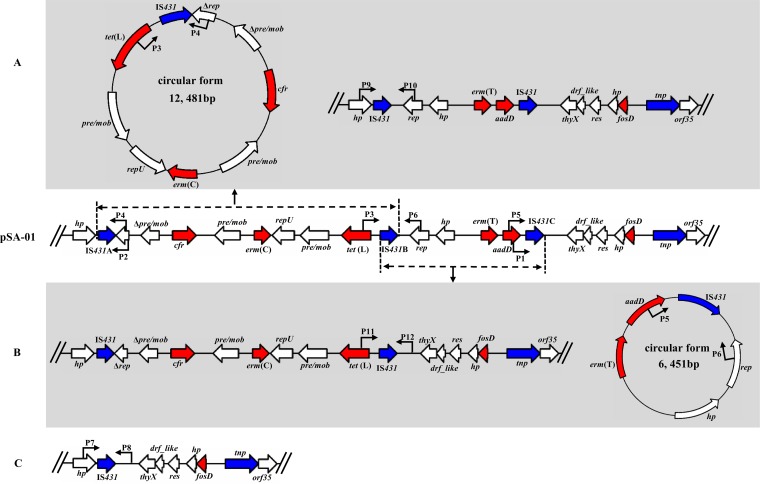
In pSA-01, the presence of multiple copies of insertion sequence (IS) (three IS431and one IS1216) were identified, which might facilitate intra- or interplasmid recombination. Three IS431 copies (named IS431A, IS431B, and IS431C based on their positions) were in the same orientation. Since direct repeats of IS may mediate dissemination of genes via formation of the circular form (1, 10, 29), inverse PCR assays (see Table S1 in the supplemental material for primers) were performed to detect whether IS431 mediated the formation of the circular form. Two circular forms of 12,481 bp and 6,451 bp were observed in SA-01 (Fig. 3). Both of the circular forms contained an intact IS431 and the region between IS431A and IS431B (Fig. 3A) or region between IS431B and IS431C (Fig. 3B). To further confirm the formation of circular forms, PCR assays (primers shown in Table S1) were performed to detect the structures that missed the corresponding region of the circular form. The results of these PCR assays matched with inverse PCR. Interestingly, although no circular form was observed between IS431A and IS431C, the structure that missed regions spanning from Δrep to IS431C was detected (Fig. 3C). This observation indicated that the two circular forms may form simultaneously. These findings might suggest that the association of resistance genes with IS431 facilitated their translocation. Besides, IS1216 might also have been involved in the recombination of pSA-01, since it has been reported to play an important role in the dissemination of antimicrobial resistance determinants (30) and in plasmid recombination (31).
FIG 3.
Formation of circular forms mediated by IS431. (A) Circular form derived from the region spanning from IS431A to IS431B and the structure that missed the corresponding region of the circular form. (B) Circular form derived from the region spanning from IS431B to IS431C and the structure that missed the corresponding region of the circular form. (C) Structure that missed the region spanning from Δrep to IS431C. The locations and orientations of primers (P1 to P12) are indicated by arrows. Primers P1 to P6 were used for inverse PCR to detect circular forms (primers P3 and P4 for the 12,481-bp circular form and P5 and P6 for the 6,451-bp circular form; inverse PCR with primers P1 and P2 produced no product); primers P7 to P12 were used to detect structures that missed the corresponding region of the circular form (P7 and P8 for structure that missed the region spanning from Δrep to IS431C; P9 and P10 for structure that missed the corresponding region of the 12,481-bp circular form; and P11 and P12 for structure that missed the corresponding region of the 6,451-bp circular form).
In conclusion, pSA-01 was a complex, hybrid multiresistance plasmid. As far as we know, the fosD gene was described in an S. arlettae plasmid for the first time. The coexistence of cfr with other resistance genes, especially fosD, will limit antimicrobial treatment options and may lead to coselection of these genes even in the absence of direct selective pressure. The structures bracketed by IS431 were unstable and could be looped out by IS-mediated recombination. The presence of IS elements might facilitate intra- or interplasmid recombination and dissemination of resistance genes. Given that the presence of the cfr–fosD-carrying multiresistance plasmid may seriously compromise the effectiveness of clinical therapy and threaten public health, its occurrence and dissemination need further surveillance.
Accession number(s).
The complete nucleotide sequence of the 63,558-bp plasmid pSA-01 characterized in this study was submitted to the GenBank database and assigned accession number KX274135.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the earmarked fund for Modern Agro-Industry Technology Research System (project no. CARS-41-K09), “973” National Basic Research Program of China (project no. 2013CB127200), Special Fund for Agro-scientific Research in the Public Interest of China (grant no. 201303044).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01388-17.
REFERENCES
- 1.He T, Wang Y, Schwarz S, Zhao Q, Shen J, Wu C. 2014. Genetic environment of the multi-resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int J Med Microbiol 304:257–261. doi: 10.1016/j.ijmm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Cuny C, Arnold P, Hermes J, Eckmanns T, Mehraj J, Schoenfelder S, Ziebuhr W, Zhao Q, Wang Y, Feßler AT. 2017. Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet Microbiol 200:88–94. doi: 10.1016/j.vetmic.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44:2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 56:1485–1490. doi: 10.1128/AAC.05827-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 8.Brenciani A, Morroni G, Pollini S, Tiberi E, Mingoia M, Varaldo PE, Rossolini GM, Giovanetti E. 2016. Characterization of novel conjugative multiresistance plasmids carrying cfr from linezolid-resistant Staphylococcus epidermidis clinical isolates from Italy. J Antimicrob Chemother 71:307–313. doi: 10.1093/jac/dkv341. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, Liu Y, He T, Shen J, Wu C. 2012. Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J Antimicrob Chemother 67:1547–1549. doi: 10.1093/jac/dks075. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob Agents Chemother 56:1650–1654. doi: 10.1128/AAC.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai L, Wu CM, Wang MG, Wang Y, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob Agents Chemother 54:3953–3955. doi: 10.1128/AAC.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J Antimicrob Chemother 54:936–939. doi: 10.1093/jac/dkh457. [DOI] [PubMed] [Google Scholar]
- 13.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. 2016. Fosfomycin. Clin Microbiol Rev 29:321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, Men S, Kong L, Ma S, Yang Y, Wang Y, Yuan Q, Cheng G, Zou W, Wang H. 2017. Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 among CTX-M-producing Escherichia coli isolates from chickens in China. Foodborne Pathog Dis 14:210–218. doi: 10.1089/fpd.2016.2230. [DOI] [PubMed] [Google Scholar]
- 15.Kitanaka H, Wachino J, Jin W, Yokoyama S, Sasano MA, Hori M, Yamada K, Kimura K, Arakawa Y. 2014. Novel integron-mediated fosfomycin resistance gene fosK. Antimicrob Agents Chemother 58:4978–4979. doi: 10.1128/AAC.03131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakaminami H, Noguchi N, Nishijima S, Kurokawa I, Sasatsu M. 2008. Characterization of the pTZ2162 encoding multidrug efflux gene qacB from Staphylococcus aureus. Plasmid 60:108–117. doi: 10.1016/j.plasmid.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 4th edition. CLSI VET01-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI VET01-S2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Liu H, Wang Y, Wu C, Schwarz S, Shen Z, Jeon B, Ding S, Zhang Q, Shen J. 2012. A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J Antimicrob Chemother 67:322–325. doi: 10.1093/jac/dkr481. [DOI] [PubMed] [Google Scholar]
- 21.Lorenzo-Diaz F, Espinosa M. 2009. Lagging-strand DNA replication origins are required for conjugal transfer of the promiscuous plasmid pMV158. J Bacteriol 191:720–727. doi: 10.1128/JB.01257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 23.Gupta A, Phung LT, Chakravarty L, Silver S. 1999. Mercury resistance in Bacillus cereus RC607: transcriptional organization and two new open reading frames. J Bacteriol 181:7080–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saltikov CW, Olson BH. 2002. Homology of Escherichia coli R773 arsA, arsB, and arsC Genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl Environ Microbiol 68:280–288. doi: 10.1128/AEM.68.1.280-288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, Werner G. 2016. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother 71:871–881. doi: 10.1093/jac/dkv420. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Sanz E, Zarazaga M, Kadlec K, Schwarz S, Torres C. 2013. Chromosomal integration of the novel plasmid pUR3912 from methicillin-susceptible Staphylococcus aureus ST398 of human origin. Clin Microbiol Infect 19:E519–E522. doi: 10.1111/1469-0691.12279. [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Sanz E, Kadlec K, Fessler AT, Billerbeck C, Zarazaga M, Schwarz S, Torres C. 2013. Analysis of a novel erm(T)- and cadDX-carrying plasmid from methicillin-susceptible Staphylococcus aureus ST398-t571 of human origin. J Antimicrob Chemother 68:471–473. doi: 10.1093/jac/dks411. [DOI] [PubMed] [Google Scholar]
- 28.Wan TW, Hung WC, Tsai JC, Lin YT, Lee H, Hsueh PR, Lee TF, Teng LJ. 2016. Novel structure of Enterococcus faecium-originated ermB-positive Tn1546-like element in Staphylococcus aureus. Antimicrob Agents Chemother 60:6108–6114. doi: 10.1128/AAC.01096-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenciani A, Morroni G, Mingoia M, Varaldo PE, Giovanetti E. 2016. Stability of the cargo regions of the cfr-carrying, multiresistance plasmid pSP01 from Staphylococcus epidermidis. Int J Med Microbiol 306:717–721. doi: 10.1016/j.ijmm.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F, Jacoby GA, Wang M. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 54:4643–4647. doi: 10.1128/AAC.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz FV, Perreten V, Teuber M. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.