ABSTRACT
In microbial biofilms, microorganisms utilize secreted signaling chemical molecules to coordinate their collective behavior. Farnesol is a quorum sensing molecule secreted by the fungal species Candida albicans and shown to play a central physiological role during fungal biofilm growth. Our pervious in vitro and in vivo studies characterized an intricate interaction between C. albicans and the bacterial pathogen Staphylococcus aureus, as these species coexist in biofilm. In this study, we aimed to investigate the impact of farnesol on S. aureus survival, biofilm formation, and response to antimicrobials. The results demonstrated that in the presence of exogenously supplemented farnesol or farnesol secreted by C. albicans in biofilm, S. aureus exhibited significantly enhanced tolerance to antimicrobials. By using gene expression studies, S. aureus mutant strains, and chemical inhibitors, the mechanism for the enhanced tolerance was attributed to upregulation of drug efflux pumps. Importantly, we showed that sequential exposure of S. aureus to farnesol generated a phenotype of high resistance to antimicrobials. Based on the presence of intracellular reactive oxygen species upon farnesol exposure, we hypothesize that antimicrobial tolerance in S. aureus may be mediated by farnesol-induced oxidative stress triggering the upregulation of efflux pumps, as part of a general stress response system. Hence, in mixed biofilms, C. albicans may influence the pathogenicity of S. aureus through acquisition of a drug-tolerant phenotype, with important therapeutic implications. Understanding interspecies signaling in polymicrobial biofilms and the specific drug resistance responses to secreted molecules may lead to the identification of novel targets for drug development.
KEYWORDS: C. albicans, S. aureus, drug resistance, microbial biofilms, quorum sensing, secreted molecules
INTRODUCTION
Polymicrobial infections caused by a combination of microorganisms are responsible for significant mortality and morbidity, particularly those associated with biofilm formation (1–3). Biofilms are structured three-dimensional communities of surface-associated microbial populations encased in a matrix of extracellular polysaccharides where microorganisms are afforded a protected environment (4–6). Biofilms are notoriously difficult to eradicate and are a source of many recalcitrant infections with important clinical repercussions (7). Although mixed fungal-bacterial biofilm-associated infections are challenging to treat, the impact of these interactions on therapy remains largely understudied.
Candida albicans is the most common fungal pathogen, causing diseases ranging from mucosal to life-threatening systemic infections (8–10). This dimorphic species is able to switch morphology between a yeast form and a hyphal form, a property crucial to its pathogenesis and ability to form biofilms (11–13). In fact, the majority of C. albicans infections are associated with biofilm formation (13–15). In various niches in the host, C. albicans coexists with various bacterial species, including Staphylococcus aureus, an important human pathogen and a cause of significant morbidity and mortality (16, 17). With the emergence of methicillin resistance, S. aureus is becoming an even greater therapeutic challenge (18–20). Although S. aureus is a poor former of biofilms, our previous studies have shown that with C. albicans, these species interact synergistically, forming a dense biofilm (21–23). Importantly, using a mouse model of oral coinfection, we identified a phenomenon of augmented pathogenesis with high morbidity and mortality (24).
In microbial biofilms, the extracellular polysaccharide matrix encasing the biofilm cells plays a crucial role in the enhanced resistance of biofilm-associated infections to antimicrobials and host defenses (5, 25, 26). We recently investigated the impact of C. albicans-S. aureus mixed biofilm growth on the response of S. aureus to antibacterial agents. Findings from the study demonstrated that the biofilm matrix, composed of secreted fungal cell wall polysaccharides, conferred on S. aureus enhanced tolerance to antimicrobials (23). Using time-lapse fluorescence confocal microscopy, we visually demonstrated impeded penetration of the drugs through the biofilm, thereby providing the bacteria with protection (23). However, findings from the study also indicated that other effectors secreted by C. albicans during biofilm growth also contribute to the mediated enhanced S. aureus tolerance to antimicrobials (23).
In microbial biofilms, and particularly in mixed-species biofilms, quorum sensing (QS), or cell-cell communication, is a crucial process mediated by small, secreted chemicals known as quorum sensing molecules. These signaling molecules released into the biofilm environment allow one species to detect and respond to the presence of another, allowing for concerted behavior in response to changing conditions. Therefore, these secreted mediators can affect cell physiology and may assume vital importance (27, 28). One of the best characterized of these molecules is farnesol, a key derivative in the sterol biosynthesis pathway in eukaryotic cells. Farnesol is endogenously generated in C. albicans by enzymatic dephosphorylation of farnesyl diphosphate (FPP) and secreted into the environment (29). This fungal QS molecule was shown to play a central role in C. albicans physiology by inhibiting hyphal formation and biofilm formation (29–31).
With C. albicans and S. aureus receiving renewed attention because of the escalating development of antimicrobial resistance and the increasing involvement of biofilms in chronic and systemic infections, coinfection with these species poses a significant therapeutic challenge (20, 32, 33). Therefore, it has become important to understand the mechanisms of their interactions in terms of therapeutic implications within the context of polymicrobial infections. To that end, in this study, we aimed to elucidate the role of the C. albicans secreted QS molecule farnesol in the S. aureus response to antibacterial agents in biofilms.
RESULTS
C. albicans spent biofilm culture medium confers to S. aureus enhanced tolerance to vancomycin.
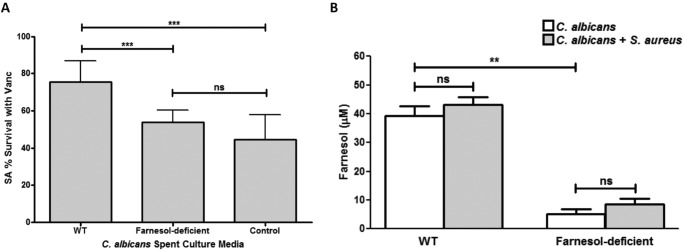
To identify the C. albicans secreted effector modulating the S. aureus response to vancomycin, spent biofilm culture medium from the wild-type (WT) C. albicans strain was used in S. aureus biofilm vancomycin susceptibility assays. Spent medium from a C. albicans strain known to be deficient in farnesol production was similarly used. Based on percent survival with vancomycin as determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium MTS assay (Fig. 1A) and CFU recovery (see Fig. S1 in the supplemental material), growth in spent medium from the farnesol-producing WT strain resulted in significantly higher (∼30%) S. aureus survival with vancomycin. In contrast, no increase in S. aureus tolerance to vancomycin was seen when it was grown in medium from the farnesol-deficient C. albicans strain.
FIG 1.
Farnesol secreted by C. albicans in biofilm confers protection against vancomycin to S. aureus. (A) S. aureus (SA) biofilms were grown in spent culture media from the farnesol-producing (SC5314) and farnesol-deficient (ATCC 10231) C. albicans strains for 24 h prior to treatment with vancomycin (Vanc) for an additional 24 h. Viability assays demonstrated that, compared to growth in control (fresh) medium, S. aureus exhibited a significant increase in tolerance to vancomycin when grown in the spent medium of the farnesol-producing WT strain but not in that of the farnesol-deficient strain. (B) HPLC analysis of spent media from single-species biofilms of both C. albicans strains as well as medium from a dual-species biofilm demonstrated a significant decrease in farnesol levels in the medium from the farnesol-deficient strain. Although some increase in farnesol levels was noted in the medium from the mixed biofilms compared to medium from the C. albicans single-species biofilm, the increase was not statistically significant (**, P < 0.01; ***, P < 0.001; ns, not significant).
Measurement of C. albicans-secreted farnesol in biofilm spent culture medium.
For comparative measurement of secreted farnesol, high-performance liquid chromatography (HPLC) analysis was performed on spent media from biofilms of both C. albicans strains. In medium from the wild-type strain, the farnesol concentration averaged approximately 40 μM, compared to less than 10 μM in medium from the farnesol-deficient strain (Fig. 1B). HPLC analysis was also performed on mixed-species biofilms with S. aureus and the two C. albicans strains. Although farnesol concentrations secreted by C. albicans were consistently higher in mixed biofilms than in C. albicans single biofilms, the difference in values did not reach statistical significance.
Exogenous farnesol modulates the S. aureus response to vancomycin.
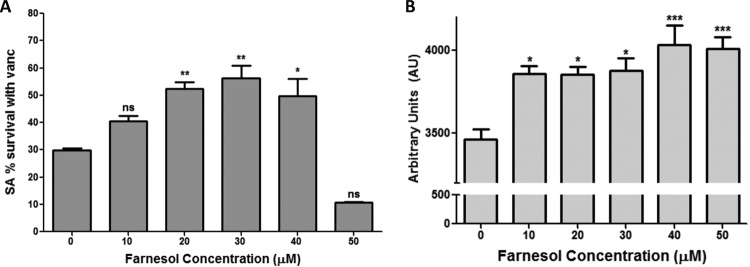
To investigate whether farnesol supplementation impacts the response to vancomycin, susceptibility assays were performed on S. aureus biofilms exogenously supplemented with farnesol at concentrations known to be physiologically secreted by C. albicans. Based on MTS assays (Fig. 2A) and CFU recovery (see Fig. S3 in the supplemental material), a significant (∼25%) farnesol dose-dependent increase in S. aureus survival with vancomycin was seen up to 40 μM farnesol. In contrast, at higher farnesol concentrations, farnesol exerted a synergistic effect with vancomycin. Susceptibility assays using farnesol alone demonstrated that farnesol did not affect S. aureus viability or biofilm formation up to 50 μM (see Fig. S2 in the supplemental material).
FIG 2.
Farnesol confers increased tolerance to vancomycin and induces intracellular ROS accumulation in S. aureus biofilms. Twenty-four-hour S. aureus biofilms grown with farnesol (0 to 50 μM) were treated with vancomycin for 24 h. (A) Based on MTS viability assays, the results demonstrated a significant increase in S. aureus survival that was proportional to the farnesol concentration up to 40 μM, above which farnesol exerted a synergistic effect with vancomycin (*, P < 0.05; **, P < 0.01; ns, not significant). (B) For ROS measurement, S. aureus biofilms treated with increasing farnesol concentrations (0 to 50 μM) were stained with DCF (ROS probe). Based on the level of fluorescence measured, intracellular ROS accumulation increased in a manner proportional to farnesol concentration (*, P < 0.05; ***, P < 0.001; ns, not significant).
Farnesol induces accumulation of intracellular ROS.
As we have previously shown that farnesol induces reactive oxygen species (ROS) production in eukaryotic cells, S. aureus biofilms treated with farnesol were analyzed for intracellular ROS accumulation using fluorescence microscopy and quantitative measurement. Microscopic images revealed an increasing presence of intracellular ROS proportional to farnesol concentrations, with the presence of dead cells at high farnesol concentrations (see Fig. S4 in the supplemental material). ROS presence was also quantified using a fluorescence plate reader. The results demonstrated an increase in ROS accumulation with increasing concentrations of farnesol (Fig. 2B).
Farnesol induces upregulation of efflux pump genes.
Since it is known that oxidative stress can be a potent inducer of efflux pump expression in S. aureus, gene expression studies were performed to examine the role of efflux pumps. RNA extracted from farnesol-treated biofilms was reverse transcribed, and quantitative reverse transcription-PCR (qRT-PCR) was performed on the most notable efflux pump genes in S. aureus, i.e., norA, norB, and norC. Gene expression was displayed as fold change, normalized to 16S rRNA gene expression. The results demonstrated that farnesol-treated biofilm cells displayed a significantly greater level of norB expression than untreated biofilm, averaging a 3-fold increase in expression (Fig. 3). Although the mean expression levels of norA and norC were higher in the presence of farnesol, these levels did not reach statistical significance.
FIG 3.
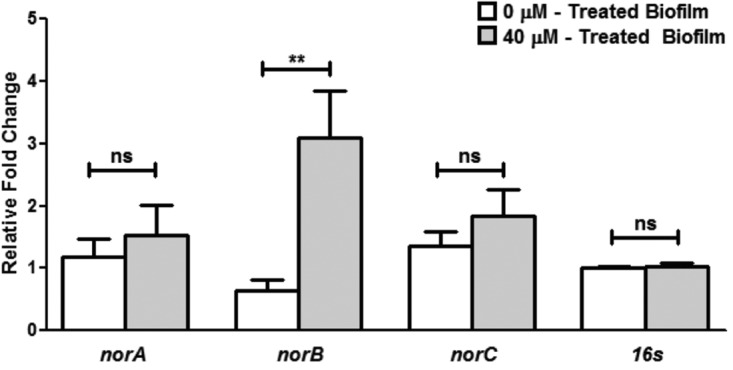
Farnesol-induced upregulation of efflux pump genes in S. aureus biofilms using qRT-PCR analysis. Based on the relative fold change, the results indicated a significant increase (3-fold) in the expression of the norB gene in farnesol-treated S. aureus biofilms. Although a similar increase was consistently seen in the norA and norC genes under these conditions, the increase did not reach statistical significance. The 16S rRNA gene was used as a housekeeping gene (**, P < 0.01; ns, not significant).
Generation of a vancomycin-resistant phenotype upon gradual repeated exposure of S. aureus cells to farnesol (sensitization).
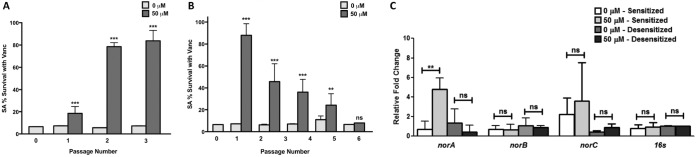
To validate the hypothesis that in mixed biofilms, S. aureus may acclimate to the farnesol increasingly secreted by C. albicans, S. aureus cells in planktonic cultures were serially passaged in medium exogenously supplemented with 50 μM farnesol for a total of 3 passages over 4 days to generate farnesol-“sensitized” cells. Simultaneously, the susceptibility of the sensitized cells to vancomycin was examined at each time point during the passaging process, where cells were recovered and used in vancomycin susceptibility assays (in the absence of farnesol). To monitor development of vancomycin tolerance over time, it was essential to test the susceptibility of these cells to vancomycin immediately upon sampling, and therefore, these assays were done using cells from the planktonic cultures. Sensitized cells did not exhibit any growth retardation (based on optical density measurements), and no defect in biofilm formation compared to that by control cells was seen (data not shown). The results demonstrated that the farnesol-sensitized cells displayed a significant increase (∼70%) in survival following treatment (Fig. 4A) and an increased MIC (10 μg/ml), based on 80% inhibition, compared to that for control cells not previously exposed to farnesol (2.5 to 5 μg/ml) (see Fig. S6 in the supplemental material). To investigate whether this phenomenon is specific to planktonic cells, susceptibility assays were also performed on S. aureus biofilms formed from the farnesol-sensitized cells using vancomycin, oxacillin, and nafcillin. The results demonstrated significant and comparable increases in tolerance to all antibiotics tested (see Fig. S7 in the supplemental material).
FIG 4.
Farnesol-presensitized S. aureus cells exhibit an increase in tolerance to vancomycin concomitant with an increase in expression of efflux pump genes. (A) S. aureus cells were serially passaged in 50 μM farnesol over 4 days for a total of 3 passages to generate cells “sensitized” to farnesol. At each time point, samples were taken and cells were planktonically tested for vancomycin susceptibility (in the absence of farnesol). Based on results from MTS viability assays and compared to control cells not exposed to farnesol, sensitized cells exhibited a gradual and significant increase in tolerance to vancomycin. (B) The farnesol-sensitized cells were serially passaged through farnesol-free medium for up to 5 passages over 6 days to generate “desensitized” cells. Vancomycin susceptibility testing performed at each time point during the passaging process (in the absence of farnesol) demonstrated a gradual loss of the acquired tolerance to vancomycin with cells fully reverting to the original susceptible phenotype after the sixth passage, where cells were considered “desensitized.” (C) Concomitant with susceptibility testing, RNA was extracted from cells after sensitization with farnesol and after desensitization for qRT-PCR analysis of the S. aureus nor efflux pump genes. Based on the fold change, the expression of the norA gene significantly increased (approximately 5-fold) in the sensitized cells but returned to baseline levels upon desensitization, similar to the case for control cells not exposed to farnesol. No significant changes in the expression of the norB and norC genes were seen. The 16S rRNA gene was used as a housekeeping gene. (**, P < 0.01; ***, P < 0.001; ns, not significant).
Examination of membrane integrity using TEM.
To rule out a potential effect for farnesol on cell structure, farnesol-sensitized cells were subjected to transmission electron microscopy (TEM) analysis. The images revealed no observable morphological changes in the cell membranes and cell walls of the sensitized cells compared to those of control cells (passaged with no farnesol) or cells from S. aureus overnight cultures (not exposed to farnesol) (see Fig. S5 in the supplemental material).
Reversion to the vancomycin-susceptible phenotype upon gradual removal of the sensitized cells from farnesol (desensitization).
To determine whether the farnesol-induced vancomycin tolerance in the sensitized cells is transient, the cells were similarly serially passaged in medium with no farnesol (desensitized) and then tested for vancomycin susceptibility at each time point. In contrast to the sensitized cells, desensitized cells exhibited a gradual loss in the acquired tolerance to vancomycin with each passaging. Following the sixth passage, the cells were considered fully desensitized, where the susceptibility to vancomycin reverted to that of control cells not previously exposed to farnesol (Fig. 4B).
Modulated expression of efflux pumps in farnesol-sensitized and desensitized cells.
To determine whether the overexpression of efflux pumps is a transient state, gene expression studies were performed on the previously farnesol-sensitized and desensitized cells (both in the absence of farnesol). The results demonstrated that sensitized cells displayed approximately 5-fold-greater expression of the norA gene, whereas desensitized cells expressed levels comparable to those in control cells not previously exposed to farnesol (Fig. 4C).
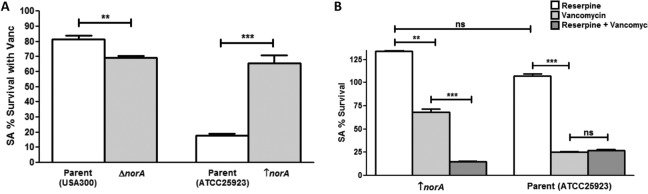
Analysis of S. aureus strains with modulated expression of norA.
To assess the role of farnesol-induced upregulation of efflux pumps in vancomycin tolerance, a norA deletion mutant and a norA-overexpressing strain were tested in vancomycin biofilm susceptibility assays. Experiments performed with the norA strains and their parent strains were performed in the absence of farnesol. The results demonstrated that whereas the mutant lacking norA displayed significantly reduced (∼10%) survival with vancomycin compared to that of its parent strain, the norA-overexpressing strain displayed significantly greater (∼45%) survival than its parent strain (Fig. 5A). Notably, the parent of the norA-overexpressing strain exhibited enhanced susceptibility compared to that of the wild-type strain (Fig. 5; see Fig. S8 in the supplemental material), which is likely due to the fact that unlike the wild-type strain, the norA-overexpressing strain and its parent strain are methicillin-susceptible S. aureus (MSSA) strains. Similar susceptibility profiles were seen when these strains were tested with other antibiotics (oxacillin and nafcillin) (Fig. S8), indicating a nonspecific role in the efflux pump-mediated protection.
FIG 5.
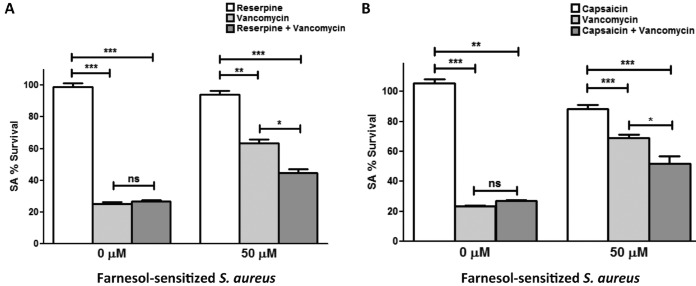
Modulation of efflux pumps impacts the S. aureus response to vancomycin. (A) S. aureus mutant strains with modulated norA expression were tested in biofilm vancomycin susceptibility testing. Based on results from MTS viability assays, the mutant strain lacking the norA gene (ΔnorA; NE1034) displayed a significant decrease in survival with vancomycin compared to that of its parent strain (USA300). In contrast, the norA-overexpressing strain (↑norA; SA1199B) displayed a significant increase in survival compared to that of its parent strain (ATCC 25923). (B) However, when a chemical inhibitor of efflux pumps (reserpine) was included in the vancomycin susceptibility testing, the initially observed increased survival of the norA-overexpressing strain was completely abolished, with no effect of the inhibitor on the susceptibility of the parent strain to vancomycin. Reserpine alone did not affect S. aureus viability (**, P < 0.01; ***, P < 0.001; ns, not significant).
Chemical inhibition of S. aureus efflux pumps using inhibitors.
To further explore the role of efflux pumps, experiments were also performed where known inhibitors of efflux pumps (reserpine and capsaicin) were incorporated in S. aureus vancomycin biofilm assays using the norA-overexpressing mutant strain as well as the farnesol-sensitized cells. The results demonstrated a significant decrease (∼50%) in survival of the norA-overexpressing strain with vancomycin in the presence of the inhibitors compared to that of the parent strain (Fig. 5B). Similarly, in the farnesol-sensitized cells, which typically displayed a significantly greater level of survival with vancomycin, the initially observed tolerance to vancomycin was significantly diminished (∼20%) in the presence of the inhibitor reserpine (Fig. 6A) or capsaicin (∼20%) (Fig. 6B). Similar results were seen when the cells were treated with the antibiotics oxacillin and nafcillin (see Fig. S9 in the supplemental material). Reserpine and capsaicin alone did not affect S. aureus viability or biofilm formation.
FIG 6.
Chemical inhibition of efflux pumps in the farnesol-sensitized S. aureus cells increases susceptibility to vancomycin. (A) Biofilms of farnesol-sensitized cells were treated with vancomycin in the absence and presence of the efflux pump inhibitor reserpine. Based on results from MTS viability assays, the biofilms displayed a significant increase in viability with vancomycin in the absence of the efflux pump inhibitor. The unsensitized S. aureus cells (0 μM) were used as a control. However, in the presence of the inhibitor, survival was significantly diminished. (B) Similar results were seen when experiments were performed using an alternate efflux pump inhibitor (capsaicin). No effect of the inhibitors on control cells not previously exposed to farnesol, in the presence or absence of vancomycin, was seen. The unsensitized S. aureus cells (0 μM) were used as a control. Reserpine and capsaicin alone did not affect S. aureus viability or biofilm formation (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
Farnesol induces efflux of intracellular vancomycin.
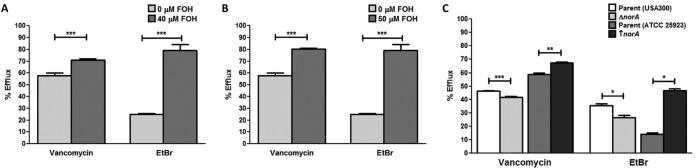
To validate the hypothesis that tolerance to vancomycin is mediated by an increase in vancomycin efflux, efflux assays were designed using a fluorescent vancomycin compound (Vanc-F). In these experiments, following internalization of Vanc-F by S. aureus cells in the biofilm, the level of fluorescent vancomycin effluxed into the supernatant as well as that retained in the biofilm cells was measured using a fluorescence microplate reader. Based on the calculated percent efflux, both farnesol-treated biofilms and biofilms of the previously farnesol-sensitized cells displayed significantly greater levels of vancomycin efflux than their respective controls (∼15% and ∼20%, respectively) (Fig. 7A and B). The role of the NorA efflux pump was further demonstrated using the norA mutant and overexpressing strains, which displayed a significant decrease (∼5%) and a significant increase (∼10%) in vancomycin efflux, respectively (Fig. 7C). Ethidium bromide (EtBr) was used as the standard efflux control, and a similar trend in ethidium bromide efflux was seen for all strains tested.
FIG 7.
Increase in efflux of intracellular vancomycin mediated by farnesol. (A and B) S. aureus biofilms were grown in the presence of farnesol (FOH) (0 or 40 μM) or using cells previously sensitized with farnesol and then treated with fluorescent vancomycin (Vanc-F) for 1 h. Following washing, PBS was added and vancomycin was allowed to efflux for 3 h. Vanc-F was then measured in the supernatant and in the biofilm cell layer. Based on calculated percent efflux, both the farnesol-treated biofilms (A) and the farnesol-sensitized biofilms (B) displayed increased efflux of Vanc-F into the supernatant compared to their respective controls. (C) The vancomycin efflux assay was also performed on the mutant strains with modulated norA expression; the norA deletion mutant strain displayed a significant decrease in efflux compared to that of its parent strain (ATCC 25923), and the norA-overexpressing strain displayed significantly increased efflux compared to that of its parent strain (USA300). The same trend in level of efflux was seen when ethidium bromide (EtBr) was used as the standard control for extracellular efflux (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In a biofilm environment, microbial species are highly interactive and employ a range of mechanisms for cell-to-cell communications (28, 34). This phenomenon for promoting collective behavior within a population is important in ensuring survival and propagation by enhancing access to nutrients and niches, as well as providing protection (3–5). The tetraprenoid farnesol is perhaps the best-characterized secreted QS molecule shown to play a central role in fungal cell physiology. We and others have shown that at high concentrations (>100 μM), farnesol impairs the growth and biofilm formation of C. albicans as well as bacteria, including staphylococci (35–38). We also demonstrated that at high concentrations, farnesol triggers a process of apoptosis in C. albicans and in human oral squamous carcinoma cells, which was preceded by ROS production in both eukaryotic cells (39–41). In S. aureus, several possibilities for farnesol's mechanism of killing have been proposed, including disruption of cell membrane integrity and leakage of K+ ions (38, 42–44). Therefore, given its toxicity, previous studies focused primarily on exploring the potential of exogenous farnesol as an antimicrobial agent. In contrast, our investigations aimed to elucidate a potential role for farnesol in orchestrating the dynamics between C. albicans and S. aureus within a mixed biofilm, focusing on the impact on responses to antimicrobials. In C. albicans biofilms, farnesol was shown to be increasingly produced with the age of the biofilm, at a reported concentration of 40 to 50 μM in 24- to 48-h-old biofilm (45, 46). These findings are important, as in our study exogenous supplementation with 30 to 40 μM farnesol conferred the highest level of tolerance to vancomycin. Most importantly, a physiological relevance was attributed to this phenomenon by the demonstration that spent culture medium recovered from C. albicans biofilm with a measured farnesol concentration of 40 μM similarly conferred on S. aureus a significant increase in survival to antimicrobials.
Based on our previous studies identifying farnesol as a redox cycling agent that induces ROS accumulation in eukaryotic cells, we explored whether a similar process occurs in S. aureus (39, 41, 45, 47). To overcome the deleterious effects of oxidative stress, staphylococci have evolved a multitude of oxidative defense strategies controlled by a complex network of regulators to detect oxidative stress (48). Therefore, it is likely that the oxidative stress exerted by farnesol may activate a protective stress response by the bacterial cell, which indirectly confers tolerance against antibacterials. In fact, an SOS response has been linked to antibiotic resistance via upregulation of drug efflux pumps (49). Efflux pumps are widespread in bacterial species and are constitutively expressed at low levels as they function in the export of bacterial products and removal of environmental substances, including toxins. Although some of these drug pumps have a narrow substrate profile, others are capable of removing many structurally distinct compounds and are therefore referred to as multidrug resistance (MDR) efflux pumps (50). S. aureus has several efflux pumps that contribute to increases in resistance to antibiotics. However, members of the Nor family (belonging to the major facilitator superfamily), particularly NorA, are the most notable (51, 52). The expression of NorA has been associated with increased tolerance to and efflux of a wide variety of structurally unrelated compounds, such as hydrophilic fluoroquinolones, benzalkonium, ethidium bromide, and rhodamine (53). Therefore, overexpression of these pumps may allow higher levels of resistance, which could be transient in the presence of an effector (54). However, the expression of the Nor family efflux pumps is not fully understood and is likely affected by a myriad of host and environmental conditions (53, 55, 56).
The involvement of the Nor efflux pumps and particularly NorA was clearly demonstrated by taking different experimental approaches. The effect of farnesol on modulation of the S. aureus NorA efflux pump gene was expected, as we had previously demonstrated that in addition to ROS accumulation, exposure to farnesol resulted in upregulation of the ABC transporters CDR1 in C. albicans and MDR1 in human oral squamous carcinoma cells (39, 40). However, the gene expression profiles in S. aureus were interesting; whereas norB was found to be significantly upregulated in the presence of farnesol, in the sensitized S. aureus cells, which exhibited high resistance to vancomycin in the absence of farnesol, norA was found to be significantly upregulated. Although NorA and NorB share several substrates, there are differences in substrate specificity, which may be related to substrate binding sites. Therefore, it is feasible to speculate that farnesol may act as a substrate for NorB, whereas the upregulation of NorA is associated with adaptation to a general stress response. In fact, in C. albicans, we showed that as a xenobiotic, farnesol conjugates with intracellular reduced glutathione, forming conjugates that act as a substrate for CDR1 (39). The upregulation of this transporter and increased extrusion of the farnesol-glutathione conjugates result in oxidative stress due to depletion of intracellular glutathione, which is an important thiol that is crucial for protection against oxidative stress (39). Interestingly, in the sensitized S. aureus cells, we found that the upregulation of norA, which was concomitant with vancomycin resistance, returned to constitutive levels, along with loss of vancomycin resistance, when the cells were gradually removed from farnesol (desensitized). These observations indicate that although the acquired resistance to vancomycin mediated by farnesol is maintained over a period of time, it is essentially transient in nature.
Perhaps the most interesting finding from analysis of the profile of susceptibility of the farnesol-sensitized cells to vancomycin was the observation that based on 80% inhibition in growth, the MIC value for the sensitized cells was approximately 10 μg/ml (see Fig. S6 in the supplemental material). These findings are of important clinical relevance, because based on the current Clinical and Laboratory Standards Institute (CLSI) breakpoints, isolates with MICs of 4 to 8 μg/ml are considered to be vancomycin-intermediate S. aureus (VISA), and those with MICs of >16 μg/ml are vancomycin resistant. Therefore, it is tempting to speculate that in terms of susceptibility to vancomycin, farnesol exposure leads to the evolution of a phenotype consistent with that for VISA strains. Although a complete understanding of the mechanisms and factors leading to vancomycin resistance in VISA strains remains elusive, based on TEM cell wall measurements, resistance was shown to go along with thickening of the cell wall due to changes in the composition of the peptidoglycan (57). However, our comparative analysis of TEM images indicated no discernible differences in cell wall thickness in the sensitized cells compared to control cells (see Fig. S5 in the supplemental material). These observations seem to indicate that although the MICs are consistent with those of VISA strains, the mechanism for the enhanced tolerance of sensitized cells to vancomycin is not associated with cell wall changes, supporting a role for efflux pumps. Nevertheless, the demonstration that the farnesol-sensitized cells are comparable to VISA isolates in vancomycin susceptibility is of significance, as VISA strains are associated with treatment failure. Additionally, transcriptomics studies had indicated that the evolution of vancomycin-susceptible S. aureus to VISA is associated with antibiotic resistance and changes likely promoting persistent infection (58). In fact, current dogma suggests that VISA represents a bacterial evolutionary state favoring persistence in the face of antibiotics and the host environment (59, 60).
The global regulator MgrA (multiple gene regulator) has been shown to regulate the transcription of the Nor family multidrug efflux pumps in S. aureus and to affect multiple genes involved in virulence and antibiotic resistance (48, 61, 62). The specific mechanism behind how MgrA regulates Nor pumps is not fully elucidated. However, findings from a study analyzing the crystal structure of the MgrA homodimer indicated the presence of a unique cysteine residue that can be oxidized in response to ROS, thereby modulating the activity of MgrA. Further, MgrA was described to utilize an oxidation-sensing mechanism and function in staphylococcal resistance to different antibiotics, including vancomycin, via regulation of transcription of several multidrug efflux pumps, including the Nor pumps (48, 63). Therefore, since MgrA acts as a regulator for the expression of Nor pumps and is modulated in response to oxidative stress, heat shock, pH level, heme stress, and other environmental conditions, it is highly likely that MgrA may sense the oxidative stress induced by farnesol and activate efflux pump regulation (63). It is interesting to note that MgrA was reported to inversely modulate the expression of norA and that of norC and norB, which may explain the observed differences in our gene expression profiles. However, it is important to note that in S. aureus, at least eight chromosomally encoded and six plasmid-encoded efflux pumps have been reported to contribute to antimicrobial tolerance, many with redundancy in substrates (51, 52). Therefore, it is doubtful that the effect of farnesol on efflux pumps is exclusive to the Nor family. Nevertheless, the goal of the study was to demonstrate that farnesol modulates the regulation of drug efflux pumps, and we focused on the Nor family members as they are the most prominent and best-characterized efflux pumps in S. aureus.
Perhaps the strongest evidence supporting the farnesol-induced vancomycin efflux phenomenon was demonstrated using the efflux assay. The results from these experiments showed that vancomycin is extruded from the cell into the milieu in the presence of farnesol and in sensitized cells (without farnesol). Applying this assay to mixed biofilms would indicate a similar role for the C. albicans secreted farnesol; however, we recently demonstrated that in mixed biofilms, C. albicans secreted polysaccharides can bind and sequester vancomycin, impeding its diffusion (23). Although the findings from our previous study strongly indicated that other C. albicans-associated secreted factors contribute to the process of enhanced antimicrobial tolerance, which is the focus of this study, the confounding effects of the C. albicans secreted polysaccharide matrix make it impossible to specifically isolate the role of vancomycin efflux in a mixed biofilm.
In conclusion, this study provides strong evidence that, in addition to the biofilm matrix, the enhanced tolerance of S. aureus in mixed biofilms involves QS effectors secreted by C. albicans. Specifically, based on the combined findings, we propose that farnesol exerts oxidative stress on the bacterial cell, which induces the expression of efflux pump genes as part of a general stress response system. This increase in drug efflux confers on S. aureus cells nonspecific transient protection against antimicrobials. With vancomycin being one of the few antibiotics that have remained effective against methicillin-resistant S. aureus (MRSA), the demonstration of enhanced S. aureus tolerance to this drug mediated by its interaction with C. albicans carries significant clinical implications, as these species are often coisolated from biofilm-associated infections.
MATERIALS AND METHODS
Reagents.
Farnesol, oxacillin sodium, nafcillin sodium, and reserpine were purchased from Sigma (St. Louis, MO), capsaicin was purchased from Millipore (Billerica, MA), the MTS proliferation assay was purchased from Promega (Madison, WI), vancomycin hydrochloride was purchased from Hospira Inc. (IL), and vancomycin bodipy Fl conjugate was purchased from Invitrogen (Grand Island, NY). Farnesol was obtained as a 3 M stock solution and diluted to a 30 mM solution in 100% methanol. Methanol was included in control experiments.
Strains and growth conditions.
The S. aureus strains used were the standard methicillin-resistant S. aureus (MRSA) strain USA300, the norA deletion strain NE1034 (Nebraska Transposon Mutant Library) (norB, norC, and double deletion strains are not available), and the methicillin-susceptible S. aureus (MSSA) norA-overexpressing strain SA1199B and its parent ATCC 25923 (64) (a MRSA norA-overexpressing strain is not available). The C. albicans strains used were reference strain SC5314 (65) and the farnesol-deficient strain ATCC 10231 (66). C. albicans strains were grown in yeast extract-peptone-dextrose (YPD) broth and S. aureus strains in tryptic soy broth (TSB), and cells were resuspended in RPMI 1640 with l-glutamine and HEPES (Invitrogen, Grand Island, NY) and used at final cell densities of 1 × 106 cells/ml.
Effect of exogenous farnesol on the S. aureus response to vancomycin in biofilm.
S. aureus biofilms were grown in 96-well flat-bottom plates; 100-μl S. aureus cell suspensions were added to the wells, and following a 90-min adhesion period, the wells were washed to remove nonadherent cells. One hundred microliters of RPMI was added to the wells and supplemented with the indicated farnesol concentrations, and biofilms were allowed to grow for 24 h. Following phosphate-buffered saline (PBS) washing, medium supplemented with vancomycin (100 μg/ml for MRSA and 25 μg/ml for MSSA) was added, and the plates were incubated for additional 24 h at 37°C. To assess viability, the MTS tetrazolium salt-based metabolic assay was used; following washing, 100 μl PBS and MTS reagent (20 μl) were added and plates incubated at 37°C. The colorimetric change at 490 nm was measured with a plate reader. Viability was also assessed based on CFU enumeration; biofilm cells recovered by sonication were diluted and plated on S. aureus-specific chromogenic medium (CHROMagar; DRG International) for determination of CFU. Drug-free wells were included as controls, and experiments were also performed using the antibiotics oxacillin (0 to 480 μg/ml) and nafcillin (0 to 160 μg/ml).
Effect of growth in C. albicans spent culture medium on the S. aureus response to vancomycin.
To investigate the role of C. albicans secreted effectors in the S. aureus response to vancomycin, cell-free C. albicans spent culture medium from wild-type (SC5314) and farnesol-deficient (ATCC 10231) strains was recovered from 48-h C. albicans biofilms grown in 10 ml of RPMI in flasks and incubated at 37°C. Spent medium was filter sterilized, supplemented 1:1 with fresh RPMI, and used to grow 24-h S. aureus biofilms. Following PBS washing, medium supplemented with vancomycin (100 μg/ml) was added and plates incubated for an additional 24 h. Viability was assessed using the MTS assay and CFU recovery as described above.
Measurement of C. albicans secreted farnesol in spent culture medium by HPLC analysis.
Recovered cell-free spent medium (from the wild-type and farnesol-deficient strains) was subjected to high-performance liquid chromatography (HPLC) analysis using an ultraperformance liquid chromatography (UPLC) H-class system with fluorescence detection (Acquity, Milford, MA) and an XBridge UPLC C18 column (4.6 by 100 mm; 3.5 μm). UV detection was done at 210 nm, and the total chromatographic run time was 35 min. The injection volume was 20.0 μl, and the farnesol retention time was 12.29 min. In addition, to examine whether the presence of S. aureus in a biofilm impacts farnesol production by C. albicans, dual-species (1 × 106 cells/ml each) biofilms were grown for 48 h in RPMI at 37°C, and spent culture medium was similarly analyzed by HPLC.
Gradual sensitization and desensitization of S. aureus to farnesol.
Farnesol-sensitized S. aureus cells were generated through sequential passages in 10 ml of TSB supplemented with 50 μM farnesol; cultures with no farnesol were simultaneously passaged as controls. This farnesol concentration was chosen because in dual-species biofilm with C. albicans, it did not exert any adverse effect on S. aureus, indicating that S. aureus can become acclimated to high concentrations of farnesol. Following 24 h of incubation at 37°C with shaking, cells were sedimented, washed with PBS, and used to inoculate a new culture supplemented with farnesol. The process was repeated 2 more times for a total of 3 passages over 4 days, generating “farnesol-sensitized” cells. At each time point, cells were recovered and immediately tested for vancomycin susceptibility and MIC determination. For desensitization, sensitized cells were similarly generated through gradual passaging in non-farnesol-containing medium for a total of 5 passages over 6 days (until fully desensitized based on vancomycin susceptibility).
Gene expression studies.
To evaluate the effect of farnesol on expression of the most notable S. aureus efflux pump genes, qRT-PCR was performed. RNA was extracted from farnesol-sensitized S. aureus cells (grown planktonically or in biofilm) using bead beating (Zymo Research) and a MiniPrep kit (Zymo Research). One microgram of DNase-treated RNA was reverse transcribed using the Omniscript RT kit (Qiagen), and 0.5 μl of cDNA was amplified using SYBR green PCR mix (Thermo Fisher) and 10 μM primers (Table 1) (67). No-template and no-RT reactions were used as controls. The relative expression ratios were calculated by using the cycle threshold (CT) of the 16S rRNA gene for each condition as the calibrator. The fold change in gene expression was calculated using the ΔΔCT method, and n-fold expression = 2ΔΔCT, where CT is the cycle number of the detection threshold.
TABLE 1.
Sequences of primers used in this study
| Gene | qRT-PCR primers |
Amplicon size (bp) | |
|---|---|---|---|
| Direction | Sequence | ||
| norA | Forward | ATCGGTTTAGTAATACCAGTCTTGC | ∼120 |
| Reverse | GCGATATAATCATTTGAGATAACGC | ||
| norB | Forward | AACCAATGATTGTGCAAATAGC | ∼110 |
| Reverse | ATGGAAAAGCCGTCAAGAGA | ||
| norC | Forward | ATGAATGAAACGTATCGCGG | ∼120 |
| Reverse | GTCTGCACCAAAACTTTGTTGTAAA | ||
| 16S rRNA gene | Forward | CCAGCAGCCGCGGTAAT | 62 |
| Reverse | CGCGCTTTACGCCCAATA | ||
Farnesol-induced accumulation of intracellular ROS.
S. aureus biofilms (supplemented with 0 to 50 μM farnesol) were stained with the ROS stain H2-DCFDA (DCF) (10 μM; Thermo Fisher) and propidium iodide (PI) (10 μM) for 1 h at 37°C. As DCF is catalyzed from a nonfluorescent form into a fluorescent form by intracellular oxidation, biofilms were not washed prior to detection. Following incubation, ROS accumulation was quantified using a fluorescence plate reader (BioTek) at excitation and emission wavelengths of 485 and 535 nm, respectively, for DCF and of 488/630 nm, respectively, for PI. Intracellular ROS were visualized using a Zeiss 710 confocal microscope. Images were obtained with LSM 5 Image Browser software with an average of 8 images per line. At least three random fields were examined, and representative images are presented.
Inhibition of S. aureus efflux pumps.
To assess the role of drug efflux pumps in response to vancomycin, two known inhibitors of efflux pumps were used. S. aureus biofilms, grown as described above, were supplemented with RPMI containing reserpine (160 to 320 μg/ml) or capsaicin (200 μg/ml). Following 24 h of incubation at 37°C, biofilms were treated with vancomycin, oxacillin, or nafcillin, and viability was assessed using the MTS assay and CFU enumeration.
Vancomycin efflux assay.
Twenty-four-hour preformed S. aureus biofilms were supplemented with 100 μl PBS containing 1 μg/ml fluorescently tagged vancomycin (Vanc-F) and incubated for 1 h at 37°C. Biofilms were washed to remove noninternalized Vanc-F, supplemented with 100 μl PBS, and incubated for 3 h at 37°C to allow for extracellular vancomycin efflux. Following the efflux period, supernatants were collected into separate wells and the biofilm layers retained in the original wells. Effluxed Vanc-F in supernatant and Vanc-F retained in biofilm were measured using a fluorescent plate reader (with excitation and emission wavelengths of 488 and 520 nm, respectively). Results were calculated as percent Vanc-F efflux from biofilms, where percent efflux = (supernatant Vanc-F/total Vanc-F) × 100. Ethidium bromide (EtBr) was used as a standard control of extracellular efflux.
TEM.
To confirm a lack of adverse effects for farnesol on cell structure, sensitized cells were processed for transmission electron microscopy (TEM). Briefly, cells were fixed and embedded in agarose, and blocks were postfixed with 1% osmium tetroxide–1.5% potassium ferrocyanide then stained with uranyl acetate. Specimens were serially dehydrated in ethanol and embedded in Spurr resin (Electron Microscopy Sciences, PA). Ultrathin sections (∼70 nm) mounted on copper grids were examined in a Tecnai T12 TEM (Thermo Fisher Scientific, Hillsboro, OR). Digital images were taken using a charge-coupled device (CCD) camera (Advanced Microscopy Techniques Corp., Woburn, MA) and AMT600 software.
Data analysis.
All experiments were performed on at least 3 separate occasions and in triplicate where applicable, and averages were used to present data. Statistical analysis was performed using GraphPad Prism 5.0 software. The Kruskal-Wallis one-way analysis of variance test was used to compare differences between multiple groups, and Dunn's multiple-comparison test was used to determine whether differences between two samples were statistically significant. Student's unpaired t test was used to compare differences between two samples. P values of <0.05 were considered to be significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Francoise Van Bambeke for providing us with the S. aureus norA-overexpressing and parent strains. We also thank Ahmed Ibrahim for assistance with HPLC analysis.
This work was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award number R01AI130170 to Mary Ann Jabra-Rizk and by the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office to Patrick Van Dijck and Mary Ann Jabra-Rizk. This work utilized an EM sample preparation instrument that was purchased with funding from a National Institutes of Health SIG grant (1S10RR26870-1) awarded to the University of Maryland, Baltimore.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01573-17.
REFERENCES
- 1.Brogden K, Guthmiller J, Taylor C. 2005. Human polymicrobial infections. Lancet 365:253–255. doi: 10.1016/S0140-6736(05)70155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters B, Jabra-Rizk MA, O'May G, Costerton J, Shirtliff M. 2012. Polymicrobial interactions in biofilms: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jabra-Rizk MA. 2011. Pathogenesis of polymicrobial biofilms. Open Mycol J 5:39–43. doi: 10.2174/1874437001105010039. [DOI] [Google Scholar]
- 4.O'Toole G, Kaplan H, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghannoum M, Roilides E, Katragkou A, Petraitis V, Walsh T. 2015. The role of echinocandins in Candida biofilm–related vascular catheter infections: in vitro and in vivo model systems. Clin Infect Dis Suppl 6:S618–S621. doi: 10.1093/cid/civ815. [DOI] [PubMed] [Google Scholar]
- 7.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jabra-Rizk MA, Kong E, Tsui C, Nguyen M, Clancy C, Fidel P, Noverr M. 2016. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun 84:2724–2739. doi: 10.1128/IAI.00469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganguly S, Mitchell A. 2011. Mucosal biofilms of Candida albicans. Curr Opin Microbiol 14:380–385. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaller M, Diekema D. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobsen ID, Wilson D, Wächtler B, Brunke S, Naglik JR, Hube B. 2012. Candida albicans dimorphism as a therapeutic target. Expert Rev Anti Infect Ther 10:85–93. doi: 10.1586/eri.11.152. [DOI] [PubMed] [Google Scholar]
- 12.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenecity mechanisms. Virulence 4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsui C, Kong E, Jabra-Rizk MA. 2016. Pathogenesis of Candida albicans biofilm. Pathog Dis doi: 10.1093/femspd/ftw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel J, Mitchell A. 2011. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tournu H, Van Dijck P. 2012. Candida biofilms and the host: models and new concepts for eradication. Int J Microbiol doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales D, Hogan D. 2010. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGavin MJ, Heinrichs DE. 2012. The staphylococci and staphylococcal pathogenesis. Front Cell Infect Microbiol doi: 10.3389/fcimb.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakoulas G, Moellering RC. 2008. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis 46:S360–S367. doi: 10.1086/533592. [DOI] [PubMed] [Google Scholar]
- 19.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong E, Johnson J, Jabra-Rizk MA. 2016. Community-associated methicillin-resistant Staphylococcus aureus: an enemy amidst us. PLoS Pathog 12:e1005837. doi: 10.1371/journal.ppat.1005837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters B, Ovchinnikova E, Schlecht L, Hoyer L, Busscher H, van der Mei H, Krom B, Jabra-Rizk MA, Shirtliff M. 2012. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiol 158:2975–2986. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters B, Jabra-Rizk MA, Scheper M, Leid J, Costerton J, Shirtliff M. 2010. Microbial interactions and differential protein expression in Staphylococcus aureus and Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol 59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong E, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 7:e01365-16. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong E, Kucharíková S, Van Dijck P, Peters B, Shirtliff M, Jabra-Rizk MA. 2015. Clinical implications of oral candidiasis: host tissue damage and disseminated bacterial disease. Infect Immun 83:604–613. doi: 10.1128/IAI.02843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taff H, Nett J, Zarnowski R, Ross K, Sanchez H, Cain M, Hamaker J, Mitchell A, Andes D. 2012. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog 8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nett JE, Sanchez H, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demuyser L, Jabra-Rizk MA, Van Dijck P. 2014. Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog Dis 70:219–230. doi: 10.1111/2049-632X.12123. [DOI] [PubMed] [Google Scholar]
- 28.Williams P. 2007. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 29.Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67:2982–2992. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Watanabe T, Mikami T, Matsumoto T. 2004. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol Pharm Bull 27:751–752. doi: 10.1248/bpb.27.751. [DOI] [PubMed] [Google Scholar]
- 31.Uppuluri P, Mekala S, Chaffin WL. 2007. Farnesol-mediated inhibition of Candida albicans yeast growth and rescue by adiacylglycerol analogue. Yeast 24:681–693. doi: 10.1002/yea.1501. [DOI] [PubMed] [Google Scholar]
- 32.Adam B, Baillie GS, Douglas LJ. 2002. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol 51:344–349. doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 33.Tawara Y, Honma K, Naito Y. 1996. Methicillin-resistant Staphylococcus aureus and Candida albicans on denture surfaces. Bull Tokyo Dent Coll 37:119–128. [PubMed] [Google Scholar]
- 34.Wargo MJ, Hogan DA. 2006. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr Opin Microbiol 9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Pammi M, Liang R, Hicks J, Barrish J, Versalovic J. 2011. Farnesol decreases biofilms of Staphylococcus epidermidis and exhibits synergy with nafcillin and vancomycin. Pediatr Res 70:578–583. doi: 10.1203/PDR.0b013e318232a984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jabra-Rizk MA, Meiller T, James C, Shirtliff M. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial resistance. Antimicrob Agents Chemother 50:1463–1469. doi: 10.1128/AAC.50.4.1463-1469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cugini C, Calfee MW, Farrow JM III, Morales DK, Pesci EC, Hogan DA. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama H, Oono T, Huh W, Yamasaki O, Ogawa S, Katsuyama M, Ichikawa H, Iwatsuki K. 2002. Actions of farnesol and xylitol against Staphylococcus aureus. Chemotherapy 48:122–128. doi: 10.1159/000064916. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Krom B, Sanglard D, Intapa C, Dawson C, Peters B, Shirtliff M, Jabra-Rizk MA. 2011. Farnesol-induced apoptosis in Candida albicans is mediated by Cdr1-p extrusion and depletion of intracellular glutathione. PLoS One 6:e28830. doi: 10.1371/journal.pone.0028830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheper MA, Shirtliff ME, Meiller TF, Peters B, Jabra-Rizk MA. 2008. Farnesol a fungal quorum sensing molecule triggers apoptosis in human oral squamous carcinoma cells. Neoplasia 10:954–963. doi: 10.1593/neo.08444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirtliff ME, Krom BP, Meijering RM, Peters BM, Zhu J, Scheper M, Jabra-Rizk MA. 2009. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother 53:2392–2401. doi: 10.1128/AAC.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerca N, Gomes F, Pereira S, Teixeira P, Oliveira R. 2012. Confocal laser scanning microscopy analysis of S. epidermidis biofilms exposed to farnesol, vancomycin and rifampicin. BMC Res Notes 16:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue Y, Togashi N, Hamashima H. 2016. Farnesol-induced disruption of the Staphylococcus aureus cytoplasmic membrane. Biol Pharm Bull 39:653–656. doi: 10.1248/bpb.b15-00416. [DOI] [PubMed] [Google Scholar]
- 44.Kaneko M, Togashi N, Hamashima H, Hirohara M, Inoue Y. 2011. Effect of farnesol on mevalonate pathway of Staphylococcus aureus. J Antibiot (Tokyo) 64:547–549. doi: 10.1038/ja.2011.49. [DOI] [PubMed] [Google Scholar]
- 45.Jabra-Rizk MA, Shirtliff M, James C, Meiller TF. 2006. Effect of farnesol on fluconazole resistance in Candida dubliniensis. FEMS Yeast Res 6:1063. doi: 10.1111/j.1567-1364.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 46.Enjalbert B, Whiteway M. 2005. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot Cell 4:1203–1210. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Intapa CBJ, Jabra-Rizk MA. 2014. Farnesol-induced apoptosis in oral squamous carcinoma cells is mediated by MRP1 extrusion and depletion of intracellular glutathione. Trends Cancer Res 10:1–6. [Google Scholar]
- 48.Gaupp R, Ledala N, Somerville G. 2012. Staphylococcal response to oxidative stress. Front Cell Infect Microbiol 2:33. doi: 10.3389/fcimb.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goerke C, Köller J, Wolz C. 2006. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother 50:171–177. doi: 10.1128/AAC.50.1.171-177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaatz G, Thyagarajan R, Seo S. 2005. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob Agents Chemother 49:161–169. doi: 10.1128/AAC.49.1.161-169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jang S. 2016. Multidrug efflux pumps in Staphylococcus aureus and their clinical implications. J Microbiol 54:1–8. doi: 10.1007/s12275-016-5159-z. [DOI] [PubMed] [Google Scholar]
- 52.Costa S, Viveiros M, Amaral L, Couto I. 2013. Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol J 7:59–71. doi: 10.2174/1874285801307010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng X, Sun F, Ji Q. 2012. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J Bacteriol 194:1753–1762. doi: 10.1128/JB.06582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez J. 2016. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 21:1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truong-Bolduc Q, Zhang X, Hooper D. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J Bacteriol 185:3127–3138. doi: 10.1128/JB.185.10.3127-3138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truong-Bolduc Q, Bolduc G, Okumura R. 2011. Implication of the NorB efflux pump in the adaptation of Staphylococcus aureus to growth at acid pH and in resistance to moxifloxacin. Antimicrob Agents Chemother 55:3214–3219. doi: 10.1128/AAC.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reipert A, Ehlert K, Kast T, Bierbaum G. 2003. Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin. Antimicrob Agents Chemother 47:568–576. doi: 10.1128/AAC.47.2.568-576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Sun X, Chang W, Dai Y, Ma X. 2015. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS One 10:e0136082. doi: 10.1371/journal.pone.0136082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howden BP, Peleg AY, Stinear TP. 2014. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol 21:575–582. doi: 10.1016/j.meegid.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 60.Howden BP, Johnson PD, Ward PB, Stinear TP, Davies JK. 2006. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 50:3039–3047. doi: 10.1128/AAC.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luong T, Dunman P, Murphy E, Projan S, Lee C. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol 188:1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Truong-Bolduc Q, Dunman P, Strahilevitz J, Projan S, Hooper D. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen P, Bae T, Williams W, Duguid E, Rice P, Schneewind O, He C. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol 2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 64.Lemaire S, Kosowska-Shick K, Appelbaum P, Glupczynski Y, Van Bambeke F, Tulkens P. 2011. Activity of moxifloxacin against intracellular community-acquired methicillin-resistant Staphylococcus aureus: comparison with clindamycin, linezolid and co-trimoxazole and attempt at defining an intracellular susceptibility breakpoint. J Antimicrob Chemother 66:596–607. doi: 10.1093/jac/dkq478. [DOI] [PubMed] [Google Scholar]
- 65.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine 5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 66.Krause J, Geginat G, Tammer I. 2015. Prostaglandin E2 from Candida albicans stimulates the growth of Staphylococcus aureus in mixed biofilms. PLoS One 10:e0135404. doi: 10.1371/journal.pone.0135404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Y, Onodera Y, Lee J, Hooper D. 2008. NorB, an efflux pump in Staphylococcus aureus strain MW2 contributes to bacterial fitness in abscesses. J Bacteriol 190:7123–7129. doi: 10.1128/JB.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.