ABSTRACT
Chemotherapy for tuberculosis (TB) is lengthy and could benefit from synergistic adjuvant therapeutics that enhance current and novel drug regimens. To identify genetic determinants of intrinsic antibiotic susceptibility in Mycobacterium tuberculosis, we applied a chemical genetic interaction (CGI) profiling approach. We screened a saturated transposon mutant library and identified mutants that exhibit altered fitness in the presence of partially inhibitory concentrations of rifampin, ethambutol, isoniazid, vancomycin, and meropenem, antibiotics with diverse mechanisms of action. This screen identified the M. tuberculosis cell envelope to be a major determinant of antibiotic susceptibility but did not yield mutants whose increase in susceptibility was due to transposon insertions in genes encoding efflux pumps. Intrinsic antibiotic resistance determinants affecting resistance to multiple antibiotics included the peptidoglycan-arabinogalactan ligase Lcp1, the mycolic acid synthase MmaA4, the protein translocase SecA2, the mannosyltransferase PimE, the cell envelope-associated protease CaeA/Hip1, and FecB, a putative iron dicitrate-binding protein. Characterization of a deletion mutant confirmed FecB to be involved in the intrinsic resistance to every antibiotic analyzed. In contrast to its predicted function, FecB was dispensable for growth in low-iron medium and instead functioned as a critical mediator of envelope integrity.
KEYWORDS: Mycobacterium tuberculosis, antibiotic resistance, cell envelope, mutational studies
INTRODUCTION
The treatment of tuberculosis (TB) is challenging, as the standard isoniazid (INH), rifampin, pyrazinamide, and ethambutol (HRZE) regimen for drug-sensitive TB takes at least 6 months, whereas more complicated cases of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB require the additional use of second-line drugs for an extended duration of 18 months and beyond (1). These lengthy treatment regimens result in toxic side effects for many patients and can lead to patient noncompliance, contributing to treatment failure and the development of drug-resistant TB. Consequently, there is a pressing need for the development of shorter therapeutic regimens, as well as a better understanding of the factors limiting the effectiveness of current drugs.
There are many reasons for the comparative difficulty of the treatment of TB relative to that of other bacterial diseases. Mycobacterium tuberculosis resides in the highly shielded niche of the host granuloma, the hallmark of TB pathology, and anti-TB drugs must diffuse across multiple layers of poorly vascularized tissue to reach the pathogen (2). The accessibility to bacterial targets by their cognate antibiotics is further hindered by the mycobacterial cell envelope, a complex multilayer structure characterized by a hydrophobic outer leaflet of long-chain mycolic acids covalently attached to an inner arabinogalactan-peptidoglycan complex (3). These permeability barriers, in combination with the development of phenotypically resistant persister populations (4) and possibly efflux pump systems (5), contribute to the recalcitrance of M. tuberculosis to chemotherapy.
Current research efforts have largely focused on the discovery of novel antibiotic targets in M. tuberculosis; however, the importance of intrinsic antibiotic resistance for the survival of M. tuberculosis during chemotherapy suggests an alternative strategy of identifying and targeting these intrinsic resistance mechanisms. This could potentiate existing antibiotic regimens and also increase the susceptibility of M. tuberculosis to antibiotics normally ineffective against this species. The use of antibiotic adjuvants in TB treatment has proven successful with the therapeutic combination of meropenem and the β-lactamase inhibitor clavulanic acid, which resulted in the efficient sterilization of M. tuberculosis in in vitro and in vivo experimental models and has shown promise in the treatment of MDR/XDR TB in preliminary clinical studies (6–10). Additionally, subinhibitory concentrations of cell wall inhibitors, such as bacitracin, vancomycin, and ethambutol, have been shown to mitigate intrinsic clarithromycin resistance in clinical M. tuberculosis strains (11), highlighting the potential of cell wall-targeting adjuvants to enhance current antibiotic therapies.
Transposon insertion sequencing (TnSeq) is an experimental methodology that uses next-generation sequencing to quantify mutant frequencies within a high-saturation transposon mutant library (12–15). TnSeq allows a high degree of genomic coverage and resolution and has previously been used for the identification of essential genes in the M. tuberculosis genome and also the genes conditionally required under multiple stress conditions and genetic backgrounds (12, 14, 16–18). Here, we used TnSeq in a chemical genetic interaction (CGI) profiling approach to identify genes involved in the intrinsic resistance of M. tuberculosis to antibiotics. We screened a saturated transposon mutant library for mutants that exhibit altered fitness in the presence of partially inhibitory concentrations of rifampin, ethambutol, isoniazid, vancomycin, and meropenem. Analysis of the change in frequency of the various transposon mutants using the TRANSIT TnSeq analysis platform (19) resulted in CGI signatures for each antibiotic. This approach successfully identified and ranked genes associated with antibiotic sensitivity in M. tuberculosis. Based on these predictions, we identified FecB to be required for the intrinsic resistance of M. tuberculosis to multiple antibiotics by controlling cell envelope permeability.
RESULTS
Identification of M. tuberculosis transposon mutants with altered antibiotic susceptibility.
To generate CGI profiles for M. tuberculosis, we constructed a saturated Mariner transposon mutant library consisting of approximately 105 unique mutants of M. tuberculosis strain H37Rv, achieving approximately 65% coverage of the possible TA dinucleotide transposon insertion sites in the H37Rv genome. We chose rifampin, ethambutol, isoniazid, vancomycin, and meropenem for selection to cover a range of clinically relevant target mechanisms (RNA polymerase and mycolic acid, arabinogalactan, and peptidoglycan biosynthesis). The library was exposed to partially inhibitory levels of these antibiotics, resulting in a 25 to 40% reduction of the overall growth rate relative to that for the antibiotic-free control library (Fig. 1A).
FIG 1.
Identification of M. tuberculosis transposon mutants with altered antibiotic susceptibility. (A) Partially inhibitory antibiotic selection of transposon mutant libraries. Growth curves are representative of the growth kinetics of the transposon mutant libraries in the presence or absence of antibiotic selection. d, days. (B) Cluster analysis of CGI profiles. Hierarchical clustering was performed on the CGI profiles derived from triplicate experiments using Pearson's correlation as the similarity metric. The mean log2 TnSeq-FC in transposon mutant frequency for each gene following antibiotic selection relative to that for the antibiotic-free control is indicated on the color scale, with increased mutant representation being indicated in red and reduced representation being indicated in blue. Genes that did not exhibit statistically significant differences (i.e., q ≥ 0.05, based on resampling testing) in transposon insertion under any of the antibiotic selection conditions tested were omitted, and the 251 remaining genes were used in this analysis.
Competitive outgrowth for approximately 6.5 generations (100-fold expansion of the population) resulted in altered frequencies of numerous mutants. We defined the antibiotic susceptibility of mutants with mutations in a particular genetic locus as a function of the log2 fold change in mutant frequency between the antibiotic-selected and antibiotic-free libraries. A negative log2 TnSeq fold change (LTnSeq-FC) indicates a decreased relative mutant frequency in the antibiotic-selected library and, thus, increased antibiotic sensitivity, whereas a positive LTnSeq-FC indicates antibiotic resistance of the corresponding mutant.
We used the TRANSIT TnSeq analysis tool (19) on experimentally independent triplicate data sets and identified 251 mutants that, when mutated, were associated with a change in fitness under antibiotic selection that was statistically significant after correction for multiple comparisons (q < 0.05) under at least one of the antibiotic selection conditions tested (see Table S1 in the supplemental material). Hierarchical cluster analysis revealed CGI profiles that were specific for each antibiotic. Unexpectedly, the vancomycin and rifampin CGI profiles clustered closely together, while the isoniazid profile was distinct from those of the other four antibiotics (Fig. 1B).
To identify genes that contribute to the intrinsic resistance of M. tuberculosis to multiple antibiotics, we determined the overlap of genes shared between two CGI profiles for mutants sensitive to different antibiotics (those with a negative LTnSeq-FC; q < 0.05) (Fig. 2A). We quantified the number of genes shared between two CGI profiles and determined the Jaccard index, which calculates the proportional overlap between the two sets [(A ∩ B)/(A ∪ B), where A and B are different sets of sensitive mutants]. Consistent with the results of cluster analysis, there was a large degree of overlap between the genes shared between the CGI profiles for the vancomycin- and rifampin-sensitive mutant subsets, with the subsets having 47 genes in common (Jaccard index = 0.43). There was slightly less overlap of shared genes between the rifampin- and meropenem-sensitive mutant subsets, which had 34 shared genes (Jaccard index = 0.32). Although both meropenem and vancomycin target peptidoglycan, the overlap of shared genes between the mutant subsets sensitive to these two antibiotics was even more modest (31 genes; proportional overlap = 0.26). The genes for the ethambutol-sensitive subset overlapped with those for the vancomycin-sensitive subset (31 genes; Jaccard index = 0.32) more than they did with the genes for the other antibiotic-sensitive subsets. In agreement with the results of cluster analysis, which revealed that the profiles for isoniazid were least related to those for the other drugs, the genes for the isoniazid-sensitive mutant subset overlapped the least with those for the other antibiotic-sensitive mutant subsets. Several mutations led to increased sensitivity to multiple antibiotics (Fig. 2B). Seventeen genes were linked to intrinsic resistance to at least 4 out of the 5 antibiotics tested, and transposon insertions in fecB were predicted to result in cross-sensitivity to all five antibiotics.
FIG 2.

Genetic determinants of intrinsic antibiotic resistance are shared between different antibiotics. (A) Overlap between CGI profiles. The overlap matrix indicates the number of genes shared between pairs of antibiotic sensitivity profiles (numbers not in parentheses). The degree of overlap is represented in the form of the Jaccard index (numbers in parentheses), i.e., the proportion of sensitivity genes shared between the two libraries over the total number of genes from both libraries [(A ∩ B)/(A ∪ B), where A and B are different sets of sensitive mutants]. (B) Venn diagram of antibiotic-sensitive mutants. Sets of significantly sensitive mutants (negative TnSeq-FC, q < 0.05) for each antibiotic are represented by the colored regions, with the number of overlapping genes in their respective zones being indicated. RIF, rifampin; INH, isoniazid; EMB, ethambutol; MERO or MER, meropenem; VAN, vancomycin.
CGI profiling ranks mutants according to their antibiotic susceptibility.
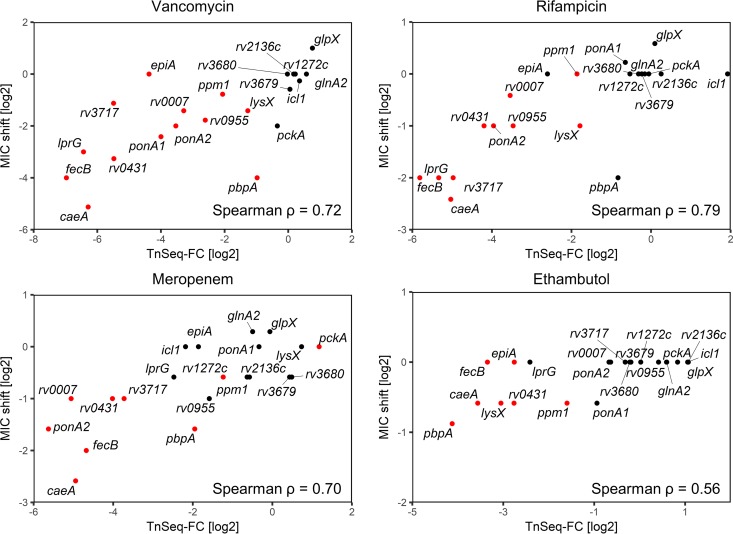
To validate the predictions of the TnSeq screen, we determined the MICs of the respective antibiotics against transposon or deletion mutants in monocultures. We selected a panel of 21 mutants, of which 15 were predicted to have significantly altered susceptibility to at least one of the drugs; 6 control mutants with no significant changes in representation in the TnSeq screen were also included (Table S2). In general, there was a good correlation between the LTnSeq-FC and the measured MIC shifts, resulting in Spearman correlation coefficients ranging from 0. 56 to 0.79 (Fig. 3), validating the ability of the TnSeq screens to predictively rank mutants in order of antibiotic sensitivity. Isoniazid was an exception: none of the 4 mutants predicted by the screen to be hypersusceptible to isoniazid showed by MIC determinations increased INH susceptibility compared to wild-type (WT) M. tuberculosis when they were assayed in single-strain cultures. Unfortunately, there were only 4 predicted isoniazid-sensitive mutants within our available validating mutant panel, which prevented a broader analysis of the correlation. It should be noted that a TnSeq screen assesses mutant fitness at a single antibiotic concentration and is not a direct measurement of the MIC. Thus, a lower mutant frequency within an antibiotic-selected transposon mutant library does not necessarily require or imply a decrease in the mutant MIC (20). To determine if this contributed to the outcome of the INH screen, we performed growth assays with various mutants in the presence of a low isoniazid concentration that affected the growth of the WT only slightly, as in the TnSeq screen (Fig. S1). We also analyzed the growth of mutants that were predicted to be hypersusceptible to ethambutol by TnSeq but showed only a minor shift in MIC. The fecB, lysX, ponA1, and ppm1 mutants exhibited stronger growth attenuation than the WT in the presence of 27 ng/ml INH. Similarly, the fecB, rv0431, lysX, rv2224c, and pbpA mutants displayed severe growth attenuation in the presence of 0.5 μg/ml of ethambutol, which affected the growth of the WT only marginally. This confirmed that some mutants have reduced fitness at partially inhibitory antibiotic concentrations, despite the absence of a substantial MIC shift compared to that for the WT.
FIG 3.
Validation of intrinsic antibiotic resistance genes with MIC measurements. The Spearman rank correlation (ρ) between the log2 fold change from the antibiotic TnSeq screen and the observed log2 MIC90 shift in knockout mutants is indicated. Log2 TnSeq-FC values are derived from triplicate experiments, while the MIC shifts shown are the means from duplicate experiments. Mutants predicted to be significantly underrepresented or enriched by the TnSeq screen (q < 0.05) are colored red.
Functional categorization of genes related to antibiotic sensitivity and resistance.
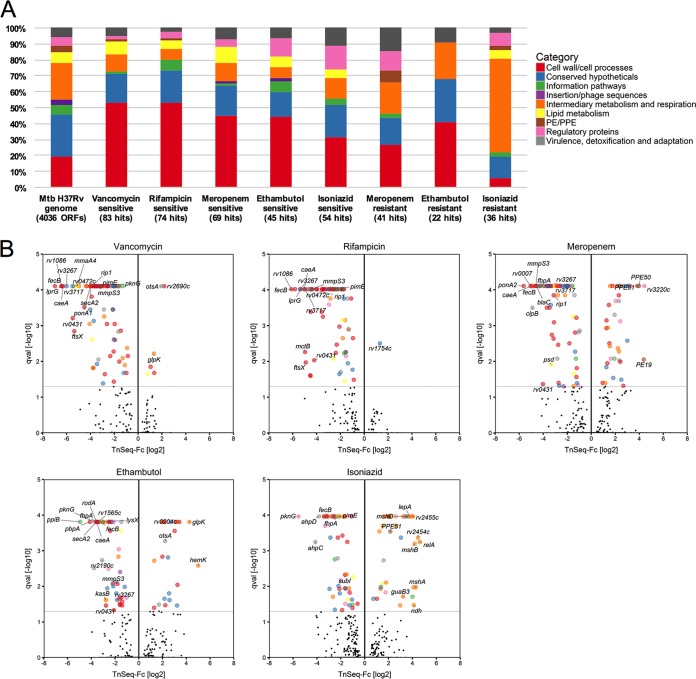
We summarized the TnSeq data in a searchable spreadsheet (Table S3). The functional classification of antibiotic susceptibility genes indicates that intrinsic antibiotic resistance is determined to a large degree by genes involved in cell wall/cell processes, comprising about 50% of the genes in the vancomycin-, rifampin-, meropenem-, and ethambutol-hypersensitive mutants and 30% of the genes in the isoniazid-sensitive mutants (Fig. 4A). Many of the proteins conferring intrinsic antibiotic resistance were shared among the groups of proteins conferring resistance to the various antibiotics (Fig. 2B and 4B). Among these are FecB, annotated as an iron dicitrate-binding periplasmic lipoprotein, Rv3267/Lcp1; a recently characterized LytR-CpsA-Psr (LCP) family protein that is capable of ligating arabinogalactan to peptidoglycan (21, 22); the protein translocase SecA2; the mannosyltransferase PimE; and Rv2224c/CaeA/Hip1, a cell envelope-associated carboxyesterase/protease important for virulence (23, 24) and associated with increased imipenem sensitivity when inactivated (25). In addition to identifying mutants with increased antibiotic susceptibility, CGI profiling also identified mutants that showed increased antibiotic resistance. While rifampin- and vancomycin-resistant mutants were infrequent, selection with the other three antibiotics enriched numerous mutants with increased resistance (Fig. 4B). Among the genes whose interruption caused increased resistance to INH were ndh, which codes for a type II NADH dehydrogenase, and the mycothiol biosynthesis genes mshA, mshB, and mshD. Mutations in these genes have previously been shown to cause INH resistance in M. smegmatis and M. bovis BCG (26–28).
FIG 4.
Identification and functional categorization of genes related to intrinsic antibiotic resistance. (A) Proportions of sensitive and resistant mutants corresponding to the different functional categories as annotated in TubercuList (57). The overall functional composition of the M. tuberculosis (Mtb) H37Rv genome is displayed to the left of the chart as a point of reference. Data for vancomycin- and rifampin-resistant mutants are not displayed due to their small number. (B) TnSeq-FC- and false discovery rate-adjusted P values (q values [qval]) from the resampling test are plotted for each genetic locus. Loci meeting the significance threshold of a q value of <0.05 are colored according to their functional categories. Selected mutants are indicated by name (see Table S3 in the supplemental material for all mutants).
Regulatory proteins linked to antibiotic cross-sensitivity include the protein kinase PknG, which was previously shown to be involved in intrinsic resistance to multiple antibiotics, including erythromycin, vancomycin, ethambutol, rifampin, and imipenem (29); the site 2 protease Rip1 (Rv2869c), which controls the transcription of multiple pathways, including mycolic acid biosynthesis (30, 31); and the uncharacterized TetR-family transcriptional regulator Rv0472c.
The transposon library contained mutant strains with insertions in 51 genes coding for putative efflux pumps (32, 33) (Table S4). Surprisingly, disruption of individual efflux pump genes did not significantly affect mutant fitness at the antibiotic concentrations used. While transposon insertions in the ABC transporter gene rv1272c were underrepresented after meropenem selection, we did not observe a decrease in the MIC of meropenem for the rv1272::Tn mutant (Fig. 3). Mutant frequencies for rv1747, encoding a probable ATP-binding protein ABC transporter, were significantly reduced after outgrowth with vancomycin, whereas transposon insertions disrupting rv1410c, encoding an efflux pump (34), were underrepresented after selection with vancomycin, rifampin, and meropenem; however, both genes have also been implicated in cell wall biosynthesis (35, 36) and may thus contribute to drug resistance in a drug efflux-independent manner.
FecB is important for M. tuberculosis resistance to multiple antibiotics.
fecB transposon mutants were significantly underrepresented in libraries grown in the presence of all antibiotics tested (Fig. 2B), suggesting that fecB has an important role in antibiotic cross-resistance. We generated a ΔfecB mutant by replacing fecB with a hygromycin resistance cassette, which we confirmed by Southern blotting (Fig. S2). On the basis of the sequence homology, FecB has been annotated as a putative iron(III) dicitrate-binding lipoprotein, similar to the substrate-binding protein in the FecBCDE iron citrate ABC transporter system in Escherichia coli (37–39). However, the other components of this system are absent from the M. tuberculosis genome, indicating that fecB may be an orphan gene fulfilling a different function. The ΔfecB mutant did not exhibit any major growth defects relative to the WT and complemented strains under a range of iron-limiting and iron-replete growth conditions (Fig. S3), showing that it is not essential for iron transport.
We tested the predictions of the TnSeq screen by MIC assays (Table 1). The MIC for the ΔfecB mutant compared to that for the WT was reduced 16-fold for vancomycin and reduced 4-fold for rifampin and meropenem, consistent with the findings of CGI profiling. We did not observe a reduction in the MICs for isoniazid and ethambutol, but the growth rate of the ΔfecB mutant relative to that of the WT strain was reduced in the presence of partially inhibitory concentrations of isoniazid and ethambutol (Fig. S1), confirming the CGI profiling predictions of reduced ΔfecB mutant fitness for growth in the presence of these antibiotics.
TABLE 1.
MICs against H37Rv WT, ΔfecB, and fecB-complemented strains
| Antibiotic | Molecular mass (g/mol) | Target | MICa (μg/ml) |
MIC ratiob | ||
|---|---|---|---|---|---|---|
| WT | ΔfecB mutant | fecB-complemented strain | ||||
| Faropenem | 285.3 | Peptidoglycan | 12.5 ± 8.2 | 1.56 ± 1.03 | 21.3 ± 20.5 | 8 |
| Meropenem | 383.5 | Peptidoglycan | 11.3 ± 4.1 | 2.81 ± 1.03 | 11.3 ± 4.1 | 4 |
| Vancomycin | 1449.3 | Peptidoglycan | 20 ± 0 | 1.25 ± 0 | 23.3 ± 8.2 | 16 |
| Fidaxomicin | 1,058.0 | RNA polymerase | 1.2 ± 0.61 | 0.098 ± 0 | 0.8 ± 0.61 | 12 |
| Rifampin | 822.9 | RNA polymerase | 0.02 ± 0 | 0.005 ± 0 | 0.02 ± 0 | 4 |
| Rifabutin | 847.0 | RNA polymerase | 0.01 ± 0 | 0.0029 ± 0.001 | 0.008 ± 0 | 3.4 |
| Ethambutol | 204.3 | Arabinogalactan | 0.91 ± 0 | 0.91 ± 0 | 1.36 ± 0 | 1 |
| Ethionamide | 166.2 | Mycolic acid | 0.55 ± 0.19 | 0.47 ± 0.24 | 0.55 ± 0.19 | 1.2 |
| Isoniazid | 137.1 | Mycolic acid | 0.05 ± 0.011 | 0.05 ± 0.011 | 0.05 ± 0.011 | 1 |
| PA-824 | 359.3 | Mycolic acid | 0.25 ± 0.21 | 0.16 ± 0 | 0.16 ± 0 | 1.6 |
| SQ109 | 330.6 | Mycolic acid | 2.5 ± 0 | 0.63 ± 0 | 2.5 ± 0 | 4 |
| Triclosan | 289.5 | Mycolic acid | 62.5 ± 0 | 62.5 ± 0 | 62.5 ± 0 | 1 |
| Moxifloxacin | 401.4 | DNA gyrase | 0.1 ± 0 | 0.1 ± 0 | 0.1 ± 0 | 1 |
| Chloramphenicol | 323.1 | Protein synthesis | 5.86 ± 0 | 5.86 ± 0 | 8.79 ± 7.18 | 1 |
| Linezolid | 337.4 | Protein synthesis | 1.37 ± 0.48 | 0.39 ± 0 | 1.37 ± 0.48 | 3.5 |
| Streptomycin | 581.6 | Protein synthesis | 0.29 ± 0 | 0.27 ± 0.06 | 0.27 ± 0.06 | 1.1 |
| Nigericin | 725.0 | Respiration | 6.25 ± 4.84 | 1.95 ± 1.28 | 6.25 ± 4.84 | 3.2 |
| Thioridazine | 370.6 | Respiration | 23.4 ± 19.1 | 15.6 ± 0 | 15.6 ± 0 | 1.5 |
| Valinomycin | 1,111.3 | Respiration | 1.56 ± 0 | 0.2 ± 0.15 | 2.34 ± 1.91 | 8 |
The mean ± SD MIC values were derived from two independent experiments with triplicate cultures. The MICs were determined as described in Materials and Methods.
MIC for WT/MIC for knockout mutant.
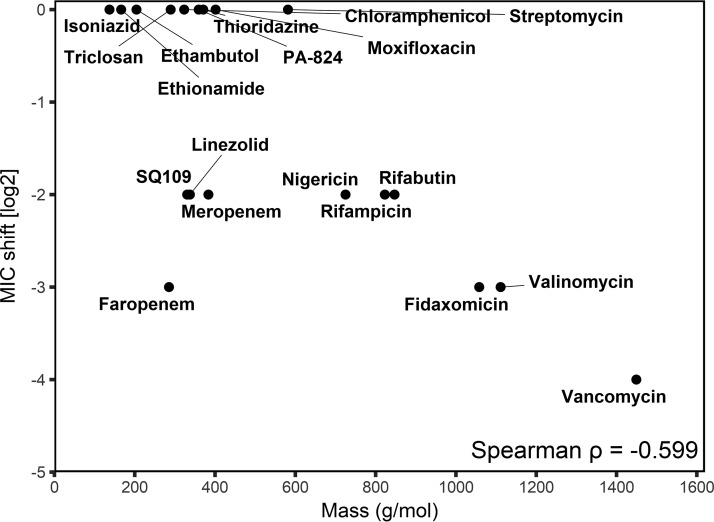
Additional MIC assays showed that the ΔfecB mutant was hypersensitive to multiple antibiotics to various degrees and that the increased antibiotic sensitivity could be complemented by the constitutive expression of fecB from an integrative plasmid (Table 1). The fold reduction in the antibiotic MIC against the ΔfecB mutant correlated with the molecular mass of the antibiotic, suggesting that the ΔfecB mutant was preferentially more sensitive than WT M. tuberculosis to higher-molecular-mass antibiotics (Fig. 5).
FIG 5.
Intrinsic drug resistance of the ΔfecB mutant correlates with the antibiotic molecular mass. A correlation plot of the reduction in the MIC against the ΔfecB mutant relative to the MIC against the H37Rv WT strain versus antibiotic molecular mass is shown.
Loss of fecB results in increased cell envelope permeability.
Given the large proportion of cell envelope processes represented in the antibiotic-sensitive mutants, as predicted by TnSeq, we hypothesized that FecB is also important in controlling the integrity of the cell envelope. We tested for increased cell envelope permeability in the ΔfecB mutant using the ethidium bromide (EtBr) uptake assay (40). The ΔfecB mutant exhibited a 4- to 5-fold increase in ethidium bromide uptake relative to the WT and complemented strains on the basis of fluorescence measurements, suggesting that FecB inactivation results in increased cell envelope permeability (Fig. 6A). To investigate the possibility that the loss of fecB may have resulted in greater EtBr accumulation due to reduced efflux, we treated the cells with the efflux pump inhibitor verapamil (Fig. 6B). All three strains showed a 50% increase in EtBr accumulation in the presence of verapamil, with the ΔfecB mutant still exhibiting a 4- to 5-fold greater accumulation than the other strains. This indicated that the increased EtBr accumulation in the ΔfecB mutant was independent of verapamil-sensitive efflux mechanisms and likely due to increased cell envelope permeability.
FIG 6.
The ΔfecB mutant exhibits increased uptake of exogenous substrates. (A, B) Uptake of ethidium bromide by the WT (black), ΔfecB (red), and complemented (Comp; blue) strains in the absence (A) and presence (B) of 100 μg/ml verapamil. The fluorescent emission at 590 nm was measured at 1-min intervals from triplicate wells, and the data shown are representative of those from two independent experiments. (C) Uptake of Bodipy-FL-tagged vancomycin. A suspension of M. tuberculosis bacteria incubated with tagged vancomycin was sampled at the time points indicated. The emission at 538 nm normalized to the OD of the suspension was measured. The data points shown are means ± SDs from triplicate measurements and are representative of those from two independent experiments.
To confirm if increased cell envelope permeability resulted in increased antibiotic uptake, we measured the uptake of fluorescence-tagged vancomycin in the various M. tuberculosis strains (Fig. 6C). The ΔfecB mutant exhibited a similar 4- to 5-fold increase in vancomycin uptake as the WT and complemented strains, implying that the observed hypersensitivity could be attributed to an increase in cell envelope permeability.
DISCUSSION
In this study, we used a TnSeq-based CGI profiling approach to identify and rank the genetic determinants of M. tuberculosis fitness under antibiotic selection pressure. MIC determinations for individual mutants validated many of the TnSeq predictions. Mutants that did not display a shift in MIC had a fitness defect at the drug concentration used in the screen, as previously observed in other studies (20, 41).
One of our goals was to determine if the mechanisms governing intrinsic antibiotic resistance are antibiotic specific or shared across the mechanisms governing resistance to different antibiotics. Transposon interrupted genes important for functions related to cell envelope processes and cell division featured predominantly among the mutants sensitive to the five antibiotics tested. This might have been due to increased cell envelope permeability and antibiotic uptake resulting from a compromised cell envelope; alternatively, the loss of function of genes involved in cell envelope processes might sensitize M. tuberculosis to normally sublethal levels of antibiotic-induced cell envelope damage. Similarly, a TnSeq screen for intrinsic resistance factors in Staphylococcus aureus identified multicomponent sensing systems that regulate the expression of cell wall biosynthetic genes and modulators of cell surface charge (42).
The CGI profile for mutants sensitive to rifampin overlapped most with that for mutants sensitive to vancomycin; this result was intriguing, given that rifampin inhibits RNA polymerase, whereas vancomycin targets peptidoglycan biosynthesis. The overlap of the genes shared by the vancomycin- and rifampin-sensitive mutants may indicate that cell envelope permeability plays an important role in intrinsic resistance to these drugs, possibly related to their large molecular sizes. These results also suggest the possibility that subinhibitory concentrations of rifampin may result in cell envelope stress that becomes synthetically lethal with the loss of function of certain genes that are important for cell envelope integrity. Recent studies have shown that point mutations in rpoB, the primary target of rifampin, result in altered fatty acid metabolism (43) and profound changes in cell wall lipid composition, suggesting that inhibition of RNA polymerase by rifampin likely affects cell envelope structure and integrity. Cell envelope damage could thus possibly be a major downstream effect of RNA polymerase inhibition, leading to bactericidality and potential synergistic combinations with rifampin. Combining rifampin with cell envelope-targeting drugs, in particular, the peptidoglycan-targeting drugs, could lead to more effective anti-TB treatment regimens. This is supported by a recent study demonstrating a synergistic effect between rifampin and carbapenem antibiotics (44).
Genes that were unique to each of the sensitivity profiles may be indicative of mechanistic pathways specific to each drug. Transposon insertions in ponA1, encoding a penicillin binding protein, were associated with increased sensitivity to vancomycin but not meropenem (see Table S2 in the supplemental material), highlighting that while both drugs inhibit peptidoglycan biosynthesis, they may affect different functional aspects of the process. Mutations in the broad-spectrum β-lactamase gene blaC specifically led to increased sensitivity to meropenem but not any of the other drugs. Transposon insertions in the mce1 locus and the PE/PPE protein genes pe19, ppe50, and ppe51 were enriched by meropenem selection but not by vancomycin or rifampin selection. Overexpression of PE19 was previously shown to result in increased cell envelope permeability (45), and we hypothesize that the loss of PE19 may specifically reduce permeability to meropenem but not to vancomycin and rifampin.
We sought to identify the main efflux pump systems contributing to intrinsic antibiotic resistance but did not observe a significant requirement for efflux pump genes in the presence of antibiotic selection. There are many possible explanations for this conspicuous absence. It may be due to redundancy between multiple efflux pump systems, which could buffer against the effects of disruption of a single efflux pump gene in the TnSeq screen. Alternatively, drug efflux may be mostly mediated by the few essential efflux pump genes (efpA, irtB, rv1463, or rv3806c), which were not detectable in our transposon mutant libraries, or the partially inhibitory antibiotic concentrations were insufficient to induce the efflux pumps. Finally, it is possible that efflux pumps do not play a key role in intrinsic resistance to the antibiotics tested. It was previously noted that treatment with the efflux pump inhibitor reserpine or verapamil did not affect the overall uptake of ofloxacin or rifampin in either replicating or nonreplicating M. tuberculosis (46, 47). The authors of these studies did, however, suggest the possibility that the efficacious induction of efflux pumps may be specific to certain drug-resistant strains and that efflux may be comparatively less important in the laboratory H37Rv strain used in the current study. We note that our study does not rule out the possibility that efflux pumps are important determinants of resistance to other antibiotics, and further screening needs to be done to address this.
The observation that certain genes affect antibiotic susceptibility in replicating M. tuberculosis cultures raises the question of whether they also contribute to antibiotic resistance in nonreplicating persister populations. Environmental stresses, such as hypoxia, oxidative stress, and nutrient deprivation, are known to induce global transcriptional changes, resulting in altered metabolism, reduced cellular replication, and structural changes to the cell envelope, leading to a state of phenotypic drug tolerance (46, 48, 49). It has previously been shown that the sulfate transport system proteins CysT, CysW, CysA1, and SubI are upregulated under starvation conditions (50); our observation that transposon insertions in subI-cysTWA1 (rv2400c-rv2397c) were associated with antibiotic cross-sensitivity suggests a possible mechanism by which the upregulation of sulfate utilization pathways may promote antibiotic tolerance through the increased synthesis of sulfolipids or other sulfated metabolites that contribute to intrinsic resistance. Further characterization of antibiotic susceptibility-determining genes may reveal novel mechanisms contributing to phenotypic drug resistance in M. tuberculosis.
Genes that are required for growth in vitro were not represented in the transposon mutant libraries used in our screen. Of the nonessential genes, those that are important for M. tuberculosis survival in vivo may still prove to be relevant chemotherapeutic targets. We thus compared the results of our antibiotic susceptibility screens with those of a previous in vivo essentiality screen (17) to identify novel targets that could also potentiate the activity of other antibiotics. We found that among the 43 mutants that were predicted to be sensitive to at least 3 out of the 5 antibiotics tested in our screen, 20 were predicted to be attenuated for growth and survival in vivo (Table S5). These included mutants with mutations in rv2224c, pknG, and fecB, the last of which was newly identified in this screen to be an important determinant of the intrinsic resistance of M. tuberculosis to antibiotics. Of note, pharmaceutical inhibition of PknG resulted in increased lysosomal localization and promoted the killing of mycobacteria in macrophages (51). As PknG is required for the intrinsic resistance of mycobacteria to multiple antibiotics (29), the use of PknG inhibitors may provide a two-pronged therapeutic approach by improving the efficacy of the host immune response as well as augmenting the activity of coadministered antibiotics (29, 51). As discussed above, many of the genes required for intrinsic antibiotic resistance and virulence have known functions in cell envelope biosynthesis and remodeling. We hypothesize that the loss of cell envelope integrity leads to pleiotropic stress phenotypes, possibly due to increased permeability to toxic agents, such as antibiotics, weak acids, reactive oxygen or nitrogen species, and antimicrobial peptides. Despite their in vitro nonessentiality, these may prove to be promising candidates whose inactivation causes growth inhibition or death in vivo and could synergize with current antibiotics.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis H37Rv (obtained from the Trudeau Institute) and derivative strains were cultured at 37°C with 5% CO2 in Middlebrook 7H9 medium (BD Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, 0.5% bovine serum albumin (Roche), 0.2% dextrose, and 0.085% NaCl.
Mutant strains.
The mutants used in this study are listed in Table S2 in the supplemental material. fecB, epiA, ponA1, and rv3717 gene deletion mutants were constructed by allelic exchange using a recombineering approach as previously described (52), and the deletions were confirmed by Southern blotting using an Amersham ECL direct nucleic acid labeling system kit (GE Healthcare) and an OptEIA tetramethylbenzidine substrate reagent set (Becton Dickinson). The ΔfecB strain was complemented by reintroducing a copy of fecB under the control of the hsp60 promoter into the attL5 site of the M. tuberculosis genome.
Transposon library construction.
An M. tuberculosis H37Rv transposon library was constructed by himar1 mutagenesis as previously described (13). Briefly, 100 ml of a mid-log-phase M. tuberculosis culture (optical density at 600 nm [OD600] = ∼0.7 to 1.0) was incubated with 1 × 1011 to 2 × 1011 PFU of ΦMycoMarT7 phage (47) at 37°C for 4 h. The cultures were then washed and plated on Middlebrook 7H9 medium with 1.5% agar supplemented with oleic acid-albumin-dextrose-catalase (BD BBL), 0.5% glycerol, and 0.05% Tween 80 and incubated for 19 days at 37°C, yielding a library of ∼105 mutants. The library was harvested by scraping and stored as frozen stocks in Middlebrook 7H9 medium with 15% glycerol at −80°C for further experiments. Library coverage of TA dinucleotide sites was determined to be ∼65% by Illumina sequencing.
Antibiotic selection of transposon libraries.
A transposon library frozen stock was thawed and incubated in Middlebrook 7H9 medium for 4 days at 37°C to allow the library to recover. This library starter culture was subcultured into 25-ml cultures at a starting OD580 of 0.01 with various antibiotic concentrations, including an antibiotic-free control. The cultures were incubated at 37°C until the OD580 was ∼1.0 in order to standardize the number of outgrowth generations between libraries at approximately 6.5 generations. A partially inhibitory concentration inhibiting the library growth rate to between 60 and 75% of that of the antibiotic-free library was determined for each antibiotic; this concentration was typically between 0.25× and 0.5× the antibiotic MIC; the selection concentrations used were 16 μg/ml for vancomycin, 4 ng/ml for rifampin, 1.2 μg/ml for meropenem, 21 to 27 ng/ml for isoniazid, and 0.4 to 0.6 μg/ml for ethambutol. Three independent library selections were performed for each antibiotic.
Sequencing of transposon mutant libraries.
Genomic DNA was extracted from selected transposon libraries, and the library mutant composition was determined by sequencing amplicons of the transposon-genome junctions as previously described (13, 53). On average, library sequencing yielded between 0.5 million and 4 million unique transposon-genome junction reads, covering between 50 and 65% of the possible TA dinucleotide insertion sites in the genome.
Mapping and quantification of transposon insertions.
Raw sequence data were processed using the TPP tool from the TRANSIT TnSeq analysis platform (19), and transposon genome junctions were mapped to the M. tuberculosis H37Rv reference genome (GenBank accession number NC_018143.1) using the Burroughs-Wheeler aligner (BWA). To account for possible PCR amplification biases, reads corresponding to the same TA site and possessing the same 7-nucleotide barcode were considered to be derived from the same template, and these duplicate reads were discarded from the final template counts. For reference, gene essentiality calls (displayed in Table S3 in the supplemental material) were assigned using a 4-state hidden Markov model (54) on the basis of the library data sets obtained from 4 independently constructed libraries outgrown on 7H9 agar plates.
Identification of genes affecting fitness under antibiotic selection.
Genes conditionally affecting fitness in the presence of antibiotic selection were identified using the resampling test module in the TRANSIT analysis platform as previously described (19). Briefly, triplicate libraries outgrown in the presence of antibiotics were compared to their corresponding antibiotic-free controls derived from the same starter cultures; differences in template counts between libraries were normalized using trimmed total reads (TTR) normalization, which normalizes data sets such that they have the same expected read counts (with minimal influence from outliers). Significant differences between the sum of the read counts under the antibiotic-free and antibiotic-selected conditions were evaluated by comparison to a resampling distribution derived by random permutation of the observed counts of TA sites within a particular genetic locus among all data sets. P values were defined as the proportion of values within 106 permutations that had a value more extreme than the observed experimental result, and these P values were adjusted for multiple comparisons (q value) using the Benjamini-Hochberg procedure. TnSeq fold changes (TnSeq-FC) were computed as log2-transformed ratios of the normalized read counts between the antibiotic-selected and antibiotic-free libraries; to facilitate logarithmic transformation, read counts of zero (no insertions) were replaced with a pseudocount corresponding to the detection limit of the sequencing data. We defined genes having a q value of <0.05 according to the permutation test to be significant determinants of fitness under antibiotic selection.
Antibiotic sensitivity assay.
The MICs of antibiotics against the various M. tuberculosis strains were determined using the broth microdilution technique. M. tuberculosis strains were grown to mid-log phase (OD580 = 0.6 to 1.0), and equal volumes of an M. tuberculosis suspension were serially diluted in Middlebrook 7H9 medium to a final OD580 of 0.01 to 0.025 per well. The OD580 in each well was measured after 10 to 14 days of outgrowth, and the MIC90 was determined to be the concentration of antibiotic at which bacterial growth was inhibited by 90% relative to the growth in the antibiotic-free control wells. Assays were repeated with a minimum of two replicates for each strain.
Permeability assay.
Cell envelope permeability was determined using the ethidium bromide (EtBr) uptake assay as previously described (40). Briefly, mid-log-phase M. tuberculosis cultures were washed once in phosphate-buffered saline (PBS) with 0.05% Tween 80 and adjusted to an OD580 of 0.8 in PBS supplemented with 0.4% glucose. One hundred microliters of bacteria was added to triplicate wells in a black 96-well plate with clear-bottomed wells (Costar), and an equal volume of 2 μg/ml EtBr in PBS with 0.4% glucose was added to each well to a final EtBr concentration of 1 μg/ml and an OD580 of 0.4. EtBr fluorescence was measured at an excitation wavelength of 530 nm and an emission wavelength of 590 nm at 1-min intervals over a course of 60 min. To investigate possible effects of efflux inhibition, the experiment was repeated with the addition of 100 μg/ml of verapamil to the test wells.
A similar assay was performed to determine permeability to fluorescence-tagged vancomycin. M. tuberculosis suspensions at an OD580 of 0.4 in PBS with 0.4% glucose were incubated with 2 μg/ml of Bodipy-FL-tagged vancomycin (Thermo Scientific), and 200-μl sample aliquots were taken at the 5-, 30-, and 60-min incubation time points, washed twice, and, finally, resuspended in 200 μl PBS. Fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength at 538 nm and normalized to the OD580 of the final suspension to account for cell loss during the washing.
Additional data analysis and representation.
The similarity between the various antibiotic sensitivity profiles was determined on the basis of average linkage clustering of uncentered Pearson correlations with the Cluster (version 3.0) program (55). Data arrays and cluster trees were visualized using Java Treeview software (56).
The overlap between antibiotic sensitivity profiles was quantified in terms of the Jaccard index [i.e., the proportion of shared genes over the total number of genes in both profiles, or (A ∩ B)/(A ∪ B)]. Venn diagrams were generated using the Venn diagrams web tool (http://bioinformatics.psb.ugent.be/webtools/Venn).
The Spearman correlation between MIC shifts and TnSeq-FCs were calculated in Prism (version 7.0) software. Numerical data and figures were prepared using Microsoft Excel, Prism (version 7.0), and R software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Namita Trikannad from the High Throughput and Spectroscopy Center at The Rockefeller University for guidance using the Tecan Freedom EVO 150 liquid dispenser. We are grateful to Eric Rubin and Christopher Sassetti and their lab members for their advice and helpful discussions.
This work was supported by National Institutes of Health (NIH) grants U19 AI107774 and U19 AI111143 (Tri-Institutional TB Research Unit).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01334-17.
REFERENCES
- 1.World Health Organization. 2010. Guidelines for treatment of tuberculosis, 4th ed World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/44165/1/9789241547833_eng.pdf. [Google Scholar]
- 2.Dartois V. 2014. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nat Rev Microbiol 12:159–167. doi: 10.1038/nrmicro3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarlier V, Nikaido H. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett 123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge BB, Fortune SM. 2014. The spectrum of drug susceptibility in mycobacteria. Microbiol Spectr 2(5):MGM2-0031-2013. doi: 10.1128/microbiolspec.MGM2-0031-2013. [DOI] [PubMed] [Google Scholar]
- 5.da Silva PEA, Von Groll A, Martin A, Palomino JC. 2011. Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol Med Microbiol 63:1–9. doi: 10.1111/j.1574-695X.2011.00831.x. [DOI] [PubMed] [Google Scholar]
- 6.De Lorenzo S, Alffenaar JW, Sotgiu G, Centis R, D'Ambrosio L, Tiberi S, Bolhuis MS, van Altena R, Viggiani P, Piana A, Spanevello A, Migliori GB. 2013. Efficacy and safety of meropenem-clavulanate added to linezolid-containing regimens in the treatment of MDR-/XDR-TB. Eur Respir J 41:1386–1392. doi: 10.1183/09031936.00124312. [DOI] [PubMed] [Google Scholar]
- 7.England K, Boshoff HIM, Arora K, Weiner D, Dayao E, Schimel D, Via LE, Barry CE. 2012. Meropenem-clavulanic acid shows activity against Mycobacterium tuberculosis in vivo. Antimicrob Agents Chemother 56:3384–3387. doi: 10.1128/AAC.05690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugonnet J-E, Tremblay LW, Boshoff HI, Barry CE, Blanchard JS. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HIM, Barry CE III. 2012. Meropenem inhibits d,d-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol 86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payen MC, De Wit S, Martin C, Sergysels R, Muylle I, Van Laethem Y, Clumeck N. 2012. Clinical use of the meropenem-clavulanate combination for extensively drug-resistant tuberculosis. Int J Tuberc Lung Dis 16:558–560. doi: 10.5588/ijtld.11.0414. [DOI] [PubMed] [Google Scholar]
- 11.Bosne-David S, Barros V, Verde SC, Portugal C. 2000. Intrinsic resistance of Mycobacterium tuberculosis to clarithromycin is effectively reversed by subinhibitory concentrations of cell wall inhibitors. J Antimicrob Chemother 46:391–395. doi: 10.1093/jac/46.3.391. [DOI] [PubMed] [Google Scholar]
- 12.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long JE, DeJesus M, Ward D, Baker RE, Ioerger T, Sassetti CM. 2015. Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol Biol 1279:79–95. doi: 10.1007/978-1-4939-2398-4_6. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog 8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieser KJ, Baranowski C, Chao MC, Long JE, Sassetti CM, Waldor MK, Sacchettini JC, Ioerger TR, Rubin EJ. 2015. Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc Natl Acad Sci U S A 112:13087–13092. doi: 10.1073/pnas.1514135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nambi S, Long JE, Mishra BB, Baker R, Murphy KC, Olive AJ, Nguyen HP, Shaffer SA, Sassetti CM. 2015. The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe 17:829–837. doi: 10.1016/j.chom.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT–a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girgis HS, Hottes AK, Tavazoie S. 2009. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One 4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison J, Lloyd G, Joe M, Lowary TL, Reynolds E, Walters-Morgan H, Bhatt A, Lovering A, Besra GS, Alderwick LJ. 2016. Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in Mycobacterium tuberculosis. mBio 7:e00972-16. doi: 10.1128/mBio.00972-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzegorzewicz AE, de Sousa-d'Auria C, McNeil MR, Huc-Claustre E, Jones V, Petit C, Angala SK, Zemanová J, Wang Q, Belardinelli JM, Gao Q, Ishizaki Y, Mikušová K, Brennan PJ, Ronning DR, Chami M, Houssin C, Jackson M. 2016. Assembling of the Mycobacterium tuberculosis cell wall core. J Biol Chem 291:18867–18879. doi: 10.1074/jbc.M116.739227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lun S, Bishai WR. 2007. Characterization of a novel cell wall-anchored protein with carboxylesterase activity required for virulence in Mycobacterium tuberculosis. J Biol Chem 282:18348–18356. doi: 10.1074/jbc.M700035200. [DOI] [PubMed] [Google Scholar]
- 24.Rengarajan JJ, Murphy EE, Park AA, Krone CLC, Hett ECE, Bloom BRB, Glimcher LHL, Rubin EJE. 2008. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A 105:264–269. doi: 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lun S, Miranda D, Kubler A, Guo H, Maiga MC, Winglee K, Pelly S, Bishai WR. 2014. Synthetic lethality reveals mechanisms of Mycobacterium tuberculosis resistance to β-lactams. mBio 5:e01767-14. doi: 10.1128/mBio.01767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR. 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol 180:2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilcheze C, Weisbrod TR, Chen B, Kremer L, Hazbon MH, Wang F, Alland D, Sacchettini JC, Jacobs WR. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Vilchèze C, Av-Gay Y, Gomez-Velasco A, Jacobs WR. 2011. Precise null deletion mutations of the mycothiol synthesis genes reveal their role in isoniazid and ethionamide resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 55:3133–3139. doi: 10.1128/AAC.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff KA, Nguyen HT, Cartabuke RH, Singh A, Ogwang S, Nguyen L. 2009. Protein kinase G is required for intrinsic antibiotic resistance in mycobacteria. Antimicrob Agents Chemother 53:3515–3519. doi: 10.1128/AAC.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider JS, Reddy SP, E HY, Evans HW, Glickman MS. 2013. Site-2 protease substrate specificity and coupling in trans by a PDZ-substrate adapter protein. Proc Natl Acad Sci U S A 110:19543–19548. doi: 10.1073/pnas.1305934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makinoshima H, Glickman MS. 2005. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature 436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black PA, Warren RM, Louw GE, van Helden PD, Victor TC, Kana BD. 2014. Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2491–2503. doi: 10.1128/AAC.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louw GE, Warren RM, Gey van Pittius NC, Leon R, Jimenez A, Hernández-Pando R, McEvoy CRE, Grobbelaar M, Murray M, van Helden PD, Victor TC. 2011. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am J Respir Crit Care Med 184:269–276. doi: 10.1164/rccm.201011-1924OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. 2009. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob Agents Chemother 53:3675–3682. doi: 10.1128/AAC.00550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spivey VL, Whalan RH, Hirst EMA, Smerdon SJ, Buxton RS. 2013. An attenuated mutant of the Rv1747 ATP-binding cassette transporter of Mycobacterium tuberculosis and a mutant of its cognate kinase, PknF, show increased expression of the efflux pump-related iniBAC operon. FEMS Microbiol Lett 347:107–115. doi: 10.1111/1574-6968.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinot AJ, Farrow M, Bai L, Layre E, Cheng T-Y, Tsai JH, Iqbal J, Annand JW, Sullivan ZA, Hussain MM, Sacchettini J, Moody DB, Seeliger JC, Rubin EJ. 2016. Mycobacterial metabolic syndrome: LprG and Rv1410 regulate triacylglyceride levels, growth rate and virulence in Mycobacterium tuberculosis. PLoS Pathog 12:e1005351. doi: 10.1371/journal.ppat.1005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staudenmaier H, Van Hove B, Yaraghi Z, Braun V. 1989. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol 171:2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun V, Herrmann C. 2007. Docking of the periplasmic FecB binding protein to the FecCD transmembrane proteins in the ferric citrate transport system of Escherichia coli. J Bacteriol 189:6913–6918. doi: 10.1128/JB.00884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee S, Paul S, Nguyen LT, Chu BCH, Vogel HJ. 2016. FecB, a periplasmic ferric-citrate transporter from E. coli, can bind different forms of ferric-citrate as well as a wide variety of metal-free and metal-loaded tricarboxylic acids. Metallomics 8:125–133. doi: 10.1039/C5MT00218D. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues L, Viveiros M, Aínsa JA. 2015. Measuring efflux and permeability in mycobacteria. Methods Mol Biol 1285:227–239. doi: 10.1007/978-1-4939-2450-9_13. [DOI] [PubMed] [Google Scholar]
- 41.Dörr T, Delgado F, Umans BD, Gerding MA, Davis BM, Waldor MK. 2016. A transposon screen identifies genetic determinants of Vibrio cholerae resistance to high-molecular-weight antibiotics. Antimicrob Agents Chemother 60:4757–4763. doi: 10.1128/AAC.00576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajagopal M, Martin MJ, Santiago M, Lee W, Kos VN, Meredith T, Gilmore MS, Walker S. 2016. Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. mBio 7:e00950-16. doi: 10.1128/mBio.00950-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.du Preez I, Loots DT. 2012. Altered fatty acid metabolism due to rifampicin-resistance conferring mutations in the rpoB gene of Mycobacterium tuberculosis: mapping the potential of pharmaco-metabolomics for global health and personalized medicine. OMICS 16:596–603. doi: 10.1089/omi.2012.0028. [DOI] [PubMed] [Google Scholar]
- 44.Kaushik A, Makkar N, Pandey P, Parrish N, Singh U, Lamichhane G. 2015. Carbapenems and rifampin exhibit synergy against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother 59:6561–6567. doi: 10.1128/AAC.01158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramakrishnan P, Aagesen AM, McKinney JD, Tischler AD. 2015. Mycobacterium tuberculosis resists stress by regulating PE19 expression. Infect Immun 84:735–746. doi: 10.1128/IAI.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarathy J, Dartois V, Dick T, Gengenbacher M. 2013. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piddock LJ, Williams KJ, Ricci V. 2000. Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J Antimicrob Chemother 45:159–165. doi: 10.1093/jac/45.2.159. [DOI] [PubMed] [Google Scholar]
- 48.Nathan C, Barry CE. 2015. TB drug development: immunology at the table. Immunol Rev 264:308–318. doi: 10.1111/imr.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gengenbacher M, Rao SPS, Pethe K, Dick T. 2010. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 50.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 51.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 52.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei J-R, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. 2012. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal 5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeJesus MA, Ioerger TR. 2013. A hidden Markov model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14:303. doi: 10.1186/1471-2105-14-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 56.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 57.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList—10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.