Abstract
Patient: Female, 14-month-old
Final Diagnosis: Acute lymphoblastic leukemia
Symptoms: Fever
Medication: —
Clinical Procedure: —
Specialty: Hematology
Objective:
Rare disease
Background:
Chromosomal translocations involving the PDGFRB gene have been reported in a broad spectrum of hematological malignancies. An ATF7IP/PDGFRB fusion was recently identified in a Philadelphia chromosome-like (Ph-like) B-progenitor acute lymphoblastic leukemia (B-ALL) patient. Here we report on a special case of a Ph-like ALL patient who had a variant ATF7IP/PDGFRB fusion.
Case Report:
In this case, a variant fusion was created between ATF7IP exon 9 (instead of exon 13) and PDGFRB exon 11, resulting in the loss of 411 nucleotides and 137 amino acids in the ATF7IP/PDGFRB fusion cDNA and its encoded chimeric protein, respectively. Interestingly, ATF7IP has also been reported as a fusion partner of the JAK2 kinase gene in Ph-like ALL, but all of the genomic breakpoints in ATF7IP in this fusion reported thus far occurred in intron 13. Enforced expression of the variant ATF7IP/PDGFRB fusion induced cytokine-independent growth and glucocorticoid resistance of BaF3 cells. Similar to the initially described ATF7IP/PDGFRB-bearing Ph-like ALL patient who was refractory to conventional chemotherapy but highly sensitive to tyrosine kinase inhibitor (TKI) monotherapy, our patient with a variant ATF7IP/PDGFRB fusion had a poor initial treatment response to chemotherapy but responded well to TKI-based therapy and is now doing well in continuous remission.
Conclusions:
Ph-like ALL patient with an ATF7IP/PDGFRB fusion is rare, but can benefit from TKI therapy.
MeSH Keywords: Oncogene Fusion, Philadelphia Chromosome, Precursor Cell Lymphoblastic Leukemia-Lymphoma
Background
Chromosomal translocations involving the PDGFRB gene have been reported in a broad spectrum of hematological malignancies, including both myeloid neoplasms and acute lymphoblastic leukemia (ALL) [1–3]. Myeloid neoplasms with PDGFRB rearrangements are genotypically and phenotypically diverse but typically present as a myeloproliferative neoplasm (MPN) with eosinophilia in the bone marrow and the peripheral blood [4]. Recently, PDGFRB rearrangements were reported in a special type of ALL named Philadelphia chromosome-like (Ph-like) ALL, which lacks the characteristic BCR-ABL1 fusion gene but possesses a similar gene expression profile to that observed in Ph+ ALL [3,5,6]. So far, over 20 fusion partners of PDGFRB have been identified, of which five genes – namely EBF1, SSBP2, TNIP1, ZEB2, and ATF7IP – were reported to be fused with PDGFRB in Ph-like ALL [2,3,7]. ATF7IP as a novel PDGFRB fusion partner was initially identified in an eight-year-old boy with B-ALL by a Japanese group in 2014 [7]. This patient had a cryptic PDGFRB translocation revealed by the conventional cytogenetics, but mRNA sequence analysis identified an in-frame fusion transcript between exon 13 of ATF7IP and exon 11 of PDGFRB.
In our case, a 14-month-old girl with B-ALL, and a completely normal karyotype was observed by conventional cytogenetics. However, fluorescence in situ hybridization (FISH) analysis, using a breakapart probe set, demonstrated rearrangement of the PDGFRB gene, and next-generation genomic DNA sequencing revealed that the ATF7IP gene was fused to PDGFRB. Unexpectedly, the breakpoint in ATF7IP was not located in intron 13 as described in the first reported case of a Ph-like ALL patient with the ATF7IP/PDGFRB fusion and as in other Ph-like ALL patients containing an ATF7IP/JAK2 fusion gene [3,7], but rather in ATF7IP intron 9. Consequently, this rearrangement created a variant ATF7IP/PDGFRB chimeric transcript in which the fusion was produced between ATF7IP exon 9 (instead of 13) and PDGFRB exon 11. This variant fusion lacks 411 nucleotides and consequently 137 amino acids compared to the originally described ATF7IP/PDGFRB chimeric cDNA and protein.
Case Report
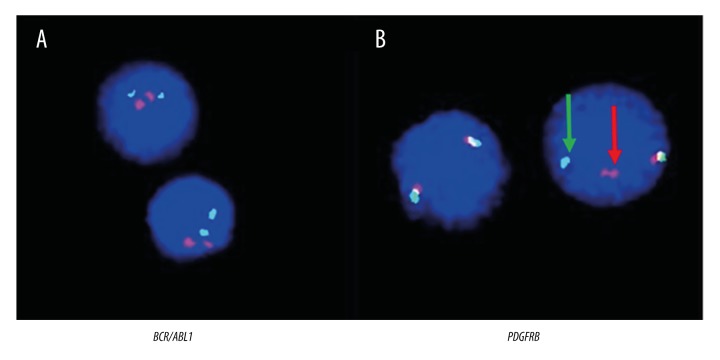
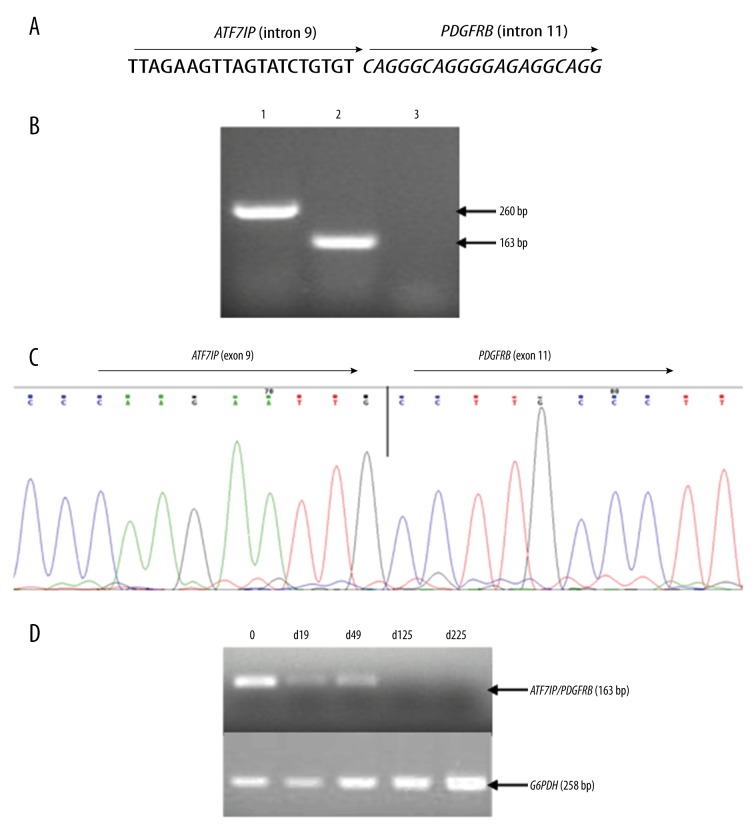
A 14-month-old female was hospitalized in October 2015 due to fever and skin petechiae. Laboratory test findings showed a white blood cell (WBC) count of 7.8×109/L with 9% immature lymphocytes, a hemoglobin concentration of 7.6 g/dL, and a platelet count of 11×109/L. Bone marrow (BM) examination was consistent with ALL-L2. Immunophenotyping results showed a pre-B cell type ALL with the presence of CD19, CD10dim, CD20, CD22, HLA-DR, cCD79a, and cIgM. Karyotyping analysis demonstrated a normal karyotype of 46 XX (data not shown). FISH analysis failed to detect the BCR/ABL1 fusion gene (Figure 1A); however, a distinct split signal was seen in the PDGFRB gene (Figure 1B). Further study with next-generation genomic DNA sequencing demonstrated fusion of the ninth intron of PDGFRB with the eleventh intron of the ATF7IP gene (Figure 2A). RT-PCR and sequence analysis of the resulting amplicon confirmed the expression of an ATF7IP/PDGFRB fusion transcript (Figure 2B, 2C).
Figure 1.
Results of FISH analysis. (A) No BCR/ABL1 fusion was detected in the interphase nuclei of leukemia cells by the BCR/ABL1 D-FISH probe (Vysis Inc., Downers Grove, IL, USA). (B) PDGFRB gene rearrangement was detected by the PDGFRB break-apart probe (Vysis). Co-localized red/green or yellow signals identify the normal PDGFRB gene. Split signals (a red and a green signal, as indicated by the red and green arrows) demonstrate PDGFRB gene translocation.
Figure 2.
Genomic and cDNA fusion junction sequences of the ATF7IP/PDGFRB fusion gene and monitoring of minimal residual disease (MRD) by RT-PCR detection of the ATF7IP/PDGFRB fusion transcript. (A) Next-generation sequencing analysis indicated the genomic breakpoint site. (B) RT-PCR test detected the ATF7IP/PDGFRB fusion transcript. First round RT-PCR using primers of ATF7IP-F1 (ACCCTACTGCCAGTGCTGCAC) and PDGFRB-R (GATGGCTGAGATCACCACCAC) yielded a product of 260 bp (lane 1) and a semi-nested PCR using primers of ATF7IP-F2 (ACAGTGGGCCCATCAGGACTC) and PDGFRB-R amplified a product of 163 bp (lane 2). Lane 3 was a negative control. (C) Sequencing analysis of the RT-PCR product indicated the fusion junction of the ATF7IP/PDGFRB cDNA. (D) MRD monitoring by RT-PCR of the ATF7IP/PDGFRB transcript indicated that no fusion gene product could be detected after 125 days of treatment.
Given these clinical and molecular findings, the patient was diagnosed with Ph-like B-ALL and treated with standard remission induction therapy of VDLP (vincristine, daunorubicin, L-asparaginase, and prednisone). At day 19, a BM examination indicated partial remission with 6.5% lymphoblasts, and a 40 mg dose of multi-targeting second generation tyrosine kinase inhibitor (TKI) dasatinib was administrated daily thereafter together with the VDLP therapy. At day 49, BM examination showed complete remission (CR) and the minimal residual disease (MRD) test by flow cytometry (FACS) was negative. However, RT-PCR analysis remained positive for the ATF7IP/PDGFRB fusion. At day 125, both the ATF7IP/PDGFRB fusion gene expression by RT-PCR testing and the MRD analysis by FACS were negative (Figure 2D). The patient is now receiving the Chinese Children’s Cancer Group (CCCG)-2015-ALL protocol plus dasatinib and remains free of molecularly detectable disease.
Discussion
Ph-like ALL is a recently recognized disease entity related to B-progenitor ALL [2,8]. It constitutes a distinct subtype of ALL characterized by a gene expression profile similar to that of BCR/ABL1 positive ALL, a diverse range of genetic alterations that activate tyrosine kinase signaling, mutations of lymphoid transcription factor genes, and a poor outcome. In contrast to the incidence of Ph-positive ALL, which is known to be lower than 5% of childhood ALL but continually increases with age, comprising 20% to 30% of adults with ALL and with a peak incidence of 50% in patients over 50 years old [9], Ph-like ALL increases in frequency from 10% among children with standard risk and 13% with high risk ALL to 21% among adolescents and 27% among young adults with ALL but then decreases significantly with more advanced age, forming a bell-shaped curve [3,10,11].
Clinically, Ph-like ALL patients have higher leukocyte counts at presentation than those with non-Ph-like ALL and non-Ph+ ALL [3,8,9]. Ph-like ALL is more commonly seen in males than females and is associated with elevated levels of MRD at the end of induction therapy [3,12,13]. Across all age groups, both the median five-year event-free survival rates and the five-year overall survival rates were reported to be inferior to those in patients with non-Ph-like ALL [3,14]. The presence of Ph-like ALL was considered to be an independent prognostic factor in all age groups [10,14].
The molecular mechanism underlying the pathogenesis of Ph-like ALL involves characteristic kinase-activating alterations that are identified in 91% of patients, including rearrangements of kinases as well as other proteins that transduce kinase signaling pathways such as ABL1, ABL2, CRLF2, CSF1R, EPOR, JAK2, NTRK3, PDGFRB, PTK2B, TSLP, or TYK2, and sequence mutations involving FLT3, IL7R, or SH2B3 [3]. Engineered overexpression of ABL-class fusions (involving ABL1, ABL2, CSF1R, or PDGFRB) and JAK fusions in cell lines results in cytokine-independent proliferation and activation of downstream signaling pathways, both of which are inhibited by TKIs, consistent with the oncogenic role of the fusion kinases in the pathogenesis of Ph-like ALL [3,10,12]. ABL-class rearrangement is more common among children and predicted to respond to ABL1 and other specific kinase inhibitors [10].
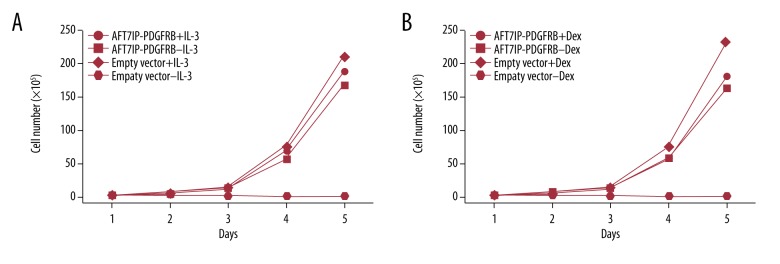
Fusion genes derived from PDGFRB play an important role in the pathogenesis of eosinophilia-associated myeloproliferative neoplasms (Eo-MPN) [1,4]. More than 20 PDGFRB fusion partners have been identified [2]. Reports indicate that the TKI imatinib is well-tolerated, and excellent long-term responses have been achieved in MPN patients with PDGFRB rearrangements, no matter the identity of the fusion partner [1]. So far, five different fusion partners have been reported in Ph-like ALL with PDGFRB rearrangements, of which only TNIP1 has been observed in both MPN and ALL [3,7]. ATF7IP was recently identified as a fusion partner of PDGFRB in a childhood Ph-like ALL case [7], and it was previously reported to be fused to JAK2 in four Ph-like ALL patients [3]. The breakpoints of ATF7IP in these five Ph-like ALL cases were all within intron 13. In our case, however, the breakpoint in ATF7IP was present in intron 9, causing a loss of 411 nucleotides from exons 9 to 13 in the variant fusion cDNA and consequently a loss of 137 amino acids in the fusion protein. Considering that in both cases the N-terminal domain of ATF7IP still contains the coiled coil structure that is known to contribute to the constitutive activation of kinase and cytokine receptor signaling, functionally there should be no difference in kinase activation and TKI responsiveness in patients with either the classic or variant ATF7IP/PDGFRB fusion. Our in vitro cell culture study indicated that enforced expression of the variant ATF7IP/PDGFRB fusion could not only transform BaF3 cells like ATF7IP/PDGFRB fusion as demonstrated by the Japanese investigators [15], but also induce the cells resistant to glucocorticoid treatment (Figure 3). Indeed, clinically both the Japanese boy with the classic ATF7IP/PDGFRB fusion and our patient showed excellent long-term responses to TKI therapy [16].
Figure 3.
Enforced expression of variant ATF7IP/PDGFRB fusion induced cytokine-independent growth and glucocorticoid resistance of BaF3 cells. (A) Compared with the parental BaF3 cells which can grow in the media with IL-3 (10 ng/mL), the cells stably transfected with variant ATF7IP/PDGFRB fusion became IL-3-independent growth. (B) The transformed BaF3 cells acquired resistance to dexamethasone treatment, even under a high concentration of the drug (10 μM).
In our case, the patient’s leukemic cells showed a normal karyotype by conventional cytogenetics. We note from the literature that a certain percentage of patients with PDGFRB fusions have a normal karyotype in MPN, MDS and ALL [2,7,17]. We suggest that FISH screening for PDGFRB rearrangement be strongly considered in such cases. As demonstrated herein in BCR-ABL1-negative B-progenitor ALL, Ph-like ALL with PDGFRB fusion is clinically refractory to conventional therapy but highly sensitive to TKI therapy.
Conclusions
Although the variant ATF7IP/PDGFRB fusion lacks 411 nucleotides and 137 amino acids compared to the originally described ATF7IP/PDGFRB chimeric cDNA and protein, in vitro cell culture study indicated that both fusion genes could transform BaF3 cells and convert the cells to GC resistance. Clinically, both of the patients with the variant or the classical fusion were refractory to conventional therapy but highly sensitive to TKI therapy. We suggest that FISH analysis for PDGFRB rearrangement be strongly considered as a supplementary screening test to identify the patients who might benefit from TKI treatment.
Acknowledgments
We thank Andy Ma and Grace Ma for English language editing.
Footnotes
Conflicts of interest
None.
References:
- 1.Cheah CY, Burbury K, Apperley JF, et al. Patients with myeloid malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood. 2014;123(23):3574–77. doi: 10.1182/blood-2014-02-555607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maccaferri M, Pierini V, Di Giacomo D, et al. The importance of cytogenetic and molecular analyses in eosinophilia-associated myeloproliferative neoplasms: An unusual case with normal karyotype and TNIP1-PDGFRB rearrangement and overview of PDGFRB partner genes. Leuk Lymphoma. 2017;58(2):489–93. doi: 10.1080/10428194.2016.1197396. [DOI] [PubMed] [Google Scholar]
- 3.Roberts KG, Li Y, Payneturner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. New Engl J Med. 2011;371(11):1005–15. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arefi M, García JL, Peñarrubia MJ, et al. Incidence and clinical characteristics of myeloproliferative neoplasms displaying a PDGFRB rearrangement. Eur J Haematol. 2012;89(1):37–41. doi: 10.1111/j.1600-0609.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 5.Lengline E, Beldjord K, Dombret H, et al. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98(11):146–48. doi: 10.3324/haematol.2013.095372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab C, Ryan SL, Chilton L, et al. EBF1-PDGFRB fusion in paediatric B-cell precursor acute lymphoblastic leukaemia (BCP-ALL): Genetic profile and clinical implications. Blood. 2016;127(18):2214–18. doi: 10.1182/blood-2015-09-670166. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi K, Mitsui K, Ichikawa H, et al. ATF7IP as a novel PDGFRB fusion partner in acute lymphoblastic leukaemia in children. Br J Haematol. 2014;165(6):836–41. doi: 10.1111/bjh.12834. [DOI] [PubMed] [Google Scholar]
- 8.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009;10(2):125–34. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu-Dumlao T, Kantarjian H, Thomas DA. Philadelphia-positive acute lymphoblastic leukemia: Current treatment options. Curr Oncol Rep. 2012;14(5):387–94. doi: 10.1007/s11912-012-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125(26):3977–87. doi: 10.1182/blood-2015-02-580043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold T, Baldus CD, Gökbuget N. Ph-like acute lymphoblastic leukemia in older adults. New Engl J Med. 2014;371(23):2235. doi: 10.1056/NEJMc1412123#SA1. [DOI] [PubMed] [Google Scholar]
- 12.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122(15):2622–29. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts KG, Pei D, Campana D, et al. Outcomes of children with BCR-ABL1 – like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014;32(27):3012–20. doi: 10.1200/JCO.2014.55.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishibashi T, Yaguchi A, Terada K, et al. Ph-like ALL-related novel fusion kinase ATF7IP-PDGFRB exhibits high sensitivity to tyrosine kinase inhibitors in murine cells. Exp Hematol. 2016;44(3):177–88. doi: 10.1016/j.exphem.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi K, Miyagawa N, Mitsui K, et al. TKI dasatinib monotherapy for a patient with Ph-like ALL bearing ATF7IP/PDGFRB translocation. Pediatr Blood Cancer. 2015;62(6):1058–60. doi: 10.1002/pbc.25327. [DOI] [PubMed] [Google Scholar]
- 17.Galimberti S, Ferreri MI, Simi P, et al. Platelet-derived growth factor beta receptor (PDGFRB) gene is rearranged in a significant percentage of myelodysplastic syndromes with normal karyotype. Br J Haematol. 2009;147(5):763–66. doi: 10.1111/j.1365-2141.2009.07878.x. [DOI] [PubMed] [Google Scholar]