ABSTRACT
Dihydroartemisinin-piperaquine (DP) has demonstrated excellent efficacy for the treatment and prevention of malaria in Uganda. However, resistance to both components of this regimen has emerged in Southeast Asia. The efficacy of artemether-lumefantrine, the first-line regimen to treat malaria in Uganda, has also been excellent, but continued pressure may select for parasites with decreased sensitivity to lumefantrine. To gain insight into current drug sensitivity patterns, ex vivo sensitivities were assessed and genotypes previously associated with altered drug sensitivity were characterized for 58 isolates collected in Tororo, Uganda, from subjects presenting in 2016 with malaria from the community or as part of a clinical trial comparing DP chemoprevention regimens. Compared to community isolates, those from trial subjects had lower sensitivities to the aminoquinolines chloroquine, monodesethyl amodiaquine, and piperaquine and greater sensitivities to lumefantrine and mefloquine, an observation consistent with DP selection pressure. Compared to results for isolates from 2010 to 2013, the sensitivities of 2016 community isolates to chloroquine, amodiaquine, and piperaquine improved (geometric mean 50% inhibitory concentrations [IC50] = 248, 76.9, and 19.1 nM in 2010 to 2013 and 33.4, 14.9, and 7.5 nM in 2016, respectively [P < 0.001 for all comparisons]), the sensitivity to lumefantrine decreased (IC50 = 3.0 nM in 2010 to 2013 and 5.4 nM in 2016 [P < 0.001]), and the sensitivity to dihydroartemisinin was unchanged (IC50 = 1.4 nM). These changes were accompanied by decreased prevalence of transporter mutations associated with aminoquinoline resistance and low prevalence of polymorphisms recently associated with resistance to artemisinins or piperaquine. Antimalarial drug sensitivities are changing in Uganda, but novel genotypes associated with DP treatment failure in Asia are not prevalent.
KEYWORDS: Plasmodium falciparum, Uganda, artemether-lumefantrine, dihydroartemisinin-piperaquine, drug resistance, ex vivo, k13, pfcrt, pfmdr1, plasmepsin 2
INTRODUCTION
With widespread resistance to older drugs, the World Health Organization currently recommends artemisinin-based combination therapies (ACTs) as first-line treatments for uncomplicated malaria in Africa, where ACTs remain highly efficacious against Plasmodium falciparum, the most lethal and prevalent of the human malaria species (1). ACTs combine a potent and fast acting artemisinin derivative with a longer-acting partner drug, notably lumefantrine, piperaquine, amodiaquine, mefloquine, or pyronaridine (2). Because the partner drugs circulate long after short-acting artemisinins have been cleared, resistance may be selected if patients are reinfected soon after therapy.
For many years, malaria was treated principally with the aminoquinoline chloroquine. Resistance to chloroquine and its analogue amodiaquine is strongly associated with mutations in two putative drug transporters: PfCRT 76T and PfMDR1 86Y and 1246Y. Treatment with chloroquine and amodiaquine selects for these same mutations (3, 4). Until recently, nearly all P. falciparum strains isolated in Uganda demonstrated chloroquine resistance, as evidenced by poor chloroquine treatment outcomes (5), high ex vivo half-maximal inhibitory (IC50) values (6, 7), and the near universal prevalence of the transporter mutations noted above (8). In 2004, Uganda established the ACT artemether-lumefantrine (AL) as the national therapy for uncomplicated malaria; implementation was slow, but AL use is currently widespread, and it has demonstrated excellent antimalarial efficacy (9, 10). AL selects for wild-type sequences at the same alleles selected for mutations by aminoquinolines, and these wild-type sequences have been associated with moderately decreased lumefantrine sensitivity (7, 11–13). Thus, aminoquinolines and lumefantrine exert opposite selective pressures on P. falciparum drug resistance mediators.
Although it is structurally related to chloroquine and amodiaquine, mediators of resistance and selective effects of the bisquinoline piperaquine, a component of the ACT dihydroartemisinin-piperaquine (DP), are less certain. In Africa, DP has shown excellent efficacy for the treatment of malaria (14, 15) and for malaria chemoprevention in children (16, 17) and pregnant women (18). For chemoprevention, DP benefits from the long half-life of piperaquine, but the impacts of this regimen on the selection of drug resistance are uncertain. Use of DP as therapy (19) or chemoprevention (20) was not associated with selection of the pfcrt and pfmdr1 mutations selected by other aminoquinolines in Burkina Faso, but in Uganda, DP treatment (12) and chemoprevention (7, 17) was followed by selection of parasites with increased prevalence of aminoquinoline resistance-mediating mutations, although selection of different mutations varied among the studies.
Artemisinin resistance emerged in Southeast Asia over the last decade, manifesting as delayed parasite clearance after therapy and causally linked to mutations in the k13 gene (PF3D7_1343700) (21–23). More recently, resistance to piperaquine also emerged in Southeast Asia, linked to an increased plasmepsin 2 (PF3D7_1408000) copy number (24, 25) and, in one study, an E415G mutation in an exonuclease (encoded by PF3D7_1362500) (24). In the same region, amplification of the pfmdr1 gene has been linked to decreased sensitivity to mefloquine and lumefantrine but increased sensitivity to aminoquinolines (26, 27). However, pfmdr1 gene amplification appears to be very uncommon in P. falciparum in Africa.
Considering recent changes in malaria treatment practices in Uganda, we investigated changes in drug sensitivity. We analyzed the drug susceptibility phenotypes and genotypes of clinical P. falciparum isolates collected from two sources in 2016 and compared them to the characteristics of isolates collected in prior years. We found marked changes in drug susceptibilities over time that were consistent with decreased selective pressure from chloroquine and/or increased selective pressure from the national treatment regimen AL.
RESULTS
P. falciparum isolates.
Isolates were collected from May to July 2016 from subjects with uncomplicated malaria from two sources: 29 children and adults from the community (mean age, 4.7 years; range, 1 to 21 years) diagnosed at Tororo District Hospital and 29 children (mean age, 1.4 years; range, 1.2 to 1.5 years) enrolled in a trial comparing two different regimens of DP (monthly or every 3 months) for the prevention of malaria.
Comparative ex vivo drug sensitivities.
We compared ex vivo drug sensitivities between samples from community members and trial subjects. Samples from trial subjects were significantly less sensitive to the aminoquinolines chloroquine, monodesethyl amodiaquine (the active metabolite of amodiaquine), and piperaquine; were significantly more sensitive to lumefantrine and mefloquine; and had no difference in sensitivity to dihydroartemisinin (DHA), atovaquone, or pyronaridine (Table 1). These results were consistent with selection by DP in trial subjects for decreased aminoquinoline sensitivity in parasites that emerged despite chemoprevention. We then compared results for samples collected in 2016 to those for samples collected at the same study site from 2010 to 2013 (7). Parasites collected from the community in 2016 were more sensitive to the tested aminoquinolines but less sensitive to lumefantrine than the samples collected earlier (Table 1).
TABLE 1.
Ex vivo drug sensitivity of P. falciparum isolates collected from 2010 to 2013 and in 2016 from trial and community patientsa
| Drug | Source and/or study period (yr) | No. of samples | Geometric mean IC50 (nM) | 95% CI (nM) | IC50 range (nM) |
P value |
||
|---|---|---|---|---|---|---|---|---|
| 2010 to 2013 vs 2016 trial | 2010 to 2013 vs 2016 community | 2016 trial vs 2016 community | ||||||
| Chloroquine | Dd2 | 7 | 209 | 180–242 | 155–240 | |||
| 3D7 | 7 | 9.2 | 7.9–10.8 | 6.7–11 | ||||
| 2010–2013 | 408 | 248 | 223–275 | 31.0–1,400 | <0.001 | <0.001 | <0.001 | |
| 2016, trial | 25 | 57.1 | 32.7–99.6 | 12.4–727 | ||||
| 2016, community | 24 | 33.4 | 19.8–56.2 | 8.7–318 | ||||
| Monodesethyl amodiaquine | Dd2 | 7 | 35.5 | 25.8–48.8 | 25–61 | |||
| 3D7 | 7 | 5.4 | 4.7–6.3 | 4.1–6.2 | ||||
| 2010–2013 | 421 | 76.9 | 70.2–84.1 | 12.5–565 | <0.001 | <0.001 | <0.001 | |
| 2016, trial | 20 | 20.6 | 15.0–28.4 | 7.5–91.8 | ||||
| 2016, community | 24 | 14.9 | 10.6–21.0 | 4.6–70.4 | ||||
| Piperaquine | Dd2 | 7 | 4.4 | 2.6–7.3 | 1.8–9.6 | |||
| 3D7 | 7 | 3.7 | 2.6–5.2 | 2–6.4 | ||||
| 2010–2013 | 381 | 19.1 | 17.1–21.4 | 3.1–189 | <0.001 | <0.001 | 0.05 | |
| 2016, trial | 25 | 8.6 | 6.5–11.2 | 1.8–26.6 | ||||
| 2016, community | 25 | 7.5 | 6.0–9.3 | 2.7–20.5 | ||||
| Lumefantrine | Dd2 | 7 | 1.8 | 1.4–2.4 | 1.3–2.7 | |||
| 3D7 | 7 | 3.4 | 3.0–3.7 | 2.7–3.7 | ||||
| 2010–2013 | 378 | 3.0 | 2.6–3.3 | 0.4–24.4 | 0.52 | <0.001 | <0.001 | |
| 2016, trial | 24 | 3.4 | 2.6–4.5 | 0.6–11.6 | ||||
| 2016, community | 25 | 5.4 | 4.3–6.9 | 1.8–23.7 | ||||
| Dihydroartemisinin | Dd2 | 7 | 0.8 | 0.5–1.3 | 0.5–2.1 | |||
| 3D7 | 7 | 0.9 | 0.6–1.5 | 0.6–2.2 | ||||
| 2010–2013 | 442 | 1.4 | 1.3–1.5 | 0.3–16.9 | 0.52 | 0.94 | 0.55 | |
| 2016, trial | 25 | 1.7 | 1.4–2.2 | 0.5–4.4 | ||||
| 2016, community | 24 | 1.4 | 1.2–1.7 | 0.4–3.9 | ||||
| Atovaquone | Dd2 | 7 | 0.2 | 0.1–0.3 | 0.1–0.3 | 0.97 | ||
| 3D7 | 7 | 0.1 | 0.1–0.2 | 0.1–0.2 | ||||
| 2016, trial | 18 | 0.5 | 0.4–0.6 | 0.2–1.0 | ||||
| 2016, community | 22 | 0.5 | 0.4–0.6 | 0.1–1.0 | ||||
| Mefloquine | Dd2 | 7 | 10.2 | 7.2–14.4 | 6.7–20 | <0.001 | ||
| 3D7 | 7 | 9.0 | 6.9–11.6 | 6.4–14 | ||||
| 2016, trial | 20 | 18.8 | 15.6–22.8 | 5.9–33.6 | ||||
| 2016, community | 24 | 21.2 | 18.5–24.4 | 8.9–41.7 | ||||
| Pyronaridine | Dd2 | 7 | 1.8 | 0.9–3.8 | 0.7–6.0 | 0.69 | ||
| 3D7 | 7 | 1.0 | 0.8–1.4 | 0.6–1.6 | ||||
| 2016, trial | 24 | 2.5 | 1.8–3.5 | 0.7–29.1 | ||||
| 2016, community | 23 | 2.2 | 1.5–3.3 | 0.2–10.1 | ||||
Geometric mean IC50s for each group were compared using t tests. Dd2 and 3D7 are laboratory control strains. Data from 2010 to 2013 were published previously (7).
Because the standard 72-h IC50 assay does not identify DHA resistance in Southeast Asian parasites (28), the ex vivo ring-stage survival assay, in which parasites are exposed to a 6-h pulse of 700 nM DHA, was performed on 16 samples collected from the community and 16 samples collected from trial subjects in 2016. Seven of the community samples had undetectable parasites at 72 h, and the remaining nine samples had median parasitemia of 0.5% that of controls. For the trial samples, seven had undetectable parasites at 72 h, and the remainder had median parasitemias that were 0.6% those of controls. Our results were similar to those for parasites collected in Kampala, Uganda, in 2014 (29) and contrasted with those for DHA-resistant parasites from Southeast Asia, in which 72-h parasitemias were much higher (median 13.5% that of controls in isolates from a region of Cambodia with frequent artemisinin resistance [30]). Thus, we did not see evidence for ex vivo artemisinin resistance in Ugandan parasites.
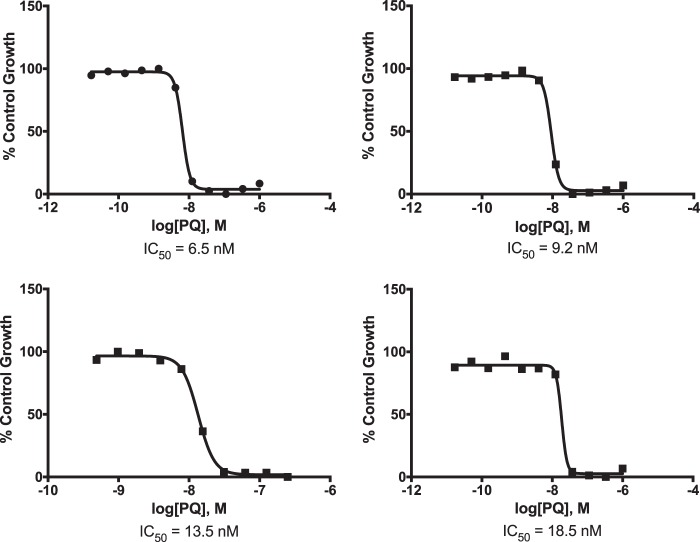
Piperaquine resistance in Southeast Asia has been accompanied by irregular dose-response curves, necessitating establishment of novel methods for determining ex vivo drug sensitivity (31). It was thus important to confirm reliable determination of piperaquine sensitivity by our methods. Standard 72-h growth inhibition assays yielded sigmoidal dose-response curves that were reproducible and readily interpretable (Fig. 1). Thus, in contrast to the case for Southeast Asian isolates resistant to piperaquine, standard IC50 determinations were deemed valid for the Ugandan isolates.
FIG 1.
Representative piperaquine (PQ) growth inhibition curves for four different P. falciparum isolates collected in 2016.
Polymorphisms in parasite drug resistance markers.
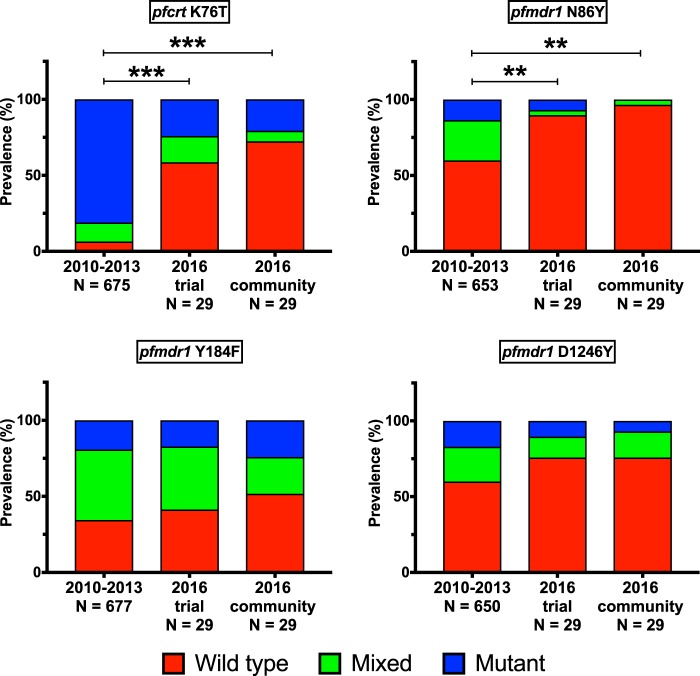
Prevalences of polymorphisms in putative transporters that are associated with sensitivity to a number of antimalarials were compared between P. falciparum isolates from 2010 to 2013 and isolates from 2016. Consistent with previous trends (7, 8, 12, 32), genotypes have changed markedly in recent years, with reversion to wild-type sequences at pfcrt K76T (P < 0.001) and pfmdr1 N86Y (P < 0.01) and D1246Y (P = 0.095) (Fig. 2). Nonsignificant trends toward greater prevalence of wild-type pfcrt K76 and pfmdr1 N86 sequences in community compared to trial samples were also seen, an observation consistent with selective pressure from DP in trial subjects. Sensitivity to some drugs is also altered by amplification of pfmdr1, but this phenomenon has been uncommon in African parasites. All 58 samples collected in 2016 had only one copy of pfmdr1.
FIG 2.
Allele prevalence over time. Wild-type, mixed, and mutant allele prevalences are indicated for the 2010 to 2013 trial and the 2016 trial and community samples. Univariate analysis using generalized estimating equations with exchangeable correlations was used to determine significance of genotype changes over time. Asterisks indicate significance of comparisons between wild-type and mixed/mutant prevalences for the indicated samples (P < 0.01 [**] and P < 0.001 [***]). Allele prevalences from 2010 to 2013 were published previously (7).
Sequencing of the propeller-encoding domain of k13, where certain mutations are strongly associated with artemisinin resistance in Southeast Asia, identified a single-nucleotide polymorphism (SNP) in 1 of 29 community samples that corresponded to an A578S mutation and a mixed wild-type/A578S genotype in 1 of 27 trial samples. The A578S mutation has been described in isolates from Uganda and other African countries, and it has not been associated with artemisinin resistance (29, 32, 33).
Recent reports have identified amplification of a gene encoding plasmepsin 2 and a SNP in an exonuclease gene (PF3D7_1362500) that encodes an E415G mutation as markers of piperaquine resistance in Southeast Asia (24, 25). We assessed the prevalence of these polymorphisms in Ugandan parasites. First, we assessed the plasmepsin 2 (PF3D7_1408000) copy number in samples collected in 2010 to 2013, when piperaquine sensitivity varied more widely in Tororo (Table 1) (7), stratifying parasites with piperaquine IC50s above or below 50 nM. Second, we assessed the plasmepsin 2 copy number in samples collected in 2016. For all three sets of samples, we identified increased copy number in 10 to 14% of samples, but there was no apparent association between increased copy number and piperaquine sensitivity (Table 2). Third, we sequenced the exonuclease gene in 29 community samples and 27 trial samples collected in 2016, as well as the same 58 samples from 2010 to 2013 for which we assessed plasmepsin 2 copy number. The exonuclease E415G mutation was not seen in any of the samples. However, two different SNPs were detected in the 2010–2013 samples, and five different SNPs were detected in the 2016 samples. Five of the seven polymorphisms were nonsynonymous, encoding (i) a D360E mutation (from 2012), (ii) a Y365C mutation (2016 community), (iii) a V352A mutation (2016 trial), (iv) a set of seven nonsynonymous SNPs (N369D, K371N, V372D, N373K, and N374V mutations; 2016 trial), and (v) a 24-base-pair insertion corresponding to 8 amino acids (DNDKVNNN), starting at position 376 (2016 community).
TABLE 2.
plasmepsin 2 copy number among samples collected from 2010 to 2013 and in 2016a
| Sample period (yr) | n | PQ IC50 (nM) | No. of samples (%) with increased plasmepsin 2 copy no. |
|---|---|---|---|
| 2010–2013 | 36 | <50 | 4 (11.1) |
| 22 | >50 | 3 (13.6) | |
| 2016, trial | 29 | <50 | 3 (10.3) |
| 2016, community | 29 | <50 | 4 (13.8) |
The copy number was considered increased when measured at >1.6 copies. n, total number of samples; PQ, piperaquine.
Associations of ex vivo drug sensitivity with transporter polymorphisms.
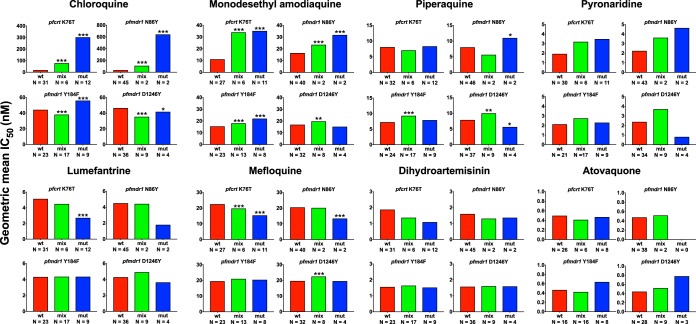
We and others have previously shown that polymorphisms in the putative transporters pfcrt and pfmdr1 are associated with altered ex vivo drug sensitivity (7, 12, 34–38). We tested for associations between pfcrt K76T and pfmdr1 N86Y, Y184F, and D1246Y polymorphisms, and the ex vivo drug sensitivities were determined for samples collected in 2016 (Fig. 3). Consistent with previous reports (7, 34, 35), decreased sensitivity to chloroquine, monodesethyl amodiaquine, and pyronaridine and increased sensitivity to lumefantrine and mefloquine were associated with the mutant pfcrt 76T and pfmdr1 86Y sequences (Fig. 3) (36). Interestingly, consistent with prior reports (7, 37), the associations seen for other aminoquinolines were not seen for piperaquine. Associations with other polymorphisms and for other drugs were mostly not significant.
FIG 3.
Associations of ex vivo geometric mean IC50s with the indicated polymorphisms. “N” represents the number of samples with wild-type (wt), mixed (mix), or mutant (mut) genotypes. The geometric mean IC50s for the wild-type genotype were compared to other genotypes using t tests, with the significance noted (P < 0.05 [*], P < 0.01 [**], and P < 0.001 [***]).
DISCUSSION
We characterized ex vivo drug sensitivities and molecular markers associated with drug sensitivity for P. falciparum isolates collected in Tororo, Uganda, in 2016. Sensitivities to aminoquinolines and other components of standard ACTs differed between isolates from the community and those receiving regular DP in a chemoprevention trial, suggesting selective pressure of piperaquine for aminoquinoline resistance. More notably, sensitivities to these drugs differed markedly compared to results for isolates collected in 2010 to 2013 (7). Specifically, sensitivities to aminoquinolines have increased and sensitivity to lumefantrine has decreased, which is consistent with the selective pressure of AL, the national malaria treatment regimen. These changes were accompanied by decreased prevalence of well-characterized transporter mutations (PfCRT 76T, PfMDR1 86Y, and 1246Y) associated with aminoquinoline resistance but not by an increased prevalence of polymorphisms recently associated with resistance to artemisinins or piperaquine. Thus, antimalarial drug sensitivities are changing in Uganda, most remarkably with reversion to aminoquinoline sensitivity, and genotypes associated with ACT treatment failure in Asia are not prevalent.
Uganda changed from chloroquine-based regimens to AL for the treatment of malaria, with implementation beginning in 2006 (39). Replacement of chloroquine by other regimens has been followed by changes in drug sensitivity in other countries. In Malawi, after discontinuation of chloroquine, the prevalence of the PfCRT 76T mutation, which is the primary mediator of chloroquine resistance (38), decreased markedly (40), followed by excellent treatment efficacy for chloroquine (41). In Uganda, ex vivo evidence of chloroquine resistance (IC50 > 50 to 100 nM) was common in isolates collected from 2006 to 2008 in Kampala (6) and from 2010 to 2013 in Tororo (7), findings consistent with a high prevalence of the PfCRT 76T mutation (3, 42). This situation changed in Tororo most notably beginning in 2012, with increasing prevalence of the pfcrt K76 wild-type genotype and decreasing chloroquine ex vivo IC50s (7, 8). Our data from 2016 show that this trend has continued, with—based on both parasitological and genetic assessments—the majority of parasites being sensitive to chloroquine and amodiaquine. The replacement of aminoquinoline-resistant parasites by sensitive parasites has been accompanied by reemergence of wild-type sequences at pfmdr1 86 and 1246 and by decreases in the ex vivo sensitivity to lumefantrine. However, the majority of malaria infections in Uganda are likely polyclonal. Minority clones may persist, allowing rapid selection of parasites with altered sensitivity when certain drugs are used, as seen in Malawi with the discontinuation of chloroquine (40) and in Uganda where treatment with artesunate-amodiaquine or AL selected for different genotypes in new infections emerging after therapy (43). The changes happening in Uganda have likely been driven by both decreased selective pressure from chloroquine and increased selective pressure from lumefantrine.
For the aminoquinolines chloroquine and amodiaquine, resistance is linked to mutations in pfcrt and, to a lesser extent, pfmdr1, and use of these drugs selects for the same mutations (3, 4, 34, 35, 44). However, associations are less straightforward for the related bisquinoline piperaquine. With the use of DP for the treatment or prevention of malaria in Uganda, selection in parasites that emerged after therapy was consistently seen for PfMDR1 86Y, but selection at other alleles was inconsistent (7, 12, 17). In the present study, samples from the DP chemoprevention trial had decreased sensitivity to chloroquine, monodesethyl amodiaquine, and piperaquine, increased sensitivity to lumefantrine and mefloquine, and an increased prevalence of the PfCRT 76T and PfMDR1 86Y mutations; all of these findings are consistent with a selective pressure of piperaquine, as seen previously for chloroquine (38) and amodiaquine (4, 45). However, in contrast to the results for chloroquine and amodiaquine, the ex vivo drug sensitivities for piperaquine were not clearly linked to the prevalences of pfcrt and pfmdr1 polymorphisms (7; the present study). The reasons for the differences in results for different aminoquinolines are uncertain; the larger size of piperaquine may lead to decreased impact of transporter mutations on sensitivity to this drug. In any event, it appears that the widespread use of DP for malaria chemoprevention will select for parasites with decreased sensitivity to aminoquinolines but increased sensitivity to lumefantrine.
Of great concern is the identification of artemisinin resistance within the last decade and of piperaquine resistance very recently in Southeast Asia (22, 24, 25, 46). Important advances from groups in Asia have identified parasitological (30, 31) and molecular (21, 23–25) markers of resistance to these drugs which, when seen together, have been accompanied by high rates of treatment failure with DP (31, 47). We utilized these new tools to characterize parasites circulating in Tororo in 2016. As seen in prior studies from Uganda, occasional k13 mutations were seen (12, 29, 33), but these were not the mutations associated with artemisinin resistance in Southeast Asia, and prevalence was not associated with drug sensitivity or recent drug pressure. In addition, as seen recently in parasites from Kampala (29), the ex vivo DHA ring survival assay did not suggest artemisinin resistance in Ugandan parasites. Similarly, an increase in plasmepsin 2 copy number, a newly identified marker associated with piperaquine resistance, was also seen occasionally in Ugandan isolates, but it was not associated with piperaquine sensitivity in recent isolates or in a comparison of older isolates with relatively high or low piperaquine sensitivity. Overall, we did not see evidence suggesting that resistance to artemisinins or piperaquine has emerged in Uganda.
Changes in antimalarial drug use have been accompanied by marked changes in the drug sensitivity of malaria parasites around the world. Our new results demonstrate marked recent changes in Ugandan parasites with, for the most part, a return to sensitivity to chloroquine and amodiaquine. Parasites have also shown some loss of sensitivity to lumefantrine, a component of the national treatment regimen, but it is unlikely that this slight change, although statistically significant, will impact AL treatment efficacy. Considering markers for resistance to artemisinins and piperaquine, our results are reassuring, without evidence for emergence of the worrisome ACT resistance seen in southeast Asia. Resistance to different antimalarial regimens has developed more slowly in Africa than in other regions, likely due to strong antimalarial immunity in Africans living in high-transmission areas, and yet it is extremely important to limit resistance spread. Our results are consistent with other recent studies in suggesting that the continued use of AL to treat malaria and the institution of DP for chemoprevention will exert opposite selective resistance pressures and thus may offer an optimal means for maintaining antimalarial treatment and chemopreventive efficacy while limiting the spread of drug resistance.
MATERIALS AND METHODS
Sample collection.
P. falciparum isolates were obtained from May to July 2016 from two sources in Tororo, Uganda. First, children and adults presenting at the Tororo District Hospital outpatient clinic with malaria (temperature ≥ 37.5°C axillary or history of fever in the previous 24 h and a positive Giemsa-stained blood smear for P. falciparum) and without signs of severe disease were enrolled after informed consent. Second, children enrolled in a clinical trial (registered at ClinicalTrials.gov [NCT02163447]) comparing monthly versus every 3-month intermittent therapy with DP to prevent malaria provided samples if they presented to the study clinic with uncomplicated malaria (defined as above). The studies were approved by the Makerere University Research and Ethics Committee, the Uganda National Council for Science and Technology, and the University of California, San Francisco Committee on Human Research. All subjects were treated with AL after sample collection.
Sample collection and parasite culture.
Blood was collected before therapy in a heparinized tube. Parasitemia was determined with Giemsa-stained thin smears. Samples containing only P. falciparum and with a parasitemia of ≥1% were selected for culture. Initiation of culture was performed as previously described (6), with slight modifications. Briefly, blood was centrifuged, plasma and buffy coat were removed, and the erythrocyte pellet was washed three times with RPMI 1640 medium prewarmed to 37°C. The pellet was resuspended in complete media (RPMI 1640 medium supplemented with 25 mM HEPES, 0.2% NaHCO3, 0.1 mM hypoxanthine, 10 μg/ml gentamicin, and 0.5% AlbuMAX II serum substitute) to produce a 50% hematocrit. Culture aliquots were spotted onto Whatman 3MM filter paper for molecular studies.
Ex vivo determination of drug susceptibilities.
Drug susceptibilities were assessed immediately or from samples stored at 4°C for a maximum of 18 h. Drug sensitivities were determined for chloroquine (Sigma-Aldrich), monodesethyl amodiaquine (BD Gentest), piperaquine (Jinan Jiaquan International Trade Co., Ltd.), lumefantrine (Sigma-Aldrich), dihydroartemisinin (DHA; TCI Tokyo Chemical Industry), mefloquine (Sigma-Aldrich), atovaquone (Sigma-Aldrich), and pyronaridine (Santa Cruz Biotechnology) using a 72-h, 96-well microplate fluorescence assay with SYBR green I detection as described previously (48). Drugs were validated by regular IC50 assessment with laboratory strains Dd2 and 3D7, which yielded results similar to those seen previously (7). Drugs were prepared as 10 mM stocks in dimethyl sulfoxide or water and stored at −20°C. For assays, fresh drug stocks were prepared in complete media by diluting chloroquine to 20 μM; piperaquine, monodesethyl amodiaquine, and mefloquine to 4 μM; lumefantrine to 0.8 μM; and dihydroartemisinin, atovaquone, and pyronaridine to 0.4 μM. Drugs were serially diluted 3-fold in complete medium in 96-well microplates, with inclusion of drug-free control wells, to final volumes of 50 μl/well. Parasite culture was added for a total volume of 200 μl/well with a 0.2% parasitemia at 2% hematocrit. Plates were maintained under a gas mixture of 5% CO2, 5% O2, and 90% N2 for 72 h at 37°C in a modular incubator (Billups Rothenberg, Del Mar, CA). Plates were frozen at −80°C and thawed, wells were mixed, and 100 μl from each well was transferred to a black 96-well plate containing 100 μl/well SYBR green lysis buffer (20 mM Tris buffer, 5 mM EDTA, 0.008% saponin, 0.08% Triton X-100, and 0.2 μl/ml SYBR green I). Plates were incubated for 1 h in the dark at room temperature, and the fluorescence was measured with a FLUOstar Omega plate reader (BMG LabTech, Inc., Cary, NC; 485-nm excitation/530-nm emission). IC50s were derived by plotting percent growth against log drug concentration and fitting the data by variable slope, sigmoidal curve fit in Prism 6.0 (GraphPad Software, San Diego, CA). DHA drug susceptibility was measured with an ex vivo ring stage survival assay as previously described (29).
Characterization of parasite polymorphisms.
Parasite DNA was extracted from filter paper blood spots using Chelex-100, genes of interest were amplified with nested PCR, and polymorphisms in pfcrt and pfmdr1 were evaluated using a ligase detection reaction-fluorescent microsphere assay, all as previously described (12, 49). pfmdr1 copy number was determined in quadruplicate using a TaqMan real-time PCR assay with 3D7 and Dd2 strain standards, as previously described (12, 50). The k13-propeller-encoding domains (codons 440 to 726; PF3D7_1343700) (21) and an exonuclease gene (PF3D7_1362500) (24) were amplified and sequenced using previously described methods and primers. For the exonuclease gene, samples that failed the initial round of PCR were amplified using nested PCR with sequencing primers as previously described (24). Sequences were aligned with the 3D7 sequence (PlasmoDB.org) using MacVector v.15 (MacVector, Inc., Apex, NC). SNPs were confirmed by inspection of individual chromatograms. The plasmepsin 2 copy number was quantified using a previously published quantitative PCR method (25). Amplification of plasmepsin 2 was defined as >1.6 copies.
Statistical methods.
Data analysis was performed using Stata v.14 (StataCorp LLC, College Station, TX). Significant differences between ex vivo IC50s were characterized with t tests. Univariate analysis using generalized estimating equations with exchangeable correlations was used to determine significance of genotype changes over time. Associations between genotype and drug sensitivity were determined by comparing wild-type and mixed/mutant genotypes using t tests. Differences were considered significant at P values of ≤0.05.
Accession number(s).
Nucleotide sequence data are available in the GenBank database under accession numbers MF477020 to MF477187.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (AI075045, AI117001, AI093784, and T37MD003407). S.A.R. and R.A.C. acknowledge travel grants from Dominican University of California.
We thank the participants in the Tororo studies, the participants' parents and guardians, the clinical study team, and our administrative staff. We are grateful for Didier Menard and Benoit Witkowski for kindly sharing the plasmepsin 2 copy number assay and to Jenny Legac for technical support.
REFERENCES
- 1.World Health Organization. 2015. Guidelines for the treatment of malaria, 3rd ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Nosten F, White NJ. 2007. Artemisinin-based combination treatment of falciparum malaria. Am J Trop Med Hyg 77:181–192. [PubMed] [Google Scholar]
- 3.Dorsey G, Kamya MR, Singh A, Rosenthal PJ. 2001. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis 183:1417–1420. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- 4.Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob Agents Chemother 51:3023–3025. doi: 10.1128/AAC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorsey G, Kamya MR, Ndeezi G, Babirye JN, Phares CR, Olson JE, Katabira ET, Rosenthal PJ. 2000. Predictors of chloroquine treatment failure in children and adults with falciparum malaria in Kampala, Uganda. Am J Trop Med Hyg 62:686–692. doi: 10.4269/ajtmh.2000.62.686. [DOI] [PubMed] [Google Scholar]
- 6.Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob Agents Chemother 54:1200–1206. doi: 10.1128/AAC.01412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumwebaze P, Conrad MD, Walakira A, LeClair N, Byaruhanga O, Nakazibwe C, Kozak B, Bloome J, Okiring J, Kakuru A, Bigira V, Kapisi J, Legac J, Gut J, Cooper RA, Kamya MR, Havlir DV, Dorsey G, Greenhouse B, Nsobya SL, Rosenthal PJ. 2015. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 59:3018–3030. doi: 10.1128/AAC.05141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, Arinaitwe E, Kamya M, Tappero J, Staedke SG, Dorsey G, Greenhouse B, Rosenthal PJ. 2014. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg 91:54–61. doi: 10.4269/ajtmh.13-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Greenhouse B, Staedke SG, Kamya MR, Dorsey G, Rosenthal PJ. 2010. Incidence of malaria and efficacy of combination antimalarial therapies over 4 years in an urban cohort of Ugandan children. PLoS One 5:e11759. doi: 10.1371/journal.pone.0011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsey G, Staedke S, Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Dokomajilar C, Kamya MR, Rosenthal PJ. 2007. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. JAMA 297:2210–2219. doi: 10.1001/jama.297.20.2210. [DOI] [PubMed] [Google Scholar]
- 11.Baliraine FN, Rosenthal PJ. 2011. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis 204:1120–1124. doi: 10.1093/infdis/jir486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, Muhindo M, Kamya MR, Tappero JW, Greenhouse B, Dorsey G, Rosenthal PJ. 2014. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis 210:344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 14.Kamya MR, Yeka A, Bukirwa H, Lugemwa M, Rwakimari JB, Staedke SG, Talisuna AO, Greenhouse B, Nosten F, Rosenthal PJ, Wabwire-Mangen F, Dorsey G. 2007. Artemether-lumefantrine versus dihydroartemisinin-piperaquine for treatment of malaria: a randomized trial. PLoS Clin Trials 2:e20. doi: 10.1371/journal.pctr.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeka A, Dorsey G, Kamya MR, Talisuna A, Lugemwa M, Rwakimari JB, Staedke SG, Rosenthal PJ, Wabwire-Mangen F, Bukirwa H. 2008. Artemether-lumefantrine versus dihydroartemisinin-piperaquine for treating uncomplicated malaria: a randomized trial to guide policy in Uganda. PLoS One 3:e2390. doi: 10.1371/journal.pone.0002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigira V, Kapisi J, Clark TD, Kinara S, Mwangwa F, Muhindo MK, Osterbauer B, Aweeka FT, Huang L, Achan J, Havlir DV, Rosenthal PJ, Kamya MR, Dorsey G. 2014. Protective efficacy and safety of three antimalarial regimens for the prevention of malaria in young Ugandan children: a randomized controlled trial. PLoS Med 11:e1001689. doi: 10.1371/journal.pmed.1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nankabirwa JI, Conrad MD, Legac J, Tukwasibwe S, Tumwebaze P, Wandera B, Brooker SJ, Staedke SG, Kamya MR, Nsobya SL, Dorsey G, Rosenthal PJ. 2016. Intermittent preventive treatment with dihydroartemisinin-piperaquine in Ugandan schoolchildren selects for Plasmodium falciparum transporter polymorphisms that modify drug sensitivity. Antimicrob Agents Chemother 60:5649–5654. doi: 10.1128/AAC.00920-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakuru A, Jagannathan P, Muhindo MK, Natureeba P, Awori P, Nakalembe M, Opira B, Olwoch P, Ategeka J, Nayebare P, Clark TD, Feeney ME, Charlebois ED, Rizzuto G, Muehlenbachs A, Havlir DV, Kamya MR, Dorsey G. 2016. Dihydroartemisinin-piperaquine for the prevention of malaria in pregnancy. N Engl J Med 374:928–939. doi: 10.1056/NEJMoa1509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Some AF, Sere YY, Dokomajilar C, Zongo I, Rouamba N, Greenhouse B, Ouedraogo JB, Rosenthal PJ. 2010. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob Agents Chemother 54:1949–1954. doi: 10.1128/AAC.01413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Some AF, Zongo I, Compaore YD, Sakande S, Nosten F, Ouedraogo JB, Rosenthal PJ. 2014. Selection of drug resistance-mediating Plasmodium falciparum genetic polymorphisms by seasonal malaria chemoprevention in Burkina Faso. Antimicrob Agents Chemother 58:3660–3665. doi: 10.1128/AAC.02406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, et al. . 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. 2015. Drug resistance: K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, Eam R, Khean C, Ken M, Loch K, Bouillon A, Domergue A, Ma L, Bouchier C, Leang R, Huy R, Nuel G, Barale JC, Legrand E, Ringwald P, Fidock DA, Mercereau-Puijalon O, Ariey F, Menard D. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, Nsobya SL, Rosenthal PJ. 2015. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother 59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witkowski B, Khim N, Chim P, Kim S, Ke S, Kloeung N, Chy S, Duong S, Leang R, Ringwald P, Dondorp AM, Tripura R, Benoit-Vical F, Berry A, Gorgette O, Ariey F, Barale JC, Mercereau-Puijalon O, Menard D. 2013. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother 57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, Ke S, Chy S, Eam R, Khean C, Loch K, Ken M, Lek D, Beghain J, Ariey F, Guerin PJ, Huy R, Mercereau-Puijalon O, Witkowski B, Menard D. 2015. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 13:305. doi: 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, Walakira A, Nankabirwa J, Yeka A, Staedke SG, Greenhouse B, Nsobya SL, Kamya MR, Dorsey G, Rosenthal PJ. 2017. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 215:631–635. doi: 10.1093/infdis/jiw614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahlstrom S, Aubouy A, Maiga-Ascofare O, Faucher JF, Wakpo A, Ezinmegnon S, Massougbodji A, Houze P, Kendjo E, Deloron P, Le Bras J, Houze S. 2014. Plasmodium falciparum polymorphisms associated with ex vivo drug susceptibility and clinical effectiveness of artemisinin-based combination therapies in Benin. Antimicrob Agents Chemother 58:1–10. doi: 10.1128/AAC.01790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyase FL, Akala HM, Ingasia L, Cheruiyot A, Omondi A, Okudo C, Juma D, Yeda R, Andagalu B, Wanja E, Kamau E, Schnabel D, Bulimo W, Waters NC, Walsh DS, Johnson JD. 2013. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008-2011. PLoS One 8:e64299. doi: 10.1371/journal.pone.0064299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madamet M, Briolant S, Amalvict R, Benoit N, Bouchiba H, Cren J, Pradines B, French National Centre for Imported Malaria Study Group. 2016. The Plasmodium falciparum chloroquine resistance transporter is associated with the ex vivo P. falciparum African parasite response to pyronaridine. Parasit Vectors 9:77. doi: 10.1186/s13071-016-1358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascual A, Madamet M, Bertaux L, Amalvict R, Benoit N, Travers D, Cren J, Taudon N, Rogier C, Parzy D, Pradines B, French National Reference Centre for Imported Malaria Study Group. 2013. In vitro piperaquine susceptibility is not associated with the Plasmodium falciparum chloroquine resistance transporter gene. Malar J 12:431. doi: 10.1186/1475-2875-12-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 39.Sears D, Kigozi R, Mpimbaza A, Kakeeto S, Sserwanga A, Staedke SG, Chang M, Kapella BK, Rubahika D, Kamya MR, Dorsey G. 2013. Anti-malarial prescription practices among outpatients with laboratory-confirmed malaria in the setting of a health facility-based sentinel site surveillance system in Uganda. Malar J 12:252. doi: 10.1186/1475-2875-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frosch AE, Laufer MK, Mathanga DP, Takala-Harrison S, Skarbinski J, Claassen CW, Dzinjalamala FK, Plowe CV. 2014. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J Infect Dis 210:1110–1114. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. 2006. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med 355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 42.Francis D, Nsobya SL, Talisuna A, Yeka A, Kamya MR, Machekano R, Dokomajilar C, Rosenthal PJ, Dorsey G. 2006. Geographic differences in antimalarial drug efficacy in Uganda are explained by differences in endemicity and not by known molecular markers of drug resistance. J Infect Dis 193:978–986. doi: 10.1086/500951. [DOI] [PubMed] [Google Scholar]
- 43.Yeka A, Kigozi R, Conrad MD, Lugemwa M, Okui P, Katureebe C, Belay K, Kapella BK, Chang MA, Kamya MR, Staedke SG, Dorsey G, Rosenthal PJ. 2016. Artesunate/amodiaquine versus artemether/lumefantrine for the treatment of uncomplicated malaria in Uganda: a randomized trial. J Infect Dis 213:1134–1142. doi: 10.1093/infdis/jiv551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmgren G, Hamrin J, Svard J, Martensson A, Gil JP, Bjorkman A. 2007. Selection of pfmdr1 mutations after amodiaquine monotherapy and amodiaquine plus artemisinin combination therapy in East Africa. Infect Genet Evol 7:562–569. doi: 10.1016/j.meegid.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Nawaz F, Nsobya SL, Kiggundu M, Joloba M, Rosenthal PJ. 2009. Selection of parasites with diminished drug susceptibility by amodiaquine-containing antimalarial regimens in Uganda. J Infect Dis 200:1650–1657. doi: 10.1086/647988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, Kim S, Witkowski B, Duru V, Domergue A, Khim N, Ringwald P, Menard D. 2015. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother 59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LeClair NP, Conrad MD, Baliraine FN, Nsanzabana C, Nsobya SL, Rosenthal PJ. 2013. Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of Plasmodium falciparum. J Clin Microbiol 51:2564–2570. doi: 10.1128/JCM.00904-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O'Neil M, Milhous W, Wirth DF, Oduola AM. 2009. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother 53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]