ABSTRACT
Pseudomonas aeruginosa biofilms contribute to its survival on biotic and abiotic surfaces and represent a major clinical threat due to their high tolerance to antibiotics. Therefore, the discovery of antibiofilm agents may hold great promise. We show that pharmacological inhibition of the P. aeruginosa quorum-sensing regulator MvfR (PqsR) using a benzamide-benzimidazole compound interferes with biofilm formation and potentiates biofilm sensitivity to antibiotics. Such a strategy could have great potential against P. aeruginosa persistence in diverse environments.
KEYWORDS: MvfR, PqsR, biofilm, antibiotic tolerance, antibiotic adjuvant, antibiotic potentiation, Pseudomonas aeruginosa, M64, antivirulence
TEXT
Pseudomonas aeruginosa, a Gram-negative bacterium, is an opportunistic human pathogen commonly isolated from soil and water. This pathogen may be problematic due to its ability to form biofilms—multicellular aggregates embedded in self-produced polymeric substances—that shield bacterial cells from antibacterial agents (such as disinfectants in water premise plumbing [1], host immune defenses [2, 3]). It is well documented that biofilms greatly contribute to the establishment of chronic infections, including chronic pneumonia in cystic fibrosis patients, relapsing and chronic wound and ear infections, as well as medical device-related infections (3). Such infections are difficult to treat because biofilms are highly tolerant to antibiotics (2, 4, 5), calling for an urgent need to develop novel approaches to combat P. aeruginosa biofilms.
One possible approach is the use of small molecules that interfere with the ability of P. aeruginosa to form biofilms. Several targets for antibiofilm drug development in P. aeruginosa have been proposed (3, 6), including the understudied MvfR quorum-sensing (QS) system. This QS system is controlled by the transcriptional regulator MvfR (also known as PqsR) that regulates the pqsABCDE operon responsible for the synthesis of ∼60 QS molecules called hydroxyl-2-alkyl-quinolines (HAQs) (7–10). The HAQ 3,4-hydroxy-2-heptyl-quinoline (PQS) as well as the enzymes PqsA and PqsD have been previously reported to play a role in biofilm formation, although their mechanism of action is not clear (11–13). We recently showed that another HAQ, 2-n-heptyl-4-hydroxyquinoline-N-oxide (HQNO), promotes biofilm formation and biofilm tolerance to antibiotics by inducing eDNA release that occurs as a result of autolysis due to self-poisoning of the respiratory chain by this molecule (14). A few studies have reported that inhibitors of PqsD or MvfR interfere with biofilm formation, but their antibiofilm potency was limited to the high micromolar range (11, 15–17). We recently described a benzamide-benzimidazole (BB) series of highly potent and cell-permeable inhibitors of MvfR (18). Here, we used one representative BB compound, M64, to evaluate the antibiofilm potential of this chemical family against P. aeruginosa and test its efficacy in potentiating antibiotic action against biofilm.
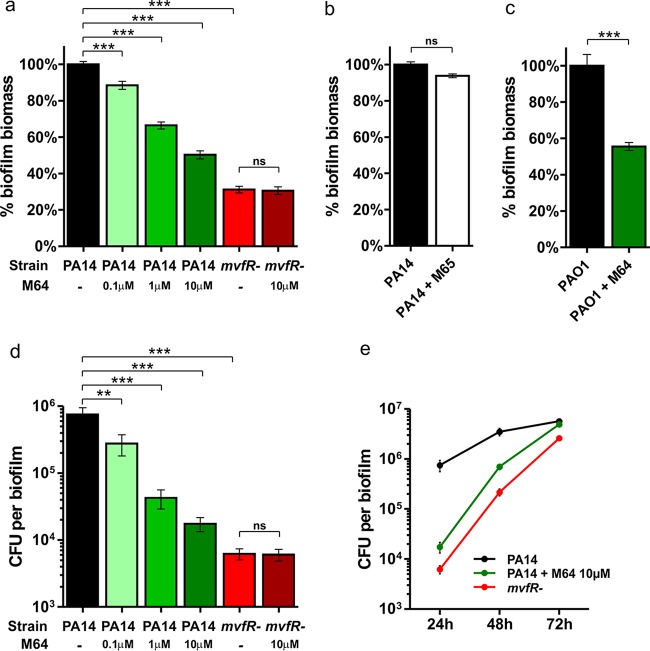
First, we sought to verify whether the transcriptional regulator MvfR itself is important in biofilm formation since MvfR directly controls several components contributing to biofilm formation. If so, the small molecules we have identified that target MvfR could be used against biofilm. To this end, we used an adaptation of the Calgary Biofilm Device (19) where biofilms are grown on peg lids in microtiter plates containing M63 minimal media supplemented with 0.2% glucose, 0.5% Casamino Acids, and 1 mM MgSO4, as previously described (14, 20, 21). Crystal violet (CV) staining and CFU counts were used to quantify biofilm biomass and viable cells, respectively, as described elsewhere (14, 20). The same setting was used throughout the study. Data presented in Fig. 1 show that the mvfR mutant cells produced a biofilm (mvfR− biofilm) at 24 h with a significantly lower biomass (P < 0.001) (Fig. 1a) and fewer viable cells (P < 0.001) (Fig. 1d) than the parental strain PA14. This demonstrates that the transcriptional regulator MvfR plays a significant role in P. aeruginosa biofilm formation, corroborating previous observations on the role of components of the MvfR QS system—PqsA, PqsD, PQS, and HQNO—in this phenomenon (11–14) and indicating that MvfR could be a promising antibiofilm target.
FIG 1.
The MvfR inhibitor M64 interferes with P. aeruginosa biofilm formation. Biofilm biomass (a, b, and c) or biofilm viable cells (d and e) were measured by crystal violet staining or CFU counts, respectively, after 24 (a to e), 48, and 72 (e) hours of growth in the presence or absence of 0.1, 1, or 10 μM M64 (a to c and e) or 10 μM M65 (b). Untreated PA14 (a, b, d, and e) or PAO1 (c) biofilms are shown in black, M64-treated biofilms in green, and mvfR− biofilms in red. Shown is the average +/− standard error of the mean (SEM) of at least 5 replicates. Statistical significance was assessed using one-way analysis of variance (ANOVA) plus Dunnett's posttest for multiple comparisons to PA14 or unpaired t test for single comparisons to mvfR− or PAO1. **, P < 0.01; ***, P < 0.001; ns, not statistically significant; -, not treated.
Next, we evaluated the potential of the MvfR inhibitor M64 in interfering with biofilm formation. Biofilms were grown for 24 h in the presence or absence of 0.1, 1, or 10 μM M64, then both biofilm biomass and biofilm viable cells were quantified as described above. Biofilm biomass quantification showed that M64 inhibited PA14 biofilm formation in a dose-dependent manner by 49.7%, 33.5%, or 11.6%, respectively, for 10 μM, 1 μM, or 0.1 μM M64 (P < 0.001) (Fig. 1a). Accordingly, we estimated the relative biofilm 50% inhibitory concentration (IC50) of M64 at 1 μM. Quantification of biofilm viable cells further support these data, as M64 also significantly reduced PA14 biofilm formation in a dose-dependent manner (P < 0.01) (Fig. 1d). Further data indicate that the M64-mediated reduction in biofilm formation is due to MvfR inhibition, as (i) M64 did not significantly modulate biofilm formation in the mvfR mutant (Fig. 1a and d) (P > 0.05), and (ii) the inactive M64 analogue M65 (18) did not interfere with PA14 biofilm formation (Fig. 1b) (P > 0.05). Biofilm formation kinetics performed over 72 h showed that 24 h was the time point when both M64-treated PA14 or mvfR− biofilms were the lowest (Fig. 1e), indicating that inhibition of the MvfR QS system significantly affects the early steps of biofilm formation and supporting the notion that MvfR inhibitors could be used preventively against the initiation of biofilm formation. M64 also interfered with PAO1 biofilm formation, indicating that its activity is not limited to only one strain (Fig. 1c) (P < 0.001). Compared to different chemical families of MvfR and PqsD inhibitors reported in several sources (11, 15–17), the BB compound M64 is at least 100 times more potent at reducing biofilm formation, and therefore the BB chemical family represents a significant step forward in the development of highly efficacious biofilm inhibitors targeting the MvfR QS system.
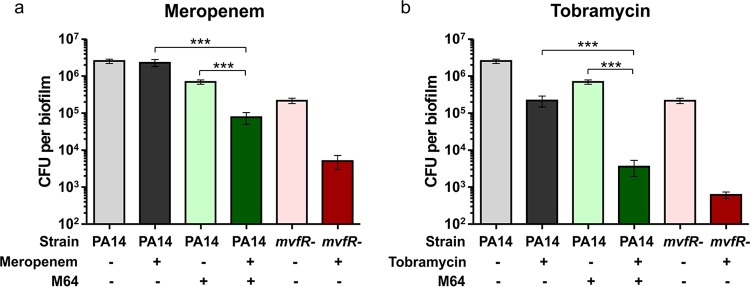
We previously reported that the MvfR QS system promotes biofilm tolerance to antibiotic via HQNO-mediated autolysis and eDNA release (14). Thus, we tested here whether chemical inhibition of the MvfR QS system could sensitize biofilms to antibiotics. To this end, biofilms were grown for 48 h in the presence or absence of 10 μM M64 then treated for 24 h with 10 μg/ml of meropenem or tobramycin, antibiotics frequently used against P. aeruginosa infections (22–24). Biofilm viable cells were quantified before and after antibiotic treatment. PA14 biofilm appeared highly tolerant to meropenem, as no biofilm cells were killed by this antibiotic (Fig. 2a). However, M64-treated or mvfR− biofilms were both significantly more susceptible to meropenem, as the number of biofilm viable cells decreased by 9.1-fold or 42.6-fold, respectively, following antibiotic treatment (Fig. 2a). A similar pattern was observed with tobramycin, as this antibiotic reduced biofilm viable cells 195-fold and 352-fold in M64-treated and mvfR− biofilms, respectively, compared to only 11.8-fold in PA14 biofilm (Fig. 2b). These data support our previous observation that the MvfR QS system is critical for the tolerance of biofilms to antibiotics (14) and demonstrate that chemical inhibition of MvfR significantly increases the ability of antibiotics to kill antibiotic-tolerant P. aeruginosa biofilms.
FIG 2.
M64 pretreatment sensitizes P. aeruginosa biofilms to the antibiotics meropenem and tobramycin. Biofilms were grown for 48 h in the presence or absence of 10 μM M64 then treated with 10 μg/ml meropenem (a) or 10 μg/ml tobramycin (b) for 24 h. Biofilm CFU were quantified preantibiotic treatment (48 h, lighter colors) and postantibiotic treatment (72 h, darker colors). PA14 biofilms are shown in gray, PA14 + M64 biofilms in green, and mvfR− biofilms in red. Data show the average +/− SEM of at least 20 replicates. Statistical significance was assessed using the Kruskal-Wallis test plus Dunn's posttest. ***, P < 0.001; +, treated; -, not treated.
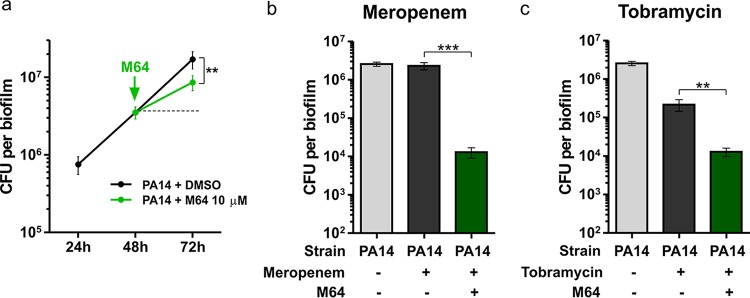
To further evaluate the utility of BB compounds, we then asked whether they could disrupt preformed biofilms. Biofilms were grown for 48 h then treated with 10 μM M64 or dimethyl sulfoxide (DMSO) vehicle control for 24 h. As shown in Fig. 3a, M64 treatment reduced the increase in biofilm CFU that occurred between 48 and 72 h by 50% relative to the DMSO control (P < 0.01). However, M64 did not reduce biofilm CFU below the level of before treatment, indicating that M64 alone can interfere with further biofilm development but does not disrupt a preformed PA14 biofilm.
FIG 3.
Treatment of preformed biofilms with M64 potentiates antibiotic-mediated biofilm disruption. (a) Biofilm CFU were quantified before (24 h or 48 h) or after (72 h) treatment with 10 μM M64. (b and c) Biofilm CFU were quantified before (48 h) or after (72 h) treatment with a combination of 10 μM M64 and 10 μg/ml antibiotic. Data show the average +/− SEM of at least 6 replicates. Statistical significance was assessed using the unpaired t test. **, P < 0.01; ***, P < 0.001; +, treated; -, not treated.
We then sought to determine whether concomitant addition of M64 and antibiotics would still increase the antibiotic efficacy against preformed biofilms, or whether M64 could be active only when added prophylactically as shown in Fig. 2. Biofilms were grown for 48 h then simultaneously treated with 10 μM M64 and 10 μg/ml of meropenem or tobramycin for 24 h. As shown in Fig. 3b, biofilm CFU were 178 times lower in the group treated with M64 and meropenem than in the group treated with meropenem alone (P < 0.001). A similar profile was observed with tobramycin, as biofilm CFU were 17 times lower in the group treated with M64 and tobramycin than in the group treated with tobramycin alone (P < 0.01) (Fig. 3c). These data indicate that when used in combination with the antibiotics meropenem and tobramycin, M64 greatly potentiates the biofilm disruption ability of both.
Altogether, this study demonstrated that the BB MvfR inhibitor M64 interferes with P. aeruginosa biofilm formation and potentiates the antibiofilm activity of antibiotics. Based on these results, M64 or M64-like compounds of the BB chemical family hold great promise as antibiofilm agents and have a great potential to be used either alone or as antibiotic adjuvants to treat established chronic infections where biofilms render antibiotic treatments particularly ineffective (2, 4). Indeed, to treat chronic infections, the dose of antibiotic is often increased but can still fail to clear the infection, frequently leading to high-level antibiotic resistance (25). The use of an efficacious anti-MvfR small molecule like M64 may therefore provide a solution to this problem in view of its ability to potentiate antibiotic efficacy against biofilms. Coating indwelling medical devices could be another application for these agents, where they may prevent P. aeruginosa biofilm buildups, which are highly problematic in catheter-associated urinary tract infections or ventilator-associated pneumonia (26, 27). Overall, this study opens new avenues for the treatment of biofilms on both biotic and abiotic surfaces and reinforces the relevance of the MvfR QS system in chronic infection and tolerance to antibiotics. It further demonstrates the clinical potential of antivirulence strategies targeting this pathway and opens possibilities for application in environmental settings, livestock, and agriculture.
ACKNOWLEDGMENTS
This work was supported by Shriners Hospital Postdoctoral Fellowship no. 84206 to D.M. and by the research grants Shriners no. 8770, Cystic Fibrosis Foundation no. 11P0, and NIAID R33AI105902 to L.G.R.
REFERENCES
- 1.Bedard E, Prevost M, Deziel E. 2016. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiologyopen 5:937–956. doi: 10.1002/mbo3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulcahy LR, Isabella VM, Lewis K. 2014. Pseudomonas aeruginosa biofilms in disease. Microb Ecol 68:1–12. doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tolker-Nielsen T. 2014. Pseudomonas aeruginosa biofilm infections: from molecular biofilm biology to new treatment possibilities. APMIS Suppl:1–51. doi: 10.1111/apm.12335. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masak J, Cejkova A, Schreiberova O, Rezanka T. 2014. Pseudomonas biofilms: possibilities of their control. FEMS Microbiol Ecol 89:1–14. doi: 10.1111/1574-6941.12344. [DOI] [PubMed] [Google Scholar]
- 7.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. 2011. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev 35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepine F, Dekimpe V, Lesic B, Milot S, Lesimple A, Mamer OA, Rahme LG, Deziel E. 2007. PqsA is required for the biosynthesis of 2,4-dihydroxyquinoline (DHQ), a newly identified metabolite produced by Pseudomonas aeruginosa and Burkholderia thailandensis. Biol Chem 388:839–845. doi: 10.1515/BC.2007.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storz MP, Maurer CK, Zimmer C, Wagner N, Brengel C, de Jong JC, Lucas S, Musken M, Haussler S, Steinbach A, Hartmann RW. 2012. Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J Am Chem Soc 134:16143–16146. doi: 10.1021/ja3072397. [DOI] [PubMed] [Google Scholar]
- 12.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 13.Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol Microbiol 74:1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x. [DOI] [PubMed] [Google Scholar]
- 14.Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, Rahme LG. 2016. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr Biol 26:195–206. doi: 10.1016/j.cub.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilangovan A, Fletcher M, Rampioni G, Pustelny C, Rumbaugh K, Heeb S, Camara M, Truman A, Chhabra SR, Emsley J, Williams P. 2013. Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLoS Pathog 9:e1003508. doi: 10.1371/journal.ppat.1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomann A, de Mello Martins AG, Brengel C, Empting M, Hartmann RW. 2016. Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chem Biol 11:1279–1286. doi: 10.1021/acschembio.6b00117. [DOI] [PubMed] [Google Scholar]
- 17.Thomann A, Brengel C, Borger C, Kail D, Steinbach A, Empting M, Hartmann RW. 2016. Structure-activity relationships of 2-sufonylpyrimidines as quorum-sensing inhibitors to tackle biofilm formation and eDNA release of Pseudomonas aeruginosa. ChemMedChem 11:2522–2533. doi: 10.1002/cmdc.201600419. [DOI] [PubMed] [Google Scholar]
- 18.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A, Rahme L. 2014. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog 10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison JJ, Stremick CA, Turner RJ, Allan ND, Olson ME, Ceri H. 2010. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc 5:1236–1254. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- 20.Maura D, Morello E, du Merle L, Bomme P, Le Bouguenec C, Debarbieux L. 2012. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ Microbiol 14:1844–1854. doi: 10.1111/j.1462-2920.2011.02644.x. [DOI] [PubMed] [Google Scholar]
- 21.Tre-Hardy M, Vanderbist F, Traore H, Devleeschouwer MJ. 2008. In vitro activity of antibiotic combinations against Pseudomonas aeruginosa biofilm and planktonic cultures. Int J Antimicrob Agents 31:329–336. doi: 10.1016/j.ijantimicag.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Pankuch GA, Lin G, Seifert H, Appelbaum PC. 2008. Activity of meropenem with and without ciprofloxacin and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother 52:333–336. doi: 10.1128/AAC.00689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratjen F, Brockhaus F, Angyalosi G. 2009. Aminoglycoside therapy against Pseudomonas aeruginosa in cystic fibrosis: a review. J Cyst Fibros 8:361–369. doi: 10.1016/j.jcf.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Maselli DJ, Keyt H, Restrepo MI. 2017. Inhaled antibiotic therapy in chronic respiratory diseases. Int J Mol Sci 18:E1062. doi: 10.3390/ijms18051062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PO, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. 2009. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health 2:101–111. doi: 10.1016/j.jiph.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Koenig SM, Truwit JD. 2006. Ventilator-associated pneumonia: diagnosis, treatment, and prevention. Clin Microbiol Rev 19:637–657. doi: 10.1128/CMR.00051-05. [DOI] [PMC free article] [PubMed] [Google Scholar]