Abstract
Since microbes were first described in the mid-1600's, we have come to appreciate that they live all around and within us with both beneficial and detrimental effects on nearly every aspect of our lives. The human gastrointestinal tract is inhabited by a dynamic community of trillions of bacteria that constantly interact with each other and their human host. The acquisition of these bacteria is not stochastic, but determined by circumstance (environment), host rules (genetics, immune state, mucus, etc), and dynamic self-selection among microbes to form stable, resilient communities that are in balance with the host. In this review, we will discuss how these factors lead to formation of the gut bacterial community and influence its interactions with the host. We will also address how gut bacteria contribute to disease and how they could potentially be targeted to prevent and treat a variety of human ailments.
Keywords: Microbiome, microbiota, colonization, commensal, niche, selection
INTRODUCTION
Microbes were first described in the mid-1600's by Robert Hooke and Antoni van Leeuwenhoek in letters to the Royal Society.1 Then, in the mid to late 1800's, Louis Pasteur and Robert Koch showed that microscopic organisms had the ability to cause disease.2 We now appreciate that microbes live all around and within us with both beneficial and detrimental effects on nearly every aspect of our modern lives. However, we are just now beginning to decipher the more complex questions of what determines the composition and function of microbial communities that colonize humans and how those communities influence health and disease in their eukaryotic hosts. In this review, we will discuss the formation of the human gastrointestinal bacterial community, interactions between the host and its gut bacteria, and the promise of disease therapies based on manipulation of the bacterial residents of the gut. We recognize that other microbial kingdoms (fungi, viruses, Archae, Protist) are also important members of the gut microbiome, but their role in the evolutionary and ecological forces that shape the microbial communities of the human gut remain poorly understood.
FORMATION OF THE GASTROINTESTINAL BACTERIAL COMMUNITY
ACQUISITION OF GUT BACTERIA
The human gastrointestinal (GI) tract is exposed to low levels of bacteria in utero, but the majority of bacterial colonization occurs after birth.3,4 The types and numbers of bacteria involved in the initial colonization are greatly influenced by the birth process.5 In healthy, full-term infants the gut is initially an aerobic environment and the earliest colonizers are aerotolerant members of the Enterobacteriaceae family.6 In vaginally delivered infants, these are primarily derived from maternal vaginal and fecal microbes. In infants delivered by Caesarean section, skin microbes are the initial gut colonizers.5,7 Within days after birth, the gut lumen becomes more anaerobic and is settled by strict anaerobes such as Bifidobacterium, Bacteroides, and Clostridium species.8 The infant milk diet further promotes colonization and proliferation of milk oligosaccharide fermenters such as Bifidobacterium, which make up an increasing proportion of the community over time.9 Later, the introduction of solid food and then weaning provides a more diverse set of microbial substrates including complex polysaccharides that are not digestible by the host. This leads to increased abundance of the genera Bacteroides, Clostridium, and Ruminococcus with decreased abundance of Bifidobacterium and Enterobacter.10–13 The ongoing development and character of the gut microbial community are further influenced by continued inputs from other individuals (caregivers, siblings, and playmates), contact with animals, ingestion of soil, and consumption of microbes with foodstuffs. Unsurprisingly, the types of microbes encountered during these interactions are highly dependent on the geographic location and lifestyle of the growing child. Whether these microbes can establish residence within the GI tract is dependent on the availability of the appropriate niche. Niche availability is dictated by the host, but is also heavily influenced by which microbes are already established in the GI tract when they attempt to infiltrate the community. Ultimately, the succession of the gut microbial community from initial colonization to adulthood is the result of changes in the anatomy and physiology of the GI tract over time, changes in the diet from milk to a variety of solid foods, and interactions among microbes within the community and between microbes and the host. At the end of this process, the gut develops a relatively stable community that has beneficial effects on digestion, host defense, and whole body development. This community interacts with the human host constantly and the outcome of these interactions determines the relative health of the 'superorganism' composed of microbes and human cells.
BACTERIAL DETERMINANTS OF GUT COLONIZATION
Microbes do not passively occupy the human GI tract, they actively colonize and establish interactions with the host and other microbes in order to remain within the GI tract, obtain nutrients, and reproduce. In order to achieve this, microbes must first be internalized by the host, which mostly occurs after birth, although some studies have suggested that this process can begin in utero.14 They also must survive the host defenses to reach an optimal colonization site. At this site the microbe or microbes must either establish an initial foothold with the host or infiltrate an existing microbial community. They then need to successfully acquire resources from the environment, avoid physical expulsion from the GI tract, avoid active host antimicrobial defenses, avoid active killing by other microbes contacting their niche, and reproduce.
Colonization factors
Bacterial residence in the gastrointestinal tract is in large part determined by cellular expression of colonization factors. These are bacterial traits that are necessary and/or sufficient for attachment to or survival within the GI tract. Colonization factors fall into two major categories: 1) factors that are required for host-microbe interactions and 2) factors that are required for microbe-microbe interactions. Thus far, most studies designed to identify colonization factors have been limited to small, defined microbial communities or even single microbes.15 Since we know that colonization is very context-dependent, these conditions likely best model early colonization of a naive host. These studies have also traditionally been limited to cultivatable organisms of clinical significance (i.e. Clostridium difficile) or those species that, by virtue of prevalence, ease of isolation, and availability of tools for genetic modification, have become commensal “model” organisms (i.e. Bacteroides thetaiotamicron). Although much is known about a relatively small number of species, it is unclear how broadly the principles governing intestinal colonization by these species can apply across microbes. Moreover, the study of fitness determinants of monocultures using traditional microbiological methods does not permit nuanced interrogation of the importance of in vivo environment or microbial community interactions. Finally, in certain contexts bacterial colonization factors may facilitate epithelial invasion or entry into the bloodstream. Therefore colonization and virulence factors should be considered a continuous, rather than a binary outcome.
The process of gastrointestinal colonization -from the microbe side
The first step in GI colonization is internalization by a potential host. Microbes from the environment or those from another eukaryotic host both have the potential to colonize the GI tract. Environmental bacteria are well adapted to conditions in the external environment, while host-associated bacteria must exit one host and survive in the external environment long enough to be re-internalized by a second host. The bacteria then enter the new host through oral ingestion in almost all cases.
After ingestion, bacteria must traverse the host defenses of the upper GI tract. This includes a number of defenses that are not specifically directed at bacteria such as pH, digestive enzymatic activity, mucus, and intestinal propulsive motility. Bacterial characteristics such as sporulation, capsule formation, and development of a sturdy cell wall permit survival during this transit process. Bacteria that survive transit must then identify a replication-permissive niche. This is often through detection of environmental characteristics, such as host metabolites, that lead to modification of bacterial characteristics. For example, Clostridium difficile spores germinate in the presence of increased taurocholate, a metabolite found in bile.16 Bacterial receptors for gastrin, somatostatin, and catecholamines have also been described and detection of these non-nutritive substrates leads to altered bacterial gene expression.17–19 Finally, evidence suggests that even genetically clonal populations of bacteria exhibit phenotypic heterogeneity, suggesting that population-level variability in gene expression facilitates survival of rapid environmental change, such as that occurring within the GI tract.20
Once bacteria reach a suitable micro-niche, they must establish themselves through physical interaction with the niche or they will be washed out of the GI tract by peristalsis and the flow of ingesta. Physical attachment to the niche can involve attachment to host tissues, host extracellular proteins, or to other microbes in the niche to form biofilms. Bacterial surface characteristics such as capsular polysaccharides or appendages such as pili and fimbrae facilitate interactions to secure bacteria to a niche.21 These surface characteristics are often only expressed when a microbe reaches a suitable niche. For example, when B. fragilis is exposed to bile salts, its cell surface is remodeled to increase expression of fimbria-like structures that facilitate attachment to epithelial cells.22,23 Host signals can also inhibit expression of colonization factors until the bacterium reaches a suitable niche. This is the case with enterohemorrhagic E. coli (EHEC) which senses and responds to biotin concentrations in the intestine. When biotin concentrations are high, as is the case in the small intestine, adherence factors are suppressed. When the bacteria pass into the large intestine, where biotin concentrations are low, they express adherence factors.24 Interactions with other microbes can also lead to modification of cell surface features to facilitate bacterial attachment. For example, co-association of B. fragilis with Klebsiella pneumoniae in mice causes B. fragilis to develop a capsule and pili.25 Expression of these bacterial attachment features can then contribute to biofilm formation and provide attachment sites for subsequent colonizers.26
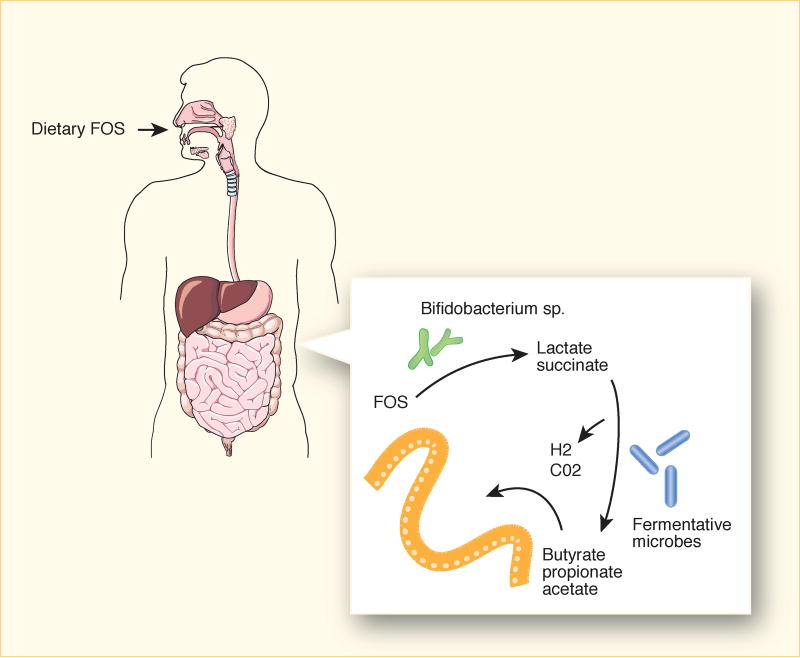
After physically securing themselves within a micro-niche, bacteria must then acquire resources for survival and reproduction from the environment. These resources include oxygen, micronutrients like iron or salts, and macronutrients like complex carbohydrates.27 Carbohydrates can be obtained from ingested foodstuffs, host tissues, or from other microbes. Some microbes, such as Bifidobacteria, thrive on complex polysaccharides, like fructo-oligosaccharides, that cannot be digested by the host (Figure 1). The end product of fructo-oligosaccharide digestion by Bifidobacteria is lactate which is then ingested by a second set of bacteria to produce short chain fatty acids such as butyrate. Butyrate is then absorbed and used as an energy source by host colonocytes. In this manner, communities of microbes within micro-niches can cooperate in metabolic chains.28–30 Iron and salt concentrations in various micro-niches also influence bacterial colonization. Enterohemorrhagic E. coli increase expression of genes for flagella and iron scavenging when exposed to bile salts.31 Salmonella enterica serovar Typhimurium exhibits iron-induced epithelial adhesion.32
Figure 1. Metabolic cross feeding during gut colonization.
Fructooligosaccharides (FOS) reach the colon largely intact where they serve as substrates for fructan-degrading bacteria such as Bifidobacteria. Utilization of down-stream products by other bacterial species, termed metabolic cross-feeding, results in production of short-chain fatty acids that are absorbed by the host. Strain variation in FOS-degradative capacity results in diet-dependent colonization or replicative capacity.
Oxygen tension is another major determinant of bacterial niche suitability. The GI tract has a radial oxygen gradient with diminishing concentrations from the epithelial surface to the gut lumen.33 This creates micro-niches of higher oxygen tension at the epithelial surface and in the epithelial-associated mucus that are colonized by facultative anaerobes such as Proteobacteria. It also creates opportunities for anaerobic bacteria that have developed mechanisms to tolerate intermittent or extremely low levels of oxygen. This includes Faecalibacterium prausnitzii, which can tolerate oxygen at the mucosal interface due to an extracellular electron shuttle utilizing flavins and thiols.34 Other bacteria, such as B. fragilis and Clostridium acetobutylicum have developed molecular switches that allow them to survive under fluctuating oxygen levels.35,36 Oxygen concentrations also sometimes define bacterial co-occurence between microaerophiles that consume oxygen and strict anaerobes that cannot grow in its presence.37 This is thought to be a factor in bacterial succession during the initial colonization of the naive infant gut.
Finally, bacteria must be able to reproduce and the progeny must re-colonize the intestine or colonization by a given organism will be short-lived. This requires availability of physical space and appropriate resources, along with access to a suitable niche before bacteria reach the end of the GI tract. Some of the progeny of resident gut bacteria will inevitably fail to find a niche within the host and will be washed out into the environment with the feces. If these microbes can adapt to the external environment, they have the potential to join new bacterial communities in that manner. However, most are adapted to the internal environment and must find a new host before they die. The majority of gut bacteria reproduce and exit the gut in a fairly constant manner. However, some pathogens employ strategies to cause diarrhea and rapidly disseminate themselves and other gut bacteria into the external environment. The classic example of this is Vibrio cholera that produces a toxin to prevent fluid re-absorption in the large intestine causing large amounts of diarrhea and bacterial shedding.38
Bacteria cooperate and compete in the GI tract
While many members of the gut bacterial community cooperate, there is also competition for space and resources within the gastrointestinal tract. Therefore, the accessibility of niches within the gut for colonization is at least somewhat dependent upon the initial colonizers of those niches.39 This suggests that there may be "keystone species" that determine the ultimate composition and function of the gut bacterial ecosystem. This concept was originally developed to explain bacterial community organization in microbial ecology and has been adopted to explain bacterial community organization in the gut.40–42 It posits that there are bacterial founders in the gut around which the rest of the community organizes. The wide differences among human bacterial communities within different individuals by taxonomic measures has led some researchers to propose that a more accurate concept may be bacterial "keystone functions" that could be performed by a number of different bacterial species. These functions would likely be those that alter the physical space, the attachment characteristics, or the resources available within a niche. They would also be expected to promote colonization by desirable bacteria and exclude bacteria likely to have detrimental effects on the established bacterial community or the host.
Competition in the gut bacterial community occurs between bacteria generally considered to be commensal and between commensal bacteria and true pathogens.43,44 Contests between commensals and pathogens are referred to as commensal exclusion or colonization resistance and encompass a number of competitive interactions for physical attachment sites and resources. Commensals and pathogens can directly compete for the same nutrients as is the case when gut resident B. thetaiomicron consumes plant-derived monosaccharides that Citrobacter rodentium requires for growth and so prevents it from gaining a foothold in the bacterial community.45 Commensal microbes can also produce antimicrobial peptides and toxins to target and eliminate bacteria competing for the same niche, whether those are other commensals or pathogens. This is exemplified by the Type VI secretion system of Bacteroides that exports antimicrobial proteins to kill competing Bacteroidales in the gut.46,47
Diversity within the gut bacterial community
The unique nature of microbial niches in the GI tract and the diverse factors that allow bacteria to occupy them explains why there is not one consistent microbial community among humans or even all along the GI tract. Micro-niches develop that represent defined communities with their own sets of host-microbe and microbe-microbe interactions.48 This diversity is primarily determined by which microbes come into contact with the host GI tract, the ability of an internalized bacterium to infiltrate any existing bacterial community and physically establish themselves within the host, and their ability to acquire resources and reproduce. Macro-niches also exist in the gut lumen that are more reliant on substrate availability, relative stasis, pH, tonicity, and other physical conditions in the gut. Many microbes can move between macro-niches in the lumen and micro-niches in the mucosa, but this ability is constrained by both microbial and host characteristics.
A secondary source of diversity is genomic modification of bacteria within the gastrointestinal tract. This occurs when bacteria undergo horizontal gene transfer to acquire new genetic material from other bacteria within the community.49 This can change the fitness of bacteria for colonization of the intestinal environment, allow them to occupy new niches, or confer virulence on otherwise non-virulent microbes. For example, a Japanese gut microbe was shown to have acquired the ability to digest complex carbohydrates in seaweed from a marine bacterium through horizontal gene transfer.50 Horizontal gene transfer has also been implicated in acquisition of antibiotic resistance genes by intestinal bacteria.51 There is extensive evidence that Bacteroides in particular undergo horizontal gene transfer to acquire antimicrobial resistance and potentially other genes of clinical significance.52–59 Therefore, the genetic composition of the gut bacterial community can change due to acquisition of new bacterial species, expulsion of previously established microbes, or genetic modification of existing microbes over the course of an individual host's lifetime.
INTERACTIONS BETWEEN A HOST AND ITS GUT BACTERIA
THE DYNAMIC NATURE OF HOST-MICROBE INTERACTIONS IN THE GUT
Bacterial colonization factors are important determinants of an organism's ability to take up residence in the human GI tract. However, there are equally important host factors, which interact with and influence the expression and characteristics of bacterial factors. Therefore, colonization is the outcome of dynamic, bidirectional interactions between the host and its microbes. These interactions modify the tissue such that the very act of colonization changes the environment encountered by the next bacterium attempting to enter a niche.
Non-bacterial specific host factors that affect bacterial colonization
The host employs mechanisms to hold growth of the bacterial population in the GI tract in check and at a distance from the epithelium while simultaneously encouraging colonization by productive partners. These mechanisms include non-bacterial specific and bacterial specific mechanisms. Non-bacterial specific mechanisms are primarily those involved in digestion. In the upper GI tract, gastric acid from the stomach, digestive enzymes from the stomach and pancreas and bile from the liver all work to create a fairly hostile environment for bacterial colonization. Reflective of this, the concentration of microbes in the stomach and proximal small intestine is fairly low relative to sites in the distal small intestine and colon. However, some bacteria have evolved to deal with bile by performing bile salt hydrolysis, hydroxyl group dehydrogenation, or 7-dehydroxylation on primary host bile acids to create new forms of bile acids with distinct functions.60
The distal GI tract has non-specific antibacterial defenses related to epithelial surface turnover. Intestinal epithelial cells are generated from stem cells in the base of the intestinal crypts. These cells then mature and migrate toward the gut lumen over the course of several days. Finally they die and are sloughed into the gut lumen where they become part of the intestinal contents.61 The epithelial surface in the small and large intestine is also coated with cell-associated and secreted mucins.62 In the small intestine the mucus layer is relatively thin and tightly packed. In the large intestine, the mucus is much thicker and is composed of a tight inner and looser outer layer. In the colon, the major secreted mucin is Muc2, which forms a gradient of decreasing density from the epithelial surface to the gut lumen and is renewed every few hours.63
The turnover of the cellular and acellular components of the epithelial surface leads to a treadmill effect with a flow away from the epithelial surface toward the gut lumen. These factors keep the majority of commensal bacteria physically segregated from the epithelial surface. The rate of epithelial turnover is at least partially dependent on gut bacteria since bacterial-produced short chain fatty acids promote epithelial turnover.64 However, responses to microbial metabolites can be very cell type specific. When intestinal stem cells, a tissue compartment that is normally isolated from short chain fatty acids, encounter butyrate, their proliferation is slowed which delays tissue repair and turnover.65 Pathogens can also affect intestinal epithelial turnover rates. Citrobacter rodentium, a murine pathogen that is used as a model for enterohemmorhagic (EHEC) and enteropathogenic (EPEC) E. coli, stimulates β-catenin signaling in stem cells, which leads to epithelial hyperplasia and an increase in crypt length.66
Gastrointestinal mucus not only limits contact between microbes and epithelial surfaces, it also serves as a food source for many gut bacteria.67 Most host epithelial surface proteins are heavily modified by the post-translational addition of a glycan moiety. This includes the surface-associated and secreted mucins in the colon. Glycan modification of mucins increases their structural integrity in the intestinal mucus layer. However, glycans are sugars and can be cleaved from the mucins by bacterial glycosidases for use as carbohydrate sources. Removal of the glycans then contributes to mucin degradation in the gut.68 Bacteria possess an astonishing diversity of glycan metabolic pathways, with the range of specific degradative enzymes varying across species.69 This means that they extensively modify host-derived glycans resulting in downstream products that influence other community members or pathogens. In an elegant example of commensal-pathogen interaction, fucose liberated from host glycans by B. thetaiotamicron is detected by a QseC sensor kinase in enterohemorrhagic Escherichia coli, resulting in fucose-concentration dependent virulence gene expression.70
Host glycan production is also influenced by gastrointestinal bacteria. Germ free mice have lower levels and different types of glycosylated proteins in their intestinal mucosa than conventionally raised mice.71 Colonization with microbes can increase glycosylation directly. This has been shown through mono-association of germ free mice with B. thetaiomicron, which directly upregulates expression of host epithelial fucosyltransferase mRNA.72 Glycans can also serve as attachment sites as is the case when the E. coli fimbrial protein FimH interacts with mannose residues on host surfaces to facilitate bacterial attachment.73,74 Finally, host protein glycosylation can influence host immune responses to microbes. Pickard et al 2014 demonstrated that systemic injection of lipopolysaccharide increased mucosal fucosylation which was in turn associated with enhanced fucose-dependent host tolerance of an enteropathogen, Citrobacter rodentium.75 Considered together, these data suggest that carbohydrates are an important currency of host-microbe exchanges, the complexity of which we are only just beginning to appreciate.
Bacterial specific host factors that affect bacterial colonization
In addition to its non-specific antibacterial defenses, the host epithelium also has a number of specific antibacterial defenses that help to determine which microbes are able to colonize and survive within the GI tract. In general, anti-bacterial factors are concentrated in the areas closest to the intestinal epithelium and are produced in response to host detection of bacteria through a number of innate and adaptive immune mechanisms.76,77 These mechanisms have been extensively reviewed elsewhere so this discussion will focus on some notable bacterial mechanisms for evasion of host anti-bacterial defenses.
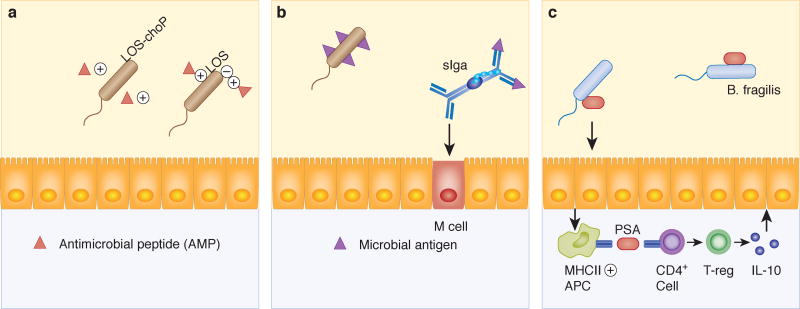
Stable colonization requires that bacteria avoid expulsion from a niche by host defenses. In order to do this they employ a number of strategies designed to avoid detection by the host. This generally leads to decreased expression of host responses detrimental to a given bacterium (Figure 2). One strategy employed by bacteria to evade host anti-bacterial defenses is modification or modulation of cell surface structures detected or targeted by the host.78,79 Antimicrobial peptides are family of proteins produced in the intestinal epithelium, which rapidly kill or inactivate microbes.80 Killing by antimicrobial peptides can be evaded by Gram negative bacteria through modification of their lipopolysaccharides (LPS) to decrease the electronegativity on their cell surface which renders them resistant to the electrostatic attraction of positively charged antimicrobial peptides.81 Gram positive bacteria can modify the lipoteichoic acids that make up their cells walls to make themselves resistant to antimicrobial peptides.82 Another strategy used by bacteria to evade host defenses is molecular mimicry. Haemophilus species are primarily respiratory pathogens, but may also cause GI disease.83 One member of this genus, Haemophilus influenzae, can harvest free host sialic acids and use them to coat and camouflage their cell surface. This prevents host recognition of pro-inflammatory molecules on the bacterial surface.84,85
Figure 2. Bacterial strategies to evade host defenses.
A) Addition of phosphorylcholine (CHOP) to bacterial lipooligosaccharide (LOS) reduces surface electronegativity and thus resistance to cationic peptides. B) Secretory IgA (sIgA) immune complexes promote M cell uptake of commensal microbial antigen. Induction of this pathway is important for induction of host-commensal homeostasis. C) Capsular polysaccharide A (PSA) from Bacteroides fragilis induces an IL-10 predominant immune response. This, in turn, is associated with protection from mucosal damage in murine models of colitis.
In contrast to immune evasion, stable colonization can also be promoted through activation of the immune system. Capsular polysaccharide A (PSA) from B. fragilis induces an immune response primarily consisting of IL-10 expressing regulatory T cells in the intestine. This leads to protection from experimental colitis in mice colonized with PSA expressing B. fragilis or treated with purified PSA.86 The presence of microbes in the gut also elicits B-cell mediated IgA responses. Immunoglobulin A (IgA) is produced by B cells within the lamina propria of the small intestine, transported across intestinal epithelial cells, and secreted into the gut lumen where it is called secretory IgA (sIgA).87 Secretory IgA likely plays multiple roles in host-microbe interactions in the gut through coating bacteria and modifying their ability to attach to the mucosal surface of the intestine and/or activate immune responses.88 Colonization with B. thetaiomicron generates sIgA responses in the gut that coat this microbe and diminish host responses to its presence in the gastrointestinal tract.89,90 Likewise, microbial degradation of IgA has been associated with increased injury in a murine colitis model.91
THE HUMAN MICROBIOME IS UNIQUELY HUMAN
Host factors share commonalities among humans, but they are uniquely modified by environmental and genetic factors so that the niches available for colonization and the ways in which they affect their resident microbes can have important differences from one host to another. Likewise, the bacteria encountered by hosts can be very different based on differences in their geography and lifestyle. These factors suggest that the gut bacterial community is uniquely assembled in each individual. This is supported by the finding that humans have huge inter-individual variability in their gut microbiomes with regard to both diversity and abundance by16S amplicon sequencing and metagenomics.92–94 This variability is likely a product of genetics and environment since even monozygotic twins are not identical in their fecal microbiomes.94–96 It also means that the gut bacterial community has host specificity and functions that make it akin to an organ system.
The unique nature of the individual human gut bacterial community also means that gut bacterial communities in model organisms are very different from their human counterparts. This has been demonstrated in studies characterizing the bacterial taxa of humans, mice, and zebrafish as well as in studies utilizing transplantation of microbes across species barriers.97–99 At high levels of taxonomic classification, such as bacterial phyla, the microbes are similar between mice and humans. However, at the genus level they are quite different, since 85% of the genera found in mice are not found in humans.100 This is reflected in conventionalization studies wherein germ-free (GF) zebrafish, mice, or rats receive transplants of gut microbes derived from other species. These studies show that the host exerts selective pressures on the transplanted microbes to reshape the community into one that is more dependent on the recipient than the donor.101–104
The specificity of microbes to their host is further exemplified by studies investigating the role of microbes in immune system maturation. Germ-free mice have well-documented defects in innate and adaptive immunity.105 When these mice are colonized by gut microbiota from specific pathogen free (SPF) mice, they undergo immune system development similar to that of conventionally raised mice.106 However, mice conventionalized with human microbiota undergo significantly different immune system maturation that also leaves them more susceptible to gastrointestinal infection with Salmonella than mice conventionalized with microbiota derived from mice.102 Defects associated with colonization by human microbiota are correctable by addition of mouse specific segmented filamentous bacteria (SFB), suggesting that SFB and other murine microbiota are necessary for full immune maturation.102 Thus, a healthy immune system is dependent on the proper match between host and microbes determined through co-evolution.
GUT BACTERIA IN HEALTH AND DISEASE
A large and growing number of studies have associated the gut bacterial community with human disease. These have primarily been single time point 16S ribosomal RNA gene sequencing-based taxonomic studies that have identified differences in community membership between healthy and disease states. However, it is not clear how to use this information to identify disease-causing bacteria since there is a large amount of inter-individual variability in community composition with extensive overlap between bacteria found in individuals with and without disease.107 It is also difficult to identify community profiles that cause disease since changes to these profiles can be the result of disease. Unraveling bacterial-related disease causation is further complicated by the fact that it is not clear whether the presence, absence, abundance, or behavior of a given bacterial species is the strongest determinant of disease.
Current sequencing based methods are also limited to characterization of the genes present in the gut bacterial community. They cannot distinguish between live and dead bacteria and bacterial characteristics must be inferred based on gene functional data generated using classical microbiological methods in model organisms. It is clear that previously elucidated concepts of pathogenicity used to identify “good” and “bad” bacteria cannot be applied to the bacterial community in the gut. This means that understanding the role of gut bacteria in human disease will require development of disease paradigms that are very different from those that explain infectious disease and Koch’s postulates will need to be re-imagined for these studies.108 There are certainly some bacteria that are obligate pathogens, but many more have the potential to promote or prevent disease in certain contexts.
It is also becoming increasingly obvious that interactions between a host and its microbes, not just the presence or absence of specific bacteria, are important in the pathophysiology of diseases related to gut bacteria. These interactions are products of genetics and the environment. Genes in both the host and bacteria dictate niche colonization, microbial behavior within those niches, and host responses to microbes. The genetic complement of the gut microbial community can change over time as microbes or genetic elements enter or leave the community. In comparison, host genetics are relatively fixed. This means that host genetic polymorphisms can potentially influence bacterial selection pressures to shape the genetic compliment of the gut microbial community in ways that contribute to disease. This is thought to be a contributing factor in Crohn's disease, one of the inflammatory bowel diseases.109 Crohn's disease is associated with polymorphisms in genes involved in several major immunological pathways and dysbiosis. The gene pathways implicated include innate microbial sensing (i.e. NOD2), autophagy (i.e. ATG16L1), the intestinal epithelial barrier (i.e. MUC1), cytokine production (i.e. IL17), and T cell functions (i.e. STAT3).110 The impact of these genetic polymorphisms on gut microbes remains to be fully characterized. However, one of the Crohn's disease-associated polymophisms, Atg16L1 (T300A), has been associated with altered appearance and function in Paneth cells.111,112 Paneth cells produce antimicrobial peptides, which are believed to be major determinants of commensal microbial assemblage. Therefore, Paneth cell dysfunction is proposed to cause and/or contribute to dysbiosis and the pathophysiology of Crohn’s disease.
Environmental factors also influence the establishment and maintenance of stable commensal relationships. They contribute to initial and ongoing colonization events and environmental disruptions can destabilize existing gut microbial communities. Disruptions include major events that cause community upheaval and minor events that cause small changes on an ongoing basis. The outcome of these events likely depends on a number of factors including the initial impact of the event on the gut bacterial community, how the community recovers after the insult, and how the host reacts to the remodeled community. Common environmental disruptions include antibiotic administration, infectious disease in the gut or elsewhere in the body, and diet changes.
Antibiotic administration can have direct effects to kill gut bacteria or indirect effects that can lead to transient or permanent changes in the bacterial community composition.113,114 In particular, antibiotic administration to young children can strongly influence the bacterial community that assembles in the gut during initial colonization events.115 Antibiotic treatment can also create opportunities for pathogens.116 For example, antibiotic-mediated depletion of Clostridia can lead to increased oxygen in the mucosal environment and aerobic expansion of Salmonella enterica serovar Typhimurium.117 Infectious diseases can also disrupt the gut bacterial community. Infection with Mycobacterium tuberculosis rapidly leads to changes in composition of the gut bacterial community and the community present after infection is distinct from the pre-infection community.118 Transient, non-colonizing bacteria can also impact the viability or function of bacteria resident in the GI tract. This principle is thought to explain the ability of pro-biotic bacteria to influence host and microbes without evidence of colonization.119
Lastly, diet is thought to be one of the strongest environmental influences on bacterial community composition and behavior.120,121 It determines the types and amounts of nutrient sources available to resident microbes, introduces new microbes associated with foodstuffs to the community, and influences host cell functions. This leads to very different gut microbiomes in animals or humans consuming high fat, high sugar Western-style diets versus those consuming plant-based diets. Additionally, studies in both mice and humans have identified fecal bile acid profiles on high fat diets that have been shown to contribute to colitis in an IL-10−/− mouse model.122,123 These microbial changes also appear to contribute to diet-induced metabolic diseases.124 Microbes play important roles in energy extraction from diets and manipulation of microbes through antibiotic administration or changes to host immune functions can promote or prevent metabolic disease.125–128 Likewise, conventionalization of GF mice with microbes from obese mice or humans can transfer phenotypic changes consistent with increased energy extraction from the diet to the recipient.129–131 Therefore, diet is likely a major contributor to disease through modification of gut bacterial community composition and/or function as well as a potential opportunity for therapeutic intervention.
THE PROMISE OF DISEASE THERAPIES BASED ON MANIPULATION OF THE BACTERIAL RESIDENTS OF THE GUT
The implication of the gut bacterial community in human disease has led to the suggestion that manipulation of the community could be used to treat disease. This idea is very attractive and has the potential to revolutionize the prevention and treatment of a large number of human diseases. However, there are a number of challenges that have made this potential difficult to realize. The first of these is the previously discussed difficulty in defining the targets for microbial manipulation. This step is essential for designing effective therapies and monitoring the efficacy of treatment. Accumulating evidence suggests that these targets are also likely to be unique to a given individual so it may require application of precision or personalized medicine principles. It is not clear whether specific bacteria or the entire bacterial community should be targeted. It is also conceptually unclear whether the goal of therapy should be to make changes to the existing bacterial community or completely eliminate the existing community and replace it with a new one. Second, a treatment would need to reach the target site and microbial population or populations of interest within the GI tract. Treatments could be delivered orally or through enemas, depending on the target area of the GI tract, but targeting specific subpopulations of bacteria would likely be difficult if not impossible. Lastly, a treatment would need to create a stable bacterial community that remains in the modified state. Whether this is achievable would presumably depend on whether the underlying cause of the dysbiosis could be addressed and the ability of the modified community to resist remodeling. Establishment of stable commensal communities requires interactions among microbes and between the microbes and the host, so both aspects would need to be considered and treated to achieve a stable modified community. Therapy targeting the gut bacterial community is in its infancy and significant challenges remain. However, several strategies have been developed and are being refined to address disease in the gut bacterial compartment of the human superorganism.
METHODS TO MANIPULATE THE GUT BACTERIAL COMMUNITY
A number of different strategies have been proposed and are in use for manipulation of gut microbes. These include antibiotics to eliminate bacteria, pro- or pre-biotics to introduce or encourage growth of bacteria, and fecal microbial transfer to theoretically replace the entire bacterial community.132 None of these strategies is completely effective, but all are in current use.
Antibiotics have been used to target pathogens in the gastrointestinal tract, often with the unintended consequences of also targeting commensal bacteria.133 The gut bacterial community composition is disrupted by antibiotic administration, but short-term treatment with narrow spectrum drugs usually leads to transient changes.134 However, longer-term treatments or use of broad-spectrum drugs can broadly or permanently change the composition of the gut bacterial community in unintended ways and potentially contribute to disease.135 Thus, it is unlikely that treatment with traditional antibiotics will be sufficient to achieve the desired changes to gut microbes.
Pro- and pre-biotics have also been used to manipulate gut bacteria. Pro-biotics are live organisms thought to have beneficial effects on the gut. Pre-biotics are food substrates intended to promote the growth of desirable microbes. Both pro- and pre-biotics have shown some efficacy for treatment of chronic gastrointestinal diseases.136 Microbial metabolites such as short chain fatty acids have also shown some limited efficacy in diseases associated with gastrointestinal bacteria such as inflammatory bowel disease.137 However, the effects associated with pro-biotics, pre-biotics, and microbial metabolites are usually small so they are generally considered adjuncts to therapy, rather than first line treatments. More recently, designer microbes and vaccinations have been proposed as strategies to manipulate gut microbes. Designer microbes are already in use commercially in yogurts containing proprietary strains of Bifidobacteria or Lactobacilli.138 Vaccinations have been developed for prevention of pathogenic bacterial infections and have the potential to work against undesirable commensal bacteria, if they can be confidently identified.139
Lastly, fecal microbial transfer (FMT) is an ancient therapy for gastrointestinal disease that has gained new interest due to its efficacy in treatment of antibiotic-induced Clostridium difficile infections.140 In this strategy, an entire microbial community is transferred from one individual to another in a procedure that has all of the elements of an organ transplant.141 The first step is preparation of the patient to receive a new bacterial community. Theoretically this would entail removal of the existing microbial community. However, it is difficult, if not impossible to completely eliminate gut resident microbes, this is essentially equivalent to excision of an organ with millions of only loosely associated cells. Several studies have shown that when FMT is performed into animals or humans with existing microbial communities, the recipient microbes persist and the community undergoes remodeling over time.142,143 The source for most FMT is freshly voided stool which can vary from individual to individual and from sample to sample. The processing of these samples can also determine which microbes survive to be transferred and low abundance or highly oxygen sensitive members of the gut bacterial community are unlikely to be transferred. Engraftment of the donor community is another factor that is difficult to monitor or control in FMT. As discussed, niche characteristics are highly dependent on the host and determine whether a donor community can colonize the recipient. Part of the appeal of FMT is the idea that relationships required for stable commensal behavior within the bacterial community might be preserved and transferred with this technique. However, selection pressures within the host and colonization by new bacteria after the transfer also have the potential to disrupt these relationships. This has led to the suggestion that the donor and recipient of fecal microbes should be matched. Indeed the major histocompatibility complex has been identified as a determinant of gut bacterial community composition, but it is unclear why this is the case or if that would be an appropriate parameter for matching a recipient and donor for fecal transplantation.144,145
Fecal microbial transfer can potentially address broad aspects of bacterial-related disease. However, it also has the potential for unexpected effects such as transfer of disease. Donor material is typically screened for evidence of common pathogens, but it is not extensively characterized generating the potential for unexpected or disease-causing effects.146 In fact, there is one case report of a patient becoming obese after receiving FMT from an obese donor.147 Although this finding has also been used to suggest that FMT could have efficacy for obesity-related diseases. Donor screening is likely to become more stringent as FMT becomes more routine. However, it is not completely clear what ideal criteria for donor feces should be based on currently available data regarding microbe functions and the potential for differences in the gut environments between donor and recipient.
Fecal microbial transfer is clearly efficacious in treatment of C. difficile-associated diarrhea, but data regarding its efficacy for treatment of chronic diseases associated with dysbiosis are less clear.148 This could be due to the underlying pathophysiology driving the disease, a failure to transfer the “right” microbial community, or persistence of the recipient's original bacteria within the community.143 Ultimately, FMT involves transfer of thousands to millions of almost completely unknown bacteria, each one with the potential to contribute to health or disease, with the hope that the community will engraft, contribute to the health of the individual, and remain stable over time.
Indigenous commensal and even dysbiotic microbiota are extremely resilient. The steady states that bacterial communities reach through self selection and the niche opportunities created by the host favor perpetuation of community stability. A single treatment, whether antibiotics, probiotics, or FMT, is not likely to have sustained effects because these manipulations are not natural to the conditions inherent to individuals. Currently, sequence tags of 16S rRNA gene profiles or metagenomic profiles are used to define health and disease states of the microbiota. However, these don't provide essential functional information, which is ultimately what is important for determining what eubiosis is and how to repair or correct dysbiosis. Finally, changing both sides of the host-microbe equation is critical to achieve effective and stable interventions for microbial-based diseases. It is difficult to change the host genetic and environmental factors that determine bacterial community assemblage. However, as technology and our knowledge base increases, many of these challenges will be overcome.
CHALLENGES FOR THE FUTURE
Commensal microbial communities are complex ecological networks in which mutualistic and antagonistic interactions are flexibly expressed. This means that in order to understand how these organisms contribute to the health and disease of a given host, they must be studied in context. The context in this case is the individual of interest, the bacterial community of interest, and their natural and complex interactions. However, this is incredibly difficult to achieve with current methods. Individuals would need to be sampled at multiple locations along the gastrointestinal tract from both the host and microbe sides over time. Then the functional characteristics of bacteria and host cells as well as indications of their interactions would have to be characterized and analyzed. This is beyond our current capabilities, but better methods and new technologies are constantly being developed to help move us closer to these goals.
In order to better understand the complex microbe-microbe and host microbe interactions in the gastrointestinal tract studies must address three key aspects of experimental design. The first of these is sampling. The majority of studies to this point have obtained a single sample at one time point, often from stool. The composition and character of the gut bacterial community vary longitudinally and radially along the GI tract and biogeography is important for identification of disease-relevant micro-niches.149 Microbes affixed to the mucosal surface of the gastrointestinal tract are presumed to be resident bacteria. The status of stool-derived microbes is less clear. These may faithfully reflect the resident gut bacteria in some cases, but are also likely to contain bacteria that were somehow unable to establish themselves within a mucosal niche. The actual status of the stool-derived bacterial population is difficult to interpret since most gut microbes cannot be easily or reliably cultured and we are currently dependent on sequencing-based techniques for most of these investigations. The character of the gut bacterial community can also vary over time, making longitudinal or multiple time point studies essential.
The second aspect of study design that must be improved is data collection. Currently available sequencing-based techniques are problematic in that they cannot differentiate between live and dead bacteria and do not provide information regarding microbial functions. Bioinformatic techniques to infer function based on functional analysis of genetically similar bacteria have been developed.150 However, this type of analysis is heavily biased by behaviors of model microbes in culture conditions or monoassociation studies and often differs widely from in vivo conditions in a complex bacterial community. In addition to the better established genomic and metagenomic techniques, metatranscriptomic techniques are now in development. The hope is that metatranscriptomics will reveal functional characteristics of gut bacteria. However, these studies are complicated by the huge number of bacterial genes in the gut and the short half-life of bacterial RNA.
Third, data analysis must be improved. Many different computational platforms have been developed to assign taxonomy to 16S sequencing data and several groups have developed software packages to infer function from bacterial genomes. As stated previously, this type of analysis relies heavily on gene characterization in model organisms. It also leads to a skewed perspective on the importance of a given microbe within the population due to the fact that available data is limited to a fairly small number of bacteria. This means that high resolution visualization platforms capable of resolving metagenomes and metatranscriptomes down to the bacterial strain level are needed.151
The ultimate goal for studies of the role of gut bacteria in human disease must be functional analysis of intact communities in humans paired with host responses to the microbes over time. Only when we achieve this goal will be able to fully define the targets for bacterially targeted therapies and understand how to manipulate those targets while also targeting host responses to establish and maintain stable host-microbe relationships that promote health across the entire organism.
Acknowledgments
DK47722, DK097268, DK42086 (DDRCC), CCFA 311120
Footnotes
Conflict of Interest: The authors declare that they have no conflicts of interest.
References
- 1.Gest H. The discovery of microorganisms by Robert Hooke and Antoni van Leeuwenhoek, Fellows of The Royal Society. Notes Rec. R. Soc. 2004;58:187–201. doi: 10.1098/rsnr.2004.0055. [DOI] [PubMed] [Google Scholar]
- 2.Karamanou M, Panayiotakopoulos G, Tsoucalas G, Kousoulis AA, Androutsos G. From miasmas to germs: a historical approach to theories of infectious disease transmission. Infez. Med. 2012;20:58–62. [PubMed] [Google Scholar]
- 3.Collado MC, et al. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romano-Keeler J, Weitkamp J-H. Maternal influences on fetal microbial colonization and immune development. Pediatr. Res. 2015;77:189–95. doi: 10.1038/pr.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: Mom matters. Trends Mol. Med. 2015;21:109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–38. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 8.Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7:e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques TM, et al. Programming infant gut microbiota: influence of dietary and environmental factors. Curr. Opin. Biotechnol. 2010;21:149–156. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Bäckhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 13.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front. Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aagaard K, et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–9. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowers MY, et al. Human gastrin: A Helicobacter pylori-specific growth factor. Gastroenterology. 1999;117:1113–1118. doi: 10.1016/s0016-5085(99)70396-3. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita K, et al. Inhibitory effect of somatostatin on Helicobacter pylori proliferation in vitro. Gastroenterology. 1998;115:1123–1130. doi: 10.1016/s0016-5085(98)70083-6. [DOI] [PubMed] [Google Scholar]
- 19.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10420–5. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 21.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial Adhesins in Host-Microbe Interactions. Cell Host Microbe. 2009;5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Pumbwe L, Skilbeck CA, Wexler HM. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach-or all three? Anaerobe. 12:211–20. doi: 10.1016/j.anaerobe.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Pumbwe L, et al. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43:78–87. doi: 10.1016/j.micpath.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, Feng L, Wang F, Wang L. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 2015;6:6592. doi: 10.1038/ncomms7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brook I, Myhal ML. Adherence of Bacteroides fragilis group species. Infect. Immun. 1991;59:742–744. doi: 10.1128/iai.59.2.742-744.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zogaj X, Bokranz W, Nimtz M, Römling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015;362:1–7. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 30.Vuyst L De, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int. J. Food Microbiol. 2011;149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Hamner S, McInnerney K, Williamson K, Franklin MJ, Ford TE. Bile Salts Affect Expression of Escherichia coli O157:H7 Genes for Virulence and Iron Acquisition, and Promote Growth under Iron Limiting Conditions. PLoS One. 2013;8:e7647. doi: 10.1371/journal.pone.0074647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortman GAM, et al. Iron-induced virulence of Salmonella enterica serovar typhimurium at the intestinal epithelial interface can be suppressed by carvacrol. Antimicrob. Agents Chemother. 2014;58:1664–1670. doi: 10.1128/AAC.02060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohland CL, Jobin C. Microbial Activities and Intestinal Homeostasis: A Delicate Balance Between Health and Disease. Cell. Mol. Gastroenterol. Hepatol. 2015;1:28–40. doi: 10.1016/j.jcmgh.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MT, et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 36.Hillmann F, Fischer R-J, Saint-Prix F, Girbal L, Bahl H. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol. Microbiol. 2008;68:848–860. doi: 10.1111/j.1365-2958.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- 37.Freilich S, et al. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–68. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reidl J, Klose KE. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol. Rev. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 39.McNally L, et al. Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:69–72. doi: 10.1098/rstb.2014.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–25. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trosvik P, de Muinck EJ. de Ecology of bacteria in the human gastrointestinal tract-identification of keystone and foundation taxa. Microbiome. 2015;3(44):1–12. doi: 10.1186/s40168-015-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 2015;194:4081–7. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–9. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell AB, et al. A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 50.Hehemann J-H, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 51.Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014;7:167–76. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for Extensive Resistance Gene Transfer among Bacteroides spp. and among Bacteroides and Other Genera in the Human Colon. Appl. Environ. Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whittle G, Shoemaker NB, Salyers AA. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 2002;59:2044–54. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whittle G, Hund BD, Shoemaker NB, Salyers AA. Characterization of the 13-Kilobase ermF Region of the Bacteroides Conjugative Transposon CTnDOT. Appl. Environ. Microbiol. 2001;67:3488–3495. doi: 10.1128/AEM.67.8.3488-3495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waters JL, Salyers AA. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. mBio. 2013;4:e00569–13. doi: 10.1128/mBio.00569-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salyers AA, Shoemaker NB, Stevens AM, Li LY. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salyers AA, Shoemaker NB. Chromosomal gene transfer elements of the Bacteroides group. Eur. J. Clin. Microbiol. Infect. Dis. 1992;11:1032–8. doi: 10.1007/BF01967795. [DOI] [PubMed] [Google Scholar]
- 58.Gupta A, Vlamakis H, Shoemaker N, Salyers AA. A new Bacteroides conjugative transposon that carries an ermB gene. Appl. Environ. Microbiol. 2003;69:6455–63. doi: 10.1128/AEM.69.11.6455-6463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu J, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2013;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 62.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 2010;12:319–30. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park J-H, et al. Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS One. 2016;11:e0156334. doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaiko GE, et al. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sellin JH, Wang Y, Singh P, Umar S. β-Catenin stabilization imparts crypt progenitor phenotype to hyperproliferating colonic epithelia. Exp. Cell Res. 2009;315:97–109. doi: 10.1016/j.yexcr.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23:1038–46. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–7. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freitas M, Axelsson L-G, Cayuela C, Midtvedt T, Trugnan G. Microbial–host interactions specifically control the glycosylation pattern in intestinal mouse mucosa. Histochem. Cell Biol. 118:149–161. doi: 10.1007/s00418-002-0432-0. [DOI] [PubMed] [Google Scholar]
- 72.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 73.Krogfelt KA, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 1990;58:1995–8. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokurenko EV, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8922–6. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pickard JM, et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 2014;514:638–41. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011;9:356–68. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 77.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008;8:411–20. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 78.Heimlich DR, Harrison A, Mason KM. Host Antimicrobial Peptides in Bacterial Homeostasis and Pathogenesis of Disease. Antibiot. 2014;3:645–76. doi: 10.3390/antibiotics3040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cole JN, Nizet V. Bacterial Evasion of Host Antimicrobial Peptide Defenses. Microbiol. Spectr. 2016;4(1):1–22. doi: 10.1128/microbiolspec.VMBF-0006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial Phosphorylcholine Decreases Susceptibility to the Antimicrobial Peptide LL-37/hCAP18 Expressed in the Upper Respiratory Tract. Infect. Immun. 2000;68:1664–1671. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saar-Dover R, et al. D-Alanylation of Lipoteichoic Acids Confers Resistance to Cationic Peptides in Group B Streptococcus by Increasing the Cell Wall Density. PLoS Pathog. 2012;8:e1002891. doi: 10.1371/journal.ppat.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mégraud F, Bébéar C, Dabernat H, Delmas C. Haemophilus species in the human gastrointestinal tract. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:437–8. doi: 10.1007/BF01962361. [DOI] [PubMed] [Google Scholar]
- 84.Severi E, et al. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 2005;58:1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 85.Bouchet V, et al. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. 2003;100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 87.Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol. Rev. 2012;245:132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 88.Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–11. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 90.Peterson DA, et al. Characterizing the Interactions between a Naturally Primed Immunoglobulin A and Its Conserved Bacteroides thetaiotaomicron Species-specific Epitope in Gnotobiotic Mice. J. Biol. Chem. 2015;290:12630–12649. doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moon C, et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature. 2015;521:90–93. doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013;29:51–58. doi: 10.1016/j.tig.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nguyen TLA, et al. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arrieta M-C, Walter J, Finlay BB. Human Microbiota-Associated Mice: A Model with Challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 99.Xiao L, et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015;33:1103–8. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 100.Ley RE, et al. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chung H, et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wos-Oxley M, et al. Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes. 3:234–49. doi: 10.4161/gmic.19934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 1998;62:1157–70. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aidy S El, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–79. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 107.Ha CWY, Lam YY, Holmes AJ. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J. Gastroenterol. 2014;20:16498–517. doi: 10.3748/wjg.v20.i44.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Byrd AL, et al. Infectious disease. Adapting Koch’s postulates. Science. 2016;351:224–6. doi: 10.1126/science.aad6753. [DOI] [PubMed] [Google Scholar]
- 109.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 110.Jianzhong H. The genetic predisposition and the interplay of host genetics and gut microbiome in Crohn disease. Clin. Lab. Med. 2014;34:763–70. doi: 10.1016/j.cll.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 113.Antonopoulos DA, et al. Reproducible Community Dynamics of the Gastrointestinal Microbiota following Antibiotic Perturbation. Infect. Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Modi SR, et al. Antibiotics and the gut microbiota. J. Clin. Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gibson MK, Crofts TS, Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 2015;27:51–6. doi: 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sekirov I, et al. Antibiotic-Induced Perturbations of the Intestinal Microbiota Alter Host Susceptibility to Enteric Infection. Infect. Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rivera-Chávez F, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winglee K, et al. Aerosol Mycobacterium tuberculosis Infection Causes Rapid Loss of Diversity in Gut Microbiota. PLoS One. 2014;9:e97048. doi: 10.1371/journal.pone.0097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 2001;73:399S–405S. doi: 10.1093/ajcn/73.2.399s. [DOI] [PubMed] [Google Scholar]
- 120.Graf D, et al. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Heal. Dis. 2015;26 doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cani PD, Everard A. Talking microbes: When gut bacteria interact with diet and host organs. Mol. Nutr. Food Res. 2016;60:58–66. doi: 10.1002/mnfr.201500406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.DiBaise JK, Frank DN, Mathur R. Impact of the Gut Microbiota on the Development of Obesity: Current Concepts. Am. J. Gastroenterol. Suppl. 2012;1:22–27. [Google Scholar]
- 125.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–81. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 126.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Turnbaugh PJ, et al. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009;1:6ra14–6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 131.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grady NG, Petrof EO, Claud EC. Microbial therapeutic interventions. Semin. Fetal Neonatal Med. 2016 doi: 10.1016/j.siny.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov. Med. 2012;13:193–9. [PubMed] [Google Scholar]
- 134.La Cochetiere MF De, et al. Resilience of the Dominant Human Fecal Microbiota upon Short-Course Antibiotic Challenge. J. Clin. Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bull MJ, Plummer NT. Part 2: Treatments for Chronic Gastrointestinal Disease and Gut Dysbiosis. Integr. Med. (Encinitas) 2015;14:25–33. [PMC free article] [PubMed] [Google Scholar]
- 137.Besten G den, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–40. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Magro DO, et al. Effect of yogurt containing polydextrose, Lactobacillus acidophilus NCFM and Bifidobacterium lactis HN019: a randomized, double-blind, controlled study in chronic constipation. Nutr. J. 2014;13:75. doi: 10.1186/1475-2891-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Eloe-Fadrosh EA, et al. Impact of Oral Typhoid Vaccination on the Human Gut Microbiota and Correlations with S. Typhi-Specific Immunological Responses. PLoS One. 2013;8:e62026. doi: 10.1371/journal.pone.0062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bakken JS, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011;9:1044–9. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Antonopoulos DA, Chang EB. Transplanting a Microbial Organ: the Good, the Bad, and the Unknown. mBio. 2016;7 doi: 10.1128/mBio.00572-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ellekilde M, et al. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci. Rep. 2014;4:1035–1045. doi: 10.1038/srep05922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li SS, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–9. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 144.Kubinak JL, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat. Commun. 2015;6:8642. doi: 10.1038/ncomms9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bolnick DI, et al. Major Histocompatibility Complex class IIb polymorphism influences gut microbiota composition and diversity. Mol. Ecol. 2014;23:4831–45. doi: 10.1111/mec.12846. [DOI] [PubMed] [Google Scholar]
- 146.Chehoud C, et al. Transfer of Viral Communities between Human Individuals during Fecal Microbiota Transplantation. mBio. 2016;7:e00322–16. doi: 10.1128/mBio.00322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Alang N, Kelly CR. Weight Gain After Fecal Microbiota Transplantation. Open Forum Infect. Dis. 2015;2:ofv004–ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fuentes S, Vos WM. De How to Manipulate the Microbiota: Fecal Microbiota Transplantation. Adv. Exp. Med. Biol. 2016;902:143–153. doi: 10.1007/978-3-319-31248-4_10. [DOI] [PubMed] [Google Scholar]
- 149.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Eren AM, et al. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015;9:968–79. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]