ABSTRACT
The biofilm life cycle is characterized by the transition of planktonic cells exhibiting high susceptibly to antimicrobial agents to a biofilm mode of growth characterized by high tolerance to antimicrobials, followed by dispersion of cells from the biofilm back into the environment. Dispersed cells, however, are not identical to planktonic cells but have been characterized as having a unique transitionary phenotype relative to biofilm and planktonic cells, with dispersed cells attaching in a manner similar to exponential-phase cells, but demonstrating gene expression patterns that are distinct from both exponential and stationary-phase planktonic cells. This raised the question whether dispersed cells are as susceptible as planktonic cells and whether the dispersion inducer or the antibiotic class affects the drug susceptibility of dispersed cells. Dispersed cells obtained in response to dispersion cues glutamate and nitric oxide (NO) were thus exposed to tobramycin and colistin. Although NO-induced dispersed cells were as susceptible to colistin and tobramycin as exponential-phase planktonic cells, glutamate-induced dispersed cells were susceptible to tobramycin but resistant to colistin. The difference in colistin susceptibility was independent of cellular c-di-GMP levels, with modulation of c-di-GMP failing to induce dispersion. Instead, drug susceptibility was inversely correlated with LPS modification system and the biofilm-specific transcriptional regulator BrlR. The susceptibility phenotype of glutamate-induced dispersed cells to colistin was found to be reversible, with dispersed cells being rendered as susceptible to colistin within 2 h postdispersion, though additional time was required for dispersed cells to display expression of genes indicative of exponential growth.
KEYWORDS: biofilm, attachment, biofilm control, colistin, dispersion, exponential phase, susceptibility, tobramycin
INTRODUCTION
Biofilms are surface-associated bacterial communities that are matrix encased and cause persistent and chronic infections in medical settings. Biofilm-related infections include chronic urinary tract infection due to catheters, chronic wounds, ventilated-associated pneumonia in intubated patients, chronic pulmonary disease in patients with cystic fibrosis or chronic obstructive lung disease. According to the National Institutes of Health, 65% of all hospital-acquired infections are due to bacteria growing as biofilms, and 80% of chronic infections are linked to biofilms. To put this in perspective, the Centers for Disease Control estimates that hospital-acquired infections account for an estimated 1.7 million infections and 99,000 associated deaths each year in American hospitals alone. The high morbidity and mortality rate is due to biofilms being extremely difficult to control in medical settings (1). In fact, conventional therapies have proven inadequate in the treatment of many (if not most) chronic biofilm infections due to the extraordinary tolerance of biofilms to antimicrobial agents relative to their planktonic counterparts (1–3) and their ability to inhibit healing (4–6). The findings underscore the need for novel approaches in treating biofilms. Recent findings have suggested a promising avenue open for biofilm control: the manipulation of the biofilm lifestyle, and more specifically, the biofilm dispersion response. Dispersion is considered an escape strategy developed by biofilm bacteria as a means of self-preservation to avoid stresses associated with biofilm growth to disseminate to new locations (7–10). During the dispersion response, surface-attached organisms liberate themselves from matrix-encased biofilms, apparent by single cells actively escaping from the biofilm, leaving behind eroded biofilms and microcolonies having central voids (7–16). This response can be induced in response to various dispersion cues including cis-2-decenoic acid, an interkingdom fatty acid signaling molecule belonging to the (B)DSF family, changing carbon source concentrations, ammonium chloride, heavy metals, and nitric oxide (NO) (9, 17). These dispersion cues function in a threshold concentration-dependent manner and have been shown to coincide with up to 80% of the biofilm biomass being removed upon induction of dispersion (11, 14, 18, 19). Dispersion furthermore coincides with the modulation of the secondary messenger molecule cyclic di-GMP (c-di-GMP). The levels of c-di-GMP are enzymatically modulated by diguanylate cyclases, proteins containing a GGDEF domain, and phosphodiesterases (PDE) harboring either an EAL or HD-GYP domain (20, 21). It is now generally accepted that c-di-GMP plays an important role in the motile-sessile transition, with low intracellular levels correlating with motility and high intracellular levels correlating with surface attachment and biofilm formation (22, 23). In agreement with the role of c-di-GMP, dispersion by Pseudomonas aeruginosa biofilms has been demonstrated to coincide with increased expression of fliC (encoding flagellin type B) (18) and dispersed cells being characterized by intracellular signaling molecule c-di-GMP at levels comparable to those found in planktonic cells (13, 14, 24). Despite the similarities, however, it is becoming increasingly apparent that dispersed cells are not identical to planktonic cells. Sauer et al. (7) demonstrated dispersed cells to differ from both planktonic and biofilm cells with respect to global protein production patterns. Li et al. (25) reported dispersion to contribute to both acute and chronic infections, with impaired dispersion capabilities coinciding with enhanced chronic infections but significantly reduced acute infections in both plant and mouse hosts. The effect of dispersion on the pathogenicity of P. aeruginosa was attributed to the finding that dispersion coincided with differences in virulence gene expression relative to planktonic and biofilm cells, with expression of genes contributing to the virulence of P. aeruginosa in cells dispersed being reduced up to 150-fold compared to planktonic cells. Moreover, a recent report indicated that dispersion of P. aeruginosa biofilms in response to NO resulted in dispersed cells that were similar to planktonic cells overproducing the Escherichia coli PDE YhjH (26, 27), suggesting dispersed cells harbor significantly reduced c-di-GMP levels compared to planktonic cells. It is thus not surprising that dispersed cells have been described as having a phenotype that is distinct from planktonic and biofilm cells (7, 25, 28).
Considering that planktonic cells are more susceptible to antimicrobial agents than their counterparts growing as a biofilm and that dispersion coincides with bacteria transitioning to the planktonic mode of growth, it has been commonly assumed that dispersed cells would also display enhanced killing by antibiotics relative to biofilm cells. However, given the apparent differences between planktonic and dispersed cells (7, 25), with recent findings linking c-di-GMP levels to the susceptibility of bacteria to antimicrobial agents (26–29), the notion of dispersed cells being as susceptible as planktonic cells needs to be challenged. Adding further uncertainty to the drug susceptibility of dispersed cells is the diverse range of structurally distinct dispersion cues used to induce dispersion. This is increasingly important considering how little is known about how dispersion cues affect antibiotic susceptibility and whether the way dispersion is induced affects the susceptibility of dispersed cells to antimicrobial agents relative to planktonic cells. In addition, it is unclear whether all antibiotic classes will exhibit enhanced activity on dispersed cells. Therefore, this study aimed at addressing some of these questions and elucidating the complexity found in this unique and important phenotype. Specifically, we focused on the susceptibility of dispersed cells to two different antibiotics, tobramycin and colistin, and sought to determine whether the manner in which the cells were dispersed from the biofilm affected the susceptibility of dispersed cells to these two antibiotics.
RESULTS
Dispersed cell susceptibility is dependent on the class of antimicrobial.
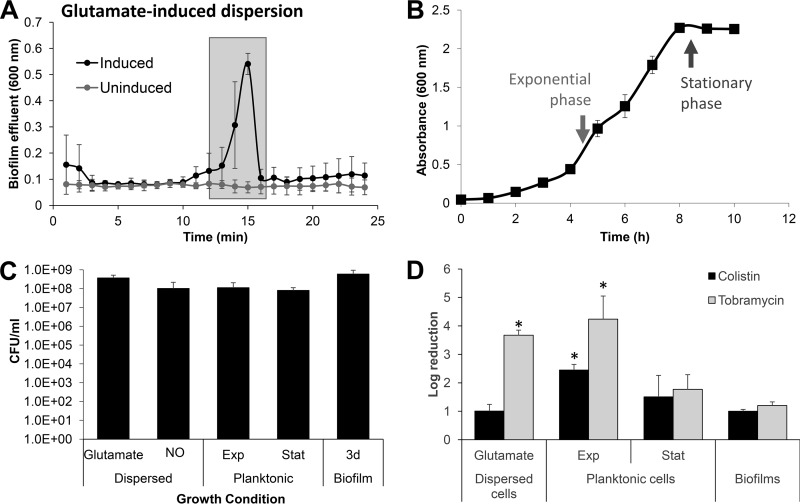
Dispersion has been characterized as an active process, which allows the bacterial cells to leave a biofilm and thus transition from the sessile, biofilm mode of growth to a motile, planktonic state (9, 10). Considering that planktonic cells are more susceptible to antimicrobial agents than their counterparts growing as a biofilm, it has been commonly assumed that dispersed cells would also display enhanced killing to antibiotics relative to biofilm cells. However, since several previous studies concluded that dispersed cells display unique properties relative to both planktonic and biofilm cells and could represent a unique phenotype (25, 28), this assumption has been brought into question. To address the question of the susceptibility phenotype of dispersed cells, we first needed to produce dispersed cells. We previously demonstrated that dispersion can be induced (9, 17) and subsequently detected by a decrease in the biofilm biomass and void formation, as determined using flow-cell grown biofilms in conjunction with microscopy, as well as by a sharp increase in the absorbance (600 nm) in the effluent within 15 to 20 min upon induction of dispersion, as determined using tube reactors (12, 14, 15, 18, 30, 31). We made use of the latter approach to ease collection of dispersed cells. P. aeruginosa biofilms were first grown for 5 days under flowing conditions in a tube reactor and then exposed to glutamate to induce dispersion. Glutamate was chosen since it simulates a dispersion in response to changes in the nutritional environment (but without being used as a carbon and energy source) (12, 18). In addition, the mechanism by which glutamate is perceived by P. aeruginosa to induce dispersion has been well characterized (12, 15). Dispersed cells were immediately collected from the biofilm effluent (Fig. 1A) and subsequently exposed to two different antibiotics, the aminoglycoside tobramycin and the cationic peptide antibiotic colistin. For comparison, planktonic cells grown to either exponential or stationary phase (4.5 or 8.5 h of growth, Fig. 1B) and 3-day-old untreated biofilms (not exposed to the dispersion cue) were used. To ensure comparable treatment conditions, all samples were grown in Vogel-Bonner citrate minimal medium (VBMM), and the OD was adjusted to 0.2. Under the conditions tested, OD-adjusted samples harbored comparable number of viable cells, as determined using viability assays and CFU counts (Fig. 1C).
FIG 1.
Susceptibility phenotype of dispersed cells obtained in response to exposure of P. aeruginosa PAO1 biofilms to glutamate. (A) Absorbance of biofilm tube reactor effluents after the induction of dispersion by glutamate. Cells that were collected for subsequent experiments and referred to as “glutamate induced” are indicated by shaded boxes. Effluents of untreated biofilms were used as control. (B) Growth curve of P. aeruginosa PAO1. Timing and average absorbance of cells referred to as “exponential-phase” and “stationary-phase” planktonic cells are indicated by arrows. (C) Average number of viable cells, expressed as CFU, present in biofilm cells, dispersed cells, and planktonic cells grown to exponential (Exp) and stationary phase (Stat) after washing and resuspension in fresh VBMM to an OD600 of 0.2. (D) Susceptibility of biofilm cells, glutamate-induced dispersed cells, and planktonic cells grown to exponential and stationary phases to tobramycin (50 μg/ml) or colistin (20 μg/ml) after exposure for 1 h at 37°C. Susceptibility was determined by log10 reduction in CFU. All experiments were repeated at least three times. Error bars indicate standard deviations. *, significantly different from PAO1 biofilms (P < 0.01 as determined by ANOVA and SigmaStat). Exp, exponential phase; Stat, stationary phase; 3d biofilm, 3-day-old biofilm.
Tobramycin treatment reduced the PAO1 biofilm cells by a 1.9-log reduction in viability relative to untreated controls. Likewise, tobramycin treatment of stationary-phase planktonic cells coincided with a 1.7-log reduction in viability (Fig. 1D). In contrast, however, the viability of exponential-phase planktonic cells was reduced on average by 4.2 logs upon exposure to tobramycin (Fig. 1D). Treatment of dispersed cells obtained in response to the dispersion cue glutamate with tobramycin resulted in a similar reduction in viability, as evidenced by a 3.7-log reduction in viability (Fig. 1D). This result is what one would expect if dispersed cells are reverting to a planktonic phenotype and thus a more antimicrobial susceptible state. Similar to tobramycin treatment, exponential-phase planktonic cells were found to be more susceptible to colistin than stationary-phase planktonic cells or biofilm cells, with colistin treatment resulting in a 2.5-log reduction in viability of exponential-phase planktonic cells, but only 1.5- and 1-log reductions in the viabilities of stationary-phase planktonic and biofilm cells, respectively (Fig. 1D). In contrast, however, colistin treatment reduced the viability of dispersed cells obtained in response to glutamate by only 1 log, a reduction that was not significantly different from those noted for biofilms (Fig. 1D). The findings suggest that although cells dispersed in response to glutamate are as susceptible to tobramycin as exponential-phase planktonic cells, dispersed cells retain the enhanced tolerance to killing by colistin similar to biofilm or stationary-phase planktonic cells.
Dispersed cells display a unique transitory phenotype.
Based on the tolerance of glutamate-derived dispersed cells to tobramycin (Fig. 1D), dispersed cells appear to be more similar to planktonic cells grown to exponential phase. However, dispersed cells were as resistant to colistin as biofilm cells or planktonic cells grown to stationary phase (Fig. 1D). To determine whether dispersed cells are more similar to planktonic cells grown to stationary or exponential phase, we made use of gene expression analysis using qRT-PCR, by focusing on genes that are differentially expressed in exponential and stationary phase. The respective genes of interest are PA1137, piv, tpiA, fdhA, and prs. These genes encode, respectively, a probable oxidoreductase, a protease, a triosephosphate isomerase, a dehydrogenase, and a pyrophosphokinase. Although the transcript abundance of PA1137, piv, and fdhA was found to be significantly increased in stationary-phase cells relative to planktonic cells grown to exponential phase, the transcript abundance of tpiA and prs was significantly reduced (Table 1). In agreement with dispersed cells demonstrating traits closely resembling those of stationary-phase or exponential-phase grown planktonic cells, dispersed cells demonstrated the tpiA, fdhA, and prs transcript abundances that were comparable to those noted for stationary-phase cells but a piv transcript abundance that was more similar to that of exponential-phase cells (Table 1). The transcript abundance of PA1137, however, was found to differ significantly from planktonic cells grown to exponential or stationary phase (Table 1). The finding suggested dispersed cells demonstrate an “intermediate” phenotype.
TABLE 1.
qRT-PCR analysis of growth phase-indicating genesa
| Sample | Mean fold change ± SEM in transcript levels relative to PAO1 grown to exponential phase |
||||
|---|---|---|---|---|---|
| PA1137 | piv | tpiA | fdhA | prs | |
| PAO1 (EP) | 1 | 1 | 1 | 1 | 1 |
| PAO1 (SP) | 3.02 ±0.84b | 31.9 ± 15.2b | 0.262 ± 0.11b | 4.31 ± 1.35b | 0.259 ± 0.88b |
| Glutamate-dispersed cells | 14.5 ± 9.4b | 6.33 ± 4.98b | 0.177 ± 0.21b | 6.11 ± 3.26b | 0.235 ± 0.14b |
| Glutamate-dispersed cells, after 2 h of recovery | 16.7 ± 10.0b | 2.60 ± 0.42b | 0.666 ± 0.15 | 7.3 ± 1.44b | 0.557 ± 0.16 |
Transcript levels of PA1137, piv, tpiA, fdhA, and prs present in P. aeruginosa PAO1 dispersed cells obtained in response to glutamate and NO, and PAO1 grown planktonically to the exponential (EP) or stationary (SP) phase. The transcript levels are relative to P. aeruginosa PAO1 grown to the exponential phase. Experiments were carried out five times. The transcript levels of mreB were used as a control.
Significantly different from PAO1 grown planktonically to exponential phase (P < 0.01 as determined by ANOVA and SigmaStat).
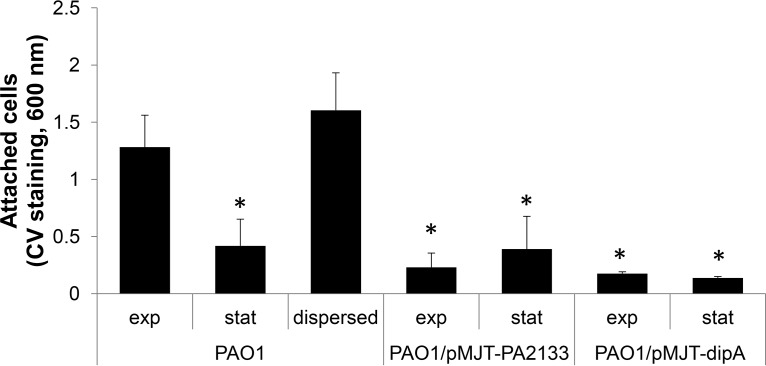
To further explore the “intermediate” phenotype of dispersed cells, we evaluated the attachment phenotype of dispersed cells. Using 96-well plate-based attachment assays, dispersed cells obtained in response to glutamate were found to attach at levels comparable to planktonic cells grown to exponential-phase after 2 h of incubation, as determined using crystal violet (CV) staining (Fig. 2). In addition, both dispersed cells and exponential-phase planktonic cells demonstrated significantly increased attachment relative to planktonic cells grown to the stationary phase (Fig. 2). It is of interest that the extent to which cells overproducing the PDEs PA2133 and DipA attached was comparable to that noted for planktonic cells grown to the stationary phase (Fig. 2). The finding not only suggested dispersed cells to be primed to reattach to a surface following egress from the biofilm mode of growth but that cells overexpressing genes encoding phosphodiesterases are not comparable to dispersed cells with respect to the attachment phenotype. Moreover, in agreement with several previous studies concluding that dispersed cells display unique properties relative to both planktonic and biofilm cells and thus could represent a unique phenotype (25, 28), our findings suggest dispersed cells display phenotypes that more closely resemble exponential-phase cells for certain traits and stationary-phase cells for others.
FIG 2.
Dispersed cells attach more than planktonic cells grown to the exponential and stationary phases or those overexpressing genes encoding phosphodiesterases. Attachment assays were carried out in 96-well plates, with each well inoculated with 200 μl of the respective bacterial culture adjusted to an OD of 0.2. Dispersed cells were obtained in response to glutamate. The adhering biomass was determined 2 h postinoculation using CV staining. Absorbance was determined at 600 nm. Experiments were carried out at least in triplicate. Error bars denote standard deviations. *, Significantly different from PAO1 grown planktonically to exponential phase (P < 0.01 as determined by ANOVA and SigmaStat). Exp, exponential phase; Stat, stationary phase.
The colistin susceptibility phenotype of dispersed cells obtained in response to glutamate is reversible.
Our findings so far illustrate that dispersion, coinciding with cells transitioning from the sessile, biofilm mode of growth to a free-living, planktonic lifestyle, is not a clear progression from one state to the other. Moreover, depending on the phenotype tested, dispersed cells resemble more closely exponential-phase cells with respect to initial attachment and susceptibility to tobramycin, or stationary-phase cells with respect to their susceptibility to colistin, whereas gene expression of select genes suggested an “intermediate” phenotype. The unique nature of the dispersed cell phenotype led us to investigate how long these cells remain in this state before fully transitioning to a planktonic, susceptible mode of growth.
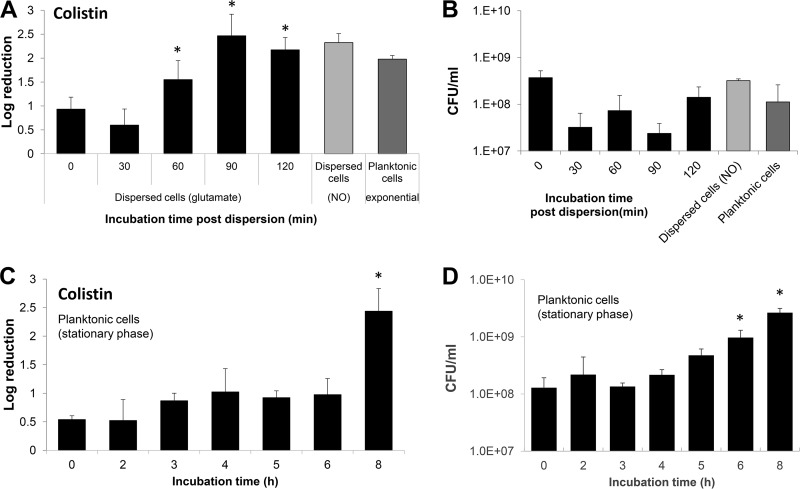
To determine the length of time required for dispersed cells to revert to a susceptible phenotype, we made use of susceptibility assays using colistin and dispersed cells obtained in response to glutamate. Freshly dispersed cells were resuspended into fresh VBMM medium, adjusted to an OD at 600 nm (OD600) of 0.2, allowed to grow for up to 2 h, and exposed at 30-min intervals to colistin (20 μg/ml) for 1 h. Relative to newly glutamate-dispersed cells, no increase in susceptibility was noted after 30 min of incubation prior to colistin treatment (Fig. 3A). Additional incubation, however, coincided with dispersed cells becoming increasingly more susceptible to colistin, as indicated by 1.5-, 2.5-, and 2.2-log reductions in viability following 1, 1.5, and 2 h of growth, respectively, prior to treatment (Fig. 3A). Moreover, after 1.5 h of incubation prior to treatment, dispersed cells were as susceptible to colistin as planktonic cells grown to exponential-phase and NO-dispersed cells. It is of interest to note that during the 2 h period of incubation, no significant increase in the overall viable cell count was noted (Fig. 3B), indicating that despite having been resuspended into fresh medium, dispersed cells experience a lag phase with respect to growth.
FIG 3.
Susceptibility of dispersed and stationary-phase cells to colistin after recovery in fresh medium under planktonic growth conditions. (A and B) Dispersed cells obtained in response to glutamate were collected, washed, resuspended in fresh VBMM, adjusted to an OD of 0.2, and subsequently allowed to grow at 22°C with aeration. At the times indicated, the cells were split into equal volumes and either left untreated or exposed to colistin (20 μg/ml) for 1 h at 37°C. (A) Susceptibility phenotype of dispersed cells and after recovery for up to 2 h under planktonic growth conditions to colistin. Colistin susceptibility is expressed as log10 reduction in CFU. (B) Average number of viable cells, expressed as CFU, present in dispersed cells at the time of recovery and over the course of 2 h under planktonic growth conditions. (C and D) Planktonic cells grown for 24 h were washed, resuspended in fresh VBMM medium, adjusted to an OD of 0.2, and subsequently allowed to grow at 22°C with aeration. At the times indicated, the cells were split into equal volumes and either left untreated or exposed to colistin (20 μg/ml) for 1 h at 37°C. (C) Susceptibility phenotype of stationary-phase cells and after growth under planktonic growth conditions in fresh VBMM to colistin. Colistin susceptibility is expressed as log10 reduction in CFU. (B) Average number of viable cells, expressed as CFU, present in stationary-phase cells at the time of recovery and over the course of 8 h under planktonic growth conditions. Experiments were repeated at least three times. Error bars indicate standard deviations. *, Significantly different from glutamate-dispersed cells at 0 min (P < 0.01 as determined by ANOVA and SigmaStat).
We furthermore sought to determine whether the 2-h time period required by dispersed cells to switch to a more susceptible phenotype (Fig. 3B) was linked to dispersed cells being similar to stationary-phase cells. We therefore repeated the susceptibility assay using stationary-phase cells. To ensure conditions comparable to those for the dispersed cells, planktonic cells were grown at room temperature for 24 h in VBMM. The resulting stationary-phase cells were resuspended into fresh VBMM medium, adjusted to an OD600 comparable to that of dispersed cells (∼ 0.2), and allowed to grow at 22°C for a given length of time before cells were exposed to colistin (20 μg/ml) for 1 h. Under the conditions tested, no significant difference in susceptibility was noted between stationary-phase cells at time zero and those reinoculated into fresh medium and allowed to grow for a period of 6 h (Fig. 3C). During this 6-h period, an increase in the viable cells was noted (Fig. 3D). Overall, stationary-phase cells were found to require additional time compared to dispersed cells, almost 8 h, to transition from a resistant to a more susceptible phenotype comparable to exponential-phase planktonic cells (Fig. 3A and C).
Our findings suggested dispersed cells require less time relative to planktonic cells grown to stationary phase, 2 h versus ∼8 h, to revert to a susceptibility phenotype that was comparable to those noted for exponential-phase planktonic cells (Fig. 3A). To determine whether the time is sufficient to fully revert dispersed cells to a phenotype that resembles exponential-phase planktonic cells, we examined the transcript abundance of genes serving as indicators for the two different planktonic growth phases in dispersed cells allowed to recover for 2 h. With the exception of PA1137 and fdhA transcript abundance, the expression of the indicator genes in recovered dispersed cells was found to be more similar to exponential-phase planktonic cells (Table 1). Together, our findings indicate that the transition from cells undergoing a dispersion response to cells resembling phenotypically planktonic cells grown to exponential phase takes approximately 2 h; however, full transition to the exponential-phase planktonic phenotype may likely require additional time.
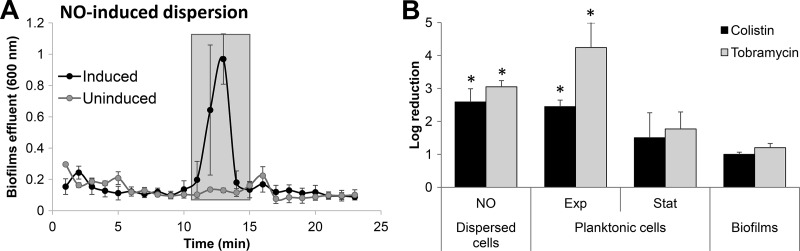
Dispersed cells obtained in response to NO are susceptible to colistin.
Given the difference in susceptibility of glutamate-induced dispersed cells to colistin and tobramycin (Fig. 1D), we furthermore sought to determine whether the susceptibility phenotype of dispersed cells is not only dependent on the antibiotic used but also on the way by which dispersed cells were obtained. To address this question, we obtained NO-induced dispersed cells by exposing biofilms to nitric oxide (NO) (Fig. 4A). It is important to note that the number of viable cells obtained upon NO-induced dispersion were comparable to those obtained in response to glutamate (Fig. 1C). Moreover, as in the case of glutamate-induced dispersed cells, NO-induced dispersed cells were immediately collected from the biofilm effluent and subsequently exposed to tobramycin or colistin. Treatment of dispersed cells obtained in response to NO coincided with a 3-log reduction in viability (Fig. 4B), with NO-induced dispersed cells being as susceptible to tobramycin as glutamate-induced dispersed cells or exponential-phase planktonic cells and more susceptible than stationary-phase and biofilm cells. Similar results were obtained upon treatment with colistin, with dispersed cells being as susceptible as exponential-phase planktonic cells but significantly more susceptible than dispersed cells obtained in response to glutamate.
FIG 4.
Susceptibility phenotype of dispersed cells obtained in response to exposure of P. aeruginosa PAO1 biofilms to NO. (A) Absorbance of biofilm tube reactor effluents after the induction of dispersion by sodium nitroprusside, with sodium nitroprusside serving as a source of nitric oxide (NO). Cells that were collected for subsequent experiments and referred to as “NO-induced dispersed cells” are indicated by shaded boxes. Effluents of untreated biofilms were used as a control. (B) Susceptibility of biofilm cells, NO-induced dispersed cells, and planktonic cells grown to exponential and stationary phases to colistin (20 μg/ml) after exposure for 1 h at 37°C. Colistin susceptibility is expressed as log10 reduction in CFU. All experiments were repeated at least 3 times. Error bars indicate standard deviations. *, Significantly different from biofilms (P < 0.01 as determined by ANOVA and SigmaStat). Exp, exponential phase; Stat, stationary phase.
The susceptibility phenotype of dispersed cell is independent of intracellular c-di-GMP levels.
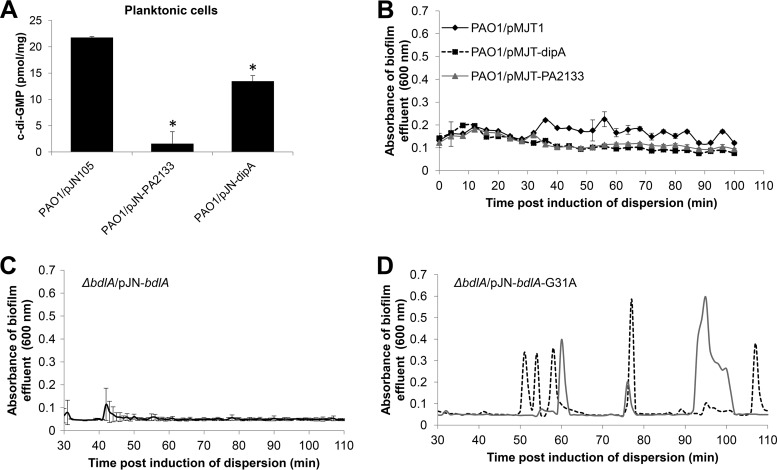
Previous studies demonstrated that dispersion coincides with a significant reduction in the intracellular levels of c-di-GMP, from the elevated levels seen in biofilm cells to the lower levels associated with the free swimming, planktonic lifestyle. Recent reports further suggested dispersed cells harbor significantly reduced c-di-GMP levels compared to planktonic cells, with dispersed P. aeruginosa cells obtained in response to NO being similar to planktonic cells overproducing the E. coli phosphodiesterase YhjH with respect to gene expression (26, 27) and susceptibility to colistin (32). Given the difference in susceptibility of glutamate- and NO-dispersed cells to colistin (Fig. 1D and 4B), we sought to determine whether differences in c-di-GMP levels affect the susceptibility of dispersed cells. We therefore first sought to obtain dispersed cells characterized by various c-di-GMP levels by overproducing phosphodiesterases, enzymes capable of degrading c-di-GMP. We selected phosphodiesterases DipA and PA2133 for several reasons. For one, overexpression of dipA and PA2133 affected c-di-GMP levels to different extents. Although overexpression of DipA coincided with a reduction in the overall cellular level of c-di-GMP present in planktonic cells, overexpression of PA2133 appeared to almost deplete planktonic cells of c-di-GMP (Fig. 5A). In addition, while the phosphodiesterase PA2133 has not been linked to dispersion, DipA was found to be required, with inactivation of dipA coinciding with impaired dispersion of mutant biofilms in response to glutamate and NO (12, 13). Moreover, the mechanism by which DipA contributes to dispersion and the modulation of c-di-GMP is fairly well understood (12). Upon perceiving dispersion cues, the chemotaxis transducer protein BdlA is activated in a process requiring phosphorylation and nonprocessive proteolytic cleavage (15). Active BdlA, in turn, induces the phosphodiesterase activity of DipA (12, 31). In addition to DipA, BdlA recruits a second phosphodiesterase, RbdA, upon induction of dispersion, and both DipA and RbdA subsequently contribute to the reduction of the intracellular pool of c-di-GMP (12).
FIG 5.
Contribution of cellular c-di-GMP levels to the biofilm dispersion response. (A) Cellular c-di-GMP level of planktonic cells harboring the empty vector pMJT1 or overexpressing PA2133 and dipA encoding phosphodiesterases. c-di-GMP levels were determined by high-pressure liquid chromatography (HPLC). “pmol/mg” refers to c-di-GMP levels (pmol) per total cell protein (in mg). *, Significantly different from PAO1 vector control (P < 0.01 as determined by ANOVA and SigmaStat). (B to D) Detection of dispersion after induction of PA2133, dipA, bdlA, and bdlA-G31A gene expression. Dispersion experiments were carried out in tube reactor-grown biofilms. Dispersion was indicated by an increase in the turbidity of biofilm tube reactor effluents. (B) Detection of dispersion after induction of PA2133 and dipA gene expression by the addition of 0.1% arabinose to the growth medium. P. aeruginosa PAO1 harboring the empty vector pMJT1 was used as a control. (C and D) Detection of dispersion after the induction of bdlA (C) and bdlA-G31A (D) gene expression upon addition of arabinose to the growth medium. The graphs are representative of two independent biofilm replicates indicated by dashed and solid lines. Dispersion was detected upon the addition of arabinose to the ΔbdlA strain carrying pJN-bdlA-G31A (D), but not to the PAO1/pMJT-1, PAO1/pMJT-PA2133, and PAO1/pMJT-dipA strains (B) or to the ΔbdlA strain carrying pJN-bdlA (C).
We made use of P. aeruginosa strains overproducing phosphodiesterases DipA and PA2133 (PAO1/pMJT-dipA and PAO1/pMJT-PA2133), with both genes being placed under the control of the arabinose-inducible PBAD promoter to enable manipulation of dipA and PA2133 expression in response to arabinose. In addition, we made use of the ΔbdlA strain carrying pJN-bdlA-G31A (31), which harbors bdlA-G31A under the control of the arabinose-inducible PBAD promoter, as positive controls. BdlA-G31A is a BdlA variant harboring an alanine substitution at amino acid residue G31. Alanine substitution of this residue was found to render P. aeruginosa biofilms hyperdispersive in the absence of dispersion cues, with biofilms of the ΔbdlA strain carrying pJN-bdlA-G31A being characterized by low cellular c-di-GMP levels and increased phosphodiesterase and DipA activity (12, 13, 15, 31). Strain PAO1/pMJT-1 harboring the empty vector and the ΔbdlA strain carrying pJN-bdlA harboring the wild-type bdlA variant were used as negative controls.
The respective strains were allowed to form wild type-like biofilms in the absence of arabinose, after which time arabinose was added to the growth medium to induce transcription (Fig. 5B to D). No dispersion events were detectable by biofilms formed by strains PAO1/pMJT-dipA and PAO1/pMJT-PA2133 or the vector control (PAO1/pMJT1 over the course of 2 h [Fig. 5B]). In agreement with previous findings (31), however, the addition of arabinose to the growth medium resulted in repeated dispersion events of the ΔbdlA strain carrying pJN-bdlA-G31A (Fig. 5D), a response that was absent in biofilms by the ΔbdlA strain complemented with the nonhyperdispersive BdlA variants (the ΔbdlA strain carrying pJN-bdlA, Fig. 5C). Our finding suggested that despite DipA and PA2133 being capable of modulating c-di-GMP levels to different extents and, in contrast to previous findings by Tolker-Nielsen and coworkers (26, 27), dispersion by P. aeruginosa biofilms is not simply inducible upon induction of phosphodiesterases encoded by P. aeruginosa.
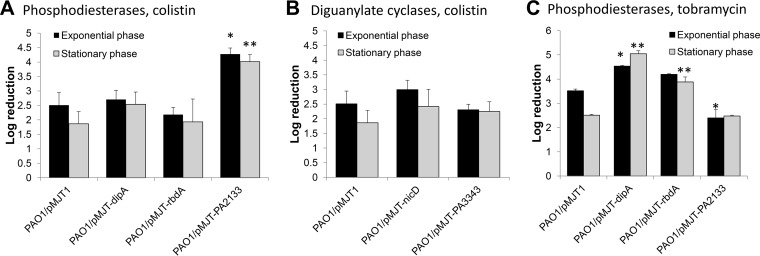
As we were unable to obtain dispersed cells with various c-di-GMP levels, we instead made use of planktonic cells overexpressing phosphodiesterases (PAO1/pMJTdipA, PAO1/pMJTrbdA, and PAO1/pMJT-PA2133). P. aeruginosa PAO1 and PAO1/pMJT1 harboring an empty vector, were used as controls. The respective strains were grown to exponential or stationary phase (see Fig. 1B), diluted to an OD600 of 0.2 in fresh medium containing arabinose, and subsequently treated with colistin (20 μg/ml) for 1 h. Overexpression of dipA or rbdA had no effect on colistin susceptibility compared to the vector control (Fig. 6A). In contrast, overexpression of PA2133 resulted in increased susceptibility to colistin compared to the vector control (Fig. 6A). Although our findings indicated a role of PA2133 in colistin resistance, the contribution of PA2133 was opposite to that observed in the dispersed cells obtained using glutamate that exhibited decreased susceptibility to colistin (Fig. 1D).
FIG 6.
Susceptibility of planktonic cells differing in c-di-GMP levels due to overexpression of genes encoding phosphodiesterases and diguanylate cyclases to colistin and tobramycin. (A) Susceptibility of planktonic cells overexpressing dipA, rbdA, and PA2133 encoding phosphodiesterases, to colistin. (B) Susceptibility of planktonic cells overexpressing nicD and PA3343 encoding diguanylate cyclases to colistin. (C) Susceptibility of planktonic cells overexpressing dipA, rbdA, and PA2133 encoding phosphodiesterases to tobramycin. Strains were grown to the exponential and stationary phases prior to treatment with colistin (20 μg/ml) and tobramycin (50 μg/ml) for 1 h at 37°C. Susceptibility is expressed as log10 reduction in CFU. All experiments were carried out at least in triplicate. Error bars denote standard deviations. *, Significantly different from PAO1/pMJT1 grown to exponential phase; **, significantly different from PAO1/pMJT1 grown to stationary phase (P < 0.01 as determined by ANOVA and SigmaStat).
We furthermore evaluated the transcript abundance of genes believed to contribute to the tolerance to cationic antimicrobial peptide colistin in response to reduced c-di-GMP levels. Resistance has been linked to spermidine, with spermidine being believed to confer stability to the bacterial outer membrane and prevent its disruption by antimicrobial peptides (33). Genes encoded by PA4773 and PA4774 play a role in spermidine production (33). Moreover, resistance has been linked to lipopolysaccharide (LPS) modification by 4-aminoarabinose. The system contributing to LPS modification is encoded by the arnBCADTEF operon (34, 35). It is of interest that no significant difference in the transcript abundance of genes arnB, arnE, and PA4774, which are involved in mediating tolerance to antimicrobial peptides, was noted upon overexpression of dipA or PA2133 (Table 2). The only significant change in expression for genes involved in spermidine biosynthesis was for PA4773 during exponential-phase growth of PAO1/pMJTdipA (Table 2). However, this change in expression was modest and did not correlate to a change in susceptibility as seen in the treatment data for similar growth conditions shown in Fig. 6A.
TABLE 2.
qRT-PCR analysis of genes involved in LPS modificationa
| Sample | Mean fold change ± SEM in transcript levels relative to PAO1/pMJT1 grown to exponential phase |
|||
|---|---|---|---|---|
| arnB | arnE | PA4773 | PA4774 | |
| PAO1 (EP) | 1 | 1 | 1 | 1 |
| PAO1 (SP) | ND | −3.74 ± 0.09b | −7.65 ± 0.03b | −2.88 ±0.05b |
| PAO1/pMJT-dipA (EP) | 1.61 ± 0.22 | 1.97 ± 0.12 | 2.81 ± 0.25b | 1.55 ± 0.26 |
| PAO1/pMJT-dipA (SP) | 1.15 ± 0.09 | 1.33 ± 0.24 | 1.59 ± 0.32) | 1.46 ± 0.09 |
| PAO1/pMJT-PA2133 (EP) | 1.11 ± 0.07 | 1.08 ± 0.05 | 1.49 ± 0.22 | 1.26 ± 0.11 |
| PAO1/pMJT-PA2133 (SP) | 1.77 ± 0.33 | 1.28 ± 0.06 | 1.46 ± 0.08 | 1.24 ± 0.15 |
Transcript levels of arnB, arnE, PA4773, and PA4774 present in P. aeruginosa PAO1 dispersed cells obtained in response to glutamate and NO, as well as wild-type and PAO1/pMJT-dipA, and PAO1/pMJT-PA2133 strains grown planktonically to exponential (EP) or stationary (SP) phase. The transcript levels are relative to P. aeruginosa PAO1 or PAO1/pMJT1 grown to the exponential phase. Experiments were carried out five times. The transcript levels of mreB were used as a control.
Significantly different from PAO1 grown planktonically to the exponential phase (P < 0.01 as determined by ANOVA and SigmaStat). ND, not determined.
To further exclude a role of c-di-GMP in the susceptibility to colistin, we also tested the effect of elevated c-di-GMP levels on colistin susceptibility, by using strains overexpressing the diguanylate cyclases encoded by nicD (12) and PA3343. No difference in susceptibility to colistin was noted in strains overexpressing nicD or PA3343 compared to the vector control (Fig. 6B). In addition, the susceptibility phenotype of P. aeruginosa strains characterized by reduced c-di-GMP levels was tested using tobramycin. While overexpression of dipA or rbdA coincided with increased susceptibility to tobramycin compared to the vector control (Fig. 6C), overexpression of PA2133 had no effect when tested under stationary-phase growth conditions or coincided with reduced susceptibility to tobramycin relative to the vector control grown to exponential phase (Fig. 6C). Taken together, our findings indicate that modulating c-di-GMP levels does not affect the susceptibility phenotype of P. aeruginosa to colistin, likely suggesting that the modulation of c-di-GMP levels alone may not be responsible for the altered susceptibility of dispersed cells to colistin.
The LPS modification system contributes to increased tolerance phenotype of glutamate-induced dispersed cells to colistin.
Proteomic analysis of dispersed and biofilm cells indicated dispersed cells are characterized by increased levels of proteins encoded by the arnBCADTEF operon, suggesting dispersion coincides with increased transcript abundance of arnBCADTEF gene expression (32). Given the difference in susceptibility of NO- and glutamate-induced dispersed cells to colistin, we sought to determine whether dispersion cues may contribute to the differential expression of genes encoded by the arn operon, thus resulting in an observed difference in tolerance to cationic antimicrobial peptides. We therefore examined the susceptibility phenotype of a mutant strain harboring a transposon insertion in arnB (referred to as ΔarnB). We reasoned that if the difference in susceptibility between NO- and glutamate-induced dispersed cells to colistin is due to differences in arnBCADTEF transcript abundance, that inactivation of arnB would eliminate the difference. If so, we anticipated finding dispersed ΔarnB cells obtained in response to glutamate to be as susceptible to colistin as NO-induced dispersed ΔarnB cells.
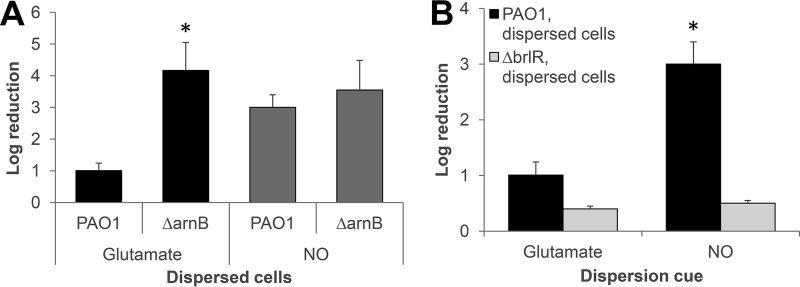
Dispersed ΔarnB cells obtained in response to glutamate exhibited increased sensitivity to colistin relative to wild-type dispersed cells (Fig. 7A). In contrast, however, insertional inactivation of arnB had no effect on the susceptibility phenotype of NO-induced dispersed cells, since no difference in colistin susceptibility was noted in dispersed cells obtained from PAO1 and the ΔarnB mutant strain biofilms in response to NO (Fig. 7A). Our data suggested dispersion cues to likely affect arn operon gene expression, with glutamate but not NO contributing to the activation of the LPS modification system in dispersed cells.
FIG 7.
The LPS modification system and BrlR contribute to the difference in the susceptibility phenotype of glutamate- and NO-induced dispersed cells to colistin. (A) Susceptibility of dispersed cells obtained from biofilms of wild-type and ΔarnB mutant strains in response to glutamate and NO to colistin (20 μg/ml) after exposure for 1 h at 37°C. (B) Inactivation of brlR coincides with dispersed cells being resistant to colistin (20 μg/ml) regardless of the dispersion cue used (glutamate, NO). Dispersed cells obtained after exposure of P. aeruginosa wild-type biofilms to glutamate or NO were used as a control. Colistin susceptibility is expressed as log10 reduction in CFU. All experiments were repeated at least three times. Error bars indicate standard deviations. *, Significantly different from PAO1 dispersed cells in response to same dispersion cue (P < 0.01 as determined by ANOVA and SigmaStat).
BrlR contributes to the difference in susceptibility of glutamate and NO-induced dispersed cells to colistin.
Considering our finding of glutamate but not NO likely contributing to the activation of the LPS modification system in dispersed cells, we sought to determine whether the two dispersion cues differentially activate the two-component regulatory systems PhoPQ and PmrAB that contribute to the expression of the arnBCADTEF operon responsible for the addition of 4-aminoarabinose to LPS (36, 37). Interestingly, the susceptibility phenotype of glutamate-induced dispersed cells, being tolerant to colistin but susceptible to tobramycin (Fig. 1D), was reminiscent of the contribution of the MerR-type transcriptional regulator BrlR in drug tolerance. This is because BrlR reciprocally contributes to colistin and tobramycin resistance in P. aeruginosa PAO1 by enhancing the expression of several multidrug efflux pumps while repressing phoP, phoQ, pmrA, and pmrB, and in turn arnBCADTEF, expression (37). Thus, expression of brlR was shown to enhance tolerance to most antimicrobial agents such as tobramycin and norfloxacin while rendering cells susceptible to colistin. In contrast, inactivation of brlR coincided with decreased tolerance to most antimicrobial agents but increased tolerance to colistin (37). We therefore sought to determine whether BrlR contributes to the difference in susceptibility to colistin of dispersed cells obtained in response to glutamate and NO, as described in Fig. 1D and 4B. We hypothesized that if BrlR contributes to the observed susceptibility of NO-dispersed cells to colistin, inactivation of brlR would coincide with dispersed cells being rendered as tolerant as glutamate-dispersed cells. P. aeruginosa PAO1 and a ΔbrlR mutant strain were allowed to form biofilms for 5 days and subsequently dispersed with either glutamate or NO. Dispersed cells were immediately collected and exposed to colistin for 1 h. As anticipated, inactivation had little effect on the susceptibility of glutamate-dispersed cells. This was supported by the finding of PAO1 and ΔbrlR strain dispersed cells being reduced in viability by 1- and ∼0.4-log reductions, respectively (Fig. 7B). Likewise, ΔbrlR strain dispersed cells obtained in response to NO were as tolerant to colistin as ΔbrlR strain dispersed cells obtained in response to glutamate, as indicated by a 0.4-log reduction in viability (Fig. 7B). This is in contrast to P. aeruginosa PAO1 dispersed cells obtained in response to NO, which were susceptible to colistin, as indicated by a 3-log reduction in the viability (Fig. 7B). Considering the previously established role of BrlR in repressing the expression of genes encoding the LPS modification system, combined with the susceptibility phenotype of ΔbrlR strain dispersed cells, our data suggested increased susceptibility to NO relative to glutamate-dispersed cells is likely due to increased brlR expression in NO-dispersed cells. This was supported by the finding of brlR transcript abundance being increased up to (8 ± 2.9)-fold in NO-dispersed cells relative to glutamate-induced cells, as determined using quantitative reverse transcription-PCR (qRT-PCR).
DISCUSSION
Dispersion has been described as a cellular release from biofilms that is distinct from a simple reversal of attachment and reversion to a planktonic mode of growth, since it involves sensing specific cues and consequent physiological alterations (9). Noted changes in the phenotype of dispersed cells relative to biofilm cells include marked differences in gene expression and protein production, reduced cellular c-di-GMP levels, increased motility, release of degradative enzymes, and virulence, with dispersed cells contributing to dissemination and a shift from chronic to an acute virulence phenotype (7–10, 18, 25, 28, 38). Although the ability to disperse biofilms is considered a promising avenue for biofilm control, the switch of dispersed cells toward an acute virulence phenotype may pose a problem. Moreover, the increase in virulence associated with dispersed cells raises the question whether the modulation of the biofilm lifestyle as a potential mechanism of infection control means trading one “biofilm problem” with another, different “dispersion problem.” The problem may be further acerbated by the uncertainty of the susceptibility phenotype of dispersed cells, considering how little is known about the response of dispersed cells to therapies and for how long the dispersion phenotype persists.
In this study, we set out to determine the susceptibility of dispersed cells and whether this susceptibility was dependent on the dispersion cues used to disperse cells from the biofilm. We anticipated finding dispersed cells to be more susceptible to antimicrobial agents, based on the notion that dispersed cells would broadly display phenotypes ascribed to the planktonic mode of growth. Although we found dispersed cells to be as susceptible to tobramycin as planktonic cells grown to exponential phase, the same was not true when dispersed cells were exposed to colistin. Instead, dispersed cells obtained in response to glutamate retained their enhanced recalcitrance to killing by colistin similar to the recalcitrance noted for biofilm cells, whereas NO-dispersed cells were more susceptible than their sessile counterparts. Thus, the susceptibility phenotype of dispersed was found to be dependent on both the dispersion cue used to induce a dispersion response and the type of antimicrobial agent used for treatment.
To elucidate the underlying mechanism contributing to the disparity in response to colistin, we explored the role of c-di-GMP in drug susceptibility. This is based on dispersion coinciding with reduction of the cellular c-di-GMP level and reported links between c-di-GMP levels and drug susceptibility. Specifically, recent findings demonstrated P. aeruginosa cells harboring reduced c-di-GMP levels were shown to be more resistant to colistin than P. aeruginosa cells with high c-di-GMP levels (32), whereas P. aeruginosa planktonic cells harboring high, biofilm-like c-di-GMP levels were found to be more resistant to tobramycin relative to those with low, planktonic-like c-di-GMP levels (29). However, we found no link in this study between c-di-GMP levels and susceptibility to colistin. Likewise, we were unable to confirm findings by Chua et al. (32) linking colistin resistance to decreased c-di-GMP levels. Instead, our findings suggest a link between specific phosphodiesterases and colistin resistance. While Chua et al. (32) used the E. coli phosphodiesterase YhjH to demonstrate that reduced levels of c-di-GMP render P. aeruginosa more resistant to colistin, we demonstrated in this study that overproduction of the phosphodiesterase PA2133, but not of the phosphodiesterases DipA or RbdA, coincides with increased susceptibility to colistin (Fig. 3). Moreover, PA2133, but not DipA or RbdA, was found to inversely affect colistin and tobramycin susceptibility. Thus, while our findings indicate that general modulation of c-di-GMP levels were not a contributing factor in colistin susceptibility, our data do support a specific link between PA2133 and drug susceptibility. However, it is also likely that in addition to the phosphodiesterase used, differences in the experimental design contribute to the observed differences in susceptibility. For one, this study made use of viability determination, whereas Chua et al. combined live/dead staining and absorbance reading to evaluate susceptibility. In addition, we made use of dispersed cells obtained from 5-day-old biofilms (grown in tube reactors under flowing conditions) in response to dispersion cue, with dispersed cells being collected from the tube reactor effluent within 20 min after exposure to dispersion cues. In contrast, Chua et al. (32) obtained dispersed cells from 1-day-old microtiter plate grown biofilms following the induction of yhiH expression with up to 1% arabinose for 5 h to induce dispersion. Regardless of the experimental design, however, the findings obtained by both us and Chua et al. (32) strongly suggest a need to evaluate the respective contributions of each phosphodiesterase to drug susceptibility separately. This appears to also be applicable for dispersion induction, since in contrast to recent findings (27, 28, 32) our study suggests that modulation of the c-di-GMP level, by employing only one of the P. aeruginosa phosphodiesterase, is not sufficient to induce dispersion by P. aeruginosa biofilms (Fig. 5).
Instead, we noted a link to BrlR and the LPS modification system encoded by the arn operon, that may explain the different response to colistin seen in NO- versus glutamate-dispersed cells. While BrlR functions as a multidrug transport activator, BrlR has been furthermore shown to act as a repressor of phoPQ expression (37), thus suppressing colistin resistance. In agreement with previous findings (37), we demonstrated here that inactivation of brlR negated the difference in susceptibility of NO-dispersed cells relative to glutamate-dispersed cells to colistin (Fig. 7B). In contrast, inactivation of arnB only coincided with increased susceptibility of glutamate-dispersed cells, but not NO-dispersed cells, to colistin (Fig. 7A). These findings are in agreement with reported differences in brlR transcript abundance and BrlR indirectly repressing arnBCADTEF gene expression, via repressing phoPQ and likely pmrAB (36, 37).
While our findings clearly indicate dispersed cells to be a problem, especially when treated with colistin, previous findings demonstrated that, despite BrlR reciprocally contributing to colistin and tobramycin resistance, differences in susceptibility to colistin and tobramycin can be eliminated by combination treatment with both antibiotics (37). Thus, the same may be applicable to dispersed cells. Moreover, our data also indicate that the “problem” with dispersed cells is not permanent, since the susceptibility phenotype of dispersed cells to colistin was found to be reversible. Under the conditions tested, glutamate-dispersed cells reverted from being resistant to being susceptible to colistin within 2 h. Although gene expression analysis of growth mode indicator genes suggested dispersed cells revert during this time period to a phenotype comparable to that of planktonic cells grown to exponential phase, the reversion was not complete. Instead, it appears that dispersed cells gradually revert, with some traits persisting for more than 2 h. In agreement with previous reports (25, 28), our findings indicate dispersed cells to have a unique transitory phenotype. However, instead of displaying a unique phenotype, dispersed cells either closely resembled exponential-phase cells, as in the case of tobramycin susceptibility and attachment those of stationary-phase cells (glutamate-induced dispersed cells and colistin susceptibility), or an “intermediate” phenotype in relation to the expression of growth mode indicator genes (Table 1). In addition, our findings suggest differences among dispersed cells, depending on which dispersion cue they were exposed to (Fig. 1 and 4). Overall, both the timing required for reversion to a truly planktonic phenotype and the differences between dispersed cells should be taken into account in future studies to better assess the dispersion problem and the applicability of dispersion as a potential mechanism of infection control.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 3. P. aeruginosa strain PAO1 was used as parental strain. All planktonic cultures were grown in flasks at 220 rpm at 37°C, unless otherwise indicated, using Vogel-Bonner citrate minimal medium (VBMM), which contains the equivalent of 1 mM Mg2+ (39). For plasmid maintenance, antibiotics were used at 250 μg/ml carbenicillin and 50 μg/ml gentamicin for P. aeruginosa.
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 tonA | Life Technologies |
| P. aeruginosa PAO1 | ||
| PAO1 | Wild type | B. H. Holloway |
| ΔbrlR strain | PAO1, ΔbrlR (PA4878) | 48 |
| ΔbdlA strain | PAO1, ΔbdlA (PA1423) | 14 |
| ΔarnB strain | PAO1, PA3552::ISphoA; Tetr | 49 |
| Plasmids | ||
| pET101D | Vector for directional cloning and high level V5/6XHis fusion protein expression; Ampr | Life Technologies |
| pMJT-1 | araC-PBAD cassette of pJN105 cloned into pUCP18; Ampr (Carbr) | 40 |
| pMJT-dipA | C-terminal V5/6×His-tagged dipA (PA5017) cloned into pMJT-1 | 13 |
| pMJT-PA2133 | P. aeruginosa PA2133 cloned into pMJT-1 | This study |
| pMJT-rbdA | C-terminal V5/6×His-tagged rbdA (PA0861) cloned into pMJT-1 | This study |
| pMJT-nicD | C-terminal V5/6×His-tagged nicD (PA4929) cloned into pMJT-1 | 12 |
| pMJT-PA3343 | P. aeruginosa PA3343 cloned into pMJT-1 | This study |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD; Gmr | 50 |
| pJN-PA2133 | P. aeruginosa PA2133 cloned into pJN105 | 51 |
| pJN-dipA | C-terminal V5/6×His-tagged dipA (PA5017) cloned into pJN105 | 13 |
| pJN-bdlA | C-terminal 6×His-tagged bdlA (PA1423) cloned into pJN105 | 31 |
| pJN-bdlA-G31A | C-terminal 6×His-tagged bdlA with G31A mutation cloned into pJN105 at NheI/SacI | 31 |
Gmr, gentamicin resistance; Carbr, carbenicillin resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance.
Strain construction.
Complementation and overexpression of P. aeruginosa genes was accomplished by inserting the respective genes into the pMJT-1 (40) vector under the control of an arabinose-inducible promoter. C-terminal V5/6×His tagging of PA3343 and RbdA (PA0861) variants was accomplished by subcloning into pET101D (Life Technologies). The tagged constructs were then cloned into pJN105 and pMJT1. The identity of all vector inserts was confirmed by PCR and sequencing. Plasmids were introduced into P. aeruginosa via electroporation or conjugation. In addition, transposon insertional inactivation of arnB was confirmed by PCR and sequencing. Primers used for strain construction and confirmation are listed in Table 4.
TABLE 4.
Primers used in this study
| Method and oligonucleotide | Sequence (5′–3′)a |
|---|---|
| qRT-PCR | |
| arnB-RT-F | ACTGGACTTTCTGCCATTCT |
| arnB-RT-R | GTTCGAGCTCCTGGTTCTT |
| arnE-RT-F | AGCTCACCGTGGAGCACT |
| arnE-RT-R | TGACCAGGACGAAGTTCA |
| PA4773-RT-F | ACTGGTCATCGAGCTGTTCG |
| PA4773-RT-R | CGTACTCCGGCCAGGTATG |
| PA4774-RT-F | CCGCCGAACCACTTCTATTTCT |
| PA4774-RT-R | GCGGTCGTAGTTGTAGGTGTC |
| PA1137-RT-F | GCGAAAATCTTCACCACGAT |
| PA1137-RT-R | CACTTCCTCGACGAAATCCT |
| piv-RT-F | GTCAACCGTCCCTACTGGAG |
| piv-RT-R | GGAGTCGGCGAAATACGATA |
| tpiA-RT-F | CCGGTAACTGGAAAATGCAC |
| tpiA-RT-R | GATGACCTGGCTGATGAACA |
| fdhA-RT-F | TACCTGCAAGGAACAGCACA |
| fdhA-RT-R | GTAGGGCACCAGCACGTATT |
| prs-RT-F | AGTTGGCAAATTCTCCGATG |
| prs-RT-R | CATCACCACCAGTTCCATCA |
| Tn-Check | |
| arnB-RT2_F | GTGGTGATCCCGGTCTAC |
| PhoA_R | CGGGTGCAGTAATATCGCCCT |
| Cloning | |
| PA2133-F-SacI | GCGCGCGCGAGCTCGTGAACGGTTCCCCACAG |
| PA2133-R-EcoRI | GCGCGCGCGAATTCTCACCCCTGGCGGC |
| PA0861_pET-for | CACCATGAGGCAGAACCGG |
| PA0861_pET-rev | CCGGAGGTTCTGTCC |
| PA0861_XbaI-for | GCGCGCTCTAGAATGAGGCAGAACCGGAC |
| PA0861_V5/His_EcoRI-rev | GCGCGCGAATTCTCAATGGTGATGGTGATG |
| PA3343_pET_for | CACCGTGTGCGTGACACA |
| PA3343_pET_rev | GGCCACCACCTGGTTACG |
| PA3343_XbaI_for | GCGCGCTCTAGAGTGTGCGTGACACAGAAG |
| PA3343_V5/His_EcoRI_rev | GCGCGCGAATTCTCAATGGTGATGGTGATG |
Restriction sites are underlined.
Planktonic growth conditions.
P. aeruginosa strains were grown planktonically in VBMM at 37°C and 220 rpm or in a test tube roller unless otherwise indicated. Fresh planktonic cultures were started from a 1% inoculum of an overnight culture grown from a frozen stock. When required, the growth medium was supplemented with antibiotics for plasmid maintenance and 1% arabinose to induce gene expression. For experiments involving a specific phase of planktonic growth, P. aeruginosa cultures were grown for 4.5 h to exponential phase (OD600 = 0.5 to 0.8) or 8.5 h to reach stationary phase, at which time an OD600 of >1.9 was reached. For stationary-phase time course studies, P. aeruginosa was grown for 24 h in VBMM at room temperature (22°C) in a test tube roller to more closely mimic growth conditions of dispersed cells. The resulting suspension was briefly spun down (16,000 × g, 1 min), resuspended in fresh VBMM, and diluted to a final OD600 of 0.2 using fresh VBMM. The suspension was subsequently grown in a test tube roller at 22°C for up to 8 h.
Antibiotic susceptibility testing of planktonic cells.
For susceptibility testing, P. aeruginosa PAO1 was grown to exponential phase (OD600 = 0.5 to 0.8), or stationary phase (OD600 > 1.9), centrifuged at 16,000 × g for 1 min, and diluted down to an OD600 of 0.2 in fresh VBMM. The cultures were subsequently split into 1 ml aliquots and either left untreated or exposed to colistin (20 μg/ml) or tobramycin (50 μg/ml) for 60 min at 37°C in a test tube roller. Cultures were then vortexed, centrifuged at 16,000 × g for 1 min, resuspended in 1 ml 0.85% saline, and serially diluted and spread-plated onto Luria-Bertani (LB) agar. Viability was determined via CFU counts. Susceptibility is expressed as log10 reduction in CFU.
For susceptibility testing of P. aeruginosa overexpressing genes encoding phosphodiesterases and diguanylate cyclases, the strains were grown in VBMM containing 250 μg/ml carbenicillin and 1% arabinose to exponential and stationary phase. The suspensions were subsequently centrifuged at 16,000 × g for 1 min, resuspended in fresh VBMM containing carbenicillin and arabinose, and split into 1-ml aliquots that were either left untreated or exposed to colistin (20 μg/ml) or tobramycin (50 μg/ml) for 60 min at 37°C in a test tube roller. Cultures were then vortexed, centrifuged at 16,000 × g for 1 min, resuspended in 1 ml of 0.85% saline, and serially diluted and spread plated onto LB agar. Viability and susceptibility were determined as described above.
The susceptibility of P. aeruginosa PAO1 grown for 24 h in VBMM to stationary phase at 22°C was determined following resuspension and dilution to a final OD600 = 0.2 in fresh VBMM (time zero) and subsequent growth (2, 3, 4, 5, 6, and 8 h) at 22°C. After the selected period of growth, cells were treated with colistin (20 μg/ml) for 60 min at 37°C. Viability and susceptibility were determined as described above.
Biofilm formation and dispersion assays.
Biofilms by P. aeruginosa PAO1 and ΔbrlR were grown in a continuous flow tube reactor system (1-m size 14 silicone tubing; Masterflex; Cole Parmer, Inc.) in 5-fold-diluted VBMM at 22°C and a flow rate of 0.2 ml/min, as previously described (7, 18, 41). After 5 days of biofilm growth, dispersion was induced by the sudden addition of l-glutamate (18 mM) or 500 μM sodium nitroprusside to the growth medium, as previously described (14, 42). Sodium nitroprusside was used as a source of NO. To evaluate whether a reduction in c-di-GMP levels coincides with dispersion, biofilms by P. aeruginosa PAO1/pMJT1, PAO1/pMJT-PA2133, PAO1/pMJT-dipA, and the ΔbdlA strain complemented with pJN-bdlA or pJN-bdlA-G31A were grown for 5 days in tube reactors under flowing conditions using 5-fold diluted VBMM medium supplemented with antibiotics (carbenicillin and gentamicin) for plasmid maintenance. No arabinose was added to the biofilm growth medium. Dispersion was subsequently induced by the addition of 0.1% arabinose to the growth medium to induce gene expression. Independent of the dispersion inducing conditions, dispersed cells were collected from the tube reactor effluent at 1-min intervals, and the OD of the biofilm effluents was assessed by spectrophotometry. Dispersion events were indicated by an increase in the turbidity measured at 600 nm relative to the effluents of biofilms not exposed to dispersion cues or, in the case of arabinose-induced dispersion, relative to biofilms by the vector control (PAO1/pMJT1).
Antibiotic susceptibility testing of biofilm and dispersed cell.
Biofilms by P. aeruginosa PAO1 were harvested following, homogenized using a tissue homogenizer, centrifuged at 16,000 × g for 1 min at 22°C, and cell pellets were resuspended in fresh VBMM to an OD600 of 0.2. The resulting suspension was exposed to colistin (20 μg/ml) or tobramycin (50 μg/ml) for 1 h at 37°C.
Newly collected dispersed cells were combined, centrifuged at 16,000 × g for 1 min at room temperature and the cell pellets resuspended in fresh VBMM to an OD600 of 0.2. The cells were then treated with colistin (20 μg/ml) or tobramycin (50 μg/ml) for 60 min at 37°C in a test tube roller, followed by serial dilutions and plating as described above.
For colistin susceptibility recovery studies, freshly glutamate-induced dispersed cells were washed and OD adjusted as described above. Then, dispersed cells were incubated at 22°C for up to 2 h in a test tube roller, followed by treatment with colistin (20 μg/ml) at 37°C for 1 h. The viability and susceptibility were determined as described above.
Assessment of attachment.
Initial attachment to a polystyrene surface was measured using the polystyrene microtiter dish assay system (96-well) as previously described (43). Briefly, each well was inoculated with 200 μl of dispersed cells or planktonic cells grown to exponential or stationary phase, adjusted to an OD600 of ∼ 0.2. The 96-well plates were then incubated for 2 h at 37°C with shaking at 220 rpm to ensure proper aeration. Arabinose (1%) was added to the growth medium to ensure the induction of strains harboring the vector pMJT1. All experiments were carried out at least in triplicate, with each repeat comprising six technical replicates.
RNA extraction and qRT-PCR.
To obtain RNA from dispersed cells, dispersed cells were collected directly into equal volumes of RNAprotect (Qiagen). To isolate RNA from planktonic cells, the respective strains were first grown in VBMM to either exponential or stationary phase. Then, ∼6-ml portions of the bacterial culture were harvested by centrifugation (10,000 × g, 5 min at 4°C) and resuspended in 1 ml of RNAprotect. Isolation of mRNA and cDNA synthesis was carried out as previously described (41, 44, 45). qRT-PCR was performed using the Bio-Rad CFX Connect real-time PCR detection system and SsoAdvanced SYBR green Supermix (Bio-Rad) with oligonucleotides listed in Table 4. mreB was used as a control. Relative transcript quantitation was accomplished using the CFX Manager Software (Bio-Rad) by first normalizing transcript abundance (based on the threshold cycle value [CT]) to mreB, followed by determining transcript abundance ratios. Melting curve analyses were used to verify specific single product amplification.
In vivo quantification of c-di-GMP from P. aeruginosa.
Cyclic di-GMP (c-di-GMP) was extracted in triplicate from wild-type and mutant strains using heat and ethanol precipitation (14) and quantitated essentially as previously described (46). Briefly, c-di-GMP was extracted in triplicate from wild-type and mutant strains grown planktonically to exponential phase or as biofilms for 6 days using heat and ethanol precipitation, followed by centrifugation. Supernatants were combined, dried using a Speed-Vac, and resuspended in water. Samples (20 μl) were analyzed using an Agilent 1100 HPLC apparatus equipped with an autosampler, degasser, and detector set to 253 nm and then separated using a reverse-phase C18 Targa column (2.1 by 40 mm; 5 μm) at a flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; and 26 to 40 min, 1% B (buffer A, 10 mM ammonium acetate; buffer B, methanol plus 10 mM ammonium acetate) (13, 47). Commercially available cyclic di-GMP was used as a reference for the identification and quantification of cyclic di-GMP in cell extracts.
Statistical analysis.
All experiments were carried out at least in triplicate. Student t test was performed for pairwise comparisons of groups, and multivariant analyses were performed using a 1-way analysis of variance (ANOVA), followed by an a posteriori test using SigmaStat software.
ACKNOWLEDGMENTS
We thank Rebecca E. Al-Feghali for her assistance in carrying out cloning and PCR-related experiments.
This study was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R01AI080710 and R01AI075257). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. [DOI] [PubMed] [Google Scholar]
- 3.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seth AK, Geringer MR, Gurjala AN, Hong SJ, Galiano RD, Leung KP, Mustoe TA. 2012. Treatment of Pseudomonas aeruginosa biofilm–infected wounds with clinical wound care strategies: a quantitative study using an in vivo rabbit ear model. Plastic Reconstruct Surg 129:262e–274e. doi: 10.1097/PRS.0b013e31823aeb3b. [DOI] [PubMed] [Google Scholar]
- 5.Seth AK, Geringer MR, Hong SJ, Leung KP, Galiano RD, Mustoe TA. 2012. Comparative analysis of single-species and polybacterial wound biofilms using a quantitative, in vivo, rabbit ear model. PLoS One 7:e42897. doi: 10.1371/journal.pone.0042897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards R, Harding KG. 2004. Bacteria and wound healing. Curr Opin Infect Dis 17:91–96. doi: 10.1097/00001432-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 9.Petrova OE, Sauer K. 2016. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr Opin Microbiol 30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies DG. 2011. Biofilm dispersion, p 1–28. In Biofilm highlights. Springer, Berlin, Germany. [Google Scholar]
- 11.Davies DG, Marques CNH. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signaling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu Roy A, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc National Acad Sci 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197 1:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques CN, Davies DG, Sauer K. 2015. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals 8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. 2009. Nitric oxide-mediated dispersal in single-and multispecies biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol 2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 22.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 24.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. 2014. BdlA, DipA, and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog 10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen LD, van Gennip M, Rybtke MT, Wu H, Chiang W-C, Alhede M, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infect Immun 81:2705–2713. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua SL, Hultqvist LD, Yuan M, Rybtke M, Nielsen TE, Givskov M, Tolker-Nielsen T, Yang L. 2015. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat Protoc 10:1165–1180. doi: 10.1038/nprot.2015.067. [DOI] [PubMed] [Google Scholar]
- 28.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyle. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 29.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. 2014. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol 92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua SL, Tan SY-Y, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. 2013. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. 2012. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 194:813–826. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPhee JB, Lewenza S, Hancock REW. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 35.Macfarlane ELA, Kwasnicka A, Hancock REW. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- 36.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FSL, Hancock REW. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol 188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers JR, Sauer K. 2013. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol 195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:e00438-13. doi: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer HP. 1991. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol 173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 44.Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol 189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allegrucci M, Sauer K. 2008. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol 190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrova OE, Sauer K. 2011. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol 193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu Roy A, Petrova OE, Sauer K. 2013. Extraction and quantification of cyclic di-GMP from Pseudomonas aeruginosa. Bio Protoc 3:e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao J, Sauer K. 2012. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol 194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 51.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]