Abstract
Nucleic acid aptamers, often termed “chemical antibodies”, are functionally comparable to traditional antibodies, but offer several advantages including their relatively small physical size, flexible structure, quick chemical production, versatile chemical modification, high stability, and lack of immunogenicity. In addition, many aptamers internalize upon binding to cellular receptors making them useful targeted delivery agents for siRNAs, microRNAs and conventional drugs. However,Ge several crucial factors, such as their inherent physicochemical characteristics and lack of safety data, have delayed the clinical translation of therapeutic aptamers. This review discusses these challenges, highlighting recent clinical developments and technological advances that have revived impetus for this promising class of therapeutics.
TOC image
Nucleic acid aptamers offer several advantages over traditional antibodies, but their clinical translation has been delayed by several factors including insufficient potency, lack of safety data and high production costs. Here, Zhou and Rossi provide an overview of aptamer generation, focusing on recent technological advances, clinical development, as well as challenges and lessons learned.
Introduction
Nucleic acid aptamers are short, single-stranded (ss) DNA or RNA molecules that are selected for binding to a specific target. In 1990, several groups isolated the first RNA aptamers. In Tuerk and Gold’s seminal report1, two high-affinity RNA ligands for the T4 DNA polymerase were enriched from an 8-nucleotide (nt) random region library using an in vitro procedure. Such a randomized library theoretically contains 4N individual sequences that can be simultaneously tested in one tube. Candidate binding sequences are then subjected to iterative selection rounds to increase high-affinity species, until they eventually dominate in the library. This procedure was thus named the “systematic evolution of ligands by exponential enrichment,” (SELEX). Around the same time, Robertson and Joyce devised a similar in vitro selection method for evolution of an RNA enzyme that specifically cleaved ssDNA2. Working independently, Ellington and Szostak also demonstrated the evolutionary power of this procedure3. Starting with a combinatorial pool of 1013 different sequences, they successfully identified RNA molecules capable of specifically binding to small organic dyes. Ellington coined the term “aptamer,” derived from the combination of the Latin word aptus (“to fit”) and the Greek word meros (“part”). Together, these elegant studies led to a generalizable method for in vitro generation of high-affinity aptamers that remains in common use today. To date, thousands of aptamers have been evolved against a wide variety of targets, including small metal ion and organic molecules, peptides, proteins, viruses, bacteria, whole cells, and even targets within live animals.
Aptamers typically contain defined structures, due to their propensity to form complementary base pairs. They can fold into various secondary structures (e.g., stem, loop, bugle, pseudoknot, G-quadruplex, and kissing hairpin)4; in turn, the collection of these secondary structures can form unique three-dimensional (3D) structures capable of specific molecular recognition of their cognate targets. These 3D interactions, including hydrophobic and electrostatic interactions, hydrogen bonding, van der Waals forces, shape complementarity, and base stacking, are essential for aptamer binding affinity and specificity5. Similar to the way antibodies bind to antigens, the specific 3D interaction drives the formation of aptamer-target complexes6. This complexity confers comparable binding affinity and specificity, and even superior advantages over antibodies (Box 1, Table 1). Aptamers are capable of distinguishing between closely related molecules, such as conformational isomers7, targets containing different functional groups8, 9, or even an amino acid mutation10.
Box 1: Nucleic acid aptamers versus protein antibodies.
Aptamers possess many attractive features. For example, compared to bulky antibodies (150–180 kDa, 15 nm), aptamers have a smaller size (6–30 kDa, 2 nm) and flexible structure, allowing them to bind to smaller targets or some hidden binding domains which are inaccessible for the larger antibodies. Larger antibodies may exhibit limited tissue penetration, particularly in solid tumors, which may compromise their therapeutic efficacy. Although their small size makes aptamers susceptible to kidney filtration and a short circulation time in vivo, rational chemical modification and conjugation can improve the pharmacokinetics profile and prolong half-life in vivo. Compared to the lengthy, laborious and expensive in vivo screening involved in antibody generation, aptamers can be isolated within days via a cost-efficient in vitro selection procedure. Because the immune response results in antibody production, its targets must trigger a strong reaction from the immune system. In principle, aptamers can be selected against an infinite spectrum of targets, including toxic and non-immunogenic molecules that antibodies cannot recognize. Furthermore, aptamers have many manufacturing benefits, particularly for production under current Good Manufacturing Practice (cGMP) standards. The production of antibodies involves animals and many different cell colonies, which offer opportunities for contamination and high batch-to-batch variation. In contrast, the manufacture of aptamers is completely in vitro, relying on a chemical synthesis procedure with no or lower risk of contamination. Moreover, the propensity of aptamers to form complementary base pairs confers additional benefits, in that the function of aptamers can be modulated in vivo using antidote oligonucleotides, which disrupt aptamer function by base pairing with the active motifs of the aptamers. In this way, a rationally designed antidote may allow precise control of the therapeutic effect. A comprehensive comparison of the advantages and disadvantages of nucleic acid aptamers versus antibodies is shown in Table 1.
Table 1.
Comparison of nucleic acid aptamers and protein antibodies
| Criteria | Aptamers | Antibody |
|---|---|---|
| Basic composition |
|
|
| Materials |
|
|
| Molecular weight/size |
|
|
| Secondary structure |
|
|
| Binding pattern/action mechanism |
|
|
| Affinity |
|
|
| Specificity |
|
|
| Potential targets |
|
|
| Generation/discovery |
|
|
| Manufacture/cost |
|
|
| Batch-to-batch variation |
|
|
| Physical/thermal stability |
|
|
| Chemical modification and conjugation |
|
|
| Tissue uptake/Penetration |
|
|
| Immunogenicity |
|
|
| Nuclease degradation |
|
|
| Kidney filtration |
|
|
| Patents/distribution |
|
|
| Development/market |
|
|
However, the development of clinically effective therapeutic aptamers has lagged far behind that of therapeutic antibodies11, 12, which still dominate the global medicine marketplace13. For example, the anti-vascular endothelial growth factor (VEGF) aptamer pegaptanib (Macugen®), the only federally approved aptamer drug14, 15, has been superseded by anti-VEGF monoclonal antibodies, such as bevacizumab and ranibizumab, which show improved therapeutic effect16, 17. In addition, the recent termination of a phase III clinical trial of an anticoagulant aptamer against Factor IXa exacerbated suspicion toward the therapeutic use of aptamers18, 19. Several crucial factors, such as their inherent physicochemical characteristics, a lack of medicinal chemistry support along with production costs, as well as reluctance to break from conventional approaches, have delayed and even suppressed the clinical translation and widespread distribution of therapeutic aptamers. Despite these limitations, recent progress in aptamer selection and formulation, and the lessons learned from nucleic acid clinical development, should encourage scientists to continue pursuit of therapeutic aptamers. Over the past decade, the demand for oligonucleotide products has driven global researchers and manufacturers to devote themselves to significantly improving approaches and technologies for oligonucleotide manufacturing (Box 2).
Box 2: Commercial efforts and support for aptamer-based products.
The advent of SELEX technology has generated a flourishing area of aptamer research for a wide range of applications, including diagnostics, biosensor technologies, affinity isolation, biomarker discovery, and in particular targeted therapeutics. Although antibody-based products still substantially dominate the global marketplace in diagnostics and therapeutics, there are a handful of biotechnology and pharmaceutical companies actively struggling against those conventional products.
The demand for oligonucleotide products has driven global researchers and manufacturers to devote themselves to significant improvements in approaches and technologies for oligonucleotide manufacturing (Table 3). Indeed, Aptagen now offer custom aptamers with prices ranging from $1–4 per microgram (for microgram quantities), $300 per gram (for milligram quantities), or even under $50 per gram (for gram quantities)211. Synthesis can be completed in two days or up to two weeks. RNA aptamers with special modifications and conjugations can be custom ordered from manufacturers such as TriLink or IDT. However, the manufacturing cost of longer RNAs (>60 nt) is typically higher than short RNAs, due to increased difficulties in synthesis and purification. For example, PAGE (polyacrylamide gel electrophoresis)-purified 2′-fluoropyrimdine-modified RNA (100-nt as an example) generally costs $2500 per milligram (in milligram scale from TriLink). In addition, SomaLogic, Inc., a powerhouse in the aptamer-based diagnostic market, has now generated SOMAmer (Slow Off-rate Modified Aptamer) reagents for more than 1,300 different protein targets critical to normal and disease biology. They are exploiting a series of SOMAmer associated array platforms, such as SOMAscan and SOMApanel, for proteomic and diagnostic applications. NOXXON has exclusive licenses for the SELEX process in discovery, manufacturing and application of Spiegelmers®, and energetically discovers and develops Spiegelmer® therapeutics using the in-house discovery platform. To date, over ten Spiegelmers® have been generated against various diseased-related targets, including three in clinical trials and four in the preclinical pipeline. Presently, diagnostic applications account for the largest aptamer market share, but therapeutic application of aptamers is estimated to grow rapidly due to increasing research activities and commercial investment.
This review will frame the current challenges and potential of therapeutic aptamers, in particular RNA aptamers. We will highlight more than a quarter-century’s worth of technological advances, clinical development, and challenges and lessons learned, with a look towards the future of aptamer-based therapeutics.
The generation of RNA aptamers
Principles of SELEX
SELEX is a gold-standard methodology for generating various DNA or RNA aptamers1–3, 20. A selection cycle typically starts with a combinatorial DNA or RNA library containing a 20~60-nt random region flanked by fixed primer regions at the 5′ and 3′ ends. An initial, unselected library theoretically consists of up to 1015 unique sequences (1–2 nmol of library), which can provide enough structural diversity to identify high-affinity binders. After iterative selection cycles consisting of binding, partitioning, recovery, and re-amplification steps, specific sequences—aptamers—are enriched and dominate the population of library species. The final product is sensitive to the selection environment, which is a combination of experimental conditions (e.g., ratio of library to target, buffer components, ion strength, pH, binding temperature/time) and target-inherent properties (e.g., pI, charge, hydrophilicity). Together, these variables contribute specific selection stringency, consequently affecting the affinity and function of the enriched aptamers.
There are some differences between the selection of RNA and DNA aptamers. In the case of DNA SELEX, a library is incubated with the target, and the bound sequences are recovered following re-amplification by PCR. In the case of RNA aptamers, they must first be reverse transcribed into double-stranded (ds)DNA to enable subsequent RNA transcription. For DNA aptamers, the resulting dsDNA library is subjected to strand separation to produce a new ssDNA library for the next selection cycle. RNA SELEX generally involves in vitro transcription. DNA and RNA aptamers are functionally similar, but each has its own benefits. DNA aptamers are inherently more stable and the related manufacturing costs are lower. RNA aptamers typically have more diverse three-dimensional conformations, and stronger intra-strand RNA-RNA interactions, which probably increase binding affinity and specificity21.
Purified protein-based SELEX
In a purified protein-based SELEX procedure22 (Figure 1A), one of the most critical steps is selection partitioning, the separation of target-bound sequences from unbound sequences in the library. Various solid-phase matrix-based segmentation strategies have been employed23, including nitrocellulose membrane, affinity column/resin chromatography, magnetic beads, and chip-based segmentation. For example, nitrocellulose membrane is a popular matrix used in SELEX due to its high protein binding affinity over nucleic acids; whereas nylon membrane favors binding of negative-charged oligonucleotide for blotting applications with nucleic acids, such as a double filter assay. When the mixture of random library and target protein is passed through a nitrocellulose membrane filter, size-exclusivity ensures that larger aptamer-protein complexes are retained on the membrane, while small, unbound nucleic acid molecules with weak affinity to the membrane freely flow through. With magnetic beads, the target protein is first immobilized on the surface of the beads, which are then incubated with the random library; the resulting mixture is subjected to magnetic precipitation to recover bound sequences. During the RNA SELEX experiment, chemically modified nucleoside triphosphate derivatives (2′-aminopyridmidine or 2′-fluoropyrimidine) are routinely incorporated into RNA to improve RNase resistance via in vitro transcription24, 25. Purified protein-based SELEX has successfully generated many high-affinity aptamers and has several advantages: the selection steps can be performed in a test tube without involving sophisticated technology, the selection conditions or evolution pressures are easily tunable, and the targets can be either intracellular or extracellular proteins26.
Figure 1. The generation of RNA aptamers.
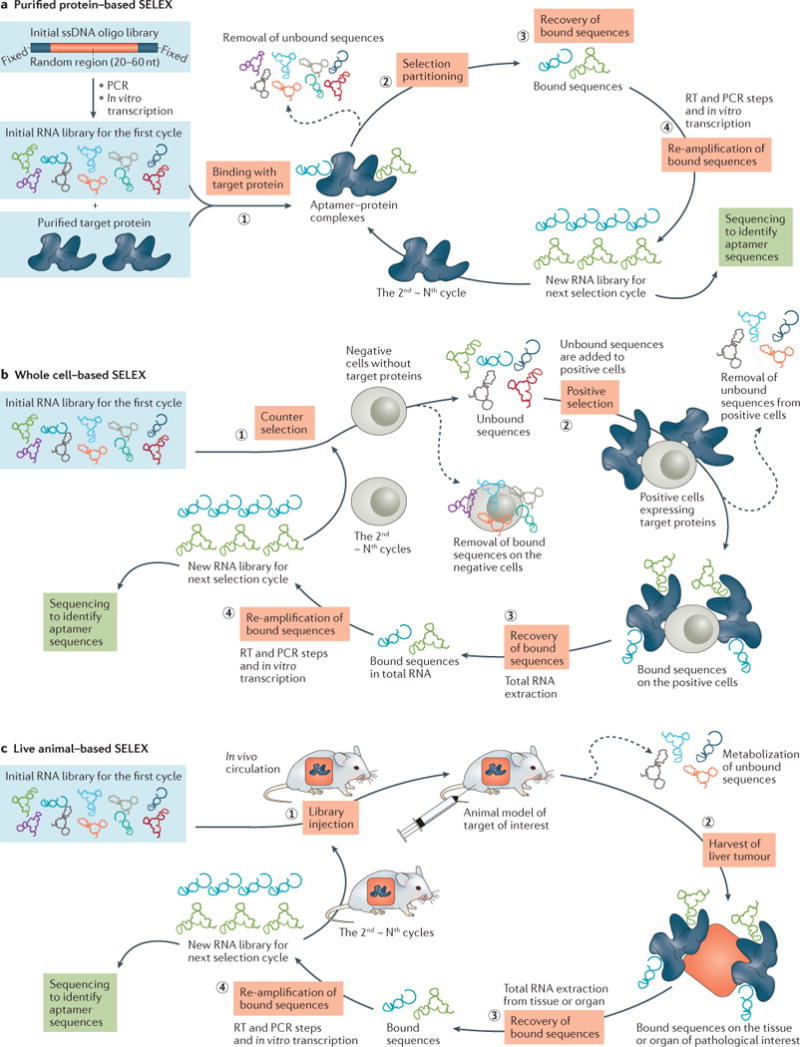
A commercially obtained ssDNA pool is used as an initial template for generating a dsDNA library by PCR, and is subsequently converted into a corresponding RNA library via in vitro transcription for the first selection cycle. A) Purified protein-based SELEX uses four key steps: (1) the RNA library is incubated with the target protein; (2) the bound species are isolated from the unbound sequences through various partitioning strategies; (3) target-bound sequences are recovered and (4) subjected to re-amplification (reverse transcription, PCR, and in vitro transcription) into a new RNA library for the next selection cycle. Through the iterative rounds, specific aptamers are enriched and identified by sequencing analysis. B) Whole cell-based SELEX, consisting of four main steps: (1) counter selection by incubating RNA library with negative cells that do not express the target protein; (2) positive selection by incubating recovered unbound sequences with positive cells expressing the target protein; (3) recovery of target-bound sequences; and (4) re-amplification of recovered species and generation of a new RNA pool for the next selection round. C) Live animal-based SELEX. After IV administration and circulation of an RNA library in the animal model, the tissue or organ of pathological interest is harvested and the bound sequences are extracted. Subsequently, the recovered RNA sequences are re-amplified to make a new RNA library for the next selection cycle.
Although protein-based SELEX may be applied to purified proteins, protein targets on the surface of whole cells, or even proteins within live animals, it is not applicable for unknown proteins, insoluble proteins, or proteins that only function in the native conformation or a multiprotein complex27. In some cases, the aptamers generated via purified protein-based SELEX fail to recognize the target protein on the cell surface28–30.
Whole cell-based SELEX
Whole cell-based SELEX offers a reasonable alternative to overcome some of the limitations of purified protein-based SELEX. It allows identification of specific aptamers that are capable of selectively binding to the target protein in its native conformation or original glycosylation pattern of the extracellular domain, and even internalizing into the target cells31–33. The basis of the selection procedure relies on the differences between the target cell population that expresses the target of interest and a negative cell population that does not. In particular, it can be applied to uncharacterized target proteins, for which there is no prior knowledge of their properties and structural features34, 35. This advantage allows whole-cell based SELEX to be exploited as a promising tool for biomarker discovery36. There are various approaches to produce negative cells (lacking targeting protein) or positive cells (overexpressing target protein). For example, gBlocks gene fragments can be used to create the corresponding cDNA containing desired target sequence. Alternatively, Cas9/CRISPR technology can be applied with positive cells to knockout target gene, thereby producing negative cells for counter selection.
A whole cell-based SELEX experiment (Figure 1B) typically involves counter selection and positive selection. The selection is performed with intact, living cells, thereby ensuring that target proteins on the cell surface keep their active conformation and functionality throughout the selection process. One caveat is that nucleic acids have a propensity for binding to dead cells in a non-specific manner, which can impede effective enrichment of target-specific sequences or even result in SELEX failure37. Modification to the process to ensure careful recovery of healthy cells (e.g. using fluorescence-activated cell sorting (FACS) technology38) expressing a high level of active receptors is critical for selection success.
Live animal-based SELEX
As aptamers selected in vitro may not necessarily function in vivo11, 39, a live animal-based SELEX procedure was developed to generate tissue-penetrating aptamers directly within a living animal model of the target disease or condition (Figure 1C). Researchers administer the RNA library intravenously (IV) to the animal, harvest the tissue or organ of pathological interest, and extract the bound sequences. The recovered RNA sequences are then re-amplified to make a new RNA library for next selection cycle. This technique has been used to identify 2′-fluoropyrimidine-modified RNA aptamers in an animal model of intrahepatic colorectal cancer metastases40. The resulting nuclease-resistant RNAs specifically localize to targets within intrahepatic tumor deposits. In addition, a 2′-fluoropyrimidine-modified RNA library has been administered to wild-type mice, followed by collection of the brain for recovery of aptamers that bind to brain capillary endothelia and penetrate into the parenchyma41. These examples demonstrate the efficacy of live animal-based SELEX as a direct method for generating RNA aptamers suitable for in vivo use.
Recent advances in SELEX technology
Although classic SELEX has proven to be a powerful and effective aptamer-selection procedure, intensive efforts have been made to streamline the methodology and optimize selection efficiency23, 42. Several important advances have improved the overall performance of SELEX, such as efficient partitioning and recovery, accurate amplification, and global analysis of sequencing data. For example, introduction of a negative-SELEX eliminates non-specific sequences that bind to environmental components43, especially to a solid-phase matrix employed for partitioning support or target immobilization. The resultant aptamers exhibit up to 10-fold higher binding affinity compared to those generated from SELEX without negative selection. A counter-SELEX excludes sequences recognizing other targets by using analogue targets, thus increasing the selectivity of the aptamers7, 8. Additionally, specialized partitioning technologies, e.g., capillary electrophoresis (CE)44–46, atomic force microscopy (AFM)47, 48, flow cytometry38, 49, microfluidics50, 51 and biacore surface plasmon resonance (SPR)52, have been successfully adopted into the classic SELEX procedure to accelerate high-affinity aptamer identification.
Previous studies have revealed that conventional PCR may limit amplification of a heterogeneous library, due to factors such as low efficiency and formation of non-specific product-product hybridizations and primer dimers53–55. In conventional PCR-driven SELEX, non-specific byproducts appear as early as the 15th amplification cycle, and all the expected products are completely converted to byproducts by the 30th cycle. With the increase of PCR cycles and iterative selection rounds, excessive accumulation of amplification artifacts hampers the enrichment of high-affinity aptamers, and may even cause selection failure54. Recently, emulsion PCR (ePCR) or droplet digital PCR (ddPCR) have been incorporated into the selection protocol to reduce the propagation of byproducts and avoid PCR bias45, 56–58. These modified techniques preserve library diversity and prevent the loss of highly structural aptamer sequences that are difficult to amplify in a conventional PCR system.
In a traditional SELEX, the final, enriched library is cloned and subjected to classic Sanger sequencing analysis for identification of individual aptamer sequences. The entire process is considered to operate within a “black box” until specific aptamers are identified at the final round. However, high throughput sequencing (HTS) technology has revolutionized the selection of aptamers by making the selection process visible with each round of selection. HTS and bioinformatics analysis combined with SELEX (HT-SELEX) not only facilitates the rapid identification of high-affinity aptamers, but also reveals a comprehensive landscape for the molecular evolution events59–61. Millions of sequence reads can be processed from each selection cycle, thus providing insight into the entire process, including primary sequences, total reads, nucleotide composition, frequency, and rate of molecular enrichment62, 63. Because HT-SELEX allows quantitative assessment of the dynamic changes in the library composition throughout selection cycles, it is capable of identifying high-affinity aptamers at a much earlier round, which is more cost-efficient and avoids the potential PCR bias associated with over-selection. Global analysis of large sequence data sets by robust bioinformatics tools can further facilitate high throughput characterization of aptamers, including structure prediction, binding affinity/specificity, functional properties, and aptamer–target interactions59.
Challenges in the development of aptamer-based therapeutics
As nucleic acid biopolymers, the in vivo therapeutic potency of aptamers is critically limited by their inherent physicochemical characteristics. These characteristics can affect pharmacokinetic properties such as metabolic instability, rapid renal filtration, rapid distribution from the plasma compartment into the tissues (e.g. liver, spleen), non-specific immune activation, and polyanionic effects. A number of chemical modifications and conjugations have therefore been developed to improve the pharmacokinetic properties of aptamer-based therapeutics64.
Nuclease degradation
Unmodified aptamers are susceptible to nuclease-mediated degradation that causes very short half-lives in vivo (less than 10 minutes)39. Most aptamers in clinical studies are chemically modified (Table 2) by replacing the 2′ position with either a fluoro- (F), amino- (NH2), or O-methyl (OCH3) group, and by capping the 3′ end with inverted thymidine to increase nuclease resistance while also enhancing binding affinity. In practice, two different strategies are applied to incorporate modified nucleotides into aptamers: in-SELEX and post-SELEX. In the first strategy, aptamers with the desired modifications are directly isolated from a DNA or RNA library containing modified nucleotides that are compatible with DNA or RNA polymerases65, 66. However, the modifications possible with this method are very limited. So far 2′-aminopyrimidines67, 2′-fluoropyrimidines68, 2′-O-methyl nucleotides69, and locked nucleic acids (LNA)70, 71 have been successfully incorporated into the SELEX protocol. Additionally, a method called SOMAmer selection (Slow Off-rate Modified Aptamer)72, 73, developed by SomaLogic, uses modified deoxyuracil carrying hydrophobic functional groups at the C5 position, which is well-tolerated by DNA polymerase (Box 2). Three clinically evaluated aptamers are Spiegelmers®, L-form RNA aptamers that are chiral inversions of their natural D-forms. Since Spiegelmers® cannot be recognized by nucleases, they display excellent in vivo biostability compared to natural nucleic acids74. However, Spiegelmer® technology requires obtaining the enantiomer of the target of interest for SELEX, thereby limiting its practicability for some biological macromolecules75.
Table 2.
Nucleic acid aptamers currently in the clinic
| Therapeutic purpose | Drug name and patent company | Target | Form and modification | Clinicaltrials.gov identifier (current status) | Condition being treated | Refs |
|---|---|---|---|---|---|---|
| Macular degeneration | Macugen® (Pegaptanib sodium) Pfizer/Eyetech |
Vascular Endothelial Growth factor isoform 165 (VEGF165) | 27-nt RNA 2′-fluoro pyrimidines, 2′-O-methyl purines, 3′-inverted dT, 40kDa PEGylated |
NCT00021736 (Phase II/III, completed); 233 NCT00040313 (Phase II, completed);234 NCT00056199 (Phase I, completed); 235 NCT00312351 (Phase 4, terminated); 236 NCT00321997 (Phase II/III, completed);237 NCT01487070 (Phase I, completed);238 Approved on Dec 2014 in the US and the EU for AMD treatment |
Age-related macular degeneration (AMD); Diabetic macular edema; Proliferative diabetic retinopathy |
14, 15, 154 |
| Zimura® (ARC1905) Ophthothech |
Complement component 5 (C5) | 38-nt RNA 2′-fluoro pyrimidines, 2′-O-methyl purines, 3′-inverted dT, 40kDa PEGylated |
NCT00709527 (Phase I, completed);239 NCT00950638 (Phase I, completed); 240 NCT02397954 (Phase II, completed);169 NCT02686658 (Phase II/III, recruiting);170 |
Age-related macular degeneration (AMD); Idiopathic Polypoidal Choroidal Vasculopathy (IPCV); Geographic Atrophy | 168 | |
| Fovista® (E10030) Opthothech/Retinal Consultants of Arizona |
Platelet-derived growth factor (PDGF) | 29-nt DNA 2′-O-methyl purines, 3′ inverted dT, 40 kDa PEGylated |
NCT00569140 (Phase I, completed);241 NCT01089517 (Phase II, completed);178 NCT02387957 (Phase II, recruiting);242 NCT02591914 (Phase I, ongoing);243 NCT01940887 (Phase III, recruiting);181 NCT01940900 (Phase III, ongoing);180 NCT01944839 (Phase III, recruiting);179 |
Age-related macular degeneration (AMD) | 174, 175 | |
| Coagulation | REG1 anticoagulation system (RB006 plus RB007) Regado Biosciences |
Coagulation factor IXa | 37-nt RNA The REG1 system consists of a drug (RB006, 37-nt RNA aptamer, 2′-ribo purine/2′-fluoro pyrimidine) and antidote (RB007, 17-nt, 40 KDa PEGylated, 2′-O-methyl antidote). |
NCT00113997 (Phase I, completed);244 NCT00715455 (Phase II, completed);189 NCT00932100 (Phase II, completed);188 NCT01872572 (Phase I, completed);190 NCT01848106 (Phase III, terminated, clinical hold due to serious anaphylactic reactions);191 |
Acute coronary syndrome (ACS); Cardiac catheterization (IV form); Coronary artery disease (CAD); Percutaneous coronary intervention (PCI) | 183–185, 187 |
| ARC1779 Archemix |
A1 domain of von Willebrand factor | 39-nt DNA 3′inverted dT, 2′-O-methyl with a single phosphorothioate linkage, 20 kDa PEGylated |
NCT00432770 (Phase I, completed);195 NCT00507338 (Phase II, terminated);198 NCT00632242 (Phase II, completed);245 NCT00694785 (Phase II, withdraw prior to enrollment);245 NCT00742612 (Phase II, terminated);197 |
Von Willebrand disease; Purpura; Thrombotic thrombocytopenic von Willebrand Disease Type-2b; Acute myocardial infarction; Percutaneous coronary intervention; Thrombosis |
192–194, 246 | |
| NU172 ARCA Biopharma |
Thrombin | 26-nt DNA Unmodified DNA |
NCT00808964 (Phase II, unknown, not verified recently);200 | Heart disease (e.g., used during cardiopulmonary bypass to maintain steady state of anticoagulation) | 199 | |
|
ARC19499 (BAX499) Baxalta US |
Tissue factor pathway inhibitor (TFPI) | 32-nt RNA 2′-O-methyl purine, 40 kDa PEGylated, 3′ inverted dT |
NCT01191372 (Phase I, terminated);203 | Hemophilia | 201, 202 | |
| Oncology | AS1411 (AGRO001) Antisoma |
Nucleolin | 26-nt DNA G-rich DNA, PEGlyated |
NCT00512083 (Phase II, completed);212 NCT00740441 (Phase II, unknown);213 NCT00881244 (Phase I, completed);211 NCT01034410 (Phase II, terminated);247 |
Acute myeloid leukemia (AML); Metastatic renal cell carcinoma; Advanced solid tumor; | 208–210 |
| NOX-A12 NOXXON Pharma |
Chemokine (C-X-C motif) ligand 12 (CXCL12) or Stromal cell-derived factor-1 (SDF-1) | 45-nt RNA (Spiegelmer®) L-Ribonucleic acid, PEGylated |
NCT00976378 (Phase I, completed);219 NCT01194934 (Phase I, completed);220 NCT01521533 (Phase II, completed);222 NCT01486797 (Phase II, ongoing, but not recruiting);221 |
Multiple myeloma (MM) and non-Hodgkin’s lymphoma (NHL); Chronic lymphocytic leukemia (CLL); Autologous stem cell transplantation; Hematopoietic stem cell transplantation; | 214, 215 | |
| Inflammation | NOX-E36 NOXXON Pharma |
Chemokine (C-C motif) ligand 2 (CCL2) | 40-nt RNA (Spiegelmer®) L-Ribonucleic acid, PEGylated |
NCT00976729 (Phase I, completed);248 NCT01085292 (Phase I/II, completed);226 NCT01372124 (Phase I, completed);249 NCT01547897 (Phase II, completed);227 |
Chronic inflammatory diseases; Type 2 diabetes mellitus; Systemic lupus erythematosus; albuminuria; Renal impairment |
74, 224, 225 |
| NOX-H94 NOXXON Pharma |
Hepcidin peptide hormone | 44-nt RNA (Spiegelmer®) L-Ribonucleic acid, PEGylated |
NCT01372137 (Phase I, completed);230 NCT01522794 (Phase I, completed);250 NCT01691040 (Phase II, completed); 231 NCT02079896 (Phase I/II, completed);232 |
Anemia; End stage renal disease; Anemia of chronic diseases; inflammation; | 228 |
In post-SELEX strategies, modifications at various positions (e.g., base, 2′-position, sugar ring, phosphate group) are introduced to pre-selected aptamers during solid-phase chemical synthesis. Sometimes multiple modifications are applied in combination for optimal performance. Since the affinity/specificity and function of an aptamer is sensitive to its structure, post-SELEX modification may affect the inherent properties and folding structures of the original aptamers, thereby compromising the binding affinity. Therefore, it is necessary to precisely tailor modifications for the desired functions76. Unfortunately, universal rules are not available for all the aptamers, and laborious evaluation/optimization is often needed77.
Renal filtration
The average diameter of aptamers (6–30 kDa) is less than 5 nm78. When non-formulated aptamers are administered into the blood stream, even using stabilizing backbone modifications, small aptamers are subject to rapid excretion through renal filtration. To overcome renal filtration and extend circulation time, aptamers are generally formulated with a bulky moiety, such as high molecular mass PEG69, cholesterol79, 80, proteins81, 82, liposomes83, organic or inorganic nanomaterials10, 84, or are multimerized85–87 to create a multivalent molecule above the cutoff threshold for the renal glomerulus (30–50 kDa). PEG is a well-studied, hydrophilic biomaterial which decreases aggregation and increases solubility of the conjugates. PEG has been widely used in many FDA-approved formulations to prolong the circulation half-life and improve in vivo bioavailability of therapeutic aptamers following IV administration. For example, PEGylation of Macugen® (see below) increased half-life to 9.3 h and 12 h in plasma after IV injection or subcutaneous injection, respectively, and even to 94 h in vitreous humor88, 89. Compared to a monovalent aptamer, rationally designed multivalent aptamers have shown an improved overall performance, including binding affinity/specificity, biological function, and circulation time. For example, tetrameric aptamer conjugates with increased retention in the circulation and improved pharmacokinetic properties have been generated90.
Toxicity
Toxicological information regarding aptamers in humans is very limited91, 92. Although aptamer-related adverse events are rare in clinical evaluations to date, potential toxicities may arise from polyanionic effects, unexpected tissue accumulation, intensive chemical modification or conjugation, and non-specific immune activation, in particular with continuous or repeated administration of aptamer therapeutics93–95. Highly negatively charged molecules (e.g. nucleic acids) are prone to non-specific binding to blood proteins, which may result in high uptake by non-target tissues and organs, thereby causing unwanted side effects and reducing therapeutic efficacy96, 97. Chemical modifications have proved to be a double-edged sword, since unnatural nucleotides may cause chemical toxic effects or become immunogenic. For example, severe hepatotoxicity was observed in LNA-modified nucleic acids98, and 2′-fluoropyridimine-modified RNAs differentially controlled the activation of pattern recognition receptors76. Thus, chemical modifications should be used cautiously, according to the desired therapeutic application of the aptamers. Adverse responses can also be associated with the formulation of therapeutic aptamers. For example, serious allergic responses to the PEG group, due to the presence of pre-existing antibodies to PEG, have been reported in the phase III study of the aptamer-based anticoagulation system, REG1 (Regado Biosciences) system (see below), raising a safety concern about the use of PEGylated agents in acutely ill patients18, 99. Highly lipophilic molecules often result in non-specific liver uptake, which may trigger potential hepatotoxicity100. Prudent formulations or administration routines are necessary for improved development of aptamer therapeutics.
Recent progress in aptamer-based therapeutics
Aptamer-based therapeutics typically exploit one of three strategies: (1) an aptamer serves as an antagonist for blocking the interaction of disease-associated targets, e.g., protein-protein, or receptor-ligand; (2) an aptamer serves as an agonist for activating the function of target receptors; or (3) a cell type-specific aptamer serves as a carrier for delivering other therapeutic agents to the target cells or tissue. This topic has been well reviewed elsewhere11, 26, 101.
Aptamers as antagonists
An inhibitory aptamer that can disrupt the function of a pathologic target protein can be used directly as a therapeutic antagonist. Currently, all the aptamers in clinical trials fall into this category (discussed in detail below). Numerous groups have developed aptamers against therapeutically relevant targets associated with a wide variety of human diseases, including ocular disorders, bleeding disorders, cancer, cardiovascular disease, autoimmune disease, degenerative neurological disease, and infectious disease26, 102. In addition, artificial RNA sequences that mimic natural RNAs have been evolved using the in vitro SELEX process. For example, an RNA decoy is designed to compete with the natural RNA sequence that is a target of an RNA-binding protein, which sequesters the interaction of the natural RNA sequence and its target protein103. In HIV-1, the trans-activator Tat protein binds to specific regulatory elements (trans-activation responsive element TAR) in the HIV-1 long-terminal repeats (LTRs) and regulates viral transcription. Synthetic TAR RNA decoys that mimic the apical loop of TAR sequence showed high affinity to Tat protein, and inhibited HIV RNA transcription and thus reduced viral replication104–106.
Despite exquisite affinity and specificity, the therapeutic potential of inhibitory aptamers may not be fully achieved due to suboptimal design or formulation. The biodistribution of aptamer conjugates varies with their actual size, charge, and other physical characteristics. Rational design and formation is crucial. Recent progress that has been made to improve design or formation is highlighted below.
Formulation with an inert antibody
The VEGF-targeted aptamer, pegaptanib, which is approved for use in ophthalmology107 (see below), was originally designed for use in cancer therapy. In vivo evaluation in a tumor xenograft model indicated its inhibitory effect on VEGF-induced vascular permeability and tumor growth108. However, its anti-cancer efficacy did not meet expectations in some early preclinical studies. To improve in vivo pharmacokinetics of the aptamer, an aptamer-antibody hybrid complex was recently generated by reacting an anti-cotinine antibody with the cotinine-conjugated pegaptanib aptamer82. This complex distributed widely throughout the tumor tissue and exhibited a prolonged half-life (8.2 h) in serum. In an A549-xenograft mouse model, systemic administration of the pegaptanib/antibody complex inhibited tumor angiogenesis with enhanced tumor tissue penetration, consequently decreasing tumor growth to a degree comparable to bevacizumab. This proof-of-principle study may represent a new formulation strategy for the use of aptamer-antibody complexes in targeted cancer therapy.
Formulation with cholesterol
Although cholesterol conjugation has been reported to reduce renal filtration and increase hepatic cell uptake via receptor-mediated endocytosis, analyses of pharmacokinetics and administration routes for cholesterol-conjugated aptamers are limited. Recently, the safety and pharmacokinetic profile of a cholesterol-conjugated RNA aptamer against the hepatitis C virus (HCV) NS5B protein was assessed in a preclinical study80. Systemic administration of the cholesterol-conjugated RNA aptamer was well-tolerated, without any abnormalities in wild-type BALB/c mice, and showed a 2-fold extended half-life compared to non-conjugated aptamer. IV administration demonstrated a longer circulation half-life (5–6-times) and higher drug exposure (2–3-times) than intraperitoneal administration, suggesting the IV route may be more suitable for cholesterol-conjugated aptamers.
Formulation with nanoparticles
A single amino acid substitution (R175H) in tumor suppressor p53 results in inactivation of wild-type p53, which impacts multiple cancer-associated pathways109. A contrast screening SELEX strategy was applied to select an exquisitely sensitive RNA aptamer capable of distinguishing p53R175H from wild-type p53 protein10. The aptamer inhibited cell migration and invasion in human lung cancer cells harboring p53R75H. To limit degradation, the aptamer was formulated with sheddable nanoparticles derived from PEGylated polyethylenimine. Following administration to a tumor xenograft model either by subcutaneous injection or tail vein IV injection, the nanoparticle-conjugated aptamer inhibited tumor growth.
Formulation with pluronic gel
Migration and proliferation of vascular smooth muscle cells (VSMC) plays a key role in the development of cardiovascular disease. Such disease is generally treated with anti-proliferative/anti-migratory drugs (e.g., paclitaxel) to directly block VSMC activation110. Through a cell-based HT-SELEX strategy, a nuclease-resistant RNA aptamer was identified to preferentially target VSMCs over endothelial cells and nonvascular myocytes, and specifically inhibited VSMC migration111. In preclinical assessment, this aptamer exhibited a half-life of ~300 h in human serum and no pathological immune response in human peripheral blood mononuclear cells. When the RNA aptamer was formulated with pluronic gel, a biphasic micellar system for transdermal drug delivery, and applied around the carotid artery in a ligation mouse model of vascular injury, neointimal formation was inhibited to levels similar to those observed with paclitaxel, the current standard of care. This confirmed effective delivery of the RNA aptamer to the vessel wall due to pluronic gel formation.
Formulation as a multimer
Multiple anticoagulant aptamers target prothrombin/thrombin or activated coagulation factor X (FXa) (see below)112. Although no individual aptamer has yet rivalled heparin, a widely-used injectable anticoagulant113, a combination of multiple aptamers targeting different coagulation enzymes can be as potent as heparin for prevention of fibrin clot formation114. Unfortunately, a cocktail formulation of multiple aptamers significantly increases the complexity and cost of clinical development115. Any unapproved individual aptamer, as well as the combination, have to be subjected to preclinical and phase I studies to ensure the safety of each ingredient. Instead, multimerization of individual aptamers into a single unit may offer an alternative strategy. Recently, by using a single bivalent aptamer that simultaneously targets two different coagulation cascade proteins, heparin-level anticoagulation potency was observed in human blood86. Notably, a single antidote rapidly reversed the bivalent aptamer-mediated anticoagulation effect in a dose-dependent manner.
Aptamers as agonists
To date, only a few aptamers have been developed to function as therapeutic agonists. This includes RNA aptamers targeting human epidermal growth factor receptor-3 (HER3/ERBB3)116, OX40 (CD134)117, 118, 4-1BB (CD137)119, CD40120, CD28121, and DNA aptamers targeting human VEGFR-2122 and the insulin receptor (IR)123. Several of the RNA aptamers targeting immune costimulatory receptors (CD28, CD40, OX40 and 4-1BB) have been engineered into multimeric versions to act as receptor agonists for improved cancer immunotherapy124. The efficient costimulation on the surface of antigen-presenting cells or T-cells induced by the interaction of a costimulatory receptor and its ligand plays a crucial role in improving anti-tumor immunity125.
4-1BB RNA aptamer
By using a complementary linker, two monomeric 4-1BB aptamers were converted to a bivalent form119 (Figure 2A). Although both monovalent and bivalent aptamer forms bound to 4-1BB expressing cells, only the bivalent aptamer was capable of costimulating T cell activation and promoting tumor rejection in vivo, with efficiency comparable to that of a previously described anti-4-1BB antibody. Since costimulatory activity is dependent on the spatial arrangement and distance of the linked aptamers, rational design is crucial. To trigger cross-linking of the 4-1BB receptor, the distance between the aptamers was formulated to be in the range of the distance (5–18 nm) between the two variable domains of the anti-4-1BB antibody. A 21-bp double-stranded linker provided a 7.1-nm distance between the two 4-1BB aptamers, and allowed the correct orientation for binding to a pair of 4-1BB receptors. This study provides a preliminary example of multivalent aptamers with agonistic function for cancer immunotherapy.
Figure 2. Schematics of bivalent RNA aptamers used as agonists.
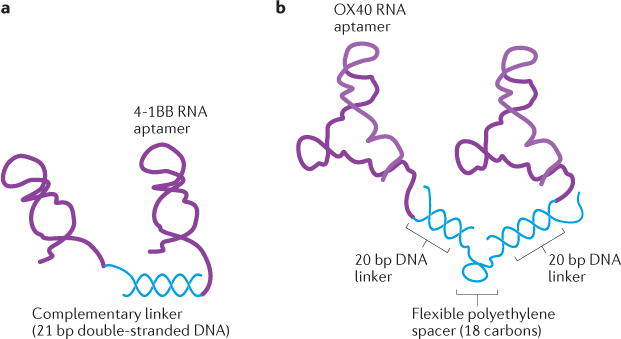
A) The bivalent 4-1BB aptamer. The 3′-terminus of two 4-1BB aptamers are attached to a 21-nt complementary linker sequence and subsequently annealed together. B) The bivalent OX40 aptamer. The 3′-end linker sequences (20 nt in length) of OX40 aptamers were annealed to a flexible DNA scaffold. A tandem repeat of 20 nt DNA oligos was connected by a flexible polyethylene spacer.
OX40 RNA aptamer
The crystal structure of the OX40 receptor-ligand complex revealed multiple binding sites for its ligand126, suggesting an OX40 aptamer would require a multivalent form to effectively stimulate OX40 function. Two copies of an anti-murine OX40 aptamer were assembled onto a flexible DNA oligonucleotide-based molecular scaffold117 (Figure 2B). The resulting bivalent aptamer activated the OX40 receptor on primed T cells in vitro, and systemic administration of this bivalent aptamer significantly enhanced anti-tumor responses generated by a dendritic cell-based vaccine in mice. Recently, two anti-human OX40 RNA aptamers containing a biotin group at the 5′-end were formulated into a bivalent via a streptavidin linker, which stimulated OX40 on human T cells, and enhanced cell proliferation as well as interferon-gamma production118.
CD28 RNA aptamer
Two RNA aptamers that specifically bind to murine CD28 have been isolated from a 2′-fluoropyrimidine-modified RNA library and have shown distinct behaviors in their monomeric and dimerized forms121. In the monomeric form, one aptamer (CD28Apt2) functioned as an antagonist to block the interaction of CD28 with its main ligand B7.2 and preclude costimulatory signaling; in contrast, the monomeric form of another (CD28Apt7) was inactive. When the two aptamers were converted into bivalent forms, both of them functioned as an agonist, providing an artificial costimulatory signal. Dimeric agonistic aptamers were engineered by either using a 21-bp double-stranded linker to fuse the two individual aptamers, or transcribing the dimeric aptamer as a single strand without any extra linker. Of the four dimeric aptamers, the CD28Apt7-dimer without extra linker significantly improved costimulatory properties and surpassed the agonistic effect of an anti-CD28 antibody. When coadministered with an idiotype vaccine, the CD28 agonistic aptamer also enhanced the cellular immune response in vivo and prolonged survival of lymphoma-bearing mice. This study demonstrates that aptamers may be rationally engineered to achieve a desired function, such as inhibiting protein-receptor interactions as an antagonist or achieving costimulation as an agonist, depending on therapeutic need. Unlike aptamers, to date, no therapeutic antibody has demonstrated such dual functionality.
CD40 RNA aptamer
Most recently, two 2′-fluoropyrimidine-modified RNA aptamers were isolated against the murine CD40 receptor120. CD40 aptamers were engineered into three constructs with different functions. The agonistic bivalent aptamer caused proliferation and activation of B lymphocytes, and accelerated recovery of bone-marrow aplasia, whereas the antagonistic monovalent aptamer reduced B-cell lymphoma proliferation and improved overall survival in BALB/c mice. Moreover, the agonistic bivalent aptamer was conjugated with an shRNA targeting SMG1, a kinase that is essential for nonsense-mediated mRNA decay (NMD) initiation. The resulting agonistic aptamer-shRNA chimera inhibited NMD, and improved tumor infiltration and overall survival in vivo.
Aptamers as delivery agents
In addition to their utility as stand-alone therapeutics, aptamers may also be used as escorts for another therapeutic. Various cell type-specific aptamers have been conjugated with therapeutic agents (e.g. siRNA, microRNA, anti-miR, therapeutic aptamer, chemotherapeutics, or toxins), or delivery vehicles (e.g., organic or inorganic nanocarriers) for targeted delivery in a cell type-specific manner. By taking advantage of the high affinity and specificity of aptamers, therapeutic compounds can be targeted to the desired cells or tissues, thereby improving their local concentration and therapeutic efficacy. A full list of cell type-specific aptamers has been published101. Several representative aptamer-mediated delivery systems are illustrated below.
Aptamer-therapeutic oligonucleotide conjugates
An initial example of an aptamer-therapeutic oligonucleotide conjugate is a 2′-fluoropyrimidine-modified aptamer targeting human prostate-specific membrane antigen (PSMA), a transmembrane protein highly expressed in human prostate cancer and the vascular endothelium127. Through either covalent fusion or physical assembly, PSMA aptamers have been successfully conjugated with various therapeutic oligonucleotides to achieve targeted delivery. In a proof of concept study, covalent PSMA aptamer-siRNA chimeras were developed128 (Figure 3A), which selectively internalized into PSMA-expressing cells and effectively suppressed the targeted tumor survival genes, such as polo-like kinase 1 (PLK1) and B-cell lymphoma 2 (BCL2), in cultured cells as well as in animal models. Subsequent efforts further optimized the design through rational modifications of both aptamer and siRNA portions129. The resulting second-generation chimeric RNA aptamer demonstrated improved circulation and bioavailability in vivo, resulting in significant regression of PSMA-expressing tumors following systemic administration. Using the same strategy, anti-NMD factor siRNAs (SMG1 and UPF2) were conjugated with the PSMA aptamer, and inhibited tumor growth after systemic administration in both subcutaneous and metastatic tumor models130. Several variants have also been developed to simplify formulation or enhance cargo-loading capability. For example, the PSMA aptamer was covalently fused with a short hairpin RNA (shRNA) against DNA-activated protein kinase (Figure 3B), which resulted in selective RNAi activity in PSMA-positive cells131. When combined with ionizing radiation, the aptamer-shRNA chimeras improved the response of the PSMA-positive tumors to ionizing radiation. Bivalent aptamer-siRNA chimeras have also been generated to increase binding affinity and cellular uptake, and to improve the inhibitory effect of siRNAs132.
Figure 3. Schematics of cell type-specific RNA aptamers used as delivery agents.
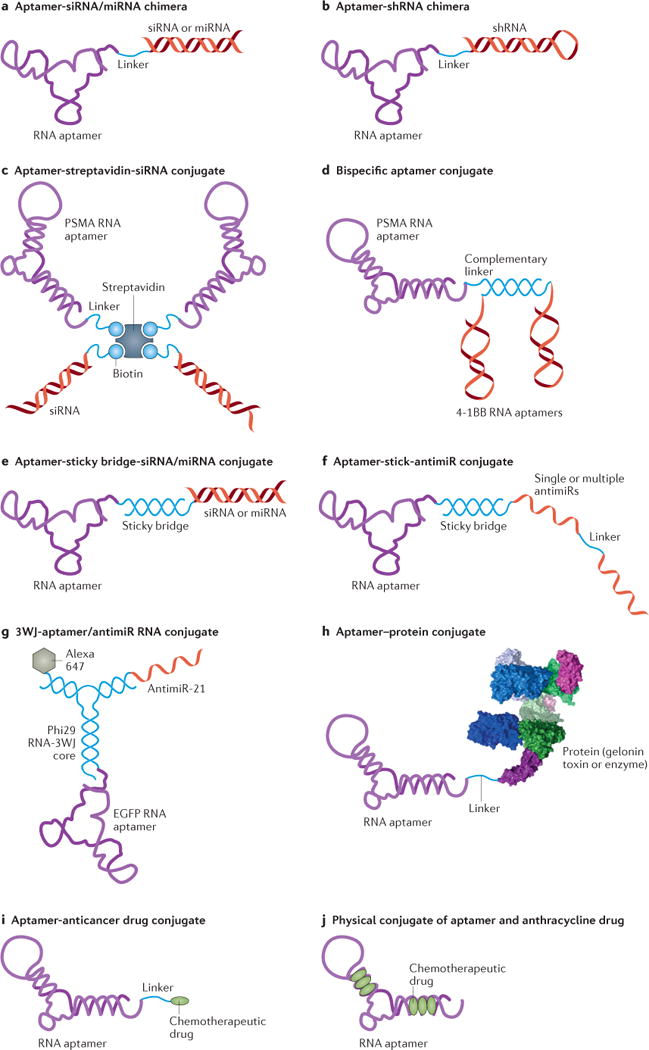
A) An aptamer-siRNA/miRNA chimera. The chimera is synthesized as two pieces followed by an annealing step to make the chimeric RNA molecule. B) An aptamer-shRNA chimera synthesized as one piece. C) An aptamer-streptavidin-siRNA conjugate. The siRNA and PSMA RNA aptamers are chemically conjugated with a biotin group. Then the two biotinylated siRNAs and two aptamers are non-covalently assembled via a streptavidin connector. D) The bispecific PSMA-4-1BB aptamer conjugate. The PSMA RNA aptamer and a bivalent 4-1BB RNA aptamer are tethered to complementary linker sequences and hybridized through Watson-Crick base pairing. E) An aptamer-sticky bridge-siRNA/miRNA conjugate. The aptamer and siRNA/miRNA are appended to complementary GC-rich bridge sequences and annealed by simple mixing that allows Watson-Crick base pairing. F) An aptamer-sticky bridge-antimiR conjugate. The single or multiple antimiR oligonucleotides are hybridized with the aptamers via a GC-rich sticky bridge. G) The 3WJ-aptamer/antimiR RNA conjugate. It contains an epidermal growth factor receptor (EGFR) aptamer as a targeting agent, an anti-miR-21 sequence as a therapeutic agent, a fluorescent dye (Alexa647) as an imaging agent, and a three-way junction (3WJ) motif as a molecular scaffold. H) An aptamer-protein conjugate. Chemical synthesis incorporates a primary amino group at the 5′-end of the PSMA aptamer, allowing chemical modification with a cross-linker agent (SPDP, N-Succinimidyl 3-[2-pyridyldithio]-propionate), and subsequent conjugation with the cysteine residue of gelonin toxin through a disulfide linkage. I) An aptamer-anticancer drug conjugate. It is made by using an acid-labile acylhydrazone linkage or formaldehyde linkage. J) Physical conjugation between an aptamer and anthracycline drug (dox) through intercalation.
PSMA aptamers can be non-covalently conjugated with therapeutic oligonucleotides via a molecular platform or bridge. For example, two copies of biotinylated PSMA aptamers and biotinylated siRNAs have been physically assembled on a streptavidin connector133 (Figure 3C). The resulting multivalent conjugate selectively delivered siRNA into PSMA-positive cells, and induced specific RNAi activity.
The PSMA aptamer can also be used to deliver therapeutic aptamers. The aforementioned bivalent 4-1BB aptamer and the PSMA aptamer have been tethered together through a complementary linker sequence, consequently resulting in a tumor-targeting costimulatory aptamer conjugate134 (Figure 3D). When administered systemically, this bispecific conjugate significantly inhibited tumor growth, and led to more effective tumor rejection in vivo than 4-1BB antibodies.
Another example of an aptamer-therapeutic oligonucleotide conjugate has been developed in the context of HIV. In HIV, the envelope glycoprotein gp120 interacts with the host cell receptor CD4 and coreceptor CCR5 during viral entry. Our own group has developed several novel dual-inhibitory gp120 aptamer-siRNA conjugates, in which both the aptamer and the siRNA portions have potent anti-HIV activities135, 136. The 2′-fluoropyrimidine-modified gp120 aptamer plays dual functions: (1) an HIV-1 inhibitor that blocks the interaction of gp120 and the CD4 receptor; and (2) a cell type-specific delivery agent for an siRNA. One design is a covalent aptamer-siRNA chimera (Figure 3A), another is a noncovalent aptamer-stick-siRNA conjugate (Figure 3E). In the latter design, the aptamer and siRNA portions are hybridized using a GC-rich sticky bridge sequence, which provides a facile approach to conjugating various aptamers or siRNAs for combination treatment. The gp120 RNA aptamer specifically delivered anti-HIV siRNA into HIV-1 infected cells and inhibited HIV-1 activity in vitro. Systemic administration of these aptamer-siRNA conjugates suppressed HIV-1 replication by several orders of magnitude and prevented CD4 T+ cell depletion in a humanized mouse model of HIV. The siRNA delivered by the gp120 aptamer was processed by Dicer and triggered specific RNAi activity137, 138. Such combinations of therapeutic siRNAs and aptamers may provide more effective treatment of highly mutated viral infection by simultaneously inhibiting multiple different targets.
Cell type-specific RNA aptamers can similarly be used to deliver anti-miR oligonucleotides139, 140 (Figure 3F, G). For example, a multifunctional RNA conjugate was recently constructed for specific delivery of an anti-miR-21 therapeutic agent140 (Figure 3G). The conjugate combined anti-miR-21 with an epidermal growth factor receptor (EGFR) aptamer as the targeting agent, a fluorescent dye (Alexa647) as an imaging agent, and a three-way junction (3WJ) motif derived from bacteriophage phi29 packaging RNA (pRNA) as a molecular scaffold. Through rational design and construction strategy, these components were efficiently assembled into a trifunctional RNA structure with an average hydrodynamic diameter of 15 nm, which showed a long half-life (18.5 h) and high thermostability. When administered systemically in a triple negative breast cancer (TNBC)-bearing mouse model, the RNA nanoparticles specifically bound and were internalized into TNBC cells, suppressed miR-21, and resulted in efficient inhibition of tumor growth. Biodistribution studies in vivo indicated that the RNA nanoparticles specifically targeted tumors, with little or no accumulation in healthy organs or tissues.
Aptamer-drug conjugates
Chemical solid-phase synthesis of aptamers offers opportunities for covalent conjugation with small organic molecules, radiopharmaceuticals, polymers, or even large proteins or enzymes (Figure 3H). For example, the PSMA aptamer has been used to deliver a toxin into PSMA-overexpressing cells141, 142. The PSMA aptamer-toxin conjugates not only showed specific cell uptake into PSMA-positive cells, but also decreased the toxicity of gelonin in non-targeted cells141. In addition, a dimeric PSMA DNA aptamer has been covalently conjugated with the antitumour chemotherapeutic drug doxorubicin, through a pH-sensitive linkage143 (Figure 3I). The resulting conjugates selectively delivered doxorubicin to PSMA-positive cells. Moreover, by taking advantage of this anthracycline drug’s ability to intercalate into the G-C-rich helical strand of the aptamer, noncovalent aptamer-drug conjugates have also been formulated without using any chemical reaction. For example, multiple doxorubicin molecules were physically loaded into the double-stranded region of the PSMA RNA aptamer144 (Figure 3J), and were selectively internalized by PSMA-positive cells, reducing cytotoxicity to PSMA-negative cells.
Similarly, target-specific radiopharmaceuticals have been developed through the conjugation of a radionuclide with a cell type-specific aptamer145. A nucleotide pre-labeled with a radionuclide can be directly incorporated into aptamers during enzymatic RNA transcription or chemical solid-phase synthesis146. Alternatively, through chemical solid-phase synthesis, desired functional groups (such as primary amines, sulfhydryls or carboxyls) can be precisely introduced into aptamers at a predefined position for covalent conjugation with radionuclide, therefore allowing more accurate and controllable conjugation than achieved with an antibody which generally contains multiple functional groups147. So far, there are many examples of radiolabeled aptamers for nuclear imaging and therapy. The most popularly used radionuclides include 99mTc, 64Cu, 68Ga, and 111In. An initial example of an aptamer-radionuclide conjugate is a RNA aptamer targeting human tenascin-C (TN-C), a large hexametric glycoprotein that is overexpressed in tumor stroma. A TN-C aptamer, TTA1148, was chemically modified with a primary amino group at its 5′-terminal end and subsequently conjugated with 99mTc via a bifunctional chelator (2-Mercaptoacetylglycylglycyl, MAG2)149. In glioblastoma and breast cancer tumor xenografts, the resultant TTA1-99mTc conjugate showed rapid tumor penetration and blood clearance, yielding a tumor-to-blood ratio of 50 within three hours. Moreover, aptamer uptake in several different human tumors was demonstrated to depend on the presence of the target TN-C protein. Although development efforts continue in this field, systemic in vivo evaluation of these systems is still lacking.
Aptamer-decorated nanomaterials as drug delivery system
In addition to the examples above, in which an aptamer is directly conjugated to a therapeutic drug, cell type-specific aptamers are also being exploited to “decorate” nanocarriers carrying multiple cargos. Multiple components are rationally assembled in one nanoscale carrier to achieve multifunctional nanomedicine150. This may include a therapeutic agent (e.g., therapeutic oligonucleotides, chemotherapy agents), an actively targeting agent (e.g., cell type-specific targeting aptamers), and an imaging component (e.g., fluorescent dyes or radioactive agents) (Figure 4). Since the first example of a PSMA aptamer-nanoparticle bioconjugate for targeted delivery of rhodamine-labeled dextran was reported in 2004151, various nanomaterials (e.g., gold nanoparticles, iron oxide nanoparticles, single-walled carbon nanotubes, quantum dots, liposomes, micelles, dendrimers, branched polymers, proteins, or viral capsids) have been assembled with cell type-specific aptamers. A recent proof-of-concept study generated aptamer-functionalized lipid nanoparticles (LNP) for siRNA delivery152. By using cell-based SELEX, DNA aptamers targeting both rat and human osteoblasts were selected and used for decoration of LNP that encapsulated osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) siRNAs. The aptamer-LNP-siRNA delivery system not only facilitated in vivo osteoblast-selective siRNA uptake (mainly via micropinocytosis), but also resulted in osteoblast-specific Plekho1 gene silencing in vivo, consequently promoting bone formation and bone microarchitecture, and enhancing mechanical properties in both osteopenic and healthy rodents. These proof-of-concept studies of aptamer-based nanomedicine provide continuing impetus for the development and clinical evaluation of therapeutic aptamers in the future.
Figure 4. Cell type-specific aptamer-functionalized nanocarriers for targeted therapy.
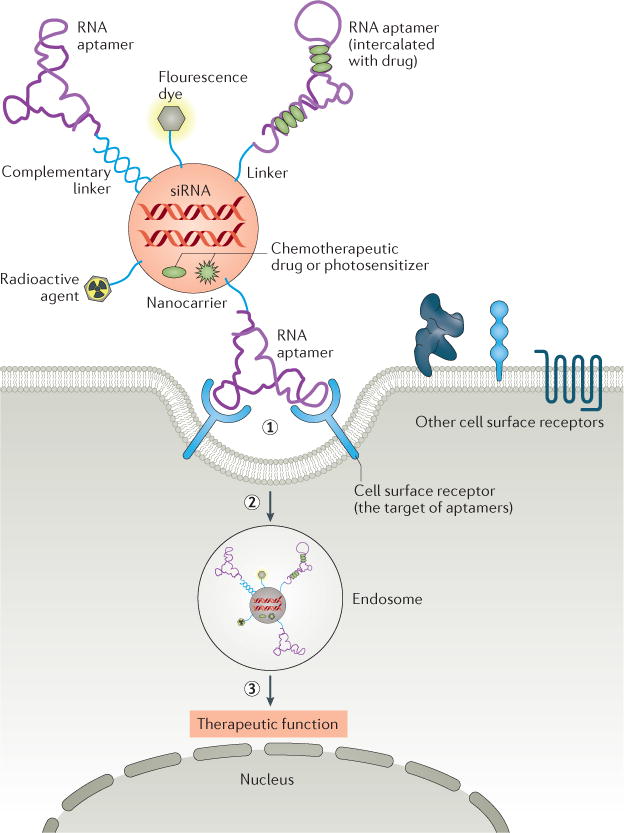
Multiple components such as therapeutics (therapeutic oligonucleotides, chemotherapy agents), actively targeting agents (cell type-specific aptamers), and imaging agents (fluorescent dyes or radioactivity agents) are rationally assembled in one nanoscale carrier to achieve multifunctional nanomedicine. (1) Upon binding of the aptamer portion of nanocarrier conjugate to the target receptor on the cell surface, (2) the conjugate is internalized into cells, probably through a receptor-mediated endocytosis pathway. (3) It is presumed that the conjugate shuttles into the endosome; subsequently, the therapeutic agents dissociate from the complex and escape the endosome. The released therapeutic agents mediated therapeutic function.
Clinical development of aptamer-based therapeutics
To date, the U.S. Food and Drug Administration (FDA) has approved one RNA aptamer and ten aptamers have undergone clinical trials for the treatment of various conditions, including macular degeneration, coagulation, oncology, and inflammation (Table 2). As indicated above, all aptamers that have entered clinical trials so far act as antagonists.
Macular degeneration
There are three aptamers designated for use in ophthalmology107, including one approved drug (Macugen®/pegaptanib), and two in late-stage development (Zimura®/ACR1905 and Fovista®/E10030). They were raised from three different target proteins, respectively: VEGF, complement component 5 (C5), and platelet-derived growth factor (PDGF).
VEGF plays an important role in neuronal development and maintenance within the central nervous system, including in the neural retina17. It is a therapeutic target for combating ocular vascular diseases: VEGF blockade reduces the growth of blood vessels, and controls vascular leakage and swelling in the eye, which would otherwise lead to loss of central vision16, 153. Pegaptanib is a 27-nt, PEGylated, modified RNA aptamer, which specifically binds to the major pathological VEGF isoform (extracellular VEGF165), inhibits VEGF binding to its receptors, and thus prevents VEGF from stimulating blood vessel growth14, 15, 154.
In 1993, the first attempt to develop VEGF aptamers started with a purified protein-based SELEX approach155, and identified unmodified RNA aptamers with low nanomolar affinity. To improve nuclease stability156, 2′-aminopyrimidines were introduced into the initial library157. The 2′-amino bases were subsequently substituted with 2′-fluoro bases68, and a novel set of inhibitory RNA aptamers with improved nuclease resistance and picomolar affinity for VEGF were identified, including the pegaptanib prototype, clone t44. Due to superior performance as an inhibitor of VEGF receptor binding and vascular permeability, clone t44 was selected for clinical translation. Further modifications, including 2′-methoxypurine, 3′-3′ inverted deoxythymidine (idT) cap, and 5′-polyethylene glycol (PEG) conjugation (40 kDa)158, 159, were incorporated to further improve inhibitory activity and plasma half-life in vivo88, 89. In 2004, the FDA approved Macugen® (pegaptanib sodium) as the world’s first therapeutic aptamer and first RNA drug for the treatment of neovascular (wet) age-related macular degeneration (AMD). Macugen® is administered by ophthalmic intravitreous injection every 6 weeks. However, pegaptanib has been largely supplanted by its protein-based competitors, monoclonal antibodies bevacizumab (Avastin®)160 and ranibizumab (Lucentis®)161, 162, and recombinant fusion protein aflibercept (Eylea®)163. Although these antibodies bind to all isoforms of VEGF and are more beneficial than pegaptanib for BVCA (Best Corrected Visual Acuity) improvement, it has been reported that blocking all VEGF activity may increase the risk of hypertension and other adverse effects164, 165. To improve benefits while limiting such risks, pegaptanib may be combined with other drugs as an adjuvant agent for long-term treatment.
Another potential approach to treat AMD involves targeting of the complement system, which plays important roles in retinal biology and AMD pathogenesis166. Several main component proteins of the complement system, such as complement component 3 (C3), C5, and the membrane attack complex (MAC), are typically expressed in the capillary vessels of the choroid and the vitreous of the human eye. These components induce VEGF expression and thus accelerate neovascular AMD. Inhibiting C5 activation can prevent inflammatory-mediated tissue injury associated with AMD167. Zimura® (ARC1905, Ophthotech) is a 38-nt modified RNA aptamer that inhibits the proteolytic activation of C5. Its original version was selected from a nuclease-resistant 2′-fluoropyrimidine RNA library via in vitro SELEX168; after truncation and a biased SELEX procedure, a new aptamer with 10-fold higher affinity was obtained. Similar to pegaptanib, ARC1905, the final anti-C5 aptamer used for clinical evaluation, contains 2′-fluoropyrimidines, 2′-methoxypurines, a 3′-idT cap, and 40 kDa-PEG. Recently, a multicenter, open-label phase IIa clinical trial (ClinicalTrials.gov identifier NCT02397954169) evaluating the safety and tolerability of Zimura® intravitreous injection in combination with anti-VEGF monotherapy for the treatment of wet AMD was completed. Zimura® was generally well-tolerated and improved visual acuity when combined with bevacizumab, ranibizumab, or aflibercept (http://www.ophthotech.com/product-candidates/arc1905/). Encouraged by the positive results, Ophthotech is recruiting participants for a phase II/III clinical trial (ClinicalTrials.gov identifier NCT02686658170) to investigate Zimura® for treatment of geographic atrophy, a severe form of dry AMD.
In vitro and animal studies have demonstrated that PDGF is also a potential therapeutic target in the treatment of AMD171, in particular when combined with anti-VEGF therapies172, 173. Fovista® (Ophthotech), an aptamer originally derived from an in vitro DNA SELEX experiment, targets PDGF174–176. The selected aptamer, which specifically inhibited the binding of the PDGF subunit B homodimer to the pericyte cell surface receptor PDGF-β, was chemically modified resulting in a 29-nt modified DNA aptamer that showed no loss of binding affinity, and a 13-fold longer half-life in plasma than the unmodified DNA aptamer177. In a randomized phase IIb clinical trial investigating the safety and efficacy of Fovista® for wet AMD therapy, intravitreous injection of Fovista® in combination with anti-VEGF monotherapy (ranibizumab) (ClinicalTrials.gov identifier NCT01089517178) exhibited a favorable safety profile and improved BCVA, representing a 62% comparative benefit from baseline (http://www.ophthotech.com/product-candidates/fovista/). Currently, several phase III clinical programs have been initiated to evaluate the safety and efficacy of Fovista® administered with bevacizumab, ranibizumab, or aflibercept, compared to anti-VEGF monotherapy (ClinicalTrials.gov identifiers NCT01944839179, NCT01940900180 and NCT01940887181). Ultimately, the combination of C5 or PDGF inhibitory aptamers with anti-VEGF monotherapy may be a promising therapeutic modality.
Coagulation
Four aptamers for coagulation therapy are currently being clinically evaluated. The aptamer-based anticoagulation system, REG1 (Regado Biosciences), comprises a 37-nt modified aptamer (RB006) and a 17-nt antidote oligonucleotide (RB007). RB006 is an inhibitory RNA aptamer targeted to coagulation factor IXa, a proximal driver of clot formation182. Starting with a 2′-fluoropyrimidine-modified RNA library, in vitro SELEX was performed against factor IXa, identifying the original aptamer (9.3) with the highest affinity183; further truncation and modifications led to RB006. RB007 is a complementary antidote sequence, which specifically binds to RB006 and rapidly reverses the anti-factor IXa activity of RE006 in a dose-dependent manner, allowing fine-tuning of the system. In preclinical studies, RB006 prevented arterial thrombosis with less bleeding than with heparin, the current standard of care. The ability to be able to rapidly reverse aptamer function by treatment with a complementary oligo is an important and underappreciated attribute of these macromolecules. The results from Phase I and II studies184–187 (ClinicalTrials.gov identifiers NCT00932100188, NCT00715455189, and NCT01872572190) indicate that the REG1 system is well-tolerated in patients. Bolus administration is feasible and controllable for anticoagulation and reversal in subjects with coronary artery disease or acute coronary syndrome during percutaneous coronary intervention (PCI) and cardiac catheterization.
Recently, Regado conducted a large-scale, randomized comparison of REG1 with bivalirudin, a specific and reversible direct thrombin inhibitor (DTI), in patients undergoing PCI (ClinicalTrials.gov identifier NCT01848106191). However, this trial was terminated prematurely due to an unacceptable risk of severe allergic reactions associated with the current formulation of REG118, 99. Additionally, there was no evidence that REG1 reduced ischemic events or bleeding complications compared with bivalirudin.
ARC1779 (Archemix Corp.) is a 39-nt modified DNA aptamer that specifically recognizes the A1 domain of von Willebrand factor (vWF). vWF plays a central role in primary hemostasis by mediating platelet adhesion to damaged vascular subendothelium and subsequent platelet aggregation192. ARC1779 was originally generated from a DNA library via SELEX, and then subjected to truncation and modifications to obtain an optimized, second-generation, PEGylated version. ARC1779 exerts antithrombotic activity by blocking the interaction between vWF A1 domain and platelet receptor glycoprotein 1B, and shows potential therapeutic benefit in acute coronary syndrome (ACS) and von Willebrand’s disease193, 194. Its evaluation in phase I and II clinical trials suggests favorable pharmacokinetic, pharmacodynamic, and safety properties in healthy individuals and patients with thrombotic thrombocytopenic purpura (TTP) (ClinicalTrials.gov identifiers NCT00432770195 and NCT00632242196). However, Archemix has terminated their phase II studies for acute myocardial infarction in patients undergoing PCI without further updates (ClinicalTrials.gov identifiers NCT00742612197 and NCT00507338198).
In contrast to therapeutic aptamers that are modified for increased nuclease resistance and prolonged half-life in vivo, NU172 is a 26-nt unmodified DNA aptamer. NU172 (ARCA biopharma, Inc.) was selected against thrombin199, and induces a short-acting anticoagulation effect. Originally identified through DNA SELEX, a truncated version was created for clinical study. In a phase Ib proof-of-concept trial, NU172 was well-tolerated without serious adverse responses, and rapidly elicited and maintained anticoagulation with a rapid return toward baseline after the infusion ended, suggesting the potential to be rapidly reversed without the need for an antidote (http://www.wikinvest.com/stock/Nuvelo_(NUVO)/Nu172#toc). ARCA initiated a phase II clinical trial to evaluate NU172 therapeutic efficacy in patients undergoing coronary artery bypass grafting surgery with median sternotomy (ClinicalTrials.gov identifier NCT00808964200), but the current status is unknown.
Unlike the previous aptamers that target the intrinsic coagulation pathway, ARC19499 (Baxter) is a 32-nt modified RNA aptamer that targets tissue factor pathway inhibitor (TFPI), the negative regulator of factor VIIa in the extrinsic tissue factor coagulation pathway201. The parent clone was originally isolated from a modified RNA library via in vitro selection; ARC19499 was generated from truncated clone variants with additional chemical modifications. By specifically inhibiting TFPI, ARC19499 enables clot initiation and propagation, thus mediating a procoagulant hemostatic effect202. In 2010, a phase I study was started to evaluate safety and efficacy of ARC19499 in hemophilia patients, but it was terminated without further updates (ClinicalTrials.gov identifier NCT01191372203).
Oncology
One DNA aptamer (AS1411) and one L-form RNA aptamer (NOX-A12) have undergone clinical trials for the treatment of cancers. AS1411, formerly named ARGO100 (Antisoma), is a 26-nt G-quadruplex DNA oligonucleotide (which was not evolved by classic SELEX204) and is the first aptamer in clinical trials for the treatment of human cancer. It was originally generated from a screen of antisense oligonucleotides with anti-proliferative function, then PEGylated to prolong its half-life for in vivo application. Due to G-quadruplex structures, AS1411 is resistant to nuclease degradation. AS1411 shows high affinity to the external domain of nucleolin205, 206, which is a B-cell lymphoma-2 (Bcl-2) mRNA binding protein involved in cell survival, growth, and proliferation which is overexpressed on the surface of tumor cells207. After binding to nucleolin, AS1411 is rapidly internalized and destabilizes Bcl-2 mRNA, probably leading to the induction of apoptosis. AS1411 can function as a tumor-targeting agent as well as a promising anti-cancer agent208. It inhibited cancer in various preclinical models209, including breast, renal, and lung cancer and in 2007, was reported to be well-tolerated without serious side-effects in an open-label phase I study in advanced solid tumors210 (ClinicalTrials.gov identifier NCT00881244211). In 2009, a phase II trial assessed the efficacy and safety of AS1411 combined with cytarabine, an anti-cancer chemotherapy drug, in the treatment of patients with primary refractory or relapsed acute myeloid leukemia (AML) (ClinicalTrials.gov identifier NCT00512083212). However, a subsequent phase II trial for renal cell carcinoma showed that AS1411 had minimal activity in unselected patients (ClinicalTrials.gov identifier NCT00740441213). Only one patient with fibroblast growth factor receptor (FGFR2) and mTOR mutations showed an excellent response, implying that future efforts may benefit from discovery of predictive biomarkers of response to AS1411 therapy.
NOX-A12214, 215 is a 45-nt L-form RNA aptamer known as a Spiegelmer®, developed against chemokine (C-X-C motif) ligand 12 (CXCL-12). Also known as stromal cell-derived factor-1 (SDF-1)216–218, CXCL-12 plays important roles in tumor proliferation, new blood vessel formation, and metastasis. It has two types of binding sites, both of which are targets of therapeutic intervention. By binding to these two key sites, NOX-A12 disrupts CXCL-12 activity and modulates the tumor microenvironment to fight tumors, including multiple myeloma and solid tumors (lung, colorectal, and brain cancer). NOX-A12 was developed by NOXXON (Box 2 and Table 3) (http://www.noxxon.com/index.php?option=com_content&view=article&id=21&Itemid=478). Spiegelmers® are mirror-image oligonucleotides that cannot be recognized by nucleases, and thus display high biostability in the bloodstream/human serum NOX-E3675. In phase I studies, NOX-A12 had a favorable safety profile when administered IV to healthy individuals (ClinicalTrials.gov identifier NCT00976378219 and NCT01194934220). Currently, NOX-A12 is being evaluated in two phase II clinical trials in combination with either chemoimmunotherapy (bendamustine and rituximab) in patients with relapsed CLL (ClinicalTrials.gov identifier NCT01486797221), or bortezomib (proteasome inhibitor) and dexamethasone (corticosteroid) in patients with relapsed multiple myeloma (ClinicalTrials.gov identifier NCT01521533222).
Table 3.
Key companies in the global aptamers market
| Company name | Founded/location | Focus and exclusive technology | Featured products and services |
|---|---|---|---|
| NOXXON Pharma AG | 1997/Berlin, Germany |
Therapeutics development: Mirror-image chemistry/Spiegelmer® (RNA or DNA-L-stereoisomer) -based therapeutics for cancer, inflammation, obesity or other diseases. |
|
| Ophthotech Corp. | 2007/New York, USA |
Therapeutics development: Modified RNA or DNA aptamer therapeutics for eye diseases: |
|
| Archemix (acquired by Baxter in 2010) |
2000/San Francisco, USA |
Therapeutics development: Modified RNA or DNA aptamers therapeutics for cardiovascular, hematology, and oncology diseases. |
|
| NeXstar (merged with Gilead) | 1999/Boulder, Colorado, USA |
Therapeutics development: SELEX license/Modified RNA or DNA aptamers therapeutics for transplant rejection and other immunological responses and aptamer-based diagnostics |
|
| SomaLogic | 1999/Boulder, Colorado, USA |
Diagnostics development: SOMAmer (Slow Off-rate Modified Aptamer) for diagnosis and proteomic analysis; |
|
| NeoVentures | 2002/Ontario, Canada |
R & D support and diagnostics development: Custom aptamer identification; diagnostics development for detecting toxins in wine, corn, beer and wheat. |
|
| AptaMatrix | 2003/New York, USA |
R & D support and diagnostics development: High throughput screening of aptamers (HTSA); diagnostics development |
|
| Aptamer Solutions (Aptasol) | 2008/York, United Kingdom |
R & D support and diagnostics development: Automated high throughput selection systems; Custom aptamer identification; diagnostics and biomarker development |
|
| Aptus Biotech | 2010/Madrid, Spain |
R & D support and diagnostics development: Custom aptamer identification; aptamer; diagnostics development |
|
| Aptagen | 2004/Jacobus, Pennsylvania, USA |
R & D support and diagnostics development: Custom aptamer identification; aptamer products and aptamer database |
|
| Aptamer Sciences (AptaSci) | 2011/Pohang, South Korea |
R & D support: Custom aptamer identification; aptamer products; |
|
| AM Biotechnologies | 2006/Houston, Texas, USA |
R & D support: X-aptamer technology; Thioaptamers (based on phosphorodithioate (PS2) DNA) |
|
| Vivonics | 2012/Sudbury, Massachusetts, USA |
R & D support: One-step rapid technique for developing aptamers (RIDA) |
|
| TriLink | 1996/San Diego, California, USA |
R & D support: Solid-phase chemical synthesis of RNA and DNA |
|
Inflammation
Two anti-inflammatory aptamers (NOX-E36 and NOX-H94) are under clinical development for the treatment of diabetic nephropathy and anemia of chronic disease, respectively. Like NOX-A12, they are PEGylated L-form RNA aptamers generated through Spiegelmer® technology by NOXXON. The 40-nt aptamer NOX-E3674, 223–225 binds human chemokine (C-C motif) ligand 2 (CCL2, also known as monocyte chemoattractant protein 1 (MCP-1)), and can specifically inhibit the chemokine’s activity, thereby preventing infiltration of pro-inflammatory cells into the kidney. CCL2 plays a key role in modulating migration and infiltration of monocytes/macrophages and it is overexpressed in several inflammatory diseases217. Evaluation in phase I and II clinical trials conducted in diabetic nephropathy patients showed that NOX-E36 was safe, well-tolerated, and showed promising efficacy for at least 12 weeks after termination of treatment (http://www.noxxon.com/index.php?option=com_content&view=article&id=20&Itemid=477) (ClinicalTrials.gov identifiers NCT01085292226 and NCT01547897227).
The 44-nt L-form RNA aptamer NOX-H94 binds human hepcidin, a regulator of iron metabolism228, thereby blocking hepcidin-induced ferroportin degradation and ferritin expression. Inhibition of hepcidin is a therapeutic approach for anemia of chronic inflammation229. In a pharmacodynamics study with healthy individuals (ClinicalTrials.gov identifier NCT01372137230), NOX-H94 blocked hepcidin-mediated effects on serum iron levels (http://www.noxxon.com/index.php?option=com_content&view=article&id=22&Itemid=479). Recently, two phase II evaluations were completed for NOX-H94 in anemic cancer patients and ESA-hyporesponsive anemia dialysis patients (ClinicalTrials.gov identifiers NCT01691040231 and NCT02079896232). Treatment with NOX-H94 induces an increase in hemoglobin (Hb) levels, suggesting hepcidin inhibition may also be an effective target for the treatment of cancer-associated anemia with functional iron deficiency.
Conclusions
The current paucity of commercial success with aptamer-based products may be due to several factors. Early exclusive intellectual property protection for SELEX technology limited initial distribution. In addition, in contrast to antibodies, with a well-developed commercial infrastructure, the development pathway for aptamers, education and investment, and related knowledge (e.g., medicinal formulation, pharmacokinetic/pharmacodynamic properties, and toxicity) are insufficient. Furthermore, challenges associated with design and formulation of RNA-based therapeutics still exist. The future success of aptamers as therapeutics relies on overcoming these challenges and taking full advantage of the unique attributes of aptamers.
Despite the limited commercial success with aptamer-based therapeutics, the lessons learned from previous failures and successful technological advancements are valuable for informing us of future opportunities for aptamer-based therapeutics. Proof-of-concept studies in aptamer technology continue to reveal their promising functionality and vast therapeutic potential. As a unique class of biomolecules, aptamers not only possess the flexibility of small molecules, allowing them to contact binding sites that may not be accessible to larger antibodies, but also possess the high specificity of antibodies, providing targeted disease therapy that small drug molecules cannot confer. These unique advantages of aptamers could fill a niche market, such as bacterial and viral treatment, cancer immunotherapy, antitoxins or targeted delivery. For example, rapid aptamer selection technology has the potential to keep up with viral emergence and mutation, and could fill the absence of fast-track vaccines. Additionally, the ability of many aptamers to internalize upon binding to cellular receptors, in a cell-specific manner, will likely improve the targeting specificity and pharmacokinetic profile of the nanocarrier, resulting in synergistic therapeutic effects when two or more therapeutic agents are administered simultaneously in a combinatorial therapy format.
The aptamer field has likely touched only the tip of the iceberg. As more researchers devote themselves to rational aptamer development, next generation aptamer-based therapeutics with superior biological function and pharmacokinetic profiles are highly anticipated. Furthermore, factors such as market demand for cost-effective treatments, technological advancements in synthesis and formulation, and expiration of the SELEX patent, provide a strong impetus for development of this promising class of therapeutics.
Key points.
Nucleic acid aptamers, often termed “chemical antibodies”, are short, single-stranded (ss) DNA or RNA molecules (20 - 100 nt) with defined structures that can specifically bind to a molecular target via three-dimensional structures.
Similar to the way antibodies bind to antigens, aptamers specifically recognize and bind to their cognate targets through unique three-dimensional structures.
SELEX (systematic evolution of ligands by exponential enrichment) is a gold-standard methodology for generating aptamers, in which an iterative selection procedure including binding, partitioning, recovery, and re-amplification steps is conducted. Specific sequences (i.e., aptamers) can be enriched and dominate the population of library species.
Aptamer-based therapeutics typically exploit one of three strategies: (1) an aptamer serves as an antagonist for blocking the interaction of disease-associated targets, e.g., receptor-ligand; (2) an aptamer serves as an agonist for activating the function of target receptors; or (3) a cell type-specific aptamer serves as a carrier for delivering other therapeutic agents to the target cells or tissue.
There are three aptamers designated for use in ophthalmology, including one FDA-approved drug (Macugen®/pegaptanib), and two in late-stage development (Zimura®/ACR1905 and Fovista®/E10030).
Six RNA and four DNA aptamers have undergone clinical trials for the treatment of various conditions, including macular degeneration, coagulation, oncology, and inflammation. All aptamers that have entered clinical trials so far act as antagonists.
Acknowledgments
This work was supported by the National Institutes of Health [grant numbers R01AI29329, R01AI42552, and R01HL07470 to J.J.R.]. Funding for open access charge: National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We would like to thank Dr. Sarah T. Wilkinson (City of Hope) for helpful advice in scientific writing.
Glossary
- Nucleic acid aptamers
short, single-stranded (ss) DNA or RNA molecules (20 - 100 nt) with defined structures that can specifically bind to a molecular target via three-dimensional structures
- SELEX (Systematic evolution of ligands by exponential enrichment)
an iterative selection procedure for aptamer generation. Each cycle consists of binding, partitioning, recovery, and re-amplification steps. Specific sequences (i.e., aptamers) can be enriched and dominate the population of library species
- Hairpin structure
A complementary region of nucleic acid that can form Watson-Crick base pairs and generally results in a stem-loop structure (hairpin)
- Kissing hairpin
If two RNA stem-loops have complementary sequences in the loop regions, the two loops will base pair to form a kissing complex
- G-quadruplex structure
A G-quadruplex structure can be formed in a guanine-rich sequence when four guanine bases are associated through hydrogen bonding
- Macugen®
trade name of pegaptanib, a modified RNA aptamer targeting vascular endothelial growth factor (VEGF), and the first federally approved aptamer drug for the treatment of wet age-related macular degeneration (AMD)
- Bevacizumab
trade name Avastin®, a recombinant humanized monoclonal antibody against VEGF, and an approved antibody drug for the treatment of certain metastatic cancers and eye diseases
- Ranibizumab
trade name Lucentis®, a monoclonal antibody fragment created from bevacizumab, and an approved antibody drug for the treatment of AMD
- Aflibercept
trade name Eylea®, a recombinant fusion protein inhibitor of VEGF, and an approved biopharmaceutical drug for the treatment of wet AMD
- Emulsion PCR (ePCR) or droplet digital PCR (ddPCR)
These systems compartmentalize and miniaturize PCR reaction by generating a water-in-oil emulsion containing numerous droplets, which creates a local homogenous amplification microenvironment
- High throughput sequencing (HTS)
a next generation sequencing technology that is capable of parallelizing the sequencing process and producing thousands or millions of sequences at once
- Zimura®
trade name of ACR1905, a modified RNA aptamer targeting complement 5 (C5) protein that has undergone clinical trials for the treatment of dry AMD
- Fovista®
trade name of E10030, a modified DNA aptamer targeting platelet-derived growth factor (PDGF) that has undergone clinical trials for wet AMD therapy
- VEGF (vascular endothelial growth factor)
a secreted protein that is capable of inducing angiogenesis and increasing vascular permeability and inflammation
- REG1 system
comprises a modified RNA aptamer RB006 (Pegnivacogin) and an antidote oligonucleotide RB007 (Anivamersen), the first aptamer-based anti-coagulation system in clinical trials
- von Willebrand factor (vWF)
a key factor in the coagulation cascade associated with platelet recruitment. A deficiency or impairment of vWF causes von Willebrand disease
- von Willebrand disease
a condition that can cause extended or excessive bleeding
- Spiegelmer®
trade name of L-ribonucleic acid aptamer developed by NOXXON; a mirror-image RNA built from natural L-ribose units
- SDF-1 (Stroma cell-derived factor-1)
also known as CXCL-12 (chemokine (C-X-C motif) ligand 12), a small cytokine protein that plays important roles in tumor proliferation, new blood vessel formation and metastasis
- Hepcidin
a small 2.8 kDa peptide thought to be the central mediator of iron homeostasis
- SOMAmer (Slow Off-rate Modified Aptamer)
chemically modified DNA aptamers highly specific for their respective cognate protein target; selected through in vitro SELEX procedure, in which modified deoxyuracil carrying hydrophic functional groups is incorporated. More than 1,300 different SOMAmer agents have been developed by SomaLogic
- Costimulatory receptors
a class of molecules expressed by T lymphocytes that regulate the activation of T cells and the generation of effector T-cell responses, including OX40, 4-1BB, CD40, CD28, and PD-1. OX40, 4-1BB, and CD40 belong to the tumor necrosis factor (TNF) family, involved in the later phase of T-cell activation, while CD28 is the member of the larger immunoglobulin superfamily, involved in triggering of cell-mediated immune response
- NMD (Non-sense mediated mRNA decay)
a translation-coupled mechanism that degrades mRNA containing premature translation-termination codons (PTC)
- RNAi (RNA interference)
a highly conserved endogenous process for post-transcriptional regulation of gene silencing, triggered by small regulatory RNAs including small interfering RNAs
- siRNA (small interfering RNA)
a class of double-stranded RNA molecules of 20–25 base-pair in length, capable of triggering sequence specific, posttranscriptional gene silencing
- shRNA (small hairpin RNA)
an artificial RNA molecule with a short hairpin turn; like siRNA, another class of RNAi trigger
- miR (microRNA)
a class of small non-coding RNA molecules of 22 nucleotides. The mechanism of miRNA-mediated silencing is repression of target mRNA translation accompanied by deadenylation and subsequent degradation of the mRNA targets
- anti-miR
a synthetic oligonucleotide designed to neutralize miRNA function
- HIV-1 gp120
HIV-1 envelope glycoprotein gp120 is exposed on the surface of virus particles and the plasma membrane of HIV-1 infected cells. The interaction of HIV-1 gp120 with the cellular CD4 receptor is a crucial step in the entry of HIV into T-cells
- Gelonin toxin
a small N-glycosidase protein of 28 kDa capable of inducing cell death
- Plekho1 (pleckstrin homology domain-containing family O member 1)
plays a role in the regulation of the actin cytoskeleton through its interactions with actin capping protein (CP)
Biographies
Dr. John J. Rossi is Chairman and Professor of Department of Molecular and Cellular Biology at Beckman Research Institute of City of Hope, and is also Associate Director of Comprehensive Cancer Center of City of Hope. He received his B.Sc degree in Biology from University of New Hampshire and Ph.D. degree in Microbial Genetics from University of Connecticut, and thereafter accepted his postdoctoral training in Brown University. Dr. Rossi has worked in health-related biomedical research at City of Hope for over 35 years. His research primary focuses on the functions and therapeutic applications of small RNAs, such as, ribozyme, siRNA, microRNA and aptamers.
Dr. Jiehua Zhou is Assistant Research Professor of Department of Molecular and Cellular Biology at Beckman Research Institute of City of Hope. She received her B.Sc degree in Chemistry and Ph.D. degree in Bioorganic Chemistry from Wuhan University, and thereafter was a postdoctoral fellow in the laboratory of Dr. John Rossi at the City of Hope. Dr. Zhou’s research encompasses the basic science and application of non-coding RNA and polymer-based nanotechnology for targeted HIV-1 and cancer therapy, including RNAi/siRNA/microRNA/mRNA, SELEX/aptamer, aptamer or nanoparticle mediated gene delivery.
Footnotes
Competing interests statement
J.J.R. and J.Z. have an issued patent entitled “Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy” [USPTO, No. US 8, 222, 226 B2, July 17, 2012]. J.J.R., J.Z., M.S.W., and K.V.M. have a patent pending on “Cell-specific internalizing RNA aptamers against human CCR5 and used therefore” [United States Patent, application number: 62/025, 368, filed on July 16, 2014].
Subject categories
Biological sciences/Drug discovery [URI/631/154]
Biological sciences/Biotechnology/Biologics/Nucleic-acid therapeutics [URI/631/61/51/391]
Biological sciences/Biotechnology/Biologics/Antibody therapy [URI/631/61/51/1568]
Biological sciences/Drug discovery/Drug delivery [URI/631/154/152]
Health sciences/Diseases/Eye diseases/Macular degeneration [URI/692/699/3161/1626]
Further information (Company website)
Ophthotech. ZimuraTM anti-C5 clinical development. http://www.ophthotech.com/product-candidates/arc1905/ (2015).
Ophthotech. FovistaTM (anti-PDGF) therapy fact sheet. http://www.ophthotech.com/product-candidates/fovista/ (2015).
ARCA. NU172 annual report. http://www.wikinvest.com/stock/Nuvelo_(NUVO)/Nu172#toc (2009).
NOXXON. Product candidates: NOX-A12. http://www.noxxon.com/index.php?option=com_content&view=article&id=21&Itemid=478 (2016).
NOXXON. Product candidates: NOX-E36. http://www.noxxon.com/index.php?option=com_content&view=article&id=20&Itemid=477 (2016).
NOXXON. Product candiates: NOX-H94. http://www.noxxon.com/index.php?option=com_content&view=article&id=22&Itemid=479 (2016).
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. One of the first three publications of the SELEX technology. A related smaller randomized library theoretically containing 48 individual sequences was used for selection of RNA ligands for T4 DNA polymerase. This procedure was thus name the “systemic evolution of ligands by exponential enrichment” (SELEX). [DOI] [PubMed] [Google Scholar]
- 2.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–8. doi: 10.1038/344467a0. One of the first three publications of the SELEX technology. The first RNA enzyme was selected via in vitro selection that could cleave single-strand DNA specifically. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. One of the first three publications of the SELEX technology. The first example of RNA aptamer specific to small organic dyes. [DOI] [PubMed] [Google Scholar]
- 4.Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48:2672–89. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 5.Gelinas AD, Davies DR, Janjic N. Embracing proteins: structural themes in aptamer-protein complexes. Curr Opin Struct Biol. 2016;36:122–32. doi: 10.1016/j.sbi.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J Biol Chem. 1995;270:13581–4. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 7.Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–36. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–9. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 9.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–3. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, et al. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc Natl Acad Sci U S A. 2015;112:10002–7. doi: 10.1073/pnas.1502159112. The first published RNA aptamer specific to single amino acid mutatation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–50. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259–71. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7:9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng EW, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–32. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqui MA, Keating GM. Pegaptanib: in exudative age-related macular degeneration. Drugs. 2005;65:1571–7. doi: 10.2165/00003495-200565110-00010. discussion 1578–9. [DOI] [PubMed] [Google Scholar]
- 16.Mousa SA, Mousa SS. Current status of vascular endothelial growth factor inhibition in age-related macular degeneration. BioDrugs. 2010;24:183–94. doi: 10.2165/11318550-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 18.Lincoff AM, et al. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. Lancet. 2016;387:349–56. doi: 10.1016/S0140-6736(15)00515-2. An recent update from REG1 anticoagulation system in a randomised clinical trial. The severe allergic reactions observed in some patients have been linked to pre-existing antibodies to PEG. [DOI] [PubMed] [Google Scholar]
- 19.Verheugt FW. An anticoagulant too good to be true for revascularisation. Lancet. 2016;387:314–5. doi: 10.1016/S0140-6736(15)00727-8. [DOI] [PubMed] [Google Scholar]
- 20.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–6. doi: 10.1038/355564a0. The first DNA aptamers targeting the protease thrombin of the blood coagulation cascade were identified through in vitro SELEX. [DOI] [PubMed] [Google Scholar]
- 21.Shu Y, et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulbachinskiy AV. Methods for selection of aptamers to protein targets. Biochemistry (Mosc) 2007;72:1505–18. doi: 10.1134/s000629790713007x. [DOI] [PubMed] [Google Scholar]
- 23.Ozer A, Pagano JM, Lis JT. New Technologies Provide Quantum Changes in the Scale, Speed, and Success of SELEX Methods and Aptamer Characterization. Mol Ther Nucleic Acids. 2014;3:e183. doi: 10.1038/mtna.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keefe AD, Cload ST. SELEX with modified nucleotides. Curr Opin Chem Biol. 2008;12:448–56. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–7. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 26.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–83. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 27.Pestourie C, et al. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides. 2006;16:323–35. doi: 10.1089/oli.2006.16.323. [DOI] [PubMed] [Google Scholar]
- 28.Cerchia L, de Franciscis V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010;28:517–25. doi: 10.1016/j.tibtech.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Cerchia L, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Liu Y, et al. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol Chem. 2009;390:137–44. doi: 10.1515/BC.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo KT, Paul A, Schichor C, Ziemer G, Wendel HP. CELL-SELEX: Novel Perspectives of Aptamer-Based Therapeutics. Int J Mol Sci. 2008;9:668–78. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohuchi S. Cell-SELEX Technology. Biores Open Access. 2012;1:265–72. doi: 10.1089/biores.2012.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. Cell-specific aptamers for targeted therapies. Methods Mol Biol. 2009;535:59–78. doi: 10.1007/978-1-59745-557-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–7. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 35.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14:1769–75. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 36.Phillips JA, Lopez-Colon D, Zhu Z, Xu Y, Tan W. Applications of aptamers in cancer cell biology. Anal Chim Acta. 2008;621:101–8. doi: 10.1016/j.aca.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Avci-Adali M, Metzger M, Perle N, Ziemer G, Wendel HP. Pitfalls of cell-systematic evolution of ligands by exponential enrichment (SELEX): existing dead cells during in vitro selection anticipate the enrichment of specific aptamers. Oligonucleotides. 2010;20:317–23. doi: 10.1089/oli.2010.0253. [DOI] [PubMed] [Google Scholar]
- 38.Raddatz MS, et al. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew Chem Int Ed Engl. 2008;47:5190–3. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 39.Healy JM, et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. 2004;21:2234–46. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 40.Mi J, et al. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6:22–4. doi: 10.1038/nchembio.277. The first pulished RNA aptamers in vivo isolated from mice bearing liver tumor. The target protein of the selected aptamer is p68, an RNA helicase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng C, Chen YH, Lennox KA, Behlke MA, Davidson BL. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol Ther Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. The first brain-penetrating RNA aptamer via in vivo SELEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv. 2015;33:1141–61. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–2. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 44.Drabovich AP, Berezovski M, Okhonin V, Krylov SN. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal Chem. 2006;78:3171–8. doi: 10.1021/ac060144h. [DOI] [PubMed] [Google Scholar]
- 45.Yufa R, et al. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal Chem. 2015;87:1411–9. doi: 10.1021/ac5044187. [DOI] [PubMed] [Google Scholar]
- 46.Mosing RK, Mendonsa SD, Bowser MT. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal Chem. 2005;77:6107–12. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 47.Miyachi Y, Shimizu N, Ogino C, Kondo A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010;38:e21. doi: 10.1093/nar/gkp1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng L, Stephens BJ, Bonin K, Cubicciotti R, Guthold M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Microsc Res Tech. 2007;70:372–81. doi: 10.1002/jemt.20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer G, et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 50.Huang CJ, Lin HI, Shiesh SC, Lee GB. Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX) Biosens Bioelectron. 2010;25:1761–6. doi: 10.1016/j.bios.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 51.Cho M, et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc Natl Acad Sci U S A. 2010;107:15373–8. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayer N, Ibrahim J, Turner K, Tahiri-Alaoui A, James W. Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem Biophys Res Commun. 2002;293:924–31. doi: 10.1016/S0006-291X(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 53.Kanagawa T. Bias and artifacts in multitemplate polymerase chain reactions (PCR) J Biosci Bioeng. 2003;96:317–23. doi: 10.1016/S1389-1723(03)90130-7. [DOI] [PubMed] [Google Scholar]
- 54.Musheev MU, Krylov SN. Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal Chim Acta. 2006;564:91–6. doi: 10.1016/j.aca.2005.09.069. [DOI] [PubMed] [Google Scholar]
- 55.Schutze T, et al. A streamlined protocol for emulsion polymerase chain reaction and subsequent purification. Anal Biochem. 2011;410:155–7. doi: 10.1016/j.ab.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 56.Shao K, et al. Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS One. 2011;6:e24910. doi: 10.1371/journal.pone.0024910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakano M, et al. Single-molecule PCR using water-in-oil emulsion. J Biotechnol. 2003;102:117–24. doi: 10.1016/s0168-1656(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 58.Ouellet E, Foley JH, Conway EM, Haynes C. Hi-Fi SELEX: A High-Fidelity Digital-PCR Based Therapeutic Aptamer Discovery Platform. Biotechnol Bioeng. 2015;112:1506–22. doi: 10.1002/bit.25581. [DOI] [PubMed] [Google Scholar]
- 59.Levay A, et al. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic Acids Res. 2015;43:e82. doi: 10.1093/nar/gkv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoon S, Zhou B, Janda KD, Brenner S, Scolnick J. Aptamer selection by high-throughput sequencing and informatic analysis. Biotechniques. 2011;51:413–6. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- 61.Hoinka J, et al. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 2015;43:5699–707. doi: 10.1093/nar/gkv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiel WH, et al. Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid Ther. 2011;21:253–63. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiel WH, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuwahara M, Sugimoto N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules. 2010;15:5423–44. doi: 10.3390/molecules15085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jager S, et al. A versatile toolbox for variable DNA functionalization at high density. J Am Chem Soc. 2005;127:15071–82. doi: 10.1021/ja051725b. [DOI] [PubMed] [Google Scholar]
- 66.Hirao I, et al. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods. 2006;3:729–35. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 67.Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 1994;22:5229–34. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruckman J, et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556–67. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 69.Burmeister PE, et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 70.Kuwahara M, Obika S. In vitro selection of BNA (LNA) aptamers. Artif DNA PNA XNA. 2013;4:39–48. doi: 10.4161/adna.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veedu RN, Wengel J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol Biosyst. 2009;5:787–92. doi: 10.1039/b905513b. [DOI] [PubMed] [Google Scholar]
- 72.Gold L, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaught JD, et al. Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc. 2010;132:4141–51. doi: 10.1021/ja908035g. [DOI] [PubMed] [Google Scholar]
- 74.Maasch C, Buchner K, Eulberg D, Vonhoff S, Klussmann S. Physicochemical stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for therapeutic applications. Nucleic Acids Symp Ser (Oxf) 2008:61–2. doi: 10.1093/nass/nrn031. [DOI] [PubMed] [Google Scholar]
- 75.Vater A, Klussmann S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr Opin Drug Discov Devel. 2003;6:253–61. [PubMed] [Google Scholar]
- 76.Lee Y, Urban JH, Xu L, Sullenger BA, Lee J. 2′Fluoro Modification Differentially Modulates the Ability of RNAs to Activate Pattern Recognition Receptors. Nucleic Acid Ther. 2016 doi: 10.1089/nat.2015.0575. RNAs containing 2′fluoro (2′F) pyrimidines differentially controlled the activation of PRR. The results demonstrate that RNAs containing 2′F pyrimidine and 5′ triphosphate (5′ppp) increased cell death and interferon-beta expression in human cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aaldering LJ, et al. Smart functional nucleic acid chimeras: enabling tissue specific RNA targeting therapy. RNA Biol. 2015;12:412–25. doi: 10.1080/15476286.2015.1017234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol. 2010;5:833–42. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rusconi CP, et al. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol. 2004;22:1423–8. doi: 10.1038/nbt1023. [DOI] [PubMed] [Google Scholar]
- 80.Lee CH, et al. Pharmacokinetics of a Cholesterol-conjugated Aptamer Against the Hepatitis C Virus (HCV) NS5B Protein. Mol Ther Nucleic Acids. 2015;4:e254. doi: 10.1038/mtna.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dougan H, et al. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27:289–97. doi: 10.1016/s0969-8051(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 82.Heo K, et al. An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies. J Control Release. 2016;229:1–9. doi: 10.1016/j.jconrel.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 83.Willis MC, et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug Chem. 1998;9:573–82. doi: 10.1021/bc980002x. [DOI] [PubMed] [Google Scholar]
- 84.Zhou J, et al. A hybrid DNA aptamer-dendrimer nanomaterial for targeted cell labeling. Macromol Biosci. 2009;9:831–5. doi: 10.1002/mabi.200900046. [DOI] [PubMed] [Google Scholar]
- 85.Musumeci D, Montesarchio D. Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol Ther. 2012;136:202–15. doi: 10.1016/j.pharmthera.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 86.Soule EE, Bompiani KM, Woodruff RS, Sullenger BA. Targeting Two Coagulation Cascade Proteases with a Bivalent Aptamer Yields a Potent and Antidote-Controllable Anticoagulant. Nucleic Acid Ther. 2016;26:1–9. doi: 10.1089/nat.2015.0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim Y, Dennis DM, Morey T, Yang L, Tan W. Engineering dendritic aptamer assemblies as superior inhibitors of protein function. Chem Asian J. 2010;5:56–9. doi: 10.1002/asia.200900421. [DOI] [PubMed] [Google Scholar]
- 88.Drolet DW, et al. Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm Res. 2000;17:1503–10. doi: 10.1023/a:1007657109012. [DOI] [PubMed] [Google Scholar]
- 89.Tucker CE, et al. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J Chromatogr B Biomed Sci Appl. 1999;732:203–12. doi: 10.1016/s0378-4347(99)00285-6. [DOI] [PubMed] [Google Scholar]
- 90.Borbas KE, Ferreira CS, Perkins A, Bruce JI, Missailidis S. Design and synthesis of mono- and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to anti-MUC1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconjug Chem. 2007;18:1205–12. doi: 10.1021/bc0700741. [DOI] [PubMed] [Google Scholar]
- 91.Choi DY, et al. Sustained elevated intraocular pressures after intravitreal injection of bevacizumab, ranibizumab, and pegaptanib. Retina. 2011;31:1028–35. doi: 10.1097/IAE.0b013e318217ffde. [DOI] [PubMed] [Google Scholar]
- 92.Steffensmeier AC, Azar AE, Fuller JJ, Muller BA, Russell SR. Vitreous injections of pegaptanib sodium triggering allergic reactions. Am J Ophthalmol. 2007;143:512–3. doi: 10.1016/j.ajo.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 93.Agrawal S, Joshi M, Christoforidis JB. Vitreous inflammation associated with intravitreal anti-VEGF pharmacotherapy. Mediators Inflamm. 2013;2013:943409. doi: 10.1155/2013/943409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Falavarjani KG, Nguyen QD. Adverse events and complications associated with intravitreal injection of anti-VEGF agents: a review of literature. Eye (Lond) 2013;27:787–94. doi: 10.1038/eye.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boyer DS, Goldbaum M, Leys AM, Starita C, Group, V.I.S.I.O.N.S. Effect of pegaptanib sodium 0.3 mg intravitreal injections (Macugen) in intraocular pressure: posthoc analysis from V.I.S.I.O.N. study. Br J Ophthalmol. 2014;98:1543–6. doi: 10.1136/bjophthalmol-2013-304075. [DOI] [PubMed] [Google Scholar]
- 96.Henry SP, et al. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther. 1997;281:810–6. [PubMed] [Google Scholar]
- 97.Farman CA, Kornbrust DJ. Oligodeoxynucleotide studies in primates: antisense and immune stimulatory indications. Toxicol Pathol. 2003;31(Suppl):119–22. doi: 10.1080/01926230390174995. [DOI] [PubMed] [Google Scholar]
- 98.Swayze EE, et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ganson NJ, et al. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J Allergy Clin Immunol. 2016;137:1610–1613 e7. doi: 10.1016/j.jaci.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Waring MJ. Lipophilicity in drug discovery. Expert Opin Drug Discov. 2010;5:235–48. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- 101.Zhou J, Rossi JJ. Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Mol Ther Nucleic Acids. 2014;3:e169. doi: 10.1038/mtna.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shum KT, Zhou J, Rossi JJ. Aptamer-based therapeutics: new approaches to combat human viral diseases. Pharmaceuticals (Basel) 2013;6:1507–42. doi: 10.3390/ph6121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kohn DB, et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–71. [PubMed] [Google Scholar]
- 104.DiGiusto DL, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Michienzi A, Li S, Zaia JA, Rossi JJ. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci U S A. 2002;99:14047–52. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Unwalla HJ, Rossi JJ. A dual function TAR Decoy serves as an anti-HIV siRNA delivery vehicle. Virol J. 2010;7:33. doi: 10.1186/1743-422X-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Drolet DW, Green LS, Gold L, Janjic N. Fit for the Eye: Aptamers in Ocular Disorders. Nucleic Acid Ther. 2016 doi: 10.1089/nat.2015.0573. A compherensive review article about the development and current progress of three aptamers in ophthalmology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eyetech Study G. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22:143–52. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 109.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 110.Axel DI, et al. Paclitaxel inhibits arterial smooth muscle cell proliferation and migration in vitro and in vivo using local drug delivery. Circulation. 1997;96:636–45. doi: 10.1161/01.cir.96.2.636. [DOI] [PubMed] [Google Scholar]
- 111.Thiel WH, et al. Smooth Muscle Cell-targeted RNA Aptamer Inhibits Neointimal Formation. Mol Ther. 2016;24:779–87. doi: 10.1038/mt.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nimjee SM, Rusconi CP, Harrington RA, Sullenger BA. The potential of aptamers as anticoagulants. Trends Cardiovasc Med. 2005;15:41–5. doi: 10.1016/j.tcm.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 113.Bompiani KM, et al. Probing the coagulation pathway with aptamers identifies combinations that synergistically inhibit blood clot formation. Chem Biol. 2014;21:935–44. doi: 10.1016/j.chembiol.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nimjee SM, et al. Synergistic effect of aptamers that inhibit exosites 1 and 2 on thrombin. RNA. 2009;15:2105–11. doi: 10.1261/rna.1240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dyke CK, et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–7. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- 116.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci U S A. 2003;100:9226–31. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dollins CM, et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15:675–82. doi: 10.1016/j.chembiol.2008.05.016. One of the first two publications of using aptamer as agonist. Bivalent aptamer was created to achieve agonic effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pratico ED, Sullenger BA, Nair SK. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013;23:35–43. doi: 10.1089/nat.2012.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McNamara JO, et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–86. doi: 10.1172/JCI33365. One of the first two publications of using aptamer as agonist. Bivalent aptamer was created to achieve agonic effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soldevilla MM, et al. 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials. 2015;67:274–85. doi: 10.1016/j.biomaterials.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 121.Pastor F, et al. CD28 aptamers as powerful immune response modulators. Mol Ther Nucleic Acids. 2013;2:e98. doi: 10.1038/mtna.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ramaswamy V, et al. DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist. Nucleic Acid Ther. 2015;25:227–34. doi: 10.1089/nat.2014.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yunn NO, et al. Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic Acids Res. 2015;43:7688–701. doi: 10.1093/nar/gkv767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gilboa E, McNamara J, 2nd, Pastor F. Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin Cancer Res. 2013;19:1054–62. doi: 10.1158/1078-0432.CCR-12-2067. [DOI] [PubMed] [Google Scholar]
- 125.Khedri M, Rafatpanah H, Abnous K, Ramezani P, Ramezani M. Cancer immunotherapy via nucleic acid aptamers. Int Immunopharmacol. 2015;29:926–36. doi: 10.1016/j.intimp.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 126.Compaan DM, Hymowitz SG. The crystal structure of the costimulatory OX40-OX40L complex. Structure. 2006;14:1321–30. doi: 10.1016/j.str.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 127.Tasch J, Gong M, Sadelain M, Heston WD. A unique folate hydrolase, prostate-specific membrane antigen (PSMA): a target for immunotherapy? Crit Rev Immunol. 2001;21:249–61. [PubMed] [Google Scholar]
- 128.McNamara JO, 2nd, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi: 10.1038/nbt1223. One of the first two publications of using PSMA aptamers for siRNA delivery. [DOI] [PubMed] [Google Scholar]
- 129.Dassie JP, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–49. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–30. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ni X, et al. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest. 2011 doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wullner U, et al. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets. 2008;8:554–65. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- 133.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pastor F, Kolonias D, McNamara JO, 2nd, Gilboa E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol Ther. 2011;19:1878–86. doi: 10.1038/mt.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou J, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–9. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou J, et al. Functional In Vivo Delivery of Multiplexed Anti-HIV-1 siRNAs via a Chemically Synthesized Aptamer With a Sticky Bridge. Mol Ther. 2013;21:192–200. doi: 10.1038/mt.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Neff CP, et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3:66ra6. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Catuogno S, Rienzo A, Di Vito A, Esposito CL, de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J Control Release. 2015;210:147–59. doi: 10.1016/j.jconrel.2015.05.276. [DOI] [PubMed] [Google Scholar]
- 140.Shu D, et al. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano. 2015;9:9731–40. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chu TC, et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–92. doi: 10.1158/0008-5472.CAN-05-4583. The first example using PSMA aptamers for toxin delivery. [DOI] [PubMed] [Google Scholar]
- 142.Hernandez LI, et al. Methods for Evaluating Cell-Specific, Cell-Internalizing RNA Aptamers. Pharmaceuticals (Basel) 2013;6:295–319. doi: 10.3390/ph6030295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Boyacioglu O, Stuart CH, Kulik G, Gmeiner WH. Dimeric DNA Aptamer Complexes for High-capacity-targeted Drug Delivery Using pH-sensitive Covalent Linkages. Mol Ther Nucleic Acids. 2013;2:e107. doi: 10.1038/mtna.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45:8149–52. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 145.Gijs M, Aerts A, Impens N, Baatout S, Luxen A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl Med Biol. 2016;43:253–71. doi: 10.1016/j.nucmedbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 146.Younes CK, Boisgard R, Tavitian B. Labelled oligonucleotides as radiopharmaceuticals: pitfalls, problems and perspectives. Curr Pharm Des. 2002;8:1451–66. doi: 10.2174/1381612023394467. [DOI] [PubMed] [Google Scholar]
- 147.Sugiura G, Kuhn H, Sauter M, Haberkorn U, Mier W. Radiolabeling strategies for tumor-targeting proteinaceous drugs. Molecules. 2014;19:2135–65. doi: 10.3390/molecules19022135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hicke BJ, et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–54. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 149.Hicke BJ, et al. Tumor targeting by an aptamer. J Nucl Med. 2006;47:668–78. [PubMed] [Google Scholar]
- 150.Lao YH, Phua KK, Leong KW. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–54. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 151.Farokhzad OC, et al. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–72. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 152.Liang C, et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat Med. 2015;21:288–94. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barakat MR, Kaiser PK. VEGF inhibitors for the treatment of neovascular age-related macular degeneration. Expert Opin Investig Drugs. 2009;18:637–46. doi: 10.1517/13543780902855316. [DOI] [PubMed] [Google Scholar]
- 154.Group, V.I.S.i.O.N.C.T. et al. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology. 2006;113:1508 e1–25. doi: 10.1016/j.ophtha.2006.02.064. [DOI] [PubMed] [Google Scholar]
- 155.Jellinek D, Lynott CK, Rifkin DB, Janjic N. High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding. Proc Natl Acad Sci U S A. 1993;90:11227–31. doi: 10.1073/pnas.90.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shaw JP, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991;19:747–50. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jellinek D, et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34:11363–72. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 158.Jaschke A, et al. Synthesis and properties of oligodeoxyribonucleotide-polyethylene glycol conjugates. Nucleic Acids Res. 1994;22:4810–7. doi: 10.1093/nar/22.22.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kawaguchi T, Asakawa H, Tashiro Y, Juni K, Sueishi T. Stability, specific binding activity, and plasma concentration in mice of an oligodeoxynucleotide modified at 5′-terminal with poly(ethylene glycol) Biol Pharm Bull. 1995;18:474–6. doi: 10.1248/bpb.18.474. [DOI] [PubMed] [Google Scholar]
- 160.Group, C.R. et al. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N Engl J Med. 2011;364:1897–908. doi: 10.1056/NEJMoa1102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brown DM, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–44. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 162.Rosenfeld PJ, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–31. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 163.Heier JS, et al. Intravitreal aflibercept (VEGF trap-eye) in wet age-related macular degeneration. Ophthalmology. 2012;119:2537–48. doi: 10.1016/j.ophtha.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 164.D’Amore PA. Vascular endothelial cell growth factor-a: not just for endothelial cells anymore. Am J Pathol. 2007;171:14–8. doi: 10.2353/ajpath.2007.070385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ratanji KD, Derrick JP, Dearman RJ, Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11:99–109. doi: 10.3109/1547691X.2013.821564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kawa MP, Machalinska A, Roginska D, Machalinski B. Complement system in pathogenesis of AMD: dual player in degeneration and protection of retinal tissue. J Immunol Res. 2014;2014:483960. doi: 10.1155/2014/483960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Baas DC, et al. The complement component 5 gene and age-related macular degeneration. Ophthalmology. 2010;117:500–11. doi: 10.1016/j.ophtha.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Biesecker G, Dihel L, Enney K, Bendele RA. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology. 1999;42:219–30. doi: 10.1016/s0162-3109(99)00020-x. [DOI] [PubMed] [Google Scholar]
- 169.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT02397954?term=NCT02397954&rank=1.
- 170.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT02686658?term=NCT02686658&rank=1.
- 171.Sadiq MA, et al. Platelet derived growth factor inhibitors: A potential therapeutic approach for ocular neovascularization. Saudi J Ophthalmol. 2015;29:287–91. doi: 10.1016/j.sjopt.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mabry R, et al. A dual-targeting PDGFRbeta/VEGF-A molecule assembled from stable antibody fragments demonstrates anti-angiogenic activity in vitro and in vivo. MAbs. 2010;2:20–34. doi: 10.4161/mabs.2.1.10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jo N, et al. Inhibition of platelet-derived growth factor B signaling enhances the efficacy of anti-vascular endothelial growth factor therapy in multiple models of ocular neovascularization. Am J Pathol. 2006;168:2036–53. doi: 10.2353/ajpath.2006.050588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Boyer DS. A Phase 2b study of FovistaTM, a platelet derived growth factor (PDGF) inhibitor in combination with a vascular endothelial growth factor (VEGF) inhibitor for neovascular age-related macular degeneration (AMD) Invest Ophthalmol Vis Sci. 2013;54 [Google Scholar]
- 175.Boyer DS. Combined inhibition of platelet derived (PDGF) and vascular endothelial (VEGF) growth factors for the treatment of neovascular age-related macular degeration (NV‑AMD) - results of a Phase I study. Investigative Ophthalmology & Visual Science. 2009;50 [Google Scholar]
- 176.Green LS, et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35:14413–24. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 177.Floege J, et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am J Pathol. 1999;154:169–79. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT01089517?term=NCT01089517&rank=1.
- 179.US National Library of Medicine. ClinicalTrials.gov. 2005 https://clinicaltrials.gov/ct2/show/NCT01944839?term=NCT01944839&rank=1.
- 180.US National Library of Medicine. ClinicalTrials.gov. 2015 https://clinicaltrials.gov/ct2/show/NCT01940900?term=NCT01940900&rank=1.
- 181.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT01940887?term=NCT01940887&rank=1.
- 182.Smiley DA, Becker RC. Factor IXa as a target for anticoagulation in thrombotic disorders and conditions. Drug Discov Today. 2014;19:1445–53. doi: 10.1016/j.drudis.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 183.Rusconi CP, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–4. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 184.Povsic TJ, et al. Use of the REG1 anticoagulation system in patients with acute coronary syndromes undergoing percutaneous coronary intervention: results from the phase II RADAR-PCI study. EuroIntervention. 2014;10:431–8. doi: 10.4244/EIJY14M06_01. [DOI] [PubMed] [Google Scholar]
- 185.Povsic TJ, et al. A Phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur Heart J. 2013;34:2481–9. doi: 10.1093/eurheartj/ehs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Povsic TJ, et al. Pegnivacogin results in near complete FIX inhibition in acute coronary syndrome patients: RADAR pharmacokinetic and pharmacodynamic substudy. Eur Heart J. 2011;32:2412–9. doi: 10.1093/eurheartj/ehr179. [DOI] [PubMed] [Google Scholar]
- 187.Cohen MG, et al. First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation. 2010;122:614–22. doi: 10.1161/CIRCULATIONAHA.109.927756. [DOI] [PubMed] [Google Scholar]
- 188.US National Library of Medicine. ClinicalTrials.gov. 2012 https://clinicaltrials.gov/ct2/show/NCT00932100?term=NCT00932100&rank=1.
- 189.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT00715455?term=NCT00715455&rank=1.
- 190.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT01872572?term=NCT01872572&rank=1.
- 191.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT01848106?term=NCT01848106&rank=1.
- 192.Diener JL, et al. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J Thromb Haemost. 2009;7:1155–62. doi: 10.1111/j.1538-7836.2009.03459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jilma B, et al. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb Haemost. 2010;104:563–70. doi: 10.1160/TH10-01-0027. [DOI] [PubMed] [Google Scholar]
- 194.Gilbert JC, et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–86. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 195.US National Library of Medicine. ClinicalTrials.gov. 2007 https://clinicaltrials.gov/ct2/show/NCT00432770?term=ARC1779&rank=6.
- 196.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00632242?term=ARC1779&rank=1.
- 197.US National Library of Medicine. ClinicalTrials.gov. 2010 https://clinicaltrials.gov/ct2/show/NCT00742612?term=ARC1779&rank=2.
- 198.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00507338?term=ARC1779&rank=5.
- 199.Waters EKR, J, Schaub RG, Kurz JC. Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole blood. J Thromb Haemost. 2009;7:683. [Google Scholar]
- 200.US National Library of Medicine. ClinicalTrials.gov. 2011 https://clinicaltrials.gov/ct2/show/NCT00808964?term=00808964&rank=1.
- 201.Waters EK, et al. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood. 2011;117:5514–22. doi: 10.1182/blood-2010-10-311936. [DOI] [PubMed] [Google Scholar]
- 202.Gorczyca ME, Jilma B, Male C, Reitter S, Gilbert JC, Pabinger I. Inhibition of tissue factor pathway inhibitor (TFPI) by ARC19499 improves clotting of hemophiliac blood. BMC Pharmacology. 2010;10 doi: 10.1111/j.1538-7836.2012.04790.x. [DOI] [PubMed] [Google Scholar]
- 203.US National Library of Medicine. ClinicalTrials.gov. 2015 https://clinicaltrials.gov/ct2/show/NCT01191372?term=NCT01191372&rank=1.
- 204.Soundararajan S, et al. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol Pharmacol. 2009;76:984–91. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J Biol Chem. 1999;274:26369–77. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 206.Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–65. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 207.Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie. 2015;113:78–85. doi: 10.1016/j.biochi.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 208.Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr Opin Mol Ther. 2010;12:107–14. [PubMed] [Google Scholar]
- 209.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–29. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86:151–64. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00881244?term=NCT00881244&rank=1.
- 212.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00512083?term=NCT00512083&rank=1.
- 213.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00740441?term=NCT00740441&rank=1.
- 214.Hoellenriegel J, et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032–9. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Marasca R, Maffei R. NOX-A12: mobilizing CLL away from home. Blood. 2014;123:952–3. doi: 10.1182/blood-2013-12-542480. [DOI] [PubMed] [Google Scholar]
- 216.Liu SC, et al. Blockade of SDF-1 after irradiation inhibits tumor recurrences of autochthonous brain tumors in rats. Neuro Oncol. 2014;16:21–8. doi: 10.1093/neuonc/not149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sayyed SG, et al. Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes. Diabetologia. 2009;52:2445–54. doi: 10.1007/s00125-009-1493-6. [DOI] [PubMed] [Google Scholar]
- 218.Burns JM, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–13. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT00976378?term=NCT00976378&rank=1.
- 220.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT01194934?term=NCT01194934&rank=1.
- 221.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT01486797?term=NCT01486797&rank=1.
- 222.US National Library of Medicine. ClinicalTrials.gov. 2015 https://clinicaltrials.gov/ct2/show/NCT01521533?term=NCT01521533&rank=1.
- 223.Oberthur D, et al. Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2. Nat Commun. 2015;6:6923. doi: 10.1038/ncomms7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ninichuk V, et al. Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36-3′PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am J Pathol. 2008;172:628–37. doi: 10.2353/ajpath.2008.070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kulkarni O, et al. Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J Am Soc Nephrol. 2007;18:2350–8. doi: 10.1681/ASN.2006121348. [DOI] [PubMed] [Google Scholar]
- 226.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT01085292?term=NCT01085292&rank=1.
- 227.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT01547897?term=NCT01547897&rank=1.
- 228.Schwoebel F, et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood. 2013;121:2311–5. doi: 10.1182/blood-2012-09-456756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003;102:783–8. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- 230.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT01372137?term=NCT01372137&rank=1.
- 231.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT01691040?term=NCT01691040&rank=1.
- 232.US National Library of Medicine. ClinicalTrials.gov. 2015 https://clinicaltrials.gov/ct2/show/NCT02079896?term=NCT02079896&rank=1.
- 233.US National Library of Medicine. ClinicalTrials.gov. 2005 https://clinicaltrials.gov/ct2/show/NCT00021736?term=NCT00021736&rank=1.
- 234.US National Library of Medicine. Clinicaltrials.gov. 2006 https://clinicaltrials.gov/ct2/show/NCT00040313?term=00040313&rank=1.
- 235.US National Library of Medicine. ClinicalTrials.gov. 2008 doi: 10.1080/15360280801989377. https://clinicaltrials.gov/ct2/show/NCT00056199?term=00056199&rank=1. [DOI] [PubMed]
- 236.US National Library of Medicine. ClinicalTrials.gov. 2007 https://clinicaltrials.gov/ct2/show/NCT00312351?term=00312351&rank=1.
- 237.US National Library of Medicine. ClinicalTrials.gov. 2006 https://clinicaltrials.gov/ct2/show/NCT00321997?term=NCT00321997&rank=1.
- 238.US National Library of Medicine. ClinicalTrials.gov. 2011 https://clinicaltrials.gov/ct2/show/NCT01487070?term=NCT01487070&rank=1.
- 239.US National Library of Medicine. ClinicalTrials.gov. 2012 https://clinicaltrials.gov/ct2/show/NCT00709527?term=NCT00709527&rank=1.
- 240.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT00950638?term=NCT00950638&rank=1.
- 241.US National Library of Medicine. ClinicalTrials.gov. 2010 https://clinicaltrials.gov/ct2/show/NCT00569140?term=NCT00569140&rank=1.
- 242.US National Library of Medicine. ClinicalTrials.gov. 2016 https://clinicaltrials.gov/ct2/show/NCT02387957?term=NCT02387957&rank=1.
- 243.US National Library of Medicine. ClinicalTrials.gov. 2005 https://clinicaltrials.gov/ct2/show/NCT02591914?term=NCT02591914&rank=1.
- 244.US National Library of Medicine. ClinicalTrials.gov. 2008 doi: 10.1080/15360280801989377. https://clinicaltrials.gov/ct2/show/NCT00113997?term=NCT00113997&rank=1. [DOI] [PubMed]
- 245.US National Library of Medicine. ClinicalTrials.gov. 2009 https://clinicaltrials.gov/ct2/show/NCT00694785?term=ARC1779&rank=3.
- 246.Jilma-Stohlawetz P, et al. Inhibition of von Willebrand factor by ARC1779 in patients with acute thrombotic thrombocytopenic purpura. Thromb Haemost. 2011;105:545–52. doi: 10.1160/TH10-08-0520. [DOI] [PubMed] [Google Scholar]
- 247.US National Library of Medicine. ClinicalTrials.gov. 2011 https://clinicaltrials.gov/ct2/show/NCT01034410?term=NCT01034410&rank=1.
- 248.US National Library of Medicine. ClinicalTrials.gov. 2013 https://clinicaltrials.gov/ct2/show/NCT00976729?term=NCT00976729&rank=1.
- 249.US National Library of Medicine. ClinicalTrials.gov. 2012 https://clinicaltrials.gov/ct2/show/NCT01372124?term=NCT01372124&rank=1.
- 250.US National Library of Medicine. ClinicalTrials.gov. 2014 https://clinicaltrials.gov/ct2/show/NCT01522794?term=NCT01522794&rank=1.


