ABSTRACT
The increasing prevalence of nosocomial infections produced by multidrug-resistant (MDR) or extensively drug-resistant (XDR) Pseudomonas aeruginosa is frequently linked to widespread international strains designated high-risk clones. In this work, we attempted to decipher the interplay between resistance profiles, high-risk clones, and virulence, testing a large (n = 140) collection of well-characterized P. aeruginosa isolates from different sources (bloodstream infections, nosocomial outbreaks, cystic fibrosis, and the environment) in a Caenorhabditis elegans infection model. Consistent with previous data, we documented a clear inverse correlation between antimicrobial resistance and virulence in the C. elegans model. Indeed, the lowest virulence was linked to XDR profiles, which were typically linked to defined high-risk clones. However, virulence varied broadly depending on the involved high-risk clone; it was high for sequence type 111 (ST111) and ST235 but very low for ST175. The highest virulence of ST235 could be attributed to its exoU+ type III secretion system (TTSS) genotype, which was found to be linked with higher virulence in our C. elegans model. Other markers, such as motility or pigment production, were not essential for virulence in the C. elegans model but seemed to be related with the higher values of the statistical normalized data. In contrast to ST235, the ST175 high-risk clone, which is widespread in Spain and France, seems to be associated with a particularly low virulence in the C. elegans model. Moreover, the previously described G154R AmpR mutation, prevalent in ST175, was found to contribute to the reduced virulence, although it was not the only factor involved. Altogether, our results provide a major step forward for understanding the interplay between P. aeruginosa resistance profiles, high-risk clones, and virulence.
KEYWORDS: Pseudomonas aeruginosa, Caenorhabditis elegans, virulence, multidrug resistant, extensively drug resistant, high-risk clones
INTRODUCTION
Pseudomonas aeruginosa, a ubiquitous microorganism, is one of the most relevant pathogens causing human opportunistic infections (1). Due to its impressive metabolic plasticity and versatility, P. aeruginosa is capable of infecting/colonizing a wide range of ecological niches, including aquatic and soil habitats, animals, and plants (2). Moreover, P. aeruginosa is a leading cause of severe nosocomial infections, particularly in critically ill and immunocompromised patients, and is the most frequent driver of chronic respiratory infections in patients suffering from cystic fibrosis (CF) or other chronic underlying diseases (3, 4).
Multiple virulence factors contribute to the pathogenesis of acute and chronic P. aeruginosa infections (1), and many of them are found to be located in the accessory genome as part of pathogenicity (PAPI) or genomic (PAGI) islands (5). One of the most relevant P. aeruginosa virulence factors is the type III secretion system (TTSS), of which ExoU determines a greater impact in bacterial virulence. In a recent multicenter study of P. aeruginosa, we showed that the TTSS genotype is a major differential factor that needs to be considered when analyzing the clinical outcome of P. aeruginosa bacteremia (6). Motility systems and pigment production are also well established as virulence determinants of P. aeruginosa (7–9).
The increasing prevalence of nosocomial infections produced by multidrug-resistant (MDR) or extensively drug-resistant (XDR) P. aeruginosa strains severely compromises the selection of appropriate treatments and is therefore associated with significant morbidity and mortality (10–12). This growing threat results from the extraordinary capacity of this pathogen for developing resistance to nearly all available antibiotics by the interplay of the selection of mutations in chromosomal genes and the increasing prevalence of transferable resistance determinants, particularly those encoding class B carbapenemases (metallo-β-lactamases [MBLs]) or extended-spectrum β-lactamases (ESBLs), which are frequently cotransferred with genes encoding aminoglycoside-modifying enzymes (13). Even more concerning are recent reports which have provided evidence of the existence of MDR/XDR clones of P. aeruginosa disseminated in multiple institutions worldwide, denominated epidemic high-risk clones (14). Among them, sequenced type 1111 (ST111), ST175, and ST235 are those likely to be most widespread (15–18).
Understanding the interplay between high-risk clones, antimicrobial resistance, and virulence is of paramount relevance for the analysis of the outcomes of P. aeruginosa infections (6, 18, 19). Indeed, resistance profiles, high-risk clones, and TTSS genotype were significantly interconnected in a previous study, having a major impact in the mortality of P. aeruginosa bloodstream infections (6). Moreover, in a recent study, we showed that the 3 likely more worldwide-relevant P. aeruginosa high-risk clones (ST111, ST175, and ST235) were associated with a defined set of biological markers that included decreased motility, pigment production, and in vitro fitness but increased biofilm formation and spontaneous mutant frequencies (20). The interplay between resistance and virulence has also been recently evaluated for a few isolates in a murine model of P. aeruginosa bacteremia (21). However, the usefulness of murine models for the analysis of large collections of isolates with different traits is limited by cost, workload, and ethical constraints. In this sense, P. aeruginosa causes lethal infections not only to mammals, but also to invertebrates, such as the nematode Caenorhabditis elegans, a powerful model organism for studying developmental biology and host-pathogen interactions (22–24). C. elegans is often used as a host due to its deep characterization and experimental simplicity (25–27). Moreover, this infection model has many practical advantages, such as being low cost, being amenable to large-scale in vivo screening, and not raising any of the ethical concerns for drug testing at the early stages of development (22, 28–31). Thus, the objective of this work was to decipher the interplay among resistance profiles, high-risk clones, and virulence, testing a large collection of well-characterized P. aeruginosa isolates from different sources (bloodstream infections, nosocomial outbreaks, CF, and the environment) in a C. elegans infection model.
RESULTS AND DISCUSSION
Inverse correlation between resistance and virulence in the C. elegans model.
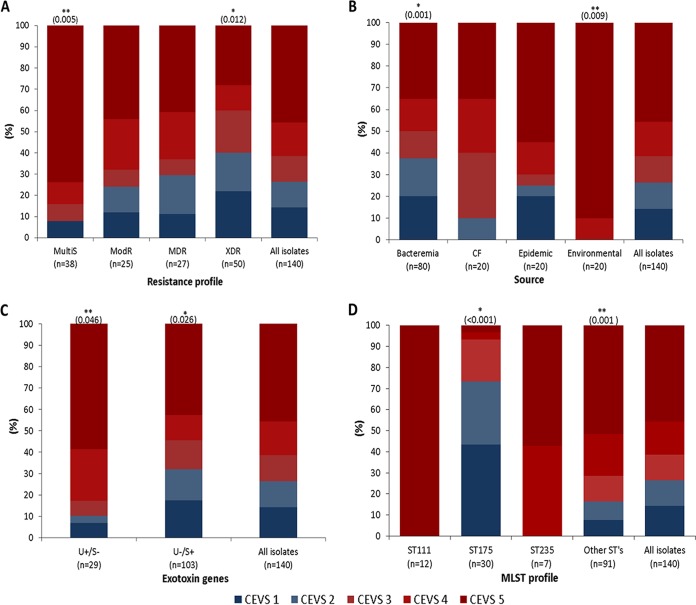
The characteristics of the 140 P. aeruginosa isolates studied are listed in Data Set S1 in the supplemental material. To establish a comparative analysis of lethality of the clinical isolates studied, we established a C. elegans virulence score (CEVS), as described in Table 1 and Materials and Methods. As shown in Fig. 1, 103 (73.6%) of the 140 studied strains were virulent (C. elegans virulence score [CEVS], 3 to 5) in the C. elegans model, whereas 37 (26.4%) were not (CEVS, 1 to 2). However, as documented in Fig. 1A, the proportion of nonvirulent strains dramatically increased according to the resistance profile, from 7.9% for isolates that were susceptible to all tested antipseudomonals (MultiS), 24% for isolates resistant to 1 or 2 antipseudomonal families (ModR), 29.6% for isolates resistant to 3 or more families (MDR), to 40% for XDR isolates (resistant to all but 1 or 2 families). Therefore, these results support previous studies suggesting that the acquisition of resistance is associated with a fitness cost and reduction in virulence (32, 33).
TABLE 1.
Caenorhabditis elegans virulence scoring
CEVS, Caenorhabditis elegans virulence score. Nonvirulent score indicates the growth of nematodes is allowed. Virulent indicates nematodes are killed.
Recorded at 72 h.
FIG 1.
Impact on C. elegans virulence score (CEVS) of resistance profiles (A), source of isolation (B), TTSS genotype (C), and MLST genotype (D). Nonvirulent strains, with a CEVS of 1 to 2, are shown in Blue tones, and virulent strains, belonging to CEVS 3 to 5, are shown in red tones. *, significantly lower virulence (P < 0.05); **, significantly higher virulence (P < 0.05).
With regard to the source (Fig. 1B), the highest virulence was documented among the environmental isolates, most of which (90%) were MultiS and none were MDR/XDR. Intriguingly, virulence in the C. elegans model was lower in blood isolates than in CF isolates. The high proportion of MDR/XDR profiles (50%) among the studied blood isolates could have partially explained these findings, but the proportion was not lower for CF isolates (85% were MDR/XDR).
exoU+ TTSS genotype is associated with enhanced virulence in the C. elegans model.
One of the most relevant virulence determinants of P. aeruginosa is the TTSS (34). This secretion system injects potent cytotoxins, including ExoS, ExoT, ExoU, or ExoY, into eukaryotic cells (35). Previous studies have indicated that exoS is present in 58 to 72% of the isolates and is typically associated with an invasive phenotype, while exoU is less frequent (28% to 42% of isolates) but associated with a highly cytotoxic phenotype (35).
The TTSS genotype was characterized for all studied isolates. The presence of exoT and exoY genes was documented in the vast majority of the isolates, in 139 (99%) and 130 (93%) isolates, respectively. Additionally, all of them were positive for either exoU (29 isolates [20.7%]) or exoS (103 isolates [73.6%]), except for 1 (0.7%) and 7 (5%) isolates that were positive or negative for both genes, respectively. As shown in Fig. 1C, exoU+ exoS-negative isolates were significantly more virulent than exoU-negative exoS+ isolates in the C. elegans model. Therefore, these results were in agreement with our previous data indicating that the exoU+ exoS-negative genotype was associated with increased early mortality in P. aeruginosa bloodstream infections (6).
ST175 high-risk clone is associated with reduced virulence in the C. elegans model.
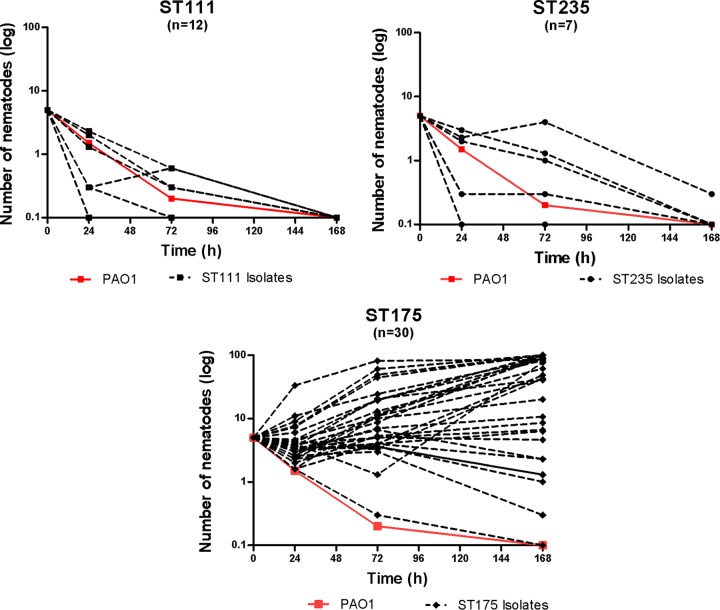
Forty-nine of the 140 isolates studied belonged to classical high-risk clones (18), including 30 ST175, 12 ST111, and 7 ST235 isolates. Figure 1D shows the association between each of the high-risk clones and CEVS. As shown, all isolates belonging to ST111 and ST235 high-risk clones were highly virulent (CEVS, 4 to 5). In contrast, 73% of ST175 isolates were nonvirulent (CEVS, 1 to 2). Moreover, most of the remaining ST175 isolates showed only moderate virulence (CEVS, 3). Figure 2 shows the dynamics of C. elegans over time in the presence of the isolates belonging to the 3 major high-risk clones, clearly illustrating the reduced virulence of ST175 isolates compared to that of ST235 or ST111.
FIG 2.
Lethality of P. aeruginosa ST111 (n = 12), ST175 (n = 30), and ST235 (n = 7) high-risk clone isolates in the C. elegans model. The mean numbers of surviving nematodes at 0, 24, 72, and 168 h are shown. PAO1 strain values (in red) are included for comparative purposes.
Motility and pigment production does not significantly impact virulence in the C. elegans model.
The motility systems and pigment production are also well established as virulence determinants of P. aeruginosa. It is widely accepted that the diverse repertoire of bacterial motility systems (swimming, twitching, and swarming) plays a pivotal role in the invasion of fluids and surfaces, including those found in the nosocomial environment or the patient's epithelial tissues (7, 36). Likewise, pyocyanin (a redox-active phenazine toxin) and pyoverdine (a siderophore) are well known to play a major role in bacterial physiology and pathogenesis (8, 9). We have previously described that P. aeruginosa high-risk clones (ST175, ST111, and ST235) were associated with a defined set of biological parameters, which included reduced motility (twitching, swimming, and swarming) and pigment production (pyoverdine and pyocyanin) (20). Therefore, we analyzed the correlations between motility and pigment production and virulence in the C. elegans model. However, statistically significant differences between virulent (CEVS, 3 to 5) and nonvirulent (CEVS, 1 to 2) isolates were not observed for any of the motility types or pigments analyzed (see Fig. S1). Thus, our results indicate that none of these factors are individually essential for virulence in the C. elegans model.
Further univariate logistic regression analysis showed that there was a real and significant association of virulence or nonvirulence (CEVS, 3 to 5 and 1 to 2, respectively) with TTSS genotype, XDR and MultiS profiles, and the ST175 high-risk clone (Table 2). So, the exoU gene (odds ratio [OR] = 4.03) and MultiS resistance profile (OR = 5.83) were positively associated with virulence, whereas the exoS gene (OR = 0.27), XDR profile (OR = 0.35), and the ST175 high-risk clone (OR = 0.06) were negatively associated with virulence. We also performed a multiple logistic regression analysis to identify factors that were independently associated with virulence. All previously identified factors were initially included in the model, and after adjustment for significant variables, the ST175 high-risk clone was the only factor that demonstrated an independent association (OR = 0.68, P ≤ 0.001) with a lack of virulence in the C. elegans model.
TABLE 2.
Univariate logistic regression analysis of predictive factors for P. aeruginosa virulence in C. elegans infection modela
| Variable | ORb | 95% CIc | P value |
|---|---|---|---|
| ST175 | 0.06 | 0.02–0.15 | ≤0.001 |
| exoS | 0.27 | 0.09–0.82 | 0.021 |
| exoU | 4.03 | 1.14–14.19 | 0.030 |
| XDR | 0.35 | 0.16–0.76 | 0.008 |
| MultiS | 5.83 | 1.67–20.34 | 0.006 |
Virulence determined as a CEVS of 3 to 5.
OR, odds ratio.
CI, confidence interval.
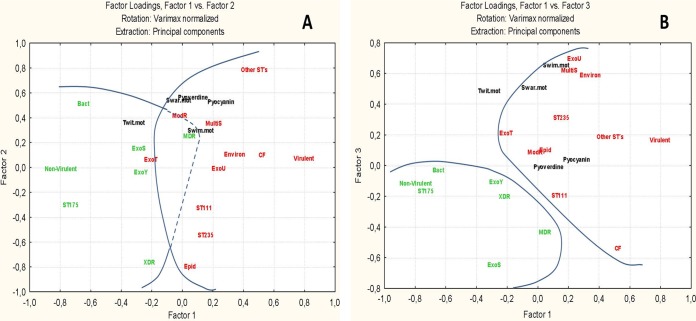
A combined analysis showed the general structure of the interrelationships among quantitative and qualitative variables in association with the virulence and nonvirulence of clones against C. elegans. Variables are well represented in the factorial space (see Table S1) despite the differences in representation in the general matrix (Data Set S1).
Figure 3 depicts the general geometric relationships established between the different studied variables, represented in a virulence gradient. Factor 1 would order the samples in a gradient according to virulence against C. elegans defined by high-risk clone ST175 and the origin (bacteremia or CF) as the main markers. Factor 2 is mainly explained by an important variation on other STs, ST235, and extensively drug-resistant (XDR) isolates; there is also an important contribution to this factor of the swarming motility and pyoverdine production. Finally, factor 3 represents mainly the variation of origin (CF or environmental), resistance profile (MultiS), secretion system (ExoU and ExoS), and the swimming motility. In this factorial space, the analysis confirms that nonvirulent strains are statistically associated with ST175, bacteremia, XDR, MDR, ExoS and ExoY secretion, and lower values of swimming and swarming motility, pyoverdine, and pyocyanin. Virulent strains are associated mainly with “other STs,” ST111, ST235, epidemic, CF, and environmental origins, ModR and MultiS profiles, ExoU and ExoT secretion (as a spurious variable), and higher values for swimming and swarming motility, pyoverdine, and pyocyanin.
FIG 3.
Relationships among variables explaining the overall differences between virulent and nonvirulent strains. (A) Defined structure according to factors 1 to 2; differences in quantitative variables are also expressed by means and standard deviations. (B) The same structure as defined by factors 1 to 3.
Characteristic G154R AmpR mutation decreases the killing of C. elegans but does not fully explain the reduced virulence of ST175.
Previous works by Balasubramanian et al. (37) and Kong et al. (38) showed that AmpR is a global transcriptional regulator connected not only to the expression of the β-lactamase AmpC but also to quorum sensing, alginate production, biofilm formation, and the expression of several other virulence factors. In this work, we analyzed whether AmpR affects bacterial virulence in the C. elegans model. Moreover, we have previously described a mutation in AmpR (G154R), detected in widespread ST175 isolates, which causes its activation as an ampC positive regulator and thus drives the hyperproduction of AmpC and β-lactam resistance (39, 40). Thus, in this work, we explored whether this specific mutation may have an impact on virulence in the C. elegans model. As shown in Fig. 4, the inactivation of AmpR significantly reduced the ability of P. aeruginosa strain PAO1 to kill C. elegans. Moreover, whereas complementation assays with wild-type ampR fully restored wild-type PAO1 virulence, complementation with the G154R ampR mutant did not. Therefore, these results indicate that AmpR is relevant for P. aeruginosa virulence in the C. elegans model and that the G154R mutation impairs this effect. We then examined the distribution of this specific mutation among the studied ST175 isolates. The G154R mutation was detected in 22 of 30 ST175 isolates studied (Data Set S1). However, the presence or absence of this mutation did not explain the within clone differences in CEVS, since the few ST175 isolates showing a higher virulence (CEVS, 4 or 5) also presented the mutation.
FIG 4.
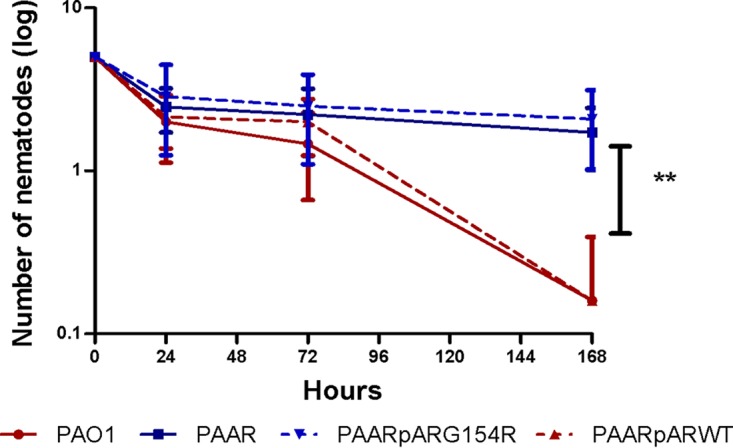
Virulence on C. elegans of strain PAO1 and ampR mutants. The mean numbers of surviving nematodes at 0, 24, 72, and 168 h are shown. Values are means ± SDs (error bars) from at least three independent experiments. A Mann-Whitney U test was performed to analyze the differences between the different mutants and PAO1. **, P < 0.001; PAAR, ampR knockout mutant of PAO1.
To search for potential mutations involved in the reduced virulence documented for most ST175 isolates, we compared the genomes of 2 nonvirulent ST175 isolates (CEVS, 1) with those of the only 2 highly virulent ST175 isolates (CEVS, 4 or 5). Up to 70 mutations present only in isolates (at least one) from one of the groups (virulent/nonvirulent) were detected (see Data Set S2). None of the mutations detected were exclusive of both nonvirulent isolates. However, 11 of them were present in both virulent isolates and neither of the nonvirulent isolates. Future studies are therefore needed to determine the potential implication of each of these 11 mutations, which include those for the pyocin S2 and the extracellular protease LepA, on the gain of virulence, if any. Likewise, further studies are needed for a full understanding of the frequent lack of virulence of ST175 isolates.
Concluding remarks.
Consistent with previous data, we documented a clear inverse correlation between antimicrobial resistance and virulence for P. aeruginosa in the C. elegans model. Indeed, the lowest virulence was linked to XDR profiles, which are typically linked to defined high-risk clones. However, virulence varied broadly depending on the involved high-risk clone; it was high for ST111 and ST235 but very low for ST175. The highest virulence of ST235 could well be attributed to its exoU+ TTSS genotype, found to be linked with higher virulence in our C. elegans model as well as in previous clinical studies (6). Moreover, ST235 appears to be associated with a particularly poor clinical outcome compared to that from other MDR/XDR strains (15, 41). In contrast to ST235, ST175 seems to be associated with a particularly low virulence in the C. elegans model. Moreover, the obtained results suggest that a specific mutation in the transcriptional regulator AmpR contributes to the reduced virulence (39). Therefore, our results are in agreement with existing data suggesting that AmpR is a global transcriptional regulator involved not only in the regulation of antibiotic resistance but also in modulating bacterial pathogenicity (37, 38). However, the presence or absence of this mutation did not explain the within clone differences in virulence. In any case, a functional analysis of the multiple ST175 genomes available will provide further insights into the drivers of the reduced virulence of the ST175 clone, despite its wide dissemination in countries such as Spain or France (40). Altogether, our results provide a major step forward for understanding the interplay between P. aeruginosa resistance profiles, high-risk clones, and virulence.
MATERIALS AND METHODS
Bacterial strains, susceptibility testing, and molecular typing.
A total of 140 P. aeruginosa isolates were evaluated. The collection comprised 80 bloodstream isolates from a Spanish multicenter study, including 20 isolates per resistance profile (XDR, MDR, ModR, and MultiS), as well as 20 epidemic XDR isolates recovered from several different outbreaks and producing diverse chromosomal and/or horizontally acquired resistance mechanisms (20). Susceptibility profiles, multilocus sequence typing (MLST) genotypes, motility, and pigment production had been already assessed in a previous study for those isolates (20). The studied collection additionally included 20 isolates from CF patients recovered during a recent multicenter study from Spain (42) and 20 environmental isolates provided by Saniconsult (Palma, Spain) recovered in 2010 from diverse sources. MICs of ticarcillin (TIC), piperacillin plus tazobactam(PIP-Tz), aztreonam (AZT), ceftazidime (CAZ), cefepime (FEP), imipenem (IMP), meropenem (MER), ciprofloxacin (CIP), tobramycin (TOB), ceftolozane plus tazobactam (TOL-TAZ), amikacin (AMI), and colistin (COL) were determined by broth microdilution following CLSI guidelines and breakpoints (43). The genotypes were documented through MLST using previously described schemes, protocols, available databases, and tools (http://pubmlst.org/paeruginosa) (44). Whole-genome sequences from selected ST175 isolates were obtained from previous studies (40). Likewise, ampR sequences for all ST175 isolates were obtained either from previous works (39, 40) or, if not available, by PCR amplification and Sanger sequencing in this study.
TTSS PCR genotyping.
The detection of exoS, exoT, exoY, and exoU genes was performed with primers and the protocol described by Feltman et al. (45). PCR assays to detect the presence of exo genes were performed on whole-DNA extracts (DNeasy tissue kit; Qiagen, Hilden, Germany) under the following conditions: denaturation for 12 min at 94°C, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, and a final extension step of 10 min at 72°C.
Motility assays. (i) Swimming motility.
Swimming medium plates (10 g/liter tryptone, 5 g/liter NaCl, and 0.3% [wt/vol] mid-resolution agarose) were inoculated with isolated colonies from an overnight culture in LB agar (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, and 1% agar) at 37°C by use of a sharp sterile toothpick, introducing it to up to half the thickness of the medium (46).
(ii) Swarming motility.
Strains were spot inoculated on swarm agar plates (M8 minimal medium supplemented with 1 mM MgSO4, 0.2% glucose, 0.5% Bacto Casamino Acids, and 0.5% agar), using 2.5-μl aliquots taken from overnight LB broth cultures (47).
(iii) Twitching motility.
Isolated colonies were inoculated with a sharp sterile toothpick inserted to the bottom of twitching medium (LB agar) plates (46). In all cases, 90-mm plates were filled with 30 ml of medium, briefly dried for 2 h, and inoculated in triplicate experiments. The plates were then wrapped with Parafilm M to prevent dehydration and incubated at 37°C for 16 h. After incubating, the zone of motility was measured. In the case of twitching medium, the agar-petri dish interface was measured. If the area to be measured was irregular, two perpendicular diameters were measured and the result was expressed as the mean of the two values.
Pigment production.
Production of pyocyanin and pyoverdine was quantified as described previously (48). Briefly, bacterial strains were grown at 37°C in Pseudomonas ACC broth for 40 h. At this time, bacteria were pelleted by centrifugation, and the amount of the blue pigment pyocyanin was evaluated by measuring the absorbance of the supernatants at 690 nm. The amount of pyoverdine was measured by fluorescence emission, by exciting the supernatants at 400 nm and measuring the emission at 460 nm. Each experiment was performed in triplicate.
C. elegans killing assay.
The assay for studying bacterial killing of C. elegans was performed as described previously (49). Briefly, a fresh culture of each bacterial strain to be tested was layered on a 55-mm-diameter plate containing 5 ml of potato dextrose agar. After spreading the bacterial culture, the plates were incubated at 37°C for 24 h to form bacterial lawns. The bacterial plates were kept overnight, and 5 worms per plate were poured on top of these bacterial lawns. The plates were incubated at 24°C and scored to detect the presence of living worms at 0 h, 24 h, 72 h, and 168 h. The nematodes were examined at ×20 and ×40 magnifications, and a worm was considered dead if it did not move spontaneously. At least three independent replicate experiments per bacterial strain were performed and the means and standard deviations (SDs) were recorded. Additionally, to determine the effect of the G154R AmpR mutation on virulence, we performed C. elegans killing assays in the wild-type PAO1 strain, the ampR knockout mutant of PAO1 (PAΔR), and the PAΔR strain complemented with either the cloned wild-type ampR (pARWT) or its G154R mutant (pARG154R). These strains were constructed in a previous study (39). To establish a comprehensive comparative analysis of the lethality of the large collection of clinical isolates tested, a C. elegans virulence score (CEVS) was developed. As described in Table 1, the isolates were classified into 5 virulence levels depending on the effect on the growth of the nematodes, including two (CEVS, 1 to 2) in which the strains were considered nonvirulent (do not kill the nematode) and three (CEVS, 3 to 5) in which the strains were considered virulent (kill the nematode).
Statistical analysis.
Quantitative variables were compared using the Mann-Whitney U test or the Student t test, as appropriate. Chi-square (χ2) and Fisher's exact tests were used to determine the association between factors and virulence. In all cases, a P value of ≤0.05 was considered statistically significant. Multivariate analyses were performed by logistic regression; variables were introduced in the models and selected using a stepwise backward process, where 0.15 was set as the limit for removal of terms. All statistical analyses were performed using GraphPad Prism 5 or IBM SPSS Statistics v22 software. Factorial analysis was performed using principal components as the method of extraction after the data were varimax normalized. The general structure of clones and variables was tested by correspondence analysis. For both factorial and correspondence analyses, the Statistica software package was used (Statistica data analysis software system, version 6; Tibco Software, Inc.).
Supplementary Material
ACKNOWLEDGMENTS
We thank all the clinical microbiologists and clinicians participating in the REIPI multicenter study of P. aeruginosa bloodstream infections (PI08/0276) and the multicenter study of cystic fibrosis respiratory infections (PI12/00734). We also thank Pilar Sanchis and Guillem Frontera from IdisBa for statistical support.
This work was supported by the Planes Nacionales de I+D+i 2008-2011/2013-2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015/0006, RD12/0015/004, RD16/0016/0004, RD16/0016/008, and RD16/0016/0011) and grants PI15/00088 and PI12/00734. This work was cofinanced by grants from the European Development Regional Fund “A way to achieve Europe” and operative program Intelligent Growth (2014-2020). I.S.-D. is the recipient of a P-FIS fellowship from Instituto de Salud Carlos III, Ministerio de Economía y Competitividad.
We declare no conflict of interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01586-17.
REFERENCES
- 1.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 2.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 3.López-Causapé C, Rojo-Molinero E, Macià MD, Oliver A. 2015. The problems of antibiotic resistance in cystic fibrosis and solutions. Expert Rev Respir Med 9:73–88. doi: 10.1586/17476348.2015.995640. [DOI] [PubMed] [Google Scholar]
- 4.Vincent JL. 2003. Nosocomial infections in adult intensive-care units. Lancet 361:2068–2077. doi: 10.1016/S0140-6736(03)13644-6. [DOI] [PubMed] [Google Scholar]
- 5.Battle SE, Meyer F, Rello J, Kung VL, Hauser AR. 2008. Hybrid pathogenicity island PAGI-5 contributes to the highly virulent phenotype of a Pseudomonas aeruginosa isolate in mammals. J Bacteriol 190:7130–7140. doi: 10.1128/JB.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peña C, Cabot G, Gómez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez-López F, Tubau F, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2015. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60:539–548. doi: 10.1093/cid/ciu866. [DOI] [PubMed] [Google Scholar]
- 7.Feinbaum RL, Urbach JM, Liberati NT, Djonovic S, Adonizio A, Carvunis AR, Ausubel FM. 2012. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog 8:e1002813. doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau GW, Hassett DJ, Ran H, Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Visca P, Imperi F, Lamont IL. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol 15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitliks SD. 1998. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med 244:379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 11.Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. 2007. Pseudomonas aeruginosa: resistance and therapeutics options in the turn of the new millennium. Clin Microbiol Infect 13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 12.Peña C, Suarez C, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Oliver A, Martínez-Martínez L, Spanish Network for Research in Infectious Diseases (REIPI). 2013. Effect of adequate single-drug vs combination antimicrobial therapy on mortality in Pseudomonas aeruginosa bloodstream infections: a post hoc analysis of a prospective cohort. Clin Infect Dis 57:208–216. doi: 10.1093/cid/cit223. [DOI] [PubMed] [Google Scholar]
- 13.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 15.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarez C, Peña C, Arch O, Dominguez MA, Tubau F, Juan C, Gavaldá L, Sora M, Oliver A, Pujol M, Ariza J. 2011. A large sustained endemic outbreak of multiresistant Pseudomonas aeruginosa: a new epidemiological scenario for nosocomial acquisition. BMC Infect Dis 11:272. doi: 10.1186/1471-2334-11-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viedma E, Juan C, Villa J, Barrado L, Orellana MA, Sanz F, Otero JR, Oliver A, Chaves F. 2012. VIM-2-producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg Infect Dis 18:1235–1241. doi: 10.3201/eid1808.111234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Juan C, Peña C, Oliver A. 2017. Host and pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Dis 215:S44–S51. doi: 10.1093/infdis/jiw299. [DOI] [PubMed] [Google Scholar]
- 20.Mulet X, Cabot G, Ocampo-Sosa AA, Domínguez MA, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C. 2013. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57:5527–5535. doi: 10.1128/AAC.01481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gómez-Zorrilla S, Juan C, Cabot G, Camoez M, Tubau F, Oliver A, Dominguez MA, Ariza J, Peña C. 2016. Impact of multidrug resistance on the pathogenicity of Pseudomonas aeruginosa: in vitro and in vivo studies. Int J Antimicrob Agents 47:368–374. doi: 10.1016/j.ijantimicag.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silverman GA, Luke CJ, Bhatia SR, Long OS, Vetica AC, Perlmutter DH, Pak SC. 2009. Modeling molecular and cellular aspects of human disease using the nematode Caenorhabditis elegans. Pediatr Res 65:10–18. doi: 10.1203/PDR.0b013e31819009b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan MW, Shapira M. 2011. Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol 13:497–507. doi: 10.1111/j.1462-5822.2011.01570.x. [DOI] [PubMed] [Google Scholar]
- 25.Lakshmanan U, Yap A, Fulwood J, Yichun L, Hoon SS, Lim J, Ting A, Sem XH, Kreisberg JF, Tan P, Tan G, Flotow H. 2014. Establishment of a novel whole-animal HTS technology platform for melioidosis drug discovery. Comb Chem High Throughput Screen 17:790–803. doi: 10.2174/1386207317666141019195031. [DOI] [PubMed] [Google Scholar]
- 26.Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. 2009. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol 4:527–533. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Reilly LP, Benson JA, Cummings EE, Perlmutter DH, Silverman GA, Pak SC. 2014. Worming our way to novel drug discovery with the Caenorhabditis elegans proteostasis network, stress response and insulin-signaling pathways. Expert Opin Drug Discov 9:1021–1032. doi: 10.1517/17460441.2014.930125. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa Caenorhabditis elegans pathogenesis model. Cell 96:47–56. doi: 10.1016/S0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 29.Tan MW, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurz CL, Ewbank JJ. 2000. Caenorhabditis elegans for the study of host-pathogen interactions. Trends Microbiol 8:142–144. doi: 10.1016/S0966-842X(99)01691-1. [DOI] [PubMed] [Google Scholar]
- 31.Tan MW, Ausubel FM. 2000. Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr Opin Microbiol 3:29–34. doi: 10.1016/S1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 32.Abdelraouf K, Kabbara S, Ledesma KR, Poole K, Tam VH. 2011. Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. J Antimicrob Chemother 66:1311–1317. doi: 10.1093/jac/dkr105. [DOI] [PubMed] [Google Scholar]
- 33.Sun Z, Jiao Peng XQ, Jiang F, Huang Y, Zhang J, Yao F. 2013. Antibiotic resistance in Pseudomonas aeruginosa is associated with decreased fitness. Cell Physiol Biochem 31:347–354. doi: 10.1159/000343372. [DOI] [PubMed] [Google Scholar]
- 34.Engel J, Balachandran P. 2009. Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 12:61–66. doi: 10.1016/j.mib.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 37.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. 2012. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob Agents Chemother 49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez MÁ, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelstein VM, Skleenova EN, Shevchenko OV, D'souza JW, Tapalski DV, Azizov IS, Sukhorukova MV, Pavlukov RA, Kozlov RS, Toleman MA, Walsh TR. 2013. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis 13:867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 42.López-Causapé C, de Dios-Caballero J, Cobo M, Escribano A, Asensio O, Oliver A, del Campo R, Cantón R, Spanish Group for the Study of Bronchopulmonary Colonisation/Infection in Cystic Fibrosis. 2017. Antibiotic resistance and population structure of cystic fibrosis Pseudomonas aeruginosa isolates from a Spanish multicentre study. Int J Antimicrob Agents 50:334–341. doi: 10.1016/j.ijantimicag.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659–2669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 46.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caiazza NC, Shanks RM, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sánchez P, Linares JF, Ruiz-Díez B, Campanario E, Navas A, Baquero F, Martínez JL. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug-resistant mutants. J Antimicrob Chemother 50:657–664. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 49.Navas A, Cobas G, Talavera M, Ayala JA, López JA, Martínez JL. 2007. Experimental validation of Haldane's hypothesis on the role of infection as an evolutionary force for metazoans. Proc Natl Acad Sci U S A 104:13728–13731. doi: 10.1073/pnas.0704497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.