Abstract
Knowledge of lectin and glycosidase specificities is fundamental to the study of glycobiology. The primary specificities of such molecules can be uncovered using well-established tools, but the complex details of their specificities are difficult to determine and describe. Here we present a language and algorithm for the analysis and description of glycan motifs with high complexity. The language uses human-readable notation and wildcards, modifiers, and logical operators to define motifs of nearly any complexity. By applying the syntax to the analysis of glycan-array data, we found that the lectin AAL had higher binding where fucose groups are displayed on separate branches. The lectin SNA showed gradations in binding based on the length of the extension displaying sialic acid and on characteristics of the opposing branches. A new algorithm to evaluate changes in lectin binding upon treatment with exoglycosidases identified the primary specificities and potential fine specificities of an α1–2-fucosidase and an α2–3,6,8-neuraminidase. The fucosidase had significantly lower action where sialic acid neighbors the fucose, and the neuraminidase showed statistically lower action where α1–2 fucose neighbors the sialic acid or is on the opposing branch. The complex features identified here would have been inaccessible to analysis using previous methods. The new language and algorithms promise to facilitate the precise determination and description of lectin and glycosidase specificities.
Protein–glycan interactions are fundamental to the biology of every organism. Glycan-binding proteins that have no enzymatic activity are called lectins and are involved in development, immune recognition, and signal transduction, among others.1 Enzymes that act upon glycans, such as glycosyltransferases and glycosidases, contribute to biosynthesis, metabolism, catabolism, and disease states.2 An important goal in the study of such proteins and enzymes is to characterize the specificities of their binding to particular glycans, which is valuable both for glycobiology research as well as for practical applications. The applications include the use of lectins to probe for specific glycans in biological samples3 and the use of glycosidases to break down glycans to enable analysis.4 Researchers also are pursuing inhibitors5 and probes6 of enzymatic activity and glycan-receptor interactions7 for therapeutic purposes.
The primary feature recognized by a lectin or glycosidase/glycosyltransferase can be uncovered using a small number of glycans in semiquantitative analyses. But the details of the interactions are usually harder to determine and describe, due to the huge range of structural features that may influence binding or activity. The amount of binding can be modulated by structural features away from the main binding site, such as substituents on neighboring monosaccharides or the length of the branch containing the binding site.8,9 Recent studies of human intelectin-110 and DC-SIGN11 provide examples of complex rules governing glycan recognition. Glycan arrays, now a standard tool for studying lectin specificities,12 have greatly helped to sort out the complexities in lectin binding, because one can obtain parallel measurements from hundreds of glycans using limited amounts of material.13 The challenge with such data is to interpret subtle differences in lectin binding between multiple, closely related glycoforms.
We previously introduced a textual system to describe patterns of substructures, or motifs, that could represent the binding determinant of a lectin.14 A system of defining motifs was important for enabling automated, statistical analyses of glycan arrays. With the text-based definitions of motifs, we could search for the presence of motifs in a set of glycans and quantify the association of lectin binding with the presence or absence of each motif. By automating the method, we performed comparative analyses of >3000 data sets.15 The textual system used at that time, however, had a limitation: it could not represent complex features that affect binding. The syntax of the language and the implementation of the software did not have the capacity to represent higher-level features that we and other researchers were finding to be important in lectin binding. Other researchers have contributed valuable tools for the analysis of glycan-array data,16−18 but these advances still would not provide the capabilities we sought.
In the research presented here, we developed a practical language for describing motifs of nearly any complexity, and we developed algorithms for testing the relationships between the motifs and the lectin binding or enzyme activity. No algorithm previously existed for the analysis of enzyme specificities from glycan-array data; here we employed a new method for finding features of glycans that associate with changes to lectin binding upon the treatment of the arrays with exoglycosidases. We show that complex features of glycans are associated with lectin binding and enzyme activity, including features that would have been inaccessible to analysis using previous methods.
Experimental Section
Preparation of the Human-Milk Oligosaccharide Arrays
The synthesis of glycans at the University of Georgia for human-milk oligosaccharide arrays was described earlier.19 A catalog of the complete set is in Figure S-1. Individual glycans were dissolved at 100 μM in a sodium phosphate buffer (50 mM, pH 9.0) and printed in replicates of six with spot volume of ∼400 pL. All compounds were printed onto NHS-activated microscope slides (Nexterion H, Schott) using a noncontact microarray printer (Scienion S3) that deposited about 460 pL per spot. The slides were printed with 24 subarrays in a 3 × 8 layout. Each subarray contained six replicate spots of each of the 60 compounds, printed in a 22 × 18 grid. After printing, the slides were incubated in a humidity chamber for 24 h and blocked for 1 h with 5 mM ethanolamine in Tris buffer (50 mM, pH 9.0). Blocked slides were rinsed with DI water, spun dry, and kept in a desiccator at room temperature until use.
Glycan-Array Data Acquisition
Screening solutions of biotinylated Aleuria aurantia lectin (AAL), Ulex europaeus agglutinin I (UEA), and Sambucus nigra agglutinin (SNA) (Vector Laboratories) were created by premixing streptavidin–AlexaFluor 635 conjugate (ThermoFisher) at a 1:3 ratio by weight of lectin/conjugate in TSM binding buffer (20 mM Tris–Cl, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween, and 1 g/L BSA, pH 7.4). Final lectin concentrations for AAL, UEA, and SNA were 1, 10, and 10 μg/mL, respectively. An amount of 100 μL of lectin solution was added to each subarray and was incubated at room temperature, in the dark, for 1 h. The slides were washed with TSM wash buffer (20 mM Tris–Cl, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, and 0.05% Tween, pH 7.4), TSM buffer (20 mM Tris–Cl, 50 mM NaCl, 2 mM CaCl2, and 2 mM MgCl2, pH 7.4), and DI water, and then spun dry as previously reported.20
Human galectin-3 and galectin-9 (R&D Systems) were dissolved at 10 and 2 μg/mL, and 3 and 0.8 μg/mL, respectively in TBS buffer (25 mM Tris, 0.15 M NaCl, pH 7.2) with 0.1% BSA and 0.05% Tween. Separately, solutions of biotinylated mouse antigalectin-3 (PeproTech) at 5 μg/mL, biotinylated mouse antigalectin-3 (PeproTech) at 5 μg/mL, and streptavidin–AlexaFluor at 5 μg/mL were each prepared in TBS buffer containing 0.1% BSA and 0.05% Tween. Each array was incubated as described above with one of the lectin solutions, followed by incubations with the corresponding secondary antibody solution and the streptavidin–AlexaFluor solution. After each incubation, the slides were washed and dried as described above. All slides were stored in the dark prior to acquiring fluorescence images using a microarray scanner (GenePix 4000B, Axon Instruments). Image analysis was performed with GenePix Pro 7 software to obtain the total fluorescence intensity of each spot. After background subtraction, the highest and lowest values of the six replicates from the same compound were removed, and the remaining values were averaged.
Additional glycan-array data were downloaded from the Consortium for Functional Glycomics (CFG). We used the data sets for AAL and UEA from array version 5.0, lectin concentration 1.0 μg/mL (IDs primscreen_4735 and primscreen_4212, respectively). The information given through the CFG and here conform to the MIRAGE standards for glycomics experiments.21 The processed data from glycan-array experiments generated for this research are available in Table S-1.
On-Chip Glycosidase Digestion
After printing the arrays and prior to blocking, the slides were placed in an 8 × 3 hybridization cassette (ArrayIt, Sunnyvale, CA) and treated with the following glycosidase conditions. An α2–3,6,8-neuraminidase from Arthrobacter ureafaciens (Roche) was prepared at 2 units per 100 μL in 50 mM sodium acetate, pH 5.5, with 5 mM CaCl2 and 1% BSA and incubated at 37 °C for 4 h. A microbial α1,2-l-fucosidase (Megazyme) was prepared at 0.5 units per 100 μL in 50 mM Tris, pH 6.5, with 1% BSA and incubated for 18 h at 37 °C. The slides were washed and dried as described above.
Software Availability and Data Preparation
The MotifFinder program was written in Matlab and is available for download at haablab.vai.org or upon request to the authors. The motifs were generated manually by text entry. Statistical analyses and graphing were done in Microsoft Excel and Graphpad Pro, and figures were prepared in Canvas.
Safety Considerations
The experimental methods presented here require standard safety precautions.
Results
A New Motif Language
The new language (Tables 1 and S-2) is expanded from a syntax introduced earlier.22 We sought to develop a system that is flexible enough to describe any binding determinant, yet human-readable and easy to use. We also wanted notation that is immediately recognizable by those familiar with glycobiology and explicit definitions rather than common terms such as “Lewis X”. Examples are given in Figure 1 of simple (Figure 1, parts A and B) and complex (Figure 1, parts D and F) motifs. Some of the motifs could not have been easily represented using previous methods, such as motifs containing noncontiguous features (Figure 1C) or exclusions of a specific number of features (Figure 1F).
Table 1. Syntax of the Motif Language.
| rule | examples | comments |
|---|---|---|
| standard glycan terms are accepted | Galb1–4(Fuca1–3)GlcNAcb | indicates the Lewis X motif |
| Fuca1–3(Galb1–4)GlcNAcb | indicates same motif; either branch may be enclosed | |
| standard abbreviations for monosaccharides | ||
| “a” and “b” for α and β, linkages separated by dash or comma | ||
| case insensitive | ||
| angled brackets specify details about the linkage or monosaccharide | Neu5AcA2-<3or6>GalB1–4<3f6f>GlcNAcB1–2<4f;6f>ManA | indicates Neu5Ac either 3′- or 6′-linked to an unbranched LacNAc |
| <T>Gal | indicates terminal galactose | |
| the “f” modifier specifies a location as unoccupied (free) | ||
| an unspecified location indicates occupied or unoccupied | ||
| the OR operator can be used to designate multiple options of linkage or monosaccharide | ||
| the XOR operator can be used to indicate multiple options of linkage, where all other options of linkage are unoccupied (free) | ||
| commas or semicolons can be included for clarity so that <3f6f>, <3f,6f>, and <3f;6f> are equivalent | ||
| the “T” modifier is shorthand for all nonreducing locations being unoccupied (<2f3f4f6f> for hexose and <8f9f> for neuraminic acids) | ||
| special monosaccharide terms are available | Any??-<3OR6>Galb1–4<3f;6f>GlcNAcb1–2<4f;6f>Mana | indicates a single monosaccharide extension linked either 3′ or 6′ to an unbranched LacNAc |
| Any??-<3OR6>Galb1–4<3f;6f>GlcNAcb1-?Mana | indicates the same but with any linkage to mannose: | |
| Fuca1-<2or3>Any?1-?Skip??-3(Fuca1-<2or3>Any?1-?Skip??-6)Galb | indicates a fucose anywhere on both a 3′ and a 6′ extension of Gal | |
| the “Any” wildcard is for unspecified monosaccharides | ||
| the “?” wildcard is for unspecified linkages | ||
| the “Skip” modifier indicates a gap of any kind | ||
| the “Skip” modifier encompasses lengths of zero, or no gap | ||
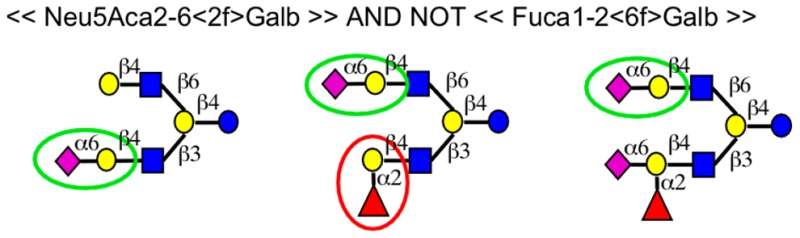
| double-angled brackets link complete terms with logical operators | ≪ <2f;3f;4f;6f>Glca ≫ OR ≪ Mana ≫ | indicates either terminal α-glucose or α-mannose |
| ≪ Fuca ≫ 2 | indicates two or more fucose | |
| ≪ Fuca1-<2or3or4>Any ≫ AND NOT ≪ Fuca ≫ 2 AND ≪ Siaa ≫ | indicates a single fucose with at least one sialic acid | |
| ≪Fuca≫ AND NOT ≪Fuca≫ 2 AND NOT k OR ≪ Fuca1–2Galb1–3GalNac ≫ ≫ | indicates a single fucose and excluding two other motifs | |
| ≪Motif1≫ AND NOT ≪Motif1≫ 2 AND NOT ≪Motif 2≫ | uses Motif IDs as shorthand | |
| embedded double-angled brackets are supported | ||
| the AND, OR, and NOT operators are supported |
Figure 1.
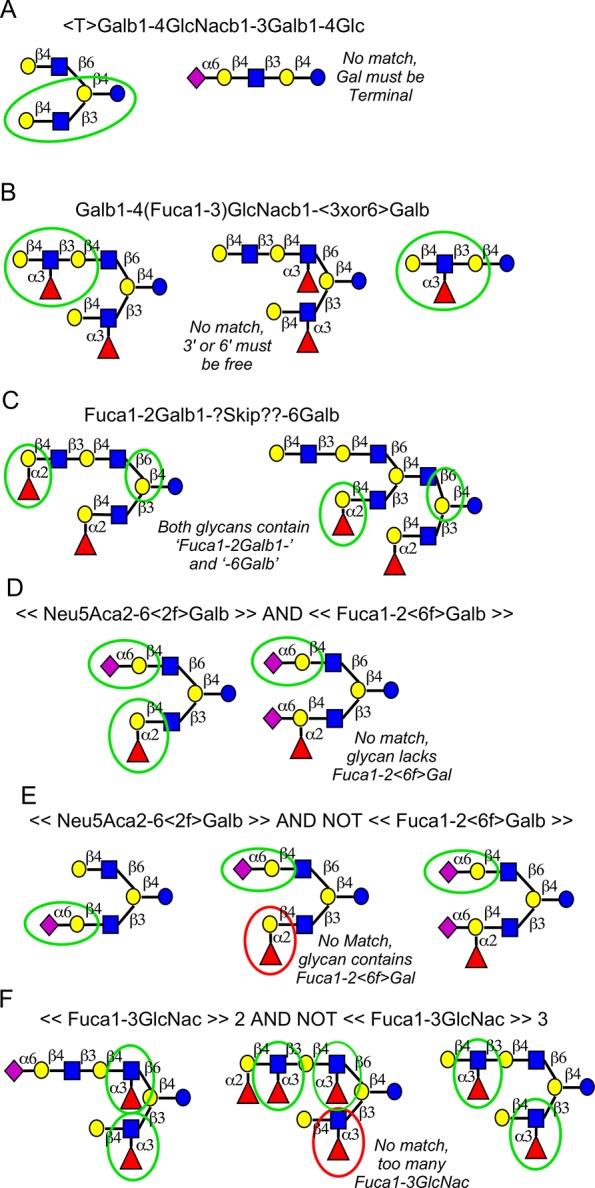
Examples of motifs and their presence or absence in glycans. In each example, the text definition of a motif is given along with the graphical representation of representative glycans. A green circle indicates the presence of the motif, and a red circle indicates a feature that precludes the motif.
An effective method to analyze glycan-array data is to identify the motifs that are present in each glycan, and then explore the relationships between lectin binding and the presence of each motif (Figure 2A), a technique we introduced earlier.14 The analysis could be partitioned into two steps (Figure 2B): (i) determine the basic motif that is nearly always present upon binding of the lectin (the primary specificity) and (ii) uncover the more complex features that modulate lectin binding (the fine specificity), such as functionalities adjacent to the primary determinant. To identify the primary binding determinant, one could compare lectin binding between glycans that contain a motif to all glycans that do not. The explore fine specificities, one could compare glycans containing a given motif to glycans containing an altered motif (Figure 2B).23,24 For the first analysis, we manually generated a “stock” set of motifs (Table S-3) that broadly covers the various types of motifs typically encountered in glycan arrays. For the second analysis, we define custom motifs to probe specific features observed from the analysis using the stock motifs.
Figure 2.
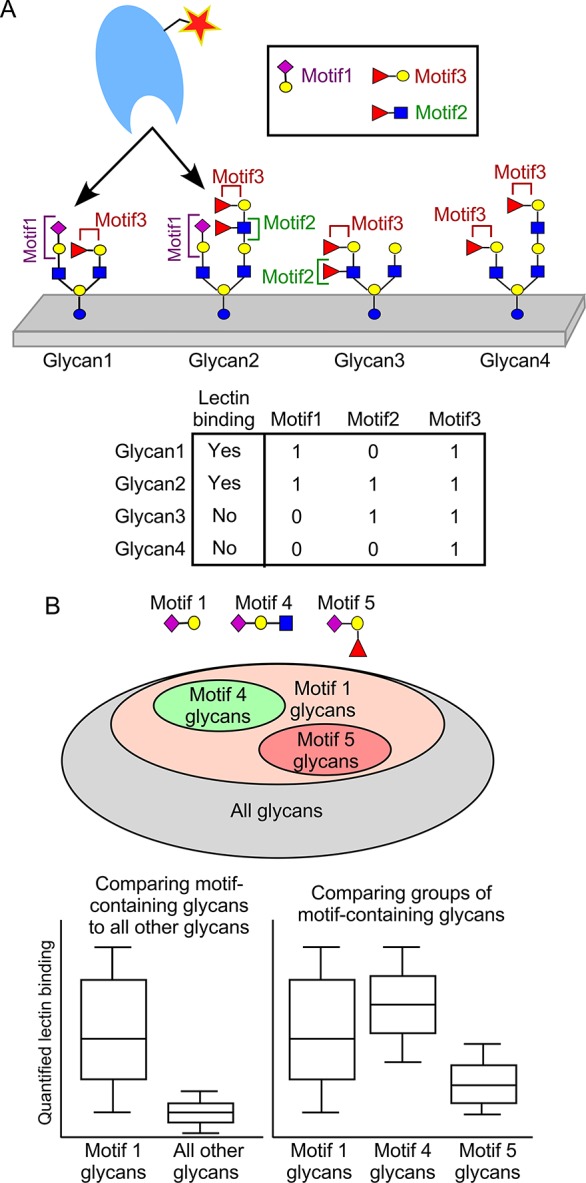
Using motifs in the analysis of glycan-array data. (A) The initial steps in the analysis of glycan-array data are to quantify the binding of a lectin to each glycan (top) and to determine the presence or absence of each motif in the glycans (bottom). (B) Adding features to a base motif defines subsets of glycans within the larger sets (top). One may compare the lectin binding between sets of glycans (bottom), such as between glycans containing a motif and all other glycans, or between subsets of glycans.
To facilitate the analysis, we developed a software program to parse the input text of glycans and motifs, search for the presence of motifs in glycans, and calculate a score for each motif indicating the strength of association between the motif and lectin binding. (See Tables S-4 and S-5 for example input and output, respectively.) The program uses various statistical measures to identify motifs that are reliably associated with binding (Figure S-2). Useful summary values are the log-transformed p-values of Welch’s t test (referred to as TScore) and the area-under-the-curve (AUC) of receiver–operator characteristic curves (see the Supporting Information for complete descriptions).
Using Motifs To Define Primary and Fine Specificities from Glycan-Array Data
The ability to define complex motifs should be especially useful when using glycan arrays with complex glycans. New libraries of glycans with controlled extensions and modifications on linear or branched oligosaccharides25 provided the opportunity to further test the capabilities of the motif language. A glycan array with 60 glycans having a variety of branching, fucosylation, and sialylation patterns19 was probed with the A. aurantia lectin (AAL), U. europaeus agglutinin (UEA), and S. nigra agglutinin (SNA). Previous analyses of glycan-array data acquired from the Consortium for Functional Glycomics (CFG) showed that the rules governing the binding of these lectins were not easy to capture using basic notation.23 Although the basic determinant of AAL is known to be α-linked fucose, other features—not readily evident—result in greater or lesser binding.
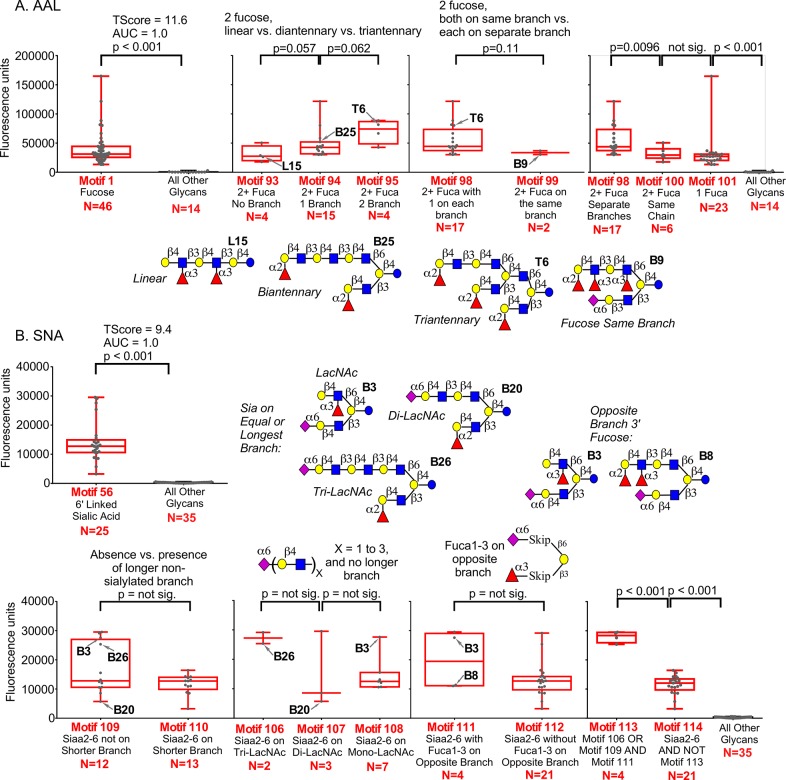
We ran the analysis with the stock set of motifs (Table S-3) to obtain the primary specificity of AAL. The top-scoring motif was “Fuca” (motif 1), TScore 11.6 and AUC 1.0. The distributions of intensities (Figure 3A) and high AUC indicate complete separation based on presence or absence of motif 1, but the distributions also showed that many glycans containing motif 1 bound AAL weakly. We examined the nonbinding glycans to identify features that potentially could explain the low binding, and then defined new motifs to test the relationships (Figure 3A and Table S-6). We found that among glycans with two or more fucoses, those with one or two branches had higher binding than linear glycans. Furthermore, if the fucoses were on different branches, binding was higher than if they were on the same branch. UEA did not show this relationship (not shown).
Figure 3.
Uncovering complex motifs in the HMO array. Each plot shows the fluorescence intensities of glycans containing the indicated motifs. Glycan arrays containing human-milk oligosaccharides were incubated with the lectins AAL (A) and SNA (B). The depicted glycans are representatives from each group (see Figure S-1 for all glycans on the array).
The top motif for SNA among the stock set of motifs was “Neu5Aca2–6Galb” (motif 56), TScore 9.4 and AUC 1.0. As with AAL, we saw complete separation of intensities based on the top motif in the initial analysis, but we also defined custom motifs that identify possible sources of variation in binding (Figure 3B and Table S-6). For example, we saw a trend toward higher binding if the sialic acid is on the longer branch, and we observed consistently higher binding when the sialic acid was at the end of a long extension. In some cases, binding was high even if the sialic acid was not on the longer branch, provided the branch opposing the sialic acid displayed 3′-linked fucose. Combining these features revealed clear distinctions in binding (Figure 3B). Sialic acid either on a long extension (tri-LacNAc), or with an opposing branch of equivalent or lesser length that displays 3′ fucose, has higher binding than all other motifs containing sialic acid. The examples demonstrate gradations of binding based on complex features of glycans. In a similar way, we found motifs associated with gradations in binding for human galectin-3 and galectin-9 (Figure S-3).We also analyzed glycan-array data from the CFG for AAL and UEA (Figure S-4 and Table S-6). The top motifs from the stock list were “Fuca” (motif 1) for AAL and “Fuca1–2Galb” (motif 4) for UEA, consistent with the known specificities. A detailed analysis showed that AAL and UEA both bind better to Fucα1–2 when the neighboring Gal is in a β1–4 linkage and when its 3′ location is free. The trends among the new motifs were reproducible at multiple concentrations of AAL and UEA, indicating the differences were not spurious features of a single array (Figure S-4). Certain features may result from the immobilization of the glycan,26 but the analysis nevertheless identified directions for further study.
Applying the Method to the Analysis of Enzyme Specificity
The characterization of glycosyltransferases and glycosidases often centers on two traits: the feature that is added or removed and the features of the glycan that modulate activity. The former trait can be fairly straightforward to determine, but the features that affect activity require empirical comparisons over many glycans. To investigate whether particular features of glycans modulate enzyme activity, we analyzed glycan arrays that in parallel were either treated or untreated by a glycosidase, and then probed with lectins (Figure 4A). Arrays treated with an α1–2-fucosidase were probed with AAL and UEA, and arrays treated with an α2–3,6,8-neuraminidase were probed with SNA. From these data, we calculated a percent change in lectin binding to each glycan.
Figure 4.
Using motifs to uncover details of exoglycosidase specificities. (A) Method of detecting changes in lectin binding upon glycosidase treatment. (B) The removal motif is the part of the glycan removed by the glycosidase, and the recognition motif is the part of the glycan that affects enzyme activity. (C) The program generates in-silico-modified glycans. If a glycan contains a recognition motif that includes the removal motif, the program eliminates the removal motif from the glycan. The program then determines, for each lectin, whether the lectin’s primary motif is present or absent in the original and modified glycans. (D) For each removal motif, the program groups the glycans according the presence or absence of the recognition motif and by the expected change in lectin binding. (E) For the fucosidase, motifs 1, 12, and 7 are examples of moderate, poor, and good recognition motifs, respectively. The graphs show the percent changes in lectin binding in each of the groups defined in panel D. Each graph contains pooled data from AAL and UEA. Fine-specificity analysis revealed statistically higher percent changes where the galactose neighboring the fucose had no sialic acid relative to where it had sialic acid at the 6′ location. (F) For the neuraminidase, motif 56 showed a very good fit to the expected changes. Fine-specificity analysis showed that α1–2-linked fucose could have an inhibitory effect.
We developed an algorithm to quantify the relationship between the motifs and the enzyme-induced changes to lectin binding (Figure 4A–D and supplementary methods in the Supporting Information). For each enzyme, we defined the feature that is removed, called the removal motif (Figure 4B). The removal motif was Fucα for the fucosidase and Siaα for the neuraminidase. We then defined recognition motifs, which are the features surrounding the removal motif that potentially influence enzyme activity. The recognition motifs initially comprised the stock set defined above. Next, for each recognition motif, we predicted the change in lectin binding to each glycan upon enzyme treatment. If a glycan contained a recognition motif that included the removal motif, we created an in-silico-modified glycan with the removal motif eliminated, and then determined the presence of the lectin-binding motif in the original and modified glycan (Figure 4C). The next step was to group the glycans by expected enzyme activity and by expected effect on lectin binding (Figure 4D).
With such groups, we could compare the experimental data to the expected trends. For each recognition motif, we performed a statistical comparison between the groups expected to show a change (groups P10 and P01) and those not expected to show a change (groups N10 and N01). A summary value of this comparison was the log-transformed p-value of the Mann–Whitney U test, referred to as the MWScore (see the Supporting Information). Another method was to calculate a “fit-to-expected” parameter based on the root-mean-squared of the differences between the expected and actual percent changes across all glycans (see the Supporting Information). The expected percent changes were 1.0 and −1.0 for groups P10 and P01, respectively, and 0 for groups N10 and N01. The fit-to-expected parameter allowed us to compare the observed patterns to the theoretical ideal (fit-to-expected value of 1).
For the fucosidase, the motif with the highest score was motif 7, Fuca1–2Galb<3or4>GlcNAc (score 0.77), which concords with the known specificity of the enzyme. In comparison, motif 1 (Fuca) and motif 12 (Fuca1–3Any) had much lower scores: 0.38 and 0, respectively. (See Table S-6 for complete scores.) Plots of the percent changes of the glycans in each group illustrate the relationships (Figure 4E). Motif 7 showed high values for nearly all group-P10 glycans and low values for nearly all the rest. (Some glycans in group P11 had the AAL-binding motif in both the original and modified glycans but showed a high change in binding.) We next investigated whether the fine specificity of the enzyme would be discernible among the glycans in group P10. By comparing percent changes between glycans containing or not containing specific recognition motifs, we found that glycans with a sialic acid neighboring the fucose had statistically lower (p = 0.015, Mann−Whitney U test) percent changes than glycans without a neighboring sialic acid (Figure 4E).
The top motif for the neuraminidase was motif 56 (Neu5Aca2–6Galb), score 0.92 (Table S-6). The known specificity of the enzyme is broader than motif 56, but we only had information about α2–6-linked sialic acid because we detected only with SNA, which binds this linkage. The use of additional lectins would provide more breadth to the analysis, but the method was robust enough to properly find a subset of the actual specificity with only one lectin. A fine-specificity analysis suggested that a fucoseα1–2 neighboring the sialic acid or on the opposing branch could reduce enzyme activity (Figure 4F).
We also compared the specificities of three neuraminidases, two that cleave all linkages (Ar. ureafaciens and Clostridium perfringens) and one that prefers to remove sialic acid in the α2–3 linkage (Salmonella typhimurium). In addition, we tested the use of lectins to detect underlying monosaccharides that would be exposed by enzymatic action, in this case Erythrinia cristagalli lectin (ECL) to detect terminal Galβ1–4GlcNAc and Bauhinia purpurea lectin (BPL) to detect terminal galactose and N-acetyl-galactosamine. The top motifs calculated by the algorithm (Table S-6) were Siaa2-<3or6>Gal (motif 175, score 0.62) for Ar. ureafaciens, <8f>Siaa2-<3or6>Galb (motif 194, score 0.63) for C. perfringens, and <8f>Siaa2–3Galb (motif 193, score 0.51) for Sa. typhimurium, thus correctly identifying the distinction between the enzymes in the linkages they cleave. The galactose-binding and sialic-acid-binding lectins contributed equivalent information (Figure S-5), confirming the value of detecting newly exposed, underlying features.
Discussion
The study of the complex details of lectin and glycosidase recognition of glycans necessitated a method to represent complex motifs and to evaluate them in experimental data. Here we demonstrated the value of a new syntax and analysis system for finding modifiers to the binding and activity of several lectins and exoglycosidases. The method identified several features that would not have been practically analyzable using manual analyses or previous modes of representing motifs. Further experiments will be required to validate the roles of the new motifs in lectin binding and glycosidase activity, but the results establish the ability to quantify features in glycan-array data that would be otherwise difficult to test. The software makes the approach accessible to non-bioinformatics researchers, and it promises to provide a useful complement to existing data-exploration tools, such as those available through the CFG. The new capabilities should have value for fundamental studies in glycobiology or for downstream use in glycan analyses. For example, one could use the output of MotifFinder to calculate the likelihood of various motifs being present in a sample, based on the binding of lectins to the sample before and after glycosidase treatment.22 In addition, the motif language may have inherent value for searching glycoinformatics databases27 or for linking data between disparate platforms, such as mass spectrometry or chromatography.
A limitation of glycan-array analysis is that it does not take into account structural and chemical information, which ultimately determine the binding between a glycan and protein. Computational glycobiologists are uncovering the chemical and structural bases for protein–glycan binding8 and are linking such analyses with results from glycan arrays.9 Both types of information are important, with the glycan arrays providing measurements of binding and the structural studies providing explanations and predictions of binding. The quantifications achieved using MotifFinder could be plugged in to the structural analyses to achieve improved accuracy.
The analyses presented here showed the necessity of complex motif definitions. The most interesting motifs were found on arrays that contained glycans with unequal extensions and substituents on each branch,19 indicating that, as the experimental data more closely model the complexity of glycobiology, the analyses will benefit from methods capable of mining that complexity. We found that AAL bound better to multiple fucoses when the fucoses were on separate branches (Figure 3A). The improved binding of AAL to fucoses on separate branches likely relates to its quaternary structure of a sixfold β-propellor, with five fucose binding sites on the edges.28,29 Multimeric structures and binding are common features among lectins,30,31 so the arrays with nonsymmetrical glycans combined with the ability to define complex motifs should be valuable to sort out the relative effects of unequal and equal modifications to branches. Similarly, with SNA we observed the influence of relationships between the branches. The relative lengths of the branches containing or not containing sialic acid seemed to modulate binding, as did the presence of α1–3-linked fucose on the opposing branch. Such relationships would need to be confirmed with more experimentation, but the results show the potential importance of complex motifs and the ability to uncover these relationships. Glycan asymmetry can be a mode of regulating glycan interactions in biology via heteromultivalent interactions, as shown with HIV-neutralizing antibodies,32 so the characterization of lectins that are influenced by unequal modifications could help to probe the regulating mechanisms.
Using the CFG data, we found that AAL and UEA bound to α1–2-linked fucose better where the galactose was β1–4-linked instead of β1–3-linked and was not substituted at the 3′ location. The former feature represents type-2 N-acetyl-lactosamine, which is the disaccharide repeat comprising glycan extensions, and the latter feature represents the H antigen (Fucα1–2Gal), in distinction to the A antigen (Fucα1–2(GalNAcα1–3)Gal) and B antigen (Fucα1–2(Galα1–3)Gal). The preference of UEA for the H antigen was noted in a previous glycan-array analysis.16 The biological effects of switching from type-2 to type-1, or switching between the A, B, and H antigens, are not known, but reducing engagement of the terminal fucose could be part of the functional mechanisms.
The use of glycan arrays to study enzyme specificities was a new challenge. The feasibility of the experiments was shown in a study of influenza neuraminidase,33 but we needed to develop an algorithm for the quantitative evaluation of enzyme preferences. Our method proved accurate in finding the primary specificities of the enzymes and showed promise for identifying fine specificities. The reduction in fucosidase action when sialic acid was near the removal motif, and the reduced action of neuraminidase in relation to fucoseα1–2, could be due to steric hindrance. But with the neuraminidase, the effect was present even when the fucose was on the opposite branch, suggesting the role not only of steric hindrance but also of engagement with the nonsialylated extension. A potential function of α1–2 fucosylation, it follows, would be to reduce neuraminidase activity. Future studies of human neuraminidases in this context could be helpful to gain insights into the functions of human A/B/O blood group antigens, which display α1–2 fucosylation.
The language and method presented here will enable deep analyses of lectins and enzymes, as well as the development of additional novel approaches in glycan analysis. Given the continual discoveries of lectins and glycan-active enzymes, researchers will need tools flexible enough to handle unusual specificities. Developers of glycan arrays have created structures that cover diverse areas of glycobiology, including mammalian glycans,13,34 microbial glycans,35,36 and various types of sialylated structures,37−39 which may be helpful in acquiring data to characterize newly discovered molecules. Another benefit of algorithms for automated interpretation is increased accessibility to researchers without expertise in glycobiology. A goal for further development will be to link software employing the motif syntax with experimental platforms that are designed to make glycan analysis routine.
Acknowledgments
We gratefully acknowledge the support of this work by the National Institute of Allergy and Infectious Diseases (NIH Common Fund Program in Glycoscience, R21AI129872) and the National Cancer Institute (Alliance of Glycobiologists for Cancer Detection, U01CA168896).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b04293.
Details of the motif score and statistical calculations, motif list preparation, and enzyme calculations, sialylated glycan-array methods, determining reliable motifs, analysis of galectins 3 and 9, glycans on the human-milk oligosaccharide array, and analysis of the specificities of three neuraminidases (PDF)
Glycan microarray data, supported modifiers, terms, and symbols, motif list, sample input of MotifFinder, sample output of MotifFinder, and top motifs in each analysis (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Sharon N. J. Biol. Chem. 2007, 282, 2753–2764. 10.1074/JBC.X600004200. [DOI] [PubMed] [Google Scholar]
- Davies G. J.; Williams S. J. Biochem. Soc. Trans. 2016, 44, 79–87. 10.1042/BST20150186. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J. Glycoconjugate J. 2004, 21, 35–40. 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Gotz L.; Abrahams J. L.; Mariethoz J.; Rudd P. M.; Karlsson N. G.; Packer N. H.; Campbell M. P.; Lisacek F. Bioinformatics 2014, 30, 3131–3133. 10.1093/bioinformatics/btu425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi Kazem Abadi S.; Tran M.; Yadav A. K.; Adabala P. J. P.; Chakladar S.; Bennet A. J. J. Am. Chem. Soc. 2017, 139, 10625–10628. 10.1021/jacs.7b05065. [DOI] [PubMed] [Google Scholar]
- Hang H. C.; Yu C.; Pratt M. R.; Bertozzi C. R. J. Am. Chem. Soc. 2004, 126, 6–7. 10.1021/ja037692m. [DOI] [PubMed] [Google Scholar]
- Hudak J. E.; Bertozzi C. R. Chem. Biol. 2014, 21, 16–37. 10.1016/j.chembiol.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. E.; Drickamer K. Glycobiology 2009, 19, 1155–1162. 10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant O. C.; Tessier M. B.; Meche L.; Mahal L. K.; Foley B. L.; Woods R. J. Glycobiology 2016, 26, 772–783. 10.1093/glycob/cww020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesener D. A.; Wangkanont K.; McBride R.; Song X.; Kraft M. B.; Hodges H. L.; Zarling L. C.; Splain R. A.; Smith D. F.; Cummings R. D.; Paulson J. C.; Forest K. T.; Kiessling L. L. Nat. Struct. Mol. Biol. 2015, 22, 603–610. 10.1038/nsmb.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs P. J.; Harrison R.; Pemberton S.; Quintero-Martinez A.; Parry S.; Haslam S. M.; Dell A.; Taylor M. E.; Drickamer K. J. Mol. Biol. 2010, 396, 685–696. 10.1016/j.jmb.2009.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan C. D.; Paulson J. C. Annu. Rev. Biochem. 2011, 80, 797–823. 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O.; Head S.; Mondala T.; Scanlan C.; Huflejt M. E.; Alvarez R.; Bryan M. C.; Fazio F.; Calarese D.; Stevens J.; Razi N.; Stevens D. J.; Skehel J. J.; van Die I.; Burton D. R.; Wilson I. A.; Cummings R.; Bovin N.; Wong C. H.; Paulson J. C. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 17033–17038. 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A.; Yue T.; Heeringa L.; Day S.; Suh E.; Haab B. B. Glycobiology 2010, 20, 369–380. 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletter D.; Singh S.; Bern M.; Haab B. B. Mol. Cell. Proteomics 2013, 12, 1026–1035. 10.1074/mcp.M112.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholleti S. R.; Agravat S.; Morris T.; Saltz J. H.; Song X.; Cummings R. D.; Smith D. F. OMICS 2012, 16, 497–512. 10.1089/omi.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan P.; Zhang Y.; Tzeng T. R.; Wan X. F.; Luo F. Glycobiology 2012, 22, 552–560. 10.1093/glycob/cwr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Cummings R. D.; Smith D. F.; Huflejt M.; Campbell C. T.; Gildersleeve J. C.; Gerlach J. Q.; Kilcoyne M.; Joshi L.; Serna S.; Reichardt N. C.; Parera Pera N.; Pieters R. J.; Eng W.; Mahal L. K. Glycobiology 2014, 24, 507–517. 10.1093/glycob/cwu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden A. R.; Liu L.; Capicciotti C. J.; Wolfert M. A.; Wang S.; Gao Z.; Meng L.; Moremen K. W.; Boons G. J. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 6954–6959. 10.1073/pnas.1701785114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg-Molinaro J.; Song X.; Smith D. F.; Cummings R. D.. Current Protocols in Protein Science; John Wiley and Sons: Hoboken, NJ, 2011; Unit12.10. DOI: 10.1002/0471140864.ps1210s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; McBride R.; Stoll M.; Palma A. S.; Silva L.; Agravat S.; Aoki-Kinoshita K. F.; Campbell M. P.; Costello C. E.; Dell A.; Haslam S. M.; Karlsson N. G.; Khoo K. H.; Kolarich D.; Novotny M. V.; Packer N. H.; Ranzinger R.; Rapp E.; Rudd P. M.; Struwe W. B.; Tiemeyer M.; Wells L.; York W. S.; Zaia J.; Kettner C.; Paulson J. C.; Feizi T.; Smith D. F. Glycobiology 2016, 27, 280–284. 10.1093/glycob/cww118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reatini B. S.; Ensink E.; Liau B.; Sinha J. Y.; Powers T. W.; Partyka K.; Bern M.; Brand R. E.; Rudd P. M.; Kletter D.; Drake R.; Haab B. B. Anal. Chem. 2016, 88, 11584–11592. 10.1021/acs.analchem.6b02998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin K. A.; Liden D.; Haab B. B. Glycobiology 2012, 22, 160–169. 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agravat S. B.; Saltz J. H.; Cummings R. D.; Smith D. F. Bioinformatics 2014, 30, 3417–3418. 10.1093/bioinformatics/btu559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Chinoy Z. S.; Ambre S. G.; Peng W.; McBride R.; de Vries R. P.; Glushka J.; Paulson J. C.; Boons G. J. Science 2013, 341, 379–383. 10.1126/science.1236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant O. C.; Smith H. M.; Firsova D.; Fadda E.; Woods R. J. Glycobiology 2014, 24, 17–25. 10.1093/glycob/cwt083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki-Kinoshita K.; Agravat S.; Aoki N. P.; Arpinar S.; Cummings R. D.; Fujita A.; Fujita N.; Hart G. M.; Haslam S. M.; Kawasaki T.; Matsubara M.; Moreman K. W.; Okuda S.; Pierce M.; Ranzinger R.; Shikanai T.; Shinmachi D.; Solovieva E.; Suzuki Y.; Tsuchiya S.; Yamada I.; York W. S.; Zaia J.; Narimatsu H. Nucleic Acids Res. 2016, 44, D1237–D1242. 10.1093/nar/gkv1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihashi M.; Peapus D. H.; Kamiya N.; Nagata Y.; Miki K. Biochemistry 2003, 42, 11093–11099. 10.1021/bi034983z. [DOI] [PubMed] [Google Scholar]
- Wimmerova M.; Mitchell E.; Sanchez J. F.; Gautier C.; Imberty A. J. Biol. Chem. 2003, 278, 27059–27067. 10.1074/jbc.M302642200. [DOI] [PubMed] [Google Scholar]
- Collins B. E.; Paulson J. C. Curr. Opin. Chem. Biol. 2004, 8, 617–625. 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Dam T. K.; Brewer C. F. Adv. Carbohydr. Chem. Biochem. 2010, 63, 139–164. 10.1016/S0065-2318(10)63005-3. [DOI] [PubMed] [Google Scholar]
- Shivatare S. S.; Chang S. H.; Tsai T. I.; Tseng S. Y.; Shivatare V. S.; Lin Y. S.; Cheng Y. Y.; Ren C. T.; Lee C. C.; Pawar S.; Tsai C. S.; Shih H. W.; Zeng Y. F.; Liang C. H.; Kwong P. D.; Burton D. R.; Wu C. Y.; Wong C. H. Nat. Chem. 2016, 8, 338–346. 10.1038/nchem.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappert M. M.; Smith D. F.; Air G. M. J. Virol 2011, 85, 12146–12159. 10.1128/JVI.05537-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui S.; Feizi T.; Galustian C.; Lawson A. M.; Chai W. Nat. Biotechnol. 2002, 20, 1011–1017. 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- Stowell S. R.; Arthur C. M.; McBride R.; Berger O.; Razi N.; Heimburg-Molinaro J.; Rodrigues L. C.; Gourdine J. P.; Noll A. J.; von Gunten S.; Smith D. F.; Knirel Y. A.; Paulson J. C.; Cummings R. D. Nat. Chem. Biol. 2014, 10, 470–476. 10.1038/nchembio.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Liu S.; Trummer B. J.; Deng C.; Wang A. Nat. Biotechnol. 2002, 20, 275–281. 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- Nycholat C. M.; McBride R.; Ekiert D. C.; Xu R.; Rangarajan J.; Peng W.; Razi N.; Gilbert M.; Wakarchuk W.; Wilson I. A.; Paulson J. C. Angew. Chem., Int. Ed. 2012, 51, 4860–4863. 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.; Yu H.; Chen X.; Lasanajak Y.; Tappert M. M.; Air G. M.; Tiwari V. K.; Cao H.; Chokhawala H. A.; Zheng H.; Cummings R. D.; Smith D. F. J. Biol. Chem. 2011, 286, 31610–31622. 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padler-Karavani V.; Song X.; Yu H.; Hurtado-Ziola N.; Huang S.; Muthana S.; Chokhawala H. A.; Cheng J.; Verhagen A.; Langereis M. A.; Kleene R.; Schachner M.; de Groot R. J.; Lasanajak Y.; Matsuda H.; Schwab R.; Chen X.; Smith D. F.; Cummings R. D.; Varki A. J. Biol. Chem. 2012, 287, 22593–22608. 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.