Abstract
Antibodies are the principal immune effectors that mediate protection against reinfection following viral infection or vaccination. Robust techniques for human mAb isolation have been developed in the last decade. The study of human mAbs isolated from subjects with prior immunity has become a mainstay for rational structure-based, next-generation vaccine development. The plethora of detailed molecular and genetic studies coupling the structure of antigen-antibody complexes with their antiviral function has begun to reveal common principles of critical interactions on which we can build better vaccines and therapeutic antibodies. This review outlines the approaches to isolating and studying human antiviral mAbs and discusses the common principles underlying the basis for their activity. This review also examines progress toward the goal of achieving a comprehensive understanding of the chemical and physical basis for molecular recognition of viral surface proteins in order to build predictive molecular models that can be used for vaccine design.
A method for generating large amounts of murine mAbs of a predefined specificity was first published in 1975 by César Milstein and Georges J.F. Köhler (Köhler and Milstein, 1975), who won the 1984 Nobel Prize in Physiology or Medicine for this advance. This laboratory method, creation of hybridoma cell lines, has revolutionized many fields of biomedical research including diagnostics and the generation of therapeutic antibodies for cancer, inflammation, immunotherapy, and infectious diseases. Increasingly, however, it has become apparent that murine antibodies are limited in their relevance for the very detailed types of structure/function studies that are needed for the design of next-generation human vaccines. Deep-sequencing studies are revealing that murine antibody variable gene repertoires are less complex and less diverse than those of humans. The average length of the most variable loop of the antigen-combining surface (the heavy-chain complementarity-determining region 3, or HCDR3) is shorter in mice than in humans (Shi et al., 2014). Long HCDR3 regions are especially important in many antiviral antibody responses. The genetics of the Ig locus of experimental animals has homology with that of humans in many cases, but the actual genomic sequences differ significantly. In some cases, animal germline gene segments do not encode the specific sequences needed to bind to certain antigens. For example, the VRC01 class of HIV CD4-binding-site-specific neutralizing antibodies is not made in mouse or even monkeys, because specific sequences of the human VH1-02 antibody variable gene segment are needed for binding that do not occur in these animals (Navis et al., 2014). The rhesus macaque VH1.23 gene segment is the closest primate ortholog to the human VH1-02 gene segment (exhibiting 92% homology to the VH1-02*02 gene). Yet even the macaque VH1.23 alleles encode only two of the three amino acids in the human gene segment that define the critical motif for VRC01-like antibodies (W50, N58 flanking the HCDR2 region, and R71).
In this review, first I will discuss the diverse laboratory techniques for isolation of human mAbs, the assays for measuring antiviral function of antibodies, and the relationship of neutralization activity to protection in vivo. Then, I present a review of the fundamental principles underlying the structural and genetic basis for development of antibodies that possess the desirable features of breadth and potency. Finally, I discuss the potential impact of our growing understanding of canonical features of protective human mAbs on the rational design of next-generation vaccines.
Diverse Approaches to Isolating Human mAbs
The first several decades of hybridoma work focused on the isolation of murine mAbs for several reasons. First, mice can be repetitively infected or hyper-immunized with foreign antigens, to increase the frequency of antigen-specific B cells. Second, animals can be brought to necropsy to collect the spleen, which is a B cell-rich tissue that typically contains about 108 cells. In contrast, a typical 50-mL human peripheral blood sample for research contains only about 50 million mononuclear cells (of which only about 5%, or 2–3 million, are B cells). Third, the non-secreting myeloma fusion partners used in fusion with B cells worked well for murine cells, but human fusion partners were lacking.
Eventually, investigators took on the task of developing techniques for efficient isolation of mAbs from human cells. First, investigators developed methods for cloning antibody variable genes by isolation of RNA followed by RT-PCR (Larrick et al., 1989). Once it was possible to obtain diverse amplicon libraries of variable genes, the logical next step was to clone and express the antibodies in phage display libraries that express the antibodies on the surface of phage particles and also package the variable gene cDNAs in phagemids inside the particles (Kang et al., 1991; McCafferty et al., 1990; Szardenings and Collins, 1990). We learned many important principles from studies in the early 1990s with human mAbs isolated from such libraries, including the surprising finding that Fabs alone could potently inhibit a virus replication program (Barbas et al., 1992; Crowe et al., 1994). Over time, however, it became apparent that the heavy- and light-chain pairing in such libraries is essentially random, and investigators sought new methods that could isolate antibodies from single cells with the natural pairing of heavy and light chains.
Techniques for physical selection of single antigen-specific B cells were developed, but initially these methods were very inefficient. Antigen labeling of B cells by binding of purified or recombinant antigens to the B cell receptor (membrane-bound surface immunoglobulin) enabled single-cell flow cytometric sorting as the sorting instruments became more precise. Sorted single cells were used directly to obtain antibody genes (Babcook et al., 1996), or sorted single cells were expanded on feeder cell layers followed by RNA extraction and RT-PCR (Weitkamp et al., 2003). The emergence of this technique was exciting, but it was difficult to label and identify antigen-specific cells when the nonspecific binding of the viral reagents to all B cells was high in the setting of low frequency of B cell precursors in peripheral blood samples of many donors. Also, in order to identify antibodies that bind to the virion particles, the antigen used to sort cells must be in the correct conformation, with a high level of structural fidelity to the form of the protein on the virus. This feature is usually not the case with soluble forms of integral membrane proteins. For instance, hundreds of human mAbs were identified using the soluble form of the dengue envelope protein as a screening target, but very few of them demonstrated neutralizing activity (Smith et al., 2012, 2013). However, a few of the antibodies that bound to soluble envelope proteins also could bind to live virus particles on immunoassay plates, suggesting that the recombinant protein was altered in some way compared to the virion protein. Subsequently, live virus particles that had been purified on density gradients (and in some cases additionally immunoaffinity purified by capture with antibodies) on ELISA plates served as a much more effective screening antigen to identify neutralizing mAbs (Smith et al., 2014).
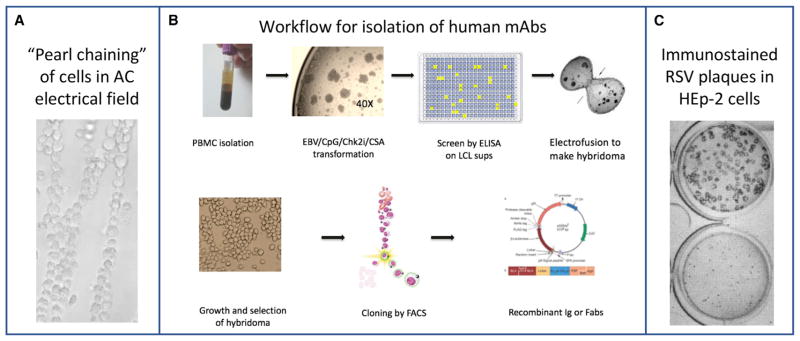
Another method for isolating antigen-specific human B cells was Epstein-Barr virus (EBV)-mediated B cell transformation, which was used to activate and transform primary B cells in culture via the EBV receptor CD21 on some B cells. Initially, however, it was found that this technique generated mostly polyreactive antibodies of the IgM isotype. Subsequently, the addition of the TLR9 agonist CPG with EBV was shown to enhance transformation of a broader array of B cells (Bernasconi et al., 2002). Addition of cyclosporine to inhibit rapidly dividing EBV-specific CD8+ cytolytic cells in the PBMC sample (that otherwise lyse EBV peptide-presenting B cells) preserves the B cell cultures longer. A number of important human mAbs to SARS (Traggiai et al., 2004), CMV (Macagno et al., 2010), paramyxoviruses (Corti et al., 2013; Wen et al., 2017), and others have been derived with this technique. The transformation protocol was modified further to include an apoptosis inhibitor, specifically a small-molecule Chk2 inhibitor (Smith and Crowe, 2015). Using this multipronged approach, a high efficiency of EBV transformation can be achieved. During this procedure, primary B cells proliferate and differentiate, secreting antibodies into the supernatants of the cell culture. EBV-transformed lymphoblastoid cell lines (LCLs) are difficult to clone biologically by passaging at the single-cell level, however. A combined transformation and hybridoma approach enables the rescue of unstable LCLs using electrofusion with a myeloma partner (Yu et al., 2008a). In this technique, the surface of LCLs and myeloma cells in suspension can be physically apposed using dielectrophoresis by delivering a low alternating current (Figure 1A), followed by direct current pulses that cause cell fusion. The human hybridoma workflow is summarized in Figure 1B.
Figure 1. Technical Approach for Isolating and Characterizing Neutralizing Human Monoclonal Antibodies.
(A) Dielectrophoresis: physical apposition of myeloma cells (large) with human B cells (small) using delivery of an alternating electrical current causes “pearl-chaining” distribution. The process is based on an electrical principle, termed dielectrophoresis, in which force is exerted on a particle when it is subjected to a non-uniform electric field.
(B) Workflow for generation of human mAbs by generating clonal hybridoma cells from primary human B cells.
(C) Immunostaining of large plaque (top) or small plaque (bottom) variants of RSV is used to quantitate residual live virus precisely in plaque reduction neutralization tests.
The resulting human hybridoma cells secreting mAbs can be cloned biologically more easily than LCLs (which often stop secreting the antibody of interest in low-cell-density culture) by limiting dilution or flow cytometric sorting. This approach has proven to be exceedingly robust, allowing isolation of very rare antigen-specific B cells from peripheral blood samples of humans infected with particular agents, decades after exposure by natural infection or immunization. For example, human mAbs that neutralized 1918 influenza A H1N1 virus were isolated from subjects who were ~100 years of age in 2007, at a time when 1918-like viruses had not circulated for about half a century (Yu et al., 2008b). The same approach has been used for isolating human mAbs to dengue virus, using the blood of U.S.-dwelling subjects who had previous dengue virus infection acquired during foreign travel decades before (Smith et al., 2012).
Additional methods for human B cell isolation and mAb generation have continued to emerge. Different methods for activation and transformation of human B cells have been reported. For example, lentivirus-mediated transduction of memory B cells with two anti-apoptotic cellular factors, BCL6 and Bcl-xL, has been used to immortalize cells for mAb discovery (Kwakkenbos et al., 2010). The TLR7/8 agonist R848 (resiquimod, an imidazo-quinoline compound) combined with IL-2 activates memory B cells (Pinna et al., 2009). IL-21 is a potent stimulator of CD40 ligand (CD154)-activated B cells, and B cell-activating factor (BAFF) enhances the stability of antibody-secreting cells. Combinations of these factors can be used to activate and maintain B cell lines. Another exciting development in the field was the discovery that antigen-specific B cells circulate as plasmablasts in the peripheral blood during a very circumscribed period following vaccination (about days 5–7) (Wrammert et al., 2008). Plasmablasts are not common in the circulation of healthy subjects, but their frequency can exceed 1% of circulating B cells a week after vaccination. This finding was useful because in this setting antigen labeling of B cells (with its attendant problems of nonspecific binding) is not required, but instead one can sort all of the circulating plasmablasts and infer that they are specific for antigens from the recent immunization or infection. This method routinely yields interesting human mAbs. However, it is unclear at this time whether the dominant antibody clones in the plasmablast population persist into the B cell memory population in humans. Finally, investigators have raised the concern that studying only the human B cells in circulation may be limiting, since the secreted antibody proteins in circulation that mediate protection derive from long-lived plasma cells in the bone marrow. Some studies suggest the repertoire of the bone marrow plasma cells and circulating memory B cells differs (Purtha et al., 2011). Various approaches are being used to address this concern, including mass spectrometry-based sequencing of antigen-specific antibody proteins captured from serum or plasma (Boutz et al., 2014) and direct study of human bone marrow specimens.
Measuring the Antiviral Activity of mAbs
Neutralization Assays
Antibody-mediated inhibition of viruses to define neutralization activity typically is measured by specific in vitro laboratory assays (although in vivo challenge studies also are used in some cases). Usually, live virus suspensions of known infectious particle count are mixed with varying concentrations of purified antibodies and incubated for a short period of time, then the mixtures are inoculated onto cell cultures to detect if any residual live virus exists. The readout is the percent reduction of infectious virus particles present in the initial inoculum. Many viruses can be detected using a plaque assay in a cell culture monolayer, such as poxviruses or paramyxoviruses like respiratory syncytial virus (RSV) (Figure 1C).
Microneutralization
This assay format is used to increase throughput and simplicity. In this approach, residual virus following incubation with antibodies is detected in cells that are inoculated in liquid medium, allowing virus to spread in culture, instead of the use of a semi-solid or agar overlay that limits local spread and causes plaque formation. In the liquid overlay assay, detection of residual replication-competent virus is performed with detection of a viral antigen and chemical substrate using a colorimetric optical density in the well, which correlates with the amount of viral antigen in the cell monolayer. Various types of microneutralization assays can be compared for benchmarking and standardization, e.g., influenza assays being tested by the international partnership CONSISE (the Consortium for the Standardization of Influenza Seroepidemiology) (Laurie et al., 2015).
Other Assays
Specific mechanistic assays have been developed to study the particular step in the virus life cycle at which the antibodies are acting. For instance, some of the most potent virus-inhibiting antibodies prevent attachment to cell surface receptors. Blocking attachment to a recombinant or purified form of cell receptor can be used in a multi-well plate assay format even when the virus cannot be propagated, such as for noroviruses that attach to histoblood group antigens (HBGAs). In this system, virus-like particles (VLPs) are used to attach to purified HBGAs, and antibodies that block binding of VLPs to HBGA are considered effectively neutralizing. Norovirus does not replicate in conventional cultured cells, although a recent report describes replication of some human noroviruses in stem cell-derived human enteroids (Ettayebi et al., 2016). The same principle of receptor blocking assay as a surrogate for neutralization also is used for hem-agglutination inhibition for viruses that bind to sialic acid, such as influenza viruses. Sialic acid molecules are common on airway epithelial cells and also on the surface of red blood cells (RBCs). Therefore, virus agglutination of RBCs can be used as a surrogate of virus attachment to respiratory epithelial cells. Antibodies that block virus-mediated agglutination of RBCs in vitro are termed hemagglutination-inhibiting (HAI) antibodies. HAI antibody titers have been validated as a correlate of protection by regulatory agencies for release of influenza vaccines (U.S. F.D.A., 2007).
Reporter Gene Systems
Variations of this technique include systems using green fluorescent protein, luciferase, or other reporter genes. These approaches are particularly helpful for standardization using validated assays and standardized reference strains in a GCLP-compliant environment, such as has been done in the HIV field with the TZM-bl assay or A3R5 assay systems (Binley et al., 2004; Sarzotti-Kelsoe et al., 2014). In the TZM-bl assay, engineered pseudovirions displaying an envelope (Env) of interest are used to deliver the tat gene to TZM-bl cells, which are grown from a CXCR4-positive HeLa cell clone that was engineered to express HIV receptor CD4 and co-receptor CCR5 (Wei et al., 2003). During the single-cycle infection that occurs in the TZM-bl cell, the viral Tat protein induces reporter gene expression in trans. Neutralization is scored as the reduction of Tat-regulated firefly luciferase reporter gene expression activity that is quantified as relative luminescence units (RLU) using a luminometer. This type of assay can be performed with large panels of reference strains in multi-well plates to achieve high-throughput capacity (deCamp et al., 2014; Mascola et al., 2005). The A3R5 cell system is similar, using expression of a Renilla luciferase reporter gene to induce expression by viral Tat protein in cis in a CEM human LCL that naturally expresses the receptor CD4 and co-receptor CXCR4 and that was engineered to express the co-receptor CCR5 (McLinden et al., 2013). In this system, engineered viruses that possess a reporter gene in the viral genome are used (Edmonds et al., 2010). This type of system allows pseudotyping of the reporter particles so that highly pathogenic organisms can be mimicked in biosafety level (BSL) 2 laboratories for agents that are typically restricted for use in BSL3 or BSL4 settings. This approach also allows conventional laboratories to study neutralization of viruses that, if possessed as a live agent, would require select agent registration or even to perform neutralization assays for viruses for which propagation is not permitted. For example, in 2012, when there was a moratorium on use of respiratory droplet-transmissible (rdt) live avian H5N1 influenza virus strains, human mAb-mediated neutralization of these viruses was studied using lentivirus-based pseudo-virions packaged with the hemagglutinin (HA) protein of the rdt viruses (Thornburg et al., 2013).
Assays for Viral Inhibition Activities Other Than Neutralization
More recently it has become apparent that many antibodies that do not exhibit neutralizing activity in the in vitro neutralization assays described above can still mediate therapeutic or protective effect in experimentally inoculated animals. Antibodies tag viruses and virus-infected cells for surveillance by cells of the immune system by interaction of cells with the fragment crystallizable (Fc) region of antigen-bound antibodies. The Fc regions of antibodies interact with a large number of Fc receptors on diverse immune cells, including macrophages, dendritic cells, neutrophils, and natural killer cells. In vitro assays to detect cell-mediated activity that is facilitated by antibodies are being performed with increasing precision. Such assays have been termed antibody-dependent cellular cytotoxicity (ADCC) assays, antibody-dependent cell-mediated virus inhibition (ADCVI) assays, and others. Surrogate assays for this type of activity using Fc dimer-binding activity and other bead-based assays have been grouped into a set of techniques that can be termed “system serology studies (Chung et al., 2015), a term designed to reflect the comprehensive and multifunctional panel of analysis that uses a discovery philosophy akin to that of the systems biology field.
Neutralization and Protection
There is a high level of interest in whether or not neutralizing assays correlate with protection in vivo. This correlation is a complex topic because the definition of in vivo protection is also complex. For instance, replication and pathogenesis of human viruses in small animals such as mice, rats, and guinea pigs may differ from that in larger animals or humans. Nonhuman primates are often the preferred animal model for virus replication and for testing therapeutic effects of human antibodies, but even these models are not perfect mimics of the human body. Genes encoding receptors on epithelial and endothelial cells and cells of the immune system differ to some extent between nonhuman primates and humans. Nevertheless, in some cases quantitative correlates have been determined for in vivo protection using neutralizing antibody assays. For instance, a quantitative level of serum neutralization titer for protection of the lower respiratory tract of small animals challenged with RSV was determined (Prince et al., 1985), and this correlate was used successfully to develop and test the RSV F protein-specific mAb palivizumab, the only mAb licensed for prevention of a virus infection. A sophisticated vocabulary about classes of correlates of immunity has been developed, principally by Plotkin, in a series of papers on the types of correlation (Heeney and Plotkin, 2006; Plotkin, 2008, 2010, 2013; Plotkin and Gilbert, 2012; Tomaras and Plotkin, 2017). This standardized nomenclature includes the term “correlate of protection” (CoP), which is a marker of immune function that statistically correlates with protection after vaccination, with two subtypes of CoP being (1) a “mechanistic correlate of protection” (mCoP) or (2) a “nonmechanistic correlate of protection” (nCoP) that does not cause protection but nevertheless predicts protection through its (partial) correlation with another immune response(s) that mechanistically protects.
Fundamental Principles of Neutralization
Many laboratories have contributed to discovery of mechanisms of human antibody-mediated neutralization. In this paper, I review the field, but focus on the function of antibodies that have been discovered in my group together with a diverse group of collaborators.
The Importance of Quaternary Structure
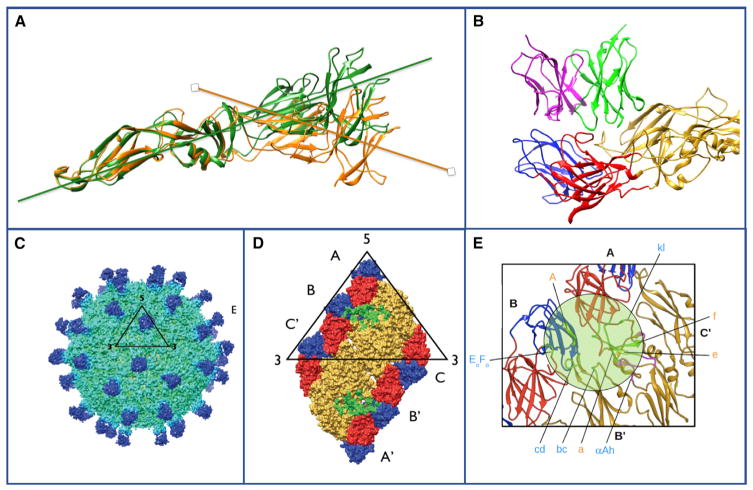
One of the most striking findings of studies in recent years is that many of the most potent neutralizing antibodies recognize complex quaternary structures on the surface of viruses. It would be convenient if linear peptides could be developed as vaccines or diagnostic epitopes for understanding antibody responses, and in fact comprehensive libraries of viral peptides have been developed as libraries that are helpful to demonstrate prior infection (Xu et al., 2015). Nevertheless, such linear peptides are rarely the target of neutralizing antibodies, and inoculating experimental animals with such peptides usually does not induce neutralizing antibodies. Therefore, it has long been known that neutralizing determinants are for the most part conformational in nature, and it was suspected that many are probably complex quaternary structures. This trend toward preferential quaternary epitope recognition is also the case for the most potent human mAbs isolated for many different pathogenic human viruses. For example, the dengue envelope protein has three domains in each protomer, and two protomers form head-to-tail dimers, then three dimers form rafts on the surface of viral particles (Kuhn et al., 2002). By large-scale isolation of human mAbs from persons with prior dengue infection, mAbs that sensitively detect each of these quaternary features have been discovered. For example, the human mAb 1F4 that is specific for dengue serotype 1 viruses (de Alwis et al., 2012) recognizes only a “bent” conformation of the domain I/II region on the dengue envelope protein (Fibriansah et al., 2014) (Figure 2A). This finding was interesting because this neutralizing antibody binds to intact dengue particles, but not to soluble extracellular recombinant forms of dengue serotype 1 envelope protein, in which the angle of the DI/II hinge is relaxed, as visualized in the atomic resolution crystal structure. Thus, there is a conformational determinant within one protomer of the dengue E protein at the DI/II hinge that is essential for recognition of this type of antibody. Next, some antibodies recognize two protomers that form the head-to-tail-oriented dimeric form of the envelope protein. For dengue virus, such antibodies have been shown in some cases to be exceptionally potent. For example, the serotype 2-specific human mAb 2D22 (de Alwis et al., 2012) recognizes the dimeric form of envelope on the surface of particles and achieves serotype 2 specificity by also interacting with DIII (Fibriansah et al., 2015a) (Figure 2B).
Figure 2. Quaternary Epitopes on Dengue Virus Particles.
(A) The domain I/II hinge angle in soluble recombinant dengue E protein differs from that in the virus. The structure of the human dengue serotype protein has been determined by crystallography (green, based on PDB: 10AN) or by cryo-EM (orange, based on EMDB: 2442). In the particle, the E protein is maintained with a “bent” conformation at the inter-domain hinge. In the crystal structure, the soluble protein achieves a more linear orientation (green).
(B) The human mAb 2D22 that is specific for serotype 2 dengue viruses engages a broad surface on the envelope protein, engaging multiple viral protein domains. The Fab is on top, with heavy chain in green and light chain in pink; the dengue E protein is on bottom with domain I in red, domain II in yellow, and domain III in blue. Based on PDB: 4UIF.
(C) Dengue virus 3-specific human mAb 5J7 recognizes a very complex quaternary epitope. The 9Å cryo-EM map of DENV3 complexed with Fab 5J7. The black triangle and numbers represent an icosahedral asymmetric unit and the icosahedral vertices, respectively.
(D) The epitope recognized by HMAb 5J7. One molecule of Fab 5J7 binds to three E proteins. Top left panel shows a raft of E proteins (containing two asymmetric units). The three individual E protein molecules in an asymmetric unit are labeled as A, B, and C, and the corresponding E proteins in the neighboring asymmetric unit as A’, B’, and C’. DI, DII, and DIII are colored in red, yellow, and blue, respectively.
(E) Enlarged view of the 5J7 epitope. The residues on the E protein that interact with heavy or light chains of the Fab are shown as green or magenta spheres, respectively. Based on PDB: 3J6U, the cryo-EM structure of dengue virus serotype 3 in complex with human antibody 5J7 Fab. All structures were determined in collaboration with the laboratory of Sheemei Lok.
An interesting class of antibody termed envelope dimer epitope (EDE) antibodies bind across the interface of two protomers in the dimer, and these antibodies can mediate cross-reactive binding and neutralization for diverse serotypes of dengue (Dejnirattisai et al., 2015). This type of quaternary epitope requires recognition of both protomers within the dimer (Rouvinski et al., 2015). Finally, a class of antibody for dengue recognizes an extremely complex quaternary epitope that spans two dimers and interacts with multiple domains within those dimers. The human mAb 5J7 is an example of this type of antibody, which only recognizes virion particles or VLPs and does not bind to recombinant envelope protein (de Alwis et al., 2012; Fibriansah et al., 2015b). Interestingly, the antibody is so sensitive to the conformation of the epitope that it recognizes only two of the six antigenic sites in each envelope protein raft (containing three dimers, and thus six E protein protomers), because of slight conformational variety in the other four sites (Figure 2C). For the related flavivirus Zika, a number of neutralizing human mAbs isolated from a subject following natural infection recognize diverse antigenic sites on the E protein (Sapparapu et al., 2016). Epitope mapping by alanine scanning mutagenesis and visualization of the structure of the antibody/virus complex by cryo-electron microscopy revealed that the most potent antibody, ZIKV-117, sits over the dimer-dimer interface on the surface of Zika particles (Hasan et al., 2017).
The Importance of Blocking Receptor Binding
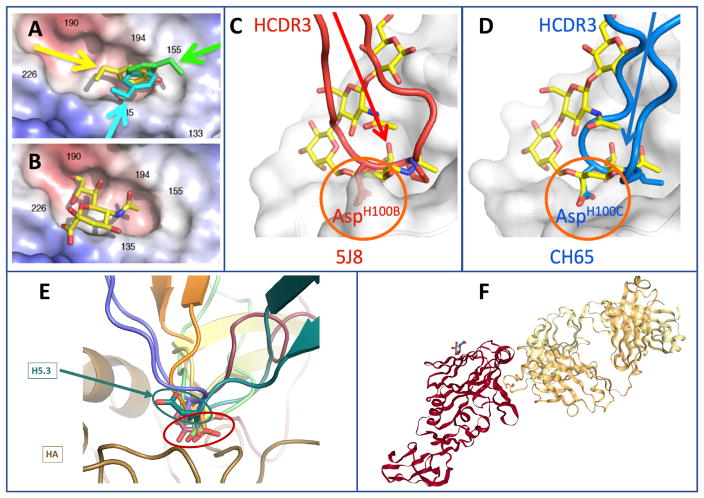
Physical blocking of virus binding to the receptor on human cells is one of the most important and common mechanisms of potent antibody-mediated virus neutralization. Antibodies that recognize the receptor binding domain (RBD) on viral surface attachment proteins can mediate very potent inhibition. For example, the most common type of strongly neutralizing antibody for influenza virus is directed to the RBD in the HA protein. Such antibodies bind near the top surface of the head domain of the HA protein that mediates attachment to the cellular receptor sialic acid. Antibodies that block receptor binding for diverse influenza type A viruses including those of subtype H1 (Krause et al., 2010, 2011a, 2011b; Tsibane et al., 2012; Xu et al., 2010; Yu et al., 2008b), H2 (Krause et al., 2012; Xu et al., 2013), H3 (Bangaru et al., 2016), H5 (Thornburg et al., 2013; Winarski et al., 2015), and H7 (Thornburg et al., 2016) have been isolated and characterized. High-resolution crystal structures of these antibodies in complex with soluble trimeric forms of HA (or recombinant HA head domain) have proven very useful for defining the chemical and physical principles underlying the molecular recognition of the RBD. Two principal protein-protein interactions have been identified as canonical in this region. First, if the antibody presents an aromatic residue at the tip of one of the hypervariable loops (CDRs), the occurrence of this residue is favorable because the aromatic amino acid, which can be phenylalanine or tyrosine, can form a pi-pi interaction with an aromatic residue in the base of the HA RBD (Xu et al., 2013). Molecular mimicry with receptor binding occurs because the insertion of a large hydrophobic amino acid into the RBD with a backbone carbonyl group makes interactions similar to those of the sialic acid carboxylate. A large number of antibodies have now been described with this feature (examples shown in Figures 3A and 3B; see also Lee and Wilson, 2015).
Figure 3. Canonical Modes of Interactions of Human Antibodies with Influenza HA Receptor Binding Domain.
(A) Prominent role for aromatic residues to mediate the interaction of human antibody CDRs with the HA RBD, in a way that mimics sialic acid interactions. The surface of the subtype H2 HA protein is shown in the region of the RBD. The angle of approach of the CDR of three different H2-reactive antibodies is indicated in yellow, cyan, or green, and the aromatic residue of the antibody on each mAb is shown.
(B) The position of the sialic acid receptor bound in the HA RBD is shown for comparison.
(C and D) Two human antibodies to influenza H1N1 virus HA RBD mimic the sialic acid receptor using an aspartate residue on the tip of a CDR loop. The sialic acid receptor is overlaid in yellow for comparison. The canonical interaction with Asp is highlighted in orange circle. (C) is based on PDB: 4M5Z, crystal structure of broadly neutralizing antibody 5J8 bound to 2009 pandemic influenza hemagglutinin, HA1 subunit. (D) is based on PDB: 5UGY, influenza hemagglutinin in complex with a neutralizing antibody.
(E) The HCDR3 of the H5 subtype-specific influenza HA RBD-reactive mAb H5.3 has an aspartate in the canonical position, but binds in an alternate fashion. H5.3 is indicated in green; conventional Asp containing antibodies is indicated in red. Figure is based on Winarski et al., 2015.
(F) Crystal structure of the broadly neutralizing antibody C05 bound to H3 influenza hemagglutinin, HA1 subunit. The antibody Fab (yellow structure, right) engages the RBD on the influenza HA protein (red structure, left), principally with a single CDR. Based on PDB: 4FP8. All structures were determined by the laboratories of Ian Wilson or Ben Spiller.
A second residue on the tip of antibody CDRs that has proven to facilitate interactions in the HA RBD is an aspartate residue, which interacts by favorable charge interactions with the HA protein. The carboxylic acid of a properly positioned aspartic acid in a CDR can mimic the carboxylate of sialic acid, and the backbone atoms mimic the acetamido groups of the receptor. A large number of antibodies with this residue have been identified and visualized in high-resolution structures as achieving important critical interactions (Figures 3C and 3D; see also Hong et al., 2013; Schmidt et al., 2013; Whittle et al., 2011; Wu and Wilson, 2017). Interestingly, although presentation of an aspartate residue in this position seems to be a rule that satisfies interaction, the neutralizing human antibody designated H5.3 has the aspartate in the right position in a CDR3, but the loop does not use the aspartate to interact in the canonical fashion (Thornburg et al., 2013; Winarski et al., 2015) (Figure 3E). Thus, although structural rules of engagement for antibodies can be identified, the structural features are not simply short sequences or single residues, but rather are elements that function as predicted only in proper context.
Interestingly, there is an antibody, F045-092 (selected from a phage-displayed library of human B cell variable genes), that possesses both the aromatic and aspartate residues in the proper position to bind to the HA RBD in mimicry of sialic acid binding (Lee et al., 2014). Crystal structures of F045-092 in complex with HAs from diverse H3N2 viruses revealed the structural basis for its neutralization breadth through insertion of its 23-residue HCDR3 into the RBD, allowing it to mimic sialic acid to a high degree (Lee et al., 2014). The carboxylate side chain of the antibody Asp100e closely aligns with the carboxylate of sialic acid that would be found in the same position and uses the same network of hydrogen bonds to bind to HA. The F045-092 HCDR3 creates an additional hydrogen bond between the Fab main chain using the Tyr100b to interact with a residue on HA.
One of the challenges for neutralizing influenza at the RBD is the high level of variability in the amino acid sequence of the surrounding rim of amino acids that encompass the approach to recessed RBD (see http://www.fludb.org). If an antibody interacts with a residue on the HA head that varies in circulating field strains, then that antibody is likely to be subtype specific, or even strain specific. This variability underlies much of the antigenic drift that occurs season to season in influenza A viruses. One engineering principle that could be used for more efficient neutralization would be to design biologics, such as antibodies, that interacted only with the base of the RBD, but did not bind to the more variable rim of the RBD. One very interesting broadly neutralizing antibody with an exceptionally long HCDR3, designated C05, essentially does achieve an interaction that focuses mostly on the base of the RBD (Ekiert et al., 2012) (Figure 3F). It is not understood at this time how rare such antibodies are, but they point the way to a possible path for rational design of new broadly neutralizing therapeutics. The HCDR3 loop of C05 possesses internal, self-stabilizing hydrogen bond interactions, similar to a β-hairpin, and provides extensive hydrogen bonding interactions with backbone atoms of the 130 loop of HA. Interesting approaches are being explored to use such stable HCDR3 loops as “mini-antibodies,” e.g., the use of computationally designed trimeric proteins mimicking C05 engagement of the HA RBD (Strauch et al., 2017).
The Generalizable Nature of Canonical Modes of Antibody-Virus Interactions
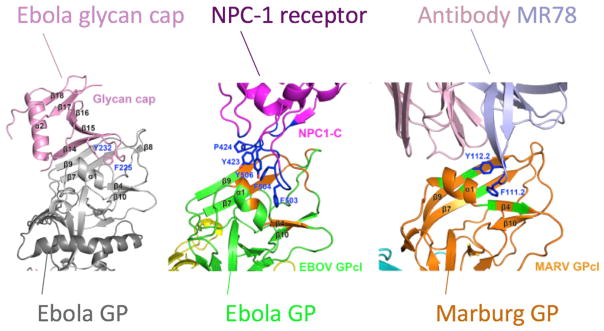
We are beginning to learn that potent neutralizing antibodies often use canonical modes of interaction that cross virus families. It is reasonable to think that the chemical and physical determinants of binding to a viral protein, such as the aromatic and aspartate residues discussed above, would be very specific to that one virus protein. However, the dominant role of aromatic residues in mediating antibody variable loops with RBDs also has proven highly pertinent for filovirus antibodies. Filoviruses, such as Marburg and Ebola viruses, bind to the Niemann-Pick disease, type C1 (NPC1) protein as a receptor in the endosome. The viruses attach to this receptor using a RBD in the surface glycoprotein (GP). Of the 50 human mAbs isolated from a single survivor of natural Marburg infection, 18 were found to be neutralizing (Flyak et al., 2015). Each of these antibodies competed for binding on the GP at a region that then was found to be the RBD when structure of the complex was determined (Hashiguchi et al., 2015). Interestingly, single-particle EM reconstruction studies of a panel of these antibodies showed that they could approach the RBD with varying angles or binding poses, but they all interacted to arrive at a common final solution, which was to place a CDR loop into the RBD. Atomic resolution structure of one of the antibodies on both Marburg and Ebola GP showed that the CDR loop presented an aromatic residue that established a pi-pi relationship with aromatic residues in the bottom of the RBD (Hashiguchi et al., 2015). Essentially, this is the same design principle as discussed above for influenza-neutralizing antibodies. Most of the isolated Marburg mAbs do not interact with the RBD of uncleaved Ebola GP, but one antibody, designated MR72, did recognize Ebola GP even in the presence of the Ebola glycan cap that partially shields the Ebola RBD (Flyak et al., 2015). Based on this canonical mode of interaction with an aromatic residue and the capacity of MR72 to bind to Ebola GP, a bispecific antibody comprising the Marburg MR72 Fab and another antibody (to NPC1 receptor or to Ebola GP glycan cap) was used to deliver a construct to the cell endosome. In that cell compartment, the Marburg antibody could recognize the RBD in the cleaved Ebola GP (which is protected by the viral glycan cap prior to cleavage) and neutralize Ebola virus (Wec et al., 2016). Even more remarkable, a three-way structural mimicry for this interaction using these principles has now been described. The Ebola virus glycan cap on the surface GP protects the RBD of Ebola virus until exposure in the endosome, using aromatic residues that interact with an aromatic amino acid in the GP RBD in the same way that the MR78 human mAb does (Hashiguchi et al., 2015). Recently, Gao and colleagues showed that the virus interacts with the NPC1 receptor using the same principles employing aromatic-rich interfaces (Wang et al., 2016) (Figure 4). It is very unusual to demonstrate a three-way structural mimicry of this type in biology; in this case a viral protein, a human antibody, and a human receptor all use the same type of interacting residues to engage the RBD of a viral protein.
Figure 4. Interactions of Ebola GP RBD in Crystal Structures of Protein Complexes Reveal a Three-Way Structural and Functional Mimicry.
The viral GP glycan cap, the human protein NPC-1 (the filovirus receptor), and the MR78 human mAb all use aromatic-rich loops to bind. Based on Wang et al., 2016.
The Search for “Universal” Epitopes Inducing Protective Antibodies
There is a strong desire in the field of immunotherapeutics to identify highly conserved epitopes in antigenic sites that can be targeted by antibodies or used as vaccine antigens. This idea has spurred a broad search for vaccine antigens that can induce broad, cross-reactive, or “universal” antibodies. For example, antibodies that equally recognized RSV fusion proteins of both antigenic subgroup A and B have been discovered, and antibodies that recognize either the pre-fusion (Mousa et al., 2017) or post-fusion (Barbas et al., 1992; Crowe et al., 1994) forms of the protein also exist. This principle is most well developed in the influenza vaccine design field. Several groups identified human mAbs that neutralized viruses of more than one subtype of influenza (H1 and H5 viruses) but did not block RBD (in other words, they did not have HAI activity) (Ekiert et al., 2009; Sui et al., 2009; Throsby et al., 2008). Crystal structures of HA/antibody complexes revealed that these antibodies were interacting with the stem domain of influenza HA protein, which is much more conserved than the head domain. Even more interesting, many of these stem-binding antibodies were found to have a very canonical mode of interaction in which only the heavy chain interacted with a hydrophobic depression in the stem protein domain, and such antibodies were encoded by a single heavy-chain variable region designated VH1-69, which is a germ-line gene segment that encodes a hydrophobic CDR. Since the initial discovery of these mAbs, it has become apparent that most humans have B cells encoding this type of antibody. Subsequently, additional, more complex interactions in the stem have been identified with even broader antibodies that recognize viruses of different major phylogenetic groups (Corti et al., 2011). Additional VH genes encoding stem antibodies have been identified, and even for VH-1-69-encoded antibodies, somatic mutation in antibody lineages can achieve novel and somewhat non-canonical interactions in this region (Pappas et al., 2014). Several of these antibodies are being tested as experimental therapeutics in humans because of their broad anti-influenza activity. Computationally designed small proteins that mimic stem antibodies also have been reported (Fleishman et al., 2011; Koday et al., 2016).
These principles can be exploited not just for antibody development, but also for engineering antigens with a view to using them as vaccines to induce this type of antibody. This field is most developed for “universal flu” vaccine antigens. Progress has been made in proof-of-principle studies, but it has been difficult to faithfully recapitulate the conformation of the HA stem simply by truncating the protein (termed a “headless” protein approach) (Steel et al., 2010). Complex engineering protocols to stabilize such stem vaccines are being pursued currently. More recently, investigators have developed chimeric HA proteins that present the conserved stem region of influenza type A or B viruses in molecules containing an avian influenza HA head domain for which immunity is not common in the human population (Ermler et al., 2017; Hai et al., 2012). The rationale is that pre-existing human memory B cells that recognize conserved epitopes in the stem will be boosted in a memory response with such chimeric proteins (while the primary B cell response to the head domain will be a less significant component of the response).
Targeting the stem region of surface GPs that are more conserved in sequence than head domains also has proven to be a useful approach for finding broadly neutralizing antibodies for viruses other than influenza. There is an entire enterprise of developing and studying HIV gp41-reactive antibodies to this region, which for HIV has been designated the membrane-proximal external region (MPER). Very broad and potent neutralizing antibodies such as 2F5 (Muster et al., 1993), 4E10 (Zwick et al., 2004), 10E8 (Huang et al., 2012), and others have been studied in great detail. Some of these antibodies appear to have some autoreactive properties, likely due to interactions of the antibodies with lipid moieties in the membrane of viruses or virus-infected cells. MPER antibodies also have been described for filoviruses, and these are very exciting because they exhibit breadth of neutralization for diverse species of Ebola viruses (Bornholdt et al., 2016; Flyak et al., 2016).
Cross-Reactive Responses
Given that naturally occurring human antibodies that recognize antigenically diverse strains for a particular virus have been isolated, it was natural to explore whether individual mAbs could be found that cross-react with different viruses within a virus family. Increasingly, we see that such broad molecular recognition is possible. For example, human mAbs that neutralize all poxviruses that are commonly pathogenic for humans, including variola (smallpox), monkeypox, cowpox, and vaccinia (“smallpox vaccine”) viruses, have been isolated (Gilchuk et al., 2016). This type of cross-reactive antibody was found within responses directed not just to one but to many different virus proteins on both of the two major structurally distinct forms of poxviruses (mature virions [MV] and enveloped virions [EV]). MAbs isolated from humans with prior chikungunya virus infection (Smith et al., 2015) included mAbs that recognize all major pathogenic alphaviruses (Fox et al., 2015). Antibodies that cross-react with the fusion proteins of the two diverse paramyxoviruses RSV and human metapneumovirus, both of which cause pediatric pneumonia and wheezing, have been reported (Corti et al., 2013; Wen et al., 2017). It has long been known that antibodies to the fusion loop antigenic site on flavivirus E proteins cross-react with most other flaviviruses, and in fact such antibodies can enhance virus replication of the second, heterologous infection (termed antibody-dependent enhancement, or ADE). However, it also has been possible to identify human mAbs that neutralize diverse flaviruses, such as dengue and Zika (Barba-Spaeth et al., 2016).
Reconstructing the Complex Nature of Polyclonal Antibodies in Serum
Although studies of the structural basis of virus neutralization by human antibodies de facto require the use of mAbs, it is important to realize that this type of study allows only a “snapshot” of the response happening in vivo. When enough antibodies are obtained from a single individual to a single agent, it becomes clear that in most cases some human B cells in the response can recognize virtually every amino acid on the surface of viral proteins. For example, mAbs for chikungunya from a single individual mapped by saturation alanine scanning mutagenesis decorate nearly the entire surface of the E2 domain of the virus (Smith et al., 2015) (Figure 5). This polyclonal response can be very beneficial if diverse antibody clones exhibit cooperative effects in neutralization or protection. For example, optimal protection against poxviruses in vivo minimally requires a cooperative mixture of 4–6 mAbs, because of the complexity of neutralizing two forms of virion particle and the use of multiple, redundant adhesion factors on each form of particle (Gilchuk et al., 2016). On the other hand, non-neutralizing mAbs that compete for binding to the site on the RSV F protein recognized by the licensed neutralizing mAb palivizumab also have been reported (Mousa et al., 2016). In principle, such antibodies could inhibit the protective effects of therapeutic antibodies. Thus, future research in this field should include studies to understand the complex biology of mixtures of antibodies.
Figure 5. Structural Analysis of Residues Important for Binding of Diverse Human mAbs from One Individual to Chikungunya E2 Protein Reveal Broad Coverage of the Protein Surface.
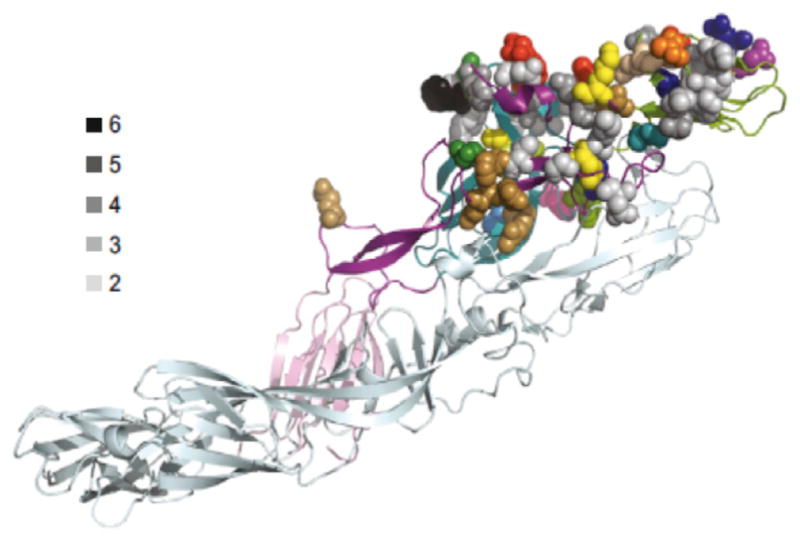
The location of residues required for mAb binding (mapped by alanine scanning mutagenesis) of 20 antibodies is shown, mapped onto the crystal structure of the mature envelope glycoprotein complex (PDB: 3N41). A side view of a ribbon trace of a single heterodimer of E1/E2 is shown, with E1 in light cyan and the domains of E2 colored. The side chains of the amino acids required for mAb binding are shown as space-filling forms and individually color coded for each of the 20 individual antibodies. Residues that bound more than one antibody are indicated by varying shades of gray. Based on Smith et al., 2015.
Genetics of Neutralizing Antibodies
Techniques for studying the genetics of antibody variable genes encoding antibodies have become increasingly sophisticated. As discussed above, it has been possible to obtain and analyze the genetic sequence encoding mAbs from single B cells or other mAb techniques using conventional Sanger sequencing since the late 1980s. More recently, large-scale, next-generation sequencing of antibody variable gene repertoires, first reported in zebrafish (Weinstein et al., 2009) and then applied to human samples, have allowed for the sequencing of millions or even billions of variable gene sequences from a human blood sample. Descriptive studies using deep sequencing revealed interesting features of antibody repertoires, including (1) the common origin of long HCDR3 antibodies in naive B cells (D3-JH6 recombinants with long N-addition regions; Briney et al., 2012a), (2) hotspot regions for insertions and deletions during somatic hypermutation (Briney et al., 2012b), (3) a relatively high frequency of VH(D-D) JH recombinants in the human peripheral blood antibody repertoire (Briney et al., 2012c), and (4) tissue-specific features of antibody repertoires (Briney et al., 2014). Coupling single B cell techniques for antigen-specific mAb discovery with next-generation sequencing of antibody variable gene repertoires of the same subject began to allow us to identify antibody clonal lineages that were diversifying by somatic hypermutation (Krause et al., 2011b). Immediately it became apparent that not only do antibody clones diversify by very robust introduction of somatic mutations within a lineage (intraclonal diversification), but also independent, genetically distinct germline-encoded B cell clones directed to a common epitope converge on common contact residues through somatic hypermutation (Krause et al., 2011b). Such studies identify specific variable genes or protein motifs that are associated with specificity. For instance, five different human mAbs identified from a single individual to influenza H1 HA were all encoded by a single VH gene, VH3-7 (Krause et al., 2011b). Deep sequencing identified “siblings” of these anti- bodies in the person’s immune repertoire. This same type of VH association has been described for influenza HA stem antibodies, as discussed above, for VH1-69 and VH3-30. In the HIV antibody field, there is an entire class of CD4-binding-site antibodies that have been designated VRC01 class antibodies, and these are typically encoded by the VH1-02 allele. This finding is particularly important, because it explains why some humans can readily make such antibodies, but experimental animals like mice and macaques cannot. Essentially, the germline genes encoding homologous antibodies in the model animals do not bind the CD4 binding site. As larger and larger numbers of antibodies are made, common sequences can be seen across individuals, which could be termed public clonotypes, using vocabulary similar to that used in studies of T cells. For example, a potent neutralizing antibody designated ZIKV-116 that recognizes DIII on the ZIKV E protein was described recently (Sapparapu et al., 2016), and subsequently, similar antibodies also encoded by the same the VH, D, and JH variable gene segments in other individuals were reported (Robbiani et al., 2017). Over time, it is expected that we will be able routinely to assign specificity for many antibodies just based on their sequence. Even more exciting, computational methods for predicting structure of antibodies using sequences are emerging. As an example, the ROSETTA modeling software suite was used to redesign the broadly neutralizing human HIV-specific mAb PG9 for increased potency and breadth. A single amino acid change in the HCDR3 that increased thermal stability without altering the contact residues converted the mode of PG9 binding to be glycan independent (Willis et al., 2015). This type of structure prediction, coupled with a novel, position-specific, structure-based scoring matrix, enabled a deep-sequencing search of antibody repertoires in HIV-naive subjects to find PG9-like antibodies that bind to and neutralize HIV (Willis et al., 2016). Such predictive techniques are not deterministic and are relatively inefficient at present, but we can see a time in the future when antibody discovery could be done simply by exploring antibody sequence databases. Technologies in the field of repertoire sequencing are evolving rapidly, allowing ever deeper sequencing. In fact, a project to sequence all of the B and T cell receptors in the human population, an effort termed the Human Immunome Program, has been initiated (Crowe and Koff, 2015).
Implication for “Reverse Vaccinology” Approaches
Understanding the “rules of engagement” for protective antibodies is beginning to help guide structure-based vaccine design (“reverse vaccinology”). The central rationale in this approach is that broadly protective potent antibodies that are naturally occurring in humans reveal antigenic sites and epitopes that could serve as components of preventative vaccines to induce that type of antibody. Proof-of-principle for this approach was first shown using the epitope for the licensed anti-RSV antibody palivizumab (termed antigenic site II) (Correia et al., 2014). Computational design of an α-helical protein scaffold that presented the correct conformation of the minimal palivizumab epitope (a small helix-loop-helix structure) discovered a structure that induced neutralizing antibodies to the epitope that bound in a similar manner to that of palivizumab (Correia et al., 2014). Progress is being made with similar approaches for HIV vaccine design, based on the epitopes for broad and potent HIV-neutralizing antibodies (Briney et al., 2016; Escolano et al., 2016; Steichen et al., 2016; Tian et al., 2016).
Conclusion
There have been major technical advances in recent years in the isolation of human mAbs to viruses. Careful studies of neutralization and other antibody-mediated antiviral functions reveal that humans can make very diverse and potent antibodies for virtually any pathogenic organism. EM and crystallography studies of antigen-antibody complexes have begun to elucidate the structural basis for molecular recognition of viruses and established some canonical modes of interaction. Deep sequencing of antibody variable gene repertoires shows the tremendous complexity of human B cell responses. As we begin to synthesize these diverse types of information, the goal of accomplishing rational development of new vaccines with reverse vaccinology principles for structure-based design seems increasingly more realistic.
Acknowledgments
The ideas presented here were birthed and shaped in a large community of scientists. First, I want to thank the many current and previous members of my laboratory who have created much of the new knowledge reviewed here. I also am deeply indebted to many collaborators and colleagues in this field who designed and performed much of the work I review here, including many current and past members of the laboratories of Ian Wilson, Andrew Ward, and Erica Saphire at Scripps; Sheemei Lok at Duke NUS Singapore; Ben Spiller and Jens Meiler at Vanderbilt; Ted Jardetsky at Stanford; Michael Diamond and Daved Fremont at Washington University in St. Louis; Michael Rossmann and Richard Kuhn at Purdue; Ben Doranz at Integral Molecular; Peter Palese, Adolfo García-Sastre, and Chris Basler at Mount Sinai, New York; and Ralph Baric and Aravinda de Silva at University of North Carolina, Chapel Hill. The work was supported by NIH grants R01 AI127828, R01 AI114816, U19 AI109711, and U19 AI117905 and NIH contract HHSN272201400024C. J.E.C. holds the Ann Scott Carell Chair at Vanderbilt University Medical Center. J.E.C. is a member of the scientific advisory boards of PaxVax, Meissa Vaccines, GigaGen, and CompuVax and has participated in advisory boards for Sanofi, Novavax, and Takeda. His laboratory has had recent sponsored research agreements with Moderna and Sanofi. His laboratory has current or recent federal consortium grants in which commercial parties are collaborators, including Inovio, Medimmune, Profectus, Arbutus, Avatar, and Moderna. Vanderbilt University has licensed materials for which he is inventor or co-inventor to GlaxoSmithKline, Takeda, Sanofi, Mapp Bio-pharmaceutical, Ridgeback Biotherapeutics, Novavax, Kerafast, and NewLink Genetics.
References
- Babcook JS, Leslie KB, Olsen OA, Salmon RA, Schrader JW. A novel strategy for generating monoclonal antibodies from single, isolated lymphocytes producing antibodies of defined specificities. Proc Natl Acad Sci USA. 1996;93:7843–7848. doi: 10.1073/pnas.93.15.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S, Nieusma T, Kose N, Thornburg NJ, Finn JA, Kaplan BS, King HG, Singh V, Lampley RM, Sapparapu G, et al. Recognition of influenza H3N2 variant virus by human neutralizing antibodies. JCI Insight. 2016;1:e86673. doi: 10.1172/jci.insight.86673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Lorière E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Crowe JE, Jr, Cababa D, Jones TM, Zebedee SL, Murphy BR, Chanock RM, Burton DR. Human monoclonal Fab fragments derived from a combinatorial library bind to respiratory syncytial virus F glycoprotein and neutralize infectivity. Proc Natl Acad Sci USA. 1992;89:10164–10168. doi: 10.1073/pnas.89.21.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, Wollen SE, et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 2016;351:1078–1083. doi: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz DR, Horton AP, Wine Y, Lavinder JJ, Georgiou G, Marcotte EM. Proteomic identification of monoclonal antibodies from serum. Anal Chem. 2014;86:4758–4766. doi: 10.1021/ac4037679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS ONE. 2012a;7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Crowe JE., Jr Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun. 2012b;13:523–529. doi: 10.1038/gene.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Hicar MD, Thomas JW, 2nd, Crowe JE., Jr Frequency and genetic characterization of V(DD)J recombinants in the human peripheral blood antibody repertoire. Immunology. 2012c;137:56–64. doi: 10.1111/j.1365-2567.2012.03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, Finn JA, McKinney BA, Crowe JE., Jr Tissue-specific expressed antibody variable gene repertoires. PLoS ONE. 2014;9:e100839. doi: 10.1371/journal.pone.0100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell. 2016;166:1459–1470. e1411. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015;163:988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- Crowe JE, Jr, Koff WC. Deciphering the human immunome. Expert Rev Vaccines. 2015;14:1421–1425. doi: 10.1586/14760584.2015.1082427. [DOI] [PubMed] [Google Scholar]
- Crowe JE, Jr, Murphy BR, Chanock RM, Williamson RA, Barbas CF, 3rd, Burton DR. Recombinant human respiratory syncytial virus (RSV) monoclonal antibody Fab is effective therapeutically when introduced directly into the lungs of RSV-infected mice. Proc Natl Acad Sci USA. 1994;91:1386–1390. doi: 10.1073/pnas.91.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2014;88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, Wieczorek L, Brown B, Polonis V, West JT, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408:1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermler ME, Kirkpatrick E, Sun W, Hai R, Amanat F, Chromikova V, Palese P, Krammer F. Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse Model. J Virol. 2017;91:e00286–e17. doi: 10.1128/JVI.00286-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458. e1412. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, et al. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015a;349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, Lok SM. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat Commun. 2015b;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen X, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, et al. Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebolavirus infection. Cell. 2016;164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MK, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell. 2015;163:1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk I, Gilchuk P, Sapparapu G, Lampley R, Singh V, Kose N, Blum DL, Hughes LJ, Satheshkumar PS, Townsend MB, et al. Cross-neutralizing and protective human antibody specificities to poxvirus infections. Cell. 2016;167:684–694. e689. doi: 10.1016/j.cell.2016.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, et al. A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat Commun. 2017;8:14722. doi: 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE, Jr, Saphire EO. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell. 2015;160:904–912. doi: 10.1016/j.cell.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney JL, Plotkin SA. Immunological correlates of protection from HIV infection and disease. Nat Immunol. 2006;7:1281–1284. doi: 10.1038/ni1206-1281. [DOI] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE, Jr, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang AS, Barbas CF, Janda KD, Benkovic SJ, Lerner RA. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc Natl Acad Sci USA. 1991;88:4363–4366. doi: 10.1073/pnas.88.10.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koday MT, Nelson J, Chevalier A, Koday M, Kalinoski H, Stewart L, Carter L, Nieusma T, Lee PS, Ward AB, et al. A computationally designed hemagglutinin stem-binding protein provides in vivo protection from influenza independent of a host immune response. PLoS Pathog. 2016;12:e1005409. doi: 10.1371/journal.ppat.1005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Krause JC, Tumpey TM, Huffman CJ, McGraw PA, Pearce MB, Tsibane T, Hai R, Basler CF, Crowe JE., Jr Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol. 2010;84:3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011a;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, Crowe JE., Jr Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011b;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, Ramos I, Fernandez-Sesma A, Edwards KM, García-Sastre A, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. 2012;86:6334–6340. doi: 10.1128/JVI.07158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrick JW, Danielsson L, Brenner CA, Abrahamson M, Fry KE, Borrebaeck CA. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun. 1989;160:1250–1256. doi: 10.1016/s0006-291x(89)80138-x. [DOI] [PubMed] [Google Scholar]
- Laurie KL, Engelhardt OG, Wood J, Heath A, Katz JM, Peiris M, Hoschler K, Hungnes O, Zhang W, Van Kerkhove MD CONSISE Laboratory Working Group participants. International laboratory comparison of influenza microneutralization assays for A(H1N1)pdm09, A(H3N2), and A(H5N1) influenza viruses by CONSISE. Clin Vaccine Immunol. 2015;22:957–964. doi: 10.1128/CVI.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, Lanzavecchia A. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, D’Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- McLinden RJ, Labranche CC, Chenine AL, Polonis VR, Eller MA, Wieczorek L, Ochsenbauer C, Kappes JC, Perfetto S, Montefiori DC, et al. Detection of HIV-1 neutralizing antibodies in a human CD4+/CXCR4+/CCR5+ T-lymphoblastoid cell assay system. PLoS ONE. 2013;8:e77756. doi: 10.1371/journal.pone.0077756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa JJ, Sauer MF, Sevy AM, Finn JA, Bates JT, Alvarado G, King HG, Loerinc LB, Fong RH, Doranz BJ, et al. Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein. Proc Natl Acad Sci USA. 2016;113:E6849–E6858. doi: 10.1073/pnas.1609449113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE., Jr A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol. 2017;2:16271. doi: 10.1038/nmicrobiol.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis M, Tran K, Bale S, Phad GE, Guenaga J, Wilson R, Soldemo M, McKee K, Sundling C, Mascola J, et al. HIV-1 receptor binding site-directed antibodies using a VH1-2 gene segment orthologue are activated by Env trimer immunization. PLoS Pathog. 2014;10:e1004337. doi: 10.1371/journal.ppat.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. Complex correlates of protection after vaccination. Clin Infect Dis. 2013;56:1458–1465. doi: 10.1093/cid/cit048. [DOI] [PubMed] [Google Scholar]
- Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. 2012;54:1615–1617. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince GA, Hemming VG, Horswood RL, Chanock RM. Immunoprophylaxis and immunotherapy of respiratory syncytial virus infection in the cotton rat. Virus Res. 1985;3:193–206. doi: 10.1016/0168-1702(85)90045-0. [DOI] [PubMed] [Google Scholar]
- Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J Exp Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, Schaefer-Babajew D, Avila-Rios S, Nogueira L, Patel R, et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell. 2017;169:597–609. e511. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Sapparapu G, Fernandez E, Kose N, Bin Cao Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Pre-configuration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Ma L, He X, Wang X, Wang P, Zhou L, Yao X. Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST. Theor Biol Med Model. 2014;11:30. doi: 10.1186/1742-4682-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Crowe JE., Jr Use of human hybridoma technology to isolate human monoclonal antibodies. Microbiol Spectr. 2015;3:AID-0027–AID-2014. doi: 10.1128/microbiolspec.AID-0027-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE., Jr Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, Crowe JE., Jr Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J Infect Dis. 2013;207:1898–1908. doi: 10.1093/infdis/jit119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE., Jr Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol. 2014;88:12233–12241. doi: 10.1128/JVI.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, Khomandiak S, Ashbrook AW, Kahle KM, Fong RH, et al. Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against chikungunya virus. Cell Host Microbe. 2015;18:86–95. doi: 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, García-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1:e00018–e10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch EM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, et al. Computational design of trimeric influenza-neutralizing proteins targeting the hemag-glutinin receptor binding site. Nat Biotechnol. 2017;35:667–671. doi: 10.1038/nbt.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szardenings M, Collins J. A phasmid optimised for protein design projects: pMAMPF. Gene. 1990;94:1–7. doi: 10.1016/0378-1119(90)90460-9. [DOI] [PubMed] [Google Scholar]
- Thornburg NJ, Nannemann DP, Blum DL, Belser JA, Tumpey TM, Deshpande S, Fritz GA, Sapparapu G, Krause JC, Lee JH, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest. 2013;123:4405–4409. doi: 10.1172/JCI69377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, Bombardi RG, Yu Y, Graham S, Branchizio A, et al. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest. 2016;126:1482–1494. doi: 10.1172/JCI85317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. Hetero-subtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Cheng C, Chen X, Duan H, Cheng HL, Dao M, Sheng Z, Kimble M, Wang L, Lin S, et al. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 2016;166:1471–1484. e1418. doi: 10.1016/j.cell.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Plotkin SA. Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol Rev. 2017;275:245–261. doi: 10.1111/imr.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Jr, Wilson IA, Basler CF. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S.F.D.A. Guidance for Industry: Clinical Data Needed to Support the Licensure of Seasonal Inactivated Influenza Vaccines. F.D.A. U.S. Department of Health and Human Services, Center for Biologics Evaluation and Research; 2007. https://www.fda.gov/biologicsbloodvaccines/guidancecomplianceregulatoryinformation/guidances/vaccines/ucm074794.htm. [Google Scholar]
- Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec AZ, Nyakatura EK, Herbert AS, Howell KA, Holtsberg FW, Bakken RR, Mittler E, Christin JR, Shulenin S, Jangra RK, et al. A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science. 2016;354:350–354. doi: 10.1126/science.aag3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, Crowe JE., Jr Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003;275:223–237. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE, Jr, Jardetzky TS. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol. 2017;2:16272. doi: 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JR, Sapparapu G, Murrell S, Julien JP, Singh V, King HG, Xia Y, Pickens JA, LaBranche CC, Slaughter JC, et al. Redesigned HIV antibodies exhibit enhanced neutralizing potency and breadth. J Clin Invest. 2015;125:2523–2531. doi: 10.1172/JCI80693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JR, Finn JA, Briney B, Sapparapu G, Singh V, King H, LaBranche CC, Montefiori DC, Meiler J, Crowe JE., Jr Long antibody HCDR3s from HIV-naïve donors presented on a PG9 neutralizing antibody background mediate HIV neutralization. Proc Natl Acad Sci USA. 2016;113:4446–4451. doi: 10.1073/pnas.1518405113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE, Jr, Spiller BW. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci USA. 2015;112:9346–9351. doi: 10.1073/pnas.1502762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Wilson IA. A perspective on the structural and functional constraints for immune evasion: Insights from influenza virus. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.06.015. Published online June 23, 2017. http://dx.doi.org/10.1016/j.jmb.2017.06.015. [DOI] [PMC free article] [PubMed]
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008a;336:142–151. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008b;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Komori HK, Stanfield RL, Church S, Wang M, Parren PW, Kunert R, Katinger H, Wilson IA, Burton DR. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J Virol. 2004;78:3155–3161. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]