LETTER
Acinetobacter species are ubiquitous pathogens that are broadly encountered in the environment, with some species, like Acinetobacter baumannii, A. pittii, and A. nosocomialis, more frequently detected in nosocomial settings (1). During the last decades, carbapenem resistance rates among such microorganisms have increased mainly because of the acquisition of carbapenem-hydrolyzing class D β-lactamase (CHDL)-encoding genes. The dissemination of blaOXA-23, blaOXA-143, and more recently blaOXA-72 is the main cause of carbapenem resistance among Brazilian A. baumannii clinical isolates (2). In contrast, blaOXA-58 has been rarely reported in clinical isolates of Acinetobacter species in Brazil (3–5). However, Cayô and colleagues recently described two OXA-58-producing A. seifertii isolates from patients hospitalized at a tertiary-care hospital in São Paulo, Brazil. Interestingly, these isolates were recovered more than 20 years ago (1993 and 1997) (3). Here, we describe an OXA-58-producing A. seifertii isolate colonizing a black-necked swan residing in the lakes of the São Paulo zoo.
During a surveillance study, 37 black-necked swans (Cygnus melanocoryphus) residing in the lakes of the São Paulo zoo were screened for colonization by carbapenem-resistant Gram-negative bacilli. Swabs of both the choana and the cloaca were collected. The swabs were streaked onto MacConkey agar supplemented with imipenem at 1 μg/ml (Sigma-Aldrich, St. Louis, MO), followed by Gram staining. We recovered a cloacal carbapenem-resistant Gram-negative coccobacillus (Ac-12.1) that was initially identified by matrix-assisted laser desorption ionization time of flight mass spectrometry as a member of the Acinetobacter calcoaceticus-baumannii complex. Subsequently, Ac-12.1 was identified to the species level as A. seifertii by rpoB sequencing (6). Antimicrobial susceptibility testing was performed by broth microdilution and interpreted according to the EUCAST guidelines (7). Ac-12.1 showed high piperacillin-tazobactam (>256 and 4 μg/ml), ampicillin-sulbactam (32 and 16 μg/ml), cefepime (16 μg/ml), ceftazidime (32 μg/ml), and ceftriaxone (64 μg/ml) MICs. It was resistant to imipenem (MIC, 16 μg/ml), meropenem (MIC, 8 μg/ml), amikacin (MIC, >256 μg/ml), and polymyxin B (MIC, 4 μg/ml). In contrast, Ac-12.1 was susceptible to gentamicin (MIC, 4 μg/ml), ciprofloxacin (MIC, 1 μg/ml), levofloxacin (MIC, 0.25 μg/ml), and sulfamethoxazole-trimethoprim (MICs, 2 and 38 μg/ml).
Screening for carbapenemase production by Blue Carba test was positive with 2 h of incubation, as previously described (8). Molecular characterization of carbapenemase-encoding genes was performed by PCR, followed by DNA sequencing with specific primers (9, 10), and demonstrated that Ac-12.1 carried the blaOXA-58 gene. No additional acquired β-lactamase-encoding genes were detected. The clonal relationship of Ac-12.1 and the two historical OXA-58-producing A. seifertii strains (Asp-70064 and Asp-1069) previously detected in Brazil (3) was analyzed by pulsed-field gel electrophoresis (PFGE) with the ApaI restriction enzyme (11). All OXA-58-producing A. seifertii isolates showed the same PFGE pattern (data not shown), despite being collected more than 20 years apart. Plasmid profiling and Southern blot hybridization were performed by the Kieser methodology and with the DIG DNA Labeling and Detection kit (Roche Diagnostics GmbH, Penzberg, Germany), respectively. Our results showed that blaOXA-58 was located on a plasmid of ∼59 kb in strain Ac12.1, which is similar to the size of plasmids carried by clinical strains Asp-70064 and Asp-1069 (3). Transfer of blaOXA-58 by transformation assays with A. baumannii ATCC 19606 as the recipient strain was unsuccessful.
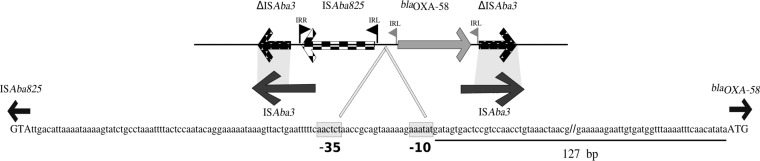
To determine the genetic surroundings of the blaOXA-58 gene, PCR, followed by sequencing by DNA walking, was performed with specific primers designed for ISAba3, as previously published (12). The sequence analyses revealed a 2.8-kb structure (GenBank accession number MF417790) in which ISAba825 was located upstream of blaOXA-58 in the opposite direction. This ISAba825–blaOXA-58 structure was located within a composite transposon formed by two truncated copies of ISAba3, as shown in Fig. 1. It has been already reported that ISAba825 was associated with blaOXA-58 overexpression (13, 14). The genetic background found in the present study was identical to those observed for plasmid pAb242, which was previously detected in an OXA-58-producing A. baumannii clinical isolate from Argentina (unpublished data; accession number KR055667.1). Distinct genetic backgrounds have been described for blaOXA-58 genes so far, demonstrating that complex transposable elements are associated with this resistance determinant (15).
FIG 1.
Genetic context of blaOXA-58 in A. seifertii isolate Ac-12.1. Genes are shown as labeled arrows; inverted repeats (IRs) of ISAba3 and ISAba825 are shown as flags (gray flags, ISAba3; black flags, ISAba825). The presence of a left IR (IRL) of ISAba3 upstream of blaOXA-58 suggests that insertion of ISAba825 occurred after transposon formation (ΔISAba3–blaOXA-58–ΔISAba3). Integral ISAba3 sequences are represented below the genetic context, and regions with similarity to ΔISAba3 are highlighted in light gray. The nucleotide sequence of the promoter region is boxed. The double bars in the127-bp nucleotide sequence represent the nucleotides omitted. Uppercase letters represent the start codons of ISAba825 and blaOXA-58.
To date, A. seifertii has been isolated in hospital and community settings (door handle and game console), illustrating its ubiquitous nature (3, 12, 16). These isolates retained susceptibility to quinolones and aminoglycosides and also showed susceptibility to most β-lactam agents when they did not carry a CHDL (3, 16). To our knowledge, this is the first report of OXA-58-producing Acinetobacter species isolated from an environmental source. The detection of an identical OXA-58-producing A. seifertii clone nearly 25 years after its first description highlights the adaptation of this fit clone in this geographic region. In addition, since only one bird was colonized by OXA-58-producing A. seifertii, it seems that this microorganism does not belong to the normal microbiota of C. melanocoryphus. Although we did not investigate the source of A. seifertii acquisition, it seems that local environmental contamination did not play an important role because other birds were not colonized by this clone. The lakes of the São Paulo zoo are often visited by migratory birds, which may have acted as vectors transmitting OXA-58-producing A. seifertii from other environmental sources. There is little evidence of the clinical relevance of A. seifertii. However, Na and colleagues (17) demonstrated that A. seifertii isolates had consistently high virulence-associated phenotypes, with no significant difference from those showed by A. baumannii (with the exception A. baumannii clone ST110) and A. nosocomialis (17). Thus, we believe that A. seifertii is capable of causing infections similar to those caused by A. baumannii. Lastly, the acquisition of a carbapenemase-encoding gene by A. seifertii is very worrisome, since this species seems to be naturally resistant to polymyxins, drastically limiting the therapeutic options available for the treatment of infections with this bacterium.
ACKNOWLEDGMENTS
We thank Ana Carolina Ramos da Silva and Lorena Cristina Correa Fehlberg for their excellent technical contribution to this study. We are also grateful to Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for supporting this work through a grant awarded to A.C.G. (process no. 2017/02258-6), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing grants to A.C.N., W.M.B.S.M., A.P.M., and R.C. (PNPD 20131991), and the National Council for Science and Technological Development (CNPq) for providing a grant to A.C.G. (process no. 305535/2014-5).
A.C.G. has recently received research funding and/or consultation fees from AstraZeneca, Bayer, BD, MSD, and Pfizer. The other authors have no conflicts of interest to declare.
REFERENCES
- 1.Al Atrouni A, Joly-Guillou ML, Hamze M, Kempf M. 2016. Reservoirs of non-baumannii Acinetobacter species. Front Microbiol 7:49. doi: 10.3389/fmicb.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasconcelos AT, Barth AL, Zavascki AP, Gales AC, Levin AS, Lucarevschi BR, Cabral BG, Brasiliense DM, Rossi F, Furtado GH, Carneiro IC, da Silva JO, Ribeiro J, Lima KV, Correa L, Britto MH, Silva MT, da Conceição ML, Moreira M, Martino MD, de Freitas MR, Oliveira MS, Dalben MF, Guzman RD, Cayô R, Morais R, Santos SA, Martins WM. 2015. The changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: a BrasNet report. Diagn Microbiol Infect Dis 83:382–385. doi: 10.1016/j.diagmicrobio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Cayô R, Rodrigues-Costa F, Pereira Matos A, Godoy Carvalhaes C, Dijkshoorn L, Gales AC. 2016. Old clinical isolates of Acinetobacter seifertii in Brazil producing OXA-58. Antimicrob Agents Chemother 60:2589–2591. doi: 10.1128/AAC.01957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueiredo DQ, Santos KR, Pereira EM, Schuenck RP, Mendonça-Souza CR, Teixeira LM, Mondino SS. 2011. First report of the bla(OXA-58) gene in a clinical isolate of Acinetobacter baumannii in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 106:368–370. doi: 10.1590/S0074-02762011000300019. [DOI] [PubMed] [Google Scholar]
- 5.de Souza Gusatti CS, Bertholdo LM, Otton LM, Marchetti DP, Ferreira AE, Corção G. 2012. First occurrence of blaOXA-58 in Acinetobacter baumannii isolated from a clinical sample in southern Brazil. Braz J Microbiol 43:243–246. doi: 10.1590/S1517-83822012000100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0, 2017. European Committee on Antimicrobial Susceptibility Testing, Basel, Switzerland: http://www.eucast.org/clinical_breakpoints/ Accessed 30 March 2017. [Google Scholar]
- 8.Pires J, Novais A, Peixe L. 2013. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins PG, Lehmann M, Seifert H. 2010. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 35:305. doi: 10.1016/j.ijantimicag.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Viana AL, Cayô R, Avelino CC, Gales AC, Franco MC, Minarini LAR. 2013. Extended-spectrum β-lactamases in Enterobacteriaceae isolated in Brazil carry distinct types of plasmid-mediated quinolone resistance genes. J Med Microbiol 62(Part 9):1326–1331. doi: 10.1099/jmm.0.055970-0. [DOI] [PubMed] [Google Scholar]
- 11.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marti S, Sánchez-Céspedes J, Blasco MD, Espinal P, Ruiz M, Alba V, Vila J. 2008. Characterization of the carbapenem-hydrolyzing oxacillinase OXA-58 in an Acinetobacter phenon 6/ct13TU clinical isolate. Diagn Microbiol Infect Dis 61:468–470. doi: 10.1016/j.diagmicrobio.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Ravasi P, Limansky AS, Rodriguez ER, Viale AM, Mussi MA. 2011. ISAba825, a functional insertion sequence modulating genomic plasticity and blaOXA-58 expression in Acinetobacter baumannii. Antimicrob Agents Chemother 55:917–920. doi: 10.1128/AAC.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes BS, Al-Hassan L, Amyes SG. 2012. ISAba825 controls the expression of the chromosomal bla(OXA-51-like) and the plasmid borne bla(OXA-58) gene in clinical isolates of Acinetobacter baumannii isolated from the USA. Clin Microbiol Infect 18:E446–E451. doi: 10.1111/j.1469-0691.2012.03979.x. [DOI] [PubMed] [Google Scholar]
- 15.Evans BA, Amyes SG. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JY, Kim Y, Ko EA, Park YK, Jheong WH, Ko G, Ko KS. 2012. Acinetobacter species isolates from a range of environments: species survey and observations of antimicrobial resistance. Diagn Microbiol Infect Dis 74:177–180. doi: 10.1016/j.diagmicrobio.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Na IY, Chung ES, Jung CY, Kim DH, Shin Y, Kang K, Kim ST, Ko KS. 2016. Comparison of the virulence-associated phenotypes of five species of Acinetobacter baumannii complex. J Microbiol Biotechnol 26:171–179. doi: 10.4014/jmb.1507.07076. [DOI] [PubMed] [Google Scholar]