Abstract
Magnetic resonance spectroscopy (MRS) is the only biomedical imaging method that can noninvasively detect endogenous signals from the neurotransmitter γ-aminobutyric acid (GABA) in the human brain. Its increasing popularity has been aided by improvements in scanner hardware and acquisition methodology, as well as by broader access to pulse sequences that can selectively detect GABA, in particular J-difference spectral editing sequences. Nevertheless, implementations of GABA-edited MRS remain diverse across research sites, making comparisons between studies challenging. This large-scale multi-vendor, multi-site study seeks to better understand the factors that impact measurement outcomes of GABA-edited MRS. An international consortium of 24 research sites was formed. Data from 272 healthy adults were acquired on scanners from the three major MRI vendors and analyzed using the Gannet processing pipeline. MRS data were acquired in the medial parietal lobe with standard GABA+ and macromolecule- (MM-) suppressed GABA editing. The coefficient of variation across the entire cohort was 12% for GABA+ measurements and 28% for MM-suppressed GABA measurements. A multilevel analysis revealed that most of the variance (72%) in the GABA+ data was accounted for by differences between participants within-site, while site-level differences accounted for comparatively more variance (20%) than vendor-level differences (8%). For MM-suppressed GABA data, the variance was distributed equally between site- (50%) and participant-level (50%) differences. The findings show that GABA+ measurements exhibit strong agreement when implemented with a standard protocol. There is, however, increased variability for MM-suppressed GABA measurements that is attributed in part to differences in site-to-site data acquisition. This study’s protocol establishes a framework for future methodological standardization of GABA-edited MRS, while the results provide valuable benchmarks for the MRS community.
Keywords: Editing, GABA, MEGA-PRESS, MRS, Multi-site study
1. Introduction
Magnetic resonance spectroscopy (MRS) is unique amongst the neuroimaging modalities in detecting endogenous signals from complex molecules in the brain noninvasively. Of particular interest is the detection and measurement of γ-aminobutyric acid (GABA), the major inhibitory neurotransmitter in the mammalian brain (McCormick, 1989). Healthy brain function relies on GABAergic inhibitory processes, and understanding GABAergic mechanisms in both healthy and pathological brain function has been one core focus of neuroscience. MRS measurements of GABA have been associated with individual differences in hemodynamic and electrophysiological signals (Donahue et al., 2010; Hu et al., 2013; Kapogiannis et al., 2013; Muthukumaraswamy et al., 2009) and a number of measures of cognition (Fujihara et al., 2015; Shibata et al., 2017; Yoon et al., 2016) and behavior (Boy et al., 2011; Greenhouse et al., 2017; Puts et al., 2011; Silveri et al., 2013). Differential levels of GABA have been observed in a number of neuropsychiatric disorders, such as schizophrenia (Kegeles et al., 2012; Öngür et al., 2010; Rowland et al., 2016; Yoon et al., 2010) and depression (Bhagwagar et al., 2008; Hasler et al., 2007; Price et al., 2009), neurodevelopmental disorders such as autism spectrum disorder (Drenthen et al., 2016; Gaetz et al., 2014; Puts et al., 2016) and attention deficit hyperactivity disorder (Bollmann et al., 2015; Edden et al., 2012a), and neurological diseases, such as Parkinson’s disease (Emir et al., 2012), amyotrophic lateral sclerosis (Foerster et al., 2012; Foerster et al., 2013) and diabetic neuropathy (Petrou et al., 2012).
The most common MRS approach for detecting the GABA signal is the Mescher– Garwood (MEGA) editing sequence (Mescher et al., 1998), a J-difference spectral editing technique that is typically implemented within a point resolved spectroscopy (PRESS) (Bottomley, 1987) acquisition. MEGA-PRESS and other spectral editing techniques exploit the known scalar coupling properties of molecules in order to separate their associated signals from the overlapping signals of other molecules. For lower-concentration metabolites such as GABA, spectral editing differentiates the weak signals of interest from the stronger, overlapping signals of higher-concentration metabolites. Difference editing techniques in particular use frequency-selective inversion pulses to achieve this (for methodological reviews, see Harris et al., 2017; Puts and Edden, 2012). The popularity of MEGA-PRESS is attributed to a number of factors, including the wide availability of the basic PRESS sequence across scanner platforms, its relatively straightforward implementation (Mullins et al., 2014), its reproducibility (Bogner et al., 2010; Brix et al., 2017; Geramita et al., 2011; Mikkelsen et al., 2016a; Near et al., 2014; O’Gorman et al., 2011; Shungu et al., 2016) and continued development of acquisition methodology and data processing tools (Chan et al., 2016; Edden et al., 2014).
However, despite these positive attributes, the diversity of implementations of MEGA-PRESS across research sites and vendors has meant that comparing data between different studies is difficult. For instance, pulse sequence parameters, and in particular pulse timings, differ between vendor-specific PRESS sequences and lead to subtle but important differences in the resolved GABA signal (Near et al., 2013b). Moreover, spectral editing of GABA is associated with a number of complexities, including TE-dependent J-evolution of the GABA spin system (Edden et al., 2012b), frequency and spatial effects of volume localization (Edden and Barker, 2007; Kaiser et al., 2008), sensitivity to B0 field frequency offsets (Edden et al., 2016; Harris et al., 2014) and contamination from co-edited macromolecules (MM) (Henry et al., 2001; Rothman et al., 1993). It is generally assumed that these factors limit the degree to which a GABA-edited measurement from one site can be compared to another at a different site.
In order to establish the extent to which site-, sequence- and vendor-specific differences impact quantitative MEGA-PRESS measurement outcomes, a multi-vendor, multi-site dataset has been assembled by an international consortium of GABA-edited MRS users. The consortium was formed with the aim of building a normative database of MEGA-PRESS data acquired on the major MRI scanner platforms at a range of imaging centers focused on neuroscience research. This dataset aims to capture some of the diversity of the sequences used, but within the framework of a standardized study design and acquisition protocol that would reflect typical MEGA-PRESS parameters. This approach reduced the number of confounding variables present within the dataset (e.g., standardizing key parameters such as TE, TR and editing pulse bandwidth), while maintaining diversity at the level of pulse sequence implementation (e.g., localization pulse waveforms/bandwidths, pulse timings and crusher gradient schemes).
This paper presents initial results from this multi-site study, focusing on how variance in creatine-referenced GABA measurements was distributed across research sites and scanner vendors and examining the influence of various acquisition- and participant-related effects. Given the complexity of this dataset, it is not possible to report on all aspects of the project in a single article, so for example, water-referenced quantification (including tissue-dependent correction factors) and site-to-site differences in voxel placement fidelity and segmentation will be presented in a future report.
2. Methods
2.1 Data collection
A consortium of 24 research institutions based in nine countries participated in this initiative, with each site contributing 5–12 datasets collected from consenting adult volunteers. Specific guidelines for each site’s participant cohort were: 18–35 years old; approximately 50:50 female/male split; no known neurological or psychiatric illness. In total, data from 272 participants were collected. Participant demographics are provided in Table 1. Scanning was conducted in accordance with ethical standards set by the institutional review board (IRB) at each site, including the sharing of anonymized data. Anonymized data files were shared securely with and analyzed by consortium members at the Johns Hopkins University School of Medicine with local IRB approval.
Table 1.
Participant demographics, displayed by site and by vendor.
| Site ID | Sample size | Age (years) (mean ± SD) | Sex (F/M) |
|---|---|---|---|
| G1 | 12 | 23.92 ± 4.81 | 7/5 |
| G2 | 12 | 26.83 ± 4.00 | 6/6 |
| G3 | 7 | 23.43 ± 5.47 | 2/5 |
| G4 | 12 | 25.58 ± 4.48 | 6/6 |
| G5 | 12 | 25.50 ± 3.73 | 5/7 |
| G6 | 12 | 24.33 ± 4.25 | 6/6 |
| G7 | 12 | 28.08 ± 4.01 | 6/6 |
| G8 | 12 | 29.67 ± 2.10 | 6/6 |
| All GE | 91 | 26.05 ± 4.43 | 44/47 |
| P1 | 12 | 25.08 ± 3.23 | 6/6 |
| P2 | 12 | 28.75 ± 3.91 | 10/2 |
| P3 | 12 | 29.25 ± 3.14 | 5/7 |
| P4 | 12 | 24.92 ± 4.29 | 7/5 |
| P5 | 8 | 23.13 ± 2.36 | 3/5 |
| P6 | 12 | 27.33 ± 3.68 | 7/5 |
| P7 | 12 | 23.58 ± 3.73 | 6/6 |
| P8 | 12 | 23.25 ± 1.96 | 5/7 |
| P9 | 12 | 25.83 ± 4.61 | 6/6 |
| All Philips | 104 | 25.78 ± 4.06 | 55/49 |
| S1 | 12 | 25.67 ± 3.65 | 6/6 |
| S2 | 5 | 40.40 ± 7.44 | 0/5 |
| S3 | 12 | 31.58 ± 3.42 | 9/3 |
| S4 | 12 | 27.67 ± 2.77 | 6/6 |
| S5 | 12 | 26.50 ± 3.68 | 6/6 |
| S6 | 12 | 24.92 ± 2.02 | 6/6 |
| S7 | 12 | 28.75 ± 3.77 | 6/6 |
| All Siemens | 77 | 28.35 ± 5.21 | 39/38 |
| Overall | 272 | 26.60 ± 4.65 | 138/134 |
2.2 Data acquisition
Each site acquired MEGA-PRESS data on a 3 T scanner by following a standard scan protocol as closely as possible. Eight sites used GE scanners, nine used Philips scanners and seven used Siemens scanners, with locally available phased-array head coils (see Table 2). Two MRS acquisitions were run: a standard GABA+-edited acquisition where ON editing pulses were placed at 1.9 ppm and OFF editing pulses were placed at 7.46 ppm; and an MM-suppressed GABA-edited acquisition where the editing pulses were placed symmetrically about the MM resonance at 1.7 ppm (ON/OFF = 1.9/1.5 ppm) (Henry et al., 2001). GE site 6 (G6) did not acquire MM-suppressed data. For the sequences used in this study, GE and Philips editing pulse offsets are calculated assuming a water frequency of 4.68 ppm and Siemens assumes 4.7 ppm. Given that GABA editing involves the use of frequency-selective editing pulses, their inversion frequency bandwidth has a significant impact on editing efficiency, determining the extent of MM co-editing in GABA+ acquisitions and the extent of GABA nulling in symmetric MM suppression (see Edden et al., 2016; Harris et al., 2014; Terpstra et al., 2002). For GE and Philips implementations where editing pulse duration is specified, editing pulse duration was set to 15 ms for the GABA+ acquisition and 20 ms for the MM-suppressed GABA acquisition. This equated to inversion bandwidths at full-width half-maximum (FWHM) of 81.7/82.5 Hz (GE/Philips) for the GABA+ acquisition and 61.3/61.9 Hz (GE/Philips) for the MM-suppressed GABA acquisition. For Siemens implementations, where the editing pulse bandwidth specified on the scanner does not correspond to the FWHM bandwidth (Lange et al., 2016), FWHM bandwidths were 82.4 Hz for the GABA+ acquisition and 61.8 Hz for the MM-suppressed GABA acquisition. The TE of the GABA+ acquisition was set to 68 ms. For the MM-suppressed acquisition, the TE was set to 80 ms on the GE and Philips platforms (Edden et al., 2012c) and to 68 ms on the Siemens platform. The higher peak B1 on some Siemens platforms makes the more selective editing pulses possible without increasing the TE. For one Siemens site (S2), the TE of the MM-suppressed acquisition was increased to 80 ms due to limited peak B1. Representative vendor-specific MEGA-PRESS pulse sequence diagrams (at TE = 68 ms) are shown in Fig. 1A. Parameters common between the two acquisitions included: TR = 2000 ms; 320 averages (i.e., 160 ON and 160 OFF transients); ~10 min scan time. Although the spectral width and number of discrete data points differed from site to site (see Table 2), in all cases the aim was to achieve a data acquisition time of ~1 s. All Philips sites except P8 addressed B0 field offsets with prospective frequency correction based on interleaved water referencing (Edden et al., 2016). Specifically, for every 40 water-suppressed acquisitions, a water-unsuppressed acquisition was performed and used to correct the center frequency in real-time. This method was only available on the Philips platform at the time of data collection. Details of B0 shimming approaches are provided in Table 2. All three vendors use a volume-localized acquisition for center frequency calibration. They differ somewhat in terms of localization method (e.g., STEAM on Siemens and semi-LASER on Philips) and acquisition resolution; both GE and Philips suppress fat signals to make algorithmic determination of center frequency more robust. GE data were saved in P-file format, Philips data were saved in SDAT/SPAR format and Siemens data were saved in TWIX format.
Table 2.
Software, hardware and acquisition parameters used to collect MEGA-PRESS data at each site.
| Site ID | Scanner vendor and model | Software release | Tx/Rx hardware | B0 shimming approach | MEGA-PRESS sequence variant | Phase cycling | Editing interleaving | TE (ms) (GABA+) | TE (ms) (MM-s GABA) | Spectral width (Hz) | Data points | Water suppression |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | GE Discovery MR750w |
DV25 | Body coil/32-ch head coil | Double-echo GRE | Interleaved sequencea | 2 steps | 2 TRs | 68 | 80 | 5000 | 4096 | CHESS |
| G2 | GE Discovery MR750 | DV24 | Body coil/8-ch head coil | Double-echo GRE | ATSM patchb | 2 steps | 1 TR | 68 | 80 | 2000 | 2048 | CHESS |
| G3 | GE Discovery MR750 | DV24 | Body coil/32-ch head coil | Double-echo GRE | ATSM patch | 8 steps | 1 TR | 68 | 80 | 2000 | 2048 | CHESS |
| G4 | GE Discovery MR750 | DV25 | Body coil/8-ch head coil | Double-echo GRE | ATSM patch | 8 steps | 1 TR | 68 | 80 | 5000 | 4096 | CHESS |
| G5 | GE Discovery MR750 | DV25 | Body coil/32-ch head coil | Double-echo GRE | ATSM patch | 8 steps | 1 TR | 68 | 80 | 2000 | 2048 | CHESS |
| G6 | GE Signa HDx | HD16 | Body coil/8-ch head coil | Double-echo GRE | ATSM patch | 2 steps | 2 TRs | 68 | – | 2000 | 2048 | CHESS |
| G7 | GE Discovery MR750 | DV24 | Body coil/8-ch head coil | Double-echo GRE | ATSM patch | 8 steps | 1 TR | 68 | 80 | 2000 | 2048 | CHESS |
| G8 | GE Discovery MR750 | DV24 | Body coil/8-ch head coil | Double-echo GRE | ATSM patch | 8 steps | 1 TR | 68 | 80 | 2000 | 2048 | CHESS |
| P1 | Philips Achieva | R5.1.7 | Body coil/32-ch head coil | PB-autoc | JHU patchd | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | VAPOR |
| P2 | Philips Achieva | R5.1.7 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | VAPOR |
| P3 | Philips Achieva | R3.2.2 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | VAPOR |
| P4 | Philips Ingenia CX | R5.1.7 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | MOIST |
| P5 | Philips Achieva TX | R5.1.7 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | MOIST |
| P6 | Philips Achieva | R3.2.3 | Body coil/8-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | MOIST |
| P7 | Philips Ingenia | R5.1.8 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | VAPOR |
| P8 | Philips Ingenia CX | R5.1.8 | Body coil/32-ch head coil | PB-auto | JHU patche | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | MOIST |
| P9 | Philips Achieva | R5.1.7 | Body coil/32-ch head coil | PB-auto | JHU patch | 16 steps | 1 TR | 68 | 80 | 2000 | 2048 | VAPOR |
| S1 | Siemens Trio | VB17 | Body coil/32-ch head coil | 3D-DESS + manual | WIP (529) | 16 steps | 1 TR | 68 | 68 | 4000f | 4096 | CHESS |
| S2 | Siemens Verio | VB17 | Body coil/32-ch head coil | 3D-DESS + manual | WIP (529) | 16 steps | 1 TR | 68 | 80 | 4000 | 4096 | CHESS |
| S3 | Siemens Prisma | VD13 | Body coil/20-ch head/neck coil | FAST(EST)MAP | WIP (859D) | 16 steps | 1 TR | 68 | 68 | 4000 | 4096 | WET |
| S4 | Siemens Prisma | VE11 | Body coil/64-ch head coil | 3D-DESS | WIP (user- modified) | 16 steps | 1 TR | 68 | 68 | 4000 | 4096 | WET |
| S5 | Siemens Trio | VB17 | Body coil/12-ch head coil | 3D-DESS | WIP (529) | 16 steps | 1 TR | 68 | 68 | 4000 | 4096 | CHESS |
| S6 | Siemens Trio | VB17 | Body coil/32-ch head coil | FAST(EST)MAP | WIP (529) | 16 steps | 1 TR | 68 | 68 | 4000 | 4096 | WET |
| S7 | Siemens Trio | VB17 | Body coil/32-ch head coil | FAST(EST)MAP | WIP (user- modified) | 16 steps | 1 TR | 68 | 68 | 2000 | 2070/2080g | CHESS |
Sequence developed by Gareth Barker, David Lythgoe (King’s College London), C. John Evans (Cardiff University) and RAEE, originally based on Dikoma Shungu’s sequence (Well Cornell Medical College).
Including source code derivatives. Sequence developed by RN.
PB-auto is a Philips pencil-beam projection-based method for automatic voxel shimming.
Sequence developed by RAEE.
Interleaved water referencing not implemented.
TWIX data are oversampled. For example, if the specified spectral width and number of discrete data points are set at 2000 Hz/2048, the data are oversampled to 4000 Hz/4096.
In the Siemens WIP, extra data points are added before and/or after the detected spin echo if the number of data points is specified as 512 or 1024. The additional points before the spin echo were removed during data processing. For the MM-suppressed GABA acquisition, the longer duration of the editing pulses prevents any extra points from being added before the echo, hence the difference in the number of data points between the two acquisitions for site S7.
ATSM, Advanced Technology Software Module; GRE, gradient echo; JHU, Johns Hopkins University; MM-s, MM-suppressed; PB, pencil beam; Rx, receive; Tx, transmit; WIP, work in progress
Fig. 1.
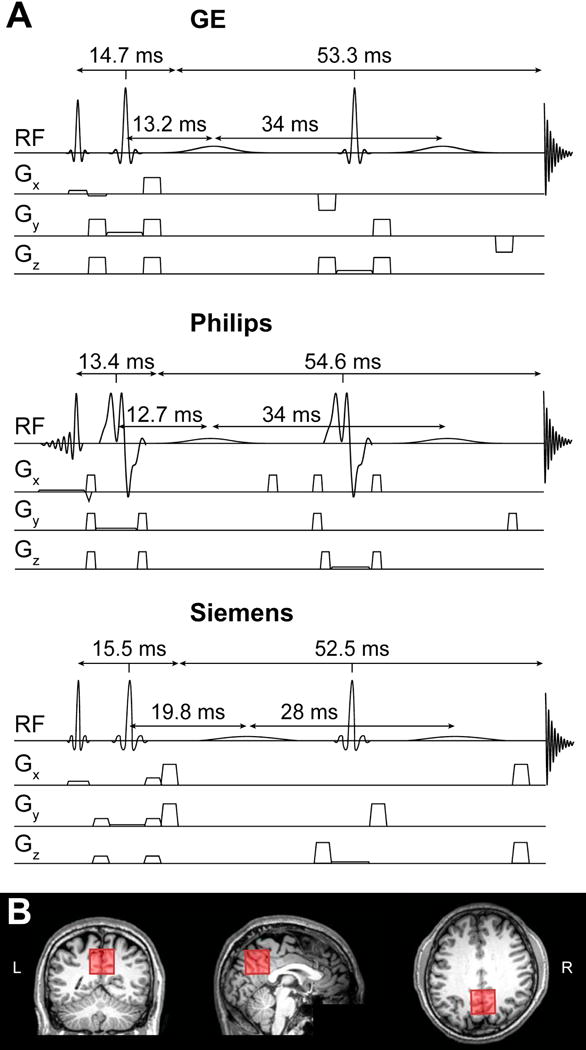
(A) Pulse sequence diagrams of vendor-specific implementations of MEGA-PRESS at TE = 68 ms. Pulse timings, including TE1/TE2, are indicated. The GE implementation employed a crusher gradient scheme based on the BASING sequence (Star-Lack et al., 1997). The Philips implementation employed non-sinc-based amplitude-modulated refocusing pulses. In the Siemens implementation, the timing between the first and second editing pulse deviates from the optimal TE/2. This slight deviation leads to the GABA signal in the ON scan being nearly, but not fully, refocused. (B) Example MRS voxel placement in the medial parietal lobe of one participant. At each research site, the voxel was rotated in the sagittal plane to be parallel with a line connecting the genu and splenium of the corpus callosum.
All MEGA-PRESS data were acquired from a 30 × 30 × 30 mm3 voxel placed in the medial parietal lobe (Fig. 1B). All sites followed the same protocol, using a guideline image, for voxel placement. Briefly, the voxel was rotated in the sagittal plane to align it with a line connecting the genu and splenium of the corpus callosum. Each site was instructed to comply with the standardized protocol, but also to avoid ventricles and/or the outer surfaces of the brain when necessary to ensure good data quality.
2.3 Data processing
Data from each site were processed in Gannet (Edden et al., 2014) using the software’s automated analysis pipeline with some in-house customization for this study. Raw time-domain data were first corrected for frequency and phase errors by spectral registration (Near et al., 2015) using the transient 10% into the acquisition (i.e., the 32nd transient) as a reference. ON/OFF transient pairs were rejected from further processing if either of their corresponding frequency/phase offset estimates were greater than 3 standard deviations (SDs) from the mean of frequency/phase offset estimates for all pre-corrected transients. A threshold of 3 SDs corresponds to 99.7% of (normally distributed) frequency/phase estimates. ON/OFF transient pairs exceeding this threshold would be expected to introduce more uncertainty into the data (Waddell et al., 2007) and were therefore removed. The data were then filtered using a 3-Hz exponential weighting function and zero-filled so as to yield a nominal spectral resolution of 0.061 Hz/point upon fast Fourier transformation. Individual ON and OFF subspectra were then averaged and subtracted to produce the edited difference (DIFF) spectrum.
Data were visually inspected for spectral artifacts, specifically lipid contamination, subtraction errors and a non-constant baseline. Individual datasets were rejected if the signal fitting routine (details below) was compromised. For instance, significant lipid contamination can distort the baseline around the 3.0 ppm GABA signal, such that the modeling algorithm converges on a clearly incorrect solution. In such cases, the data were removed from further analysis. Quantitative data quality metrics were also measured, including N-acetylaspartate (NAA) and GABA signal-to-noise ratios (SNR), linewidth and average center frequency offset . SNR estimates were measured as the amplitude of the given modeled signal (either NAA in the averaged OFF spectrum, fit with a Lorentzian function, or GABA in the DIFF spectrum) divided by twice the SD of the noise signal. Estimating noise using a consistent methodology across the whole dataset proved surprisingly challenging. Examination of the downfield portion (> 8 ppm) of the frequency-domain data revealed signal artifacts in some datasets, likely a result of suboptimal water suppression. Therefore, the following algorithm was employed to estimate artifact-free noise. First, two independent segments of the OFF or DIFF spectrum, 10–11 ppm and 11–12 ppm, were detrended using a second-order polynomial function and the SD of each detrended segment was then calculated. Detrending is required to remove baseline artifacts (often related to the water signal). The lesser of the two residuals was assumed to be the better estimate of noise in each spectrum. The NAA and GABA signal amplitudes were then divided by twice the respective SD of noise. This approach ensured that variations in baseline and signal-related artifacts did not bias SNR measurements. Linewidth was measured as the FWHM of the modeled NAA signal. was calculated as the mean (over the course of the acquisition) difference between the observed frequency of the residual water signal in the pre-frequency-corrected subspectra and the nominal water frequency at δ0 4.68 ppm. It should be noted that using the mean of offset differences does not fully characterize center frequency offsets but is a useful heuristic.
2.4 Quantification
The DIFF spectrum was modeled between 2.79 and 4.10 ppm with a three-Gaussian function with a nonlinear baseline to quantify the 3.0 ppm GABA signal and 3.75 ppm glutamate + glutamine (Glx) signals using nonlinear least-squares fitting. The OFF spectrum was modeled between 2.6 and 3.6 ppm with a two-Lorentzian model to quantify creatine (Cr) as an internal reference signal. GABA measurements derived from the GABA+ and MM-suppressed GABA acquisitions were quantified as signal integral ratios: IGABA/ICr, where IGABA is the integral of the modeled 3.0 ppm GABA signal and ICr is the integral of the modeled 3.0 ppm Cr signal. No signal scaling factors were applied. Measurements are denoted GABA+/Cr and MM-suppressed GABA/Cr. Fit quality for each model (εGABA, εCr) was assessed by normalizing the SD of the model residuals to the amplitude of the respective modeled signal. For GABA, the residuals were limited to the frequency range between 2.79 and 3.55 ppm. Overall fit error was then defined as .
2.5 Statistical analysis
The data had a nested structure. That is, each participant was scanned at one site and each site had a scanner manufactured by one of the three vendors. Therefore, a multilevel model (Hayes, 2006; Peugh, 2010; Snijders and Bosker, 2012) was used for the primary statistical analysis. This approach involves the use of a linear mixed-effects model, an extension of the well-known general linear model, but one which explicitly takes into account systematic effects ascribed to the hierarchical structure of data.
The principal aim of this study was to examine vendor-, site- and participant-related effects on measurement outcomes of GABA-edited MRS. This was achieved by fitting a three-level unconditional linear mixed-effects model to the GABA+ and MM-suppressed GABA data:
| [1] |
where yijk is the observed GABA measurement for participant i at site j on a scanner manufactured by vendor k, β0 is the model intercept (the grand mean), v0k is the level-3 random effect of vendor, s0jk is the level-2 random effect of site and pijk is the level-1 random effect of participant (the residual error). The random effects are assumed to follow a normal distribution with zero mean and constant variance. Since the total variance in the model is equal to the sum of the variance attributed to the three effects, it follows that vendor-, site- and participant-level variance partition coefficients (VPCs) can be respectively calculated as:
| [2] |
| [3] |
| [4] |
Each VPC represents the proportion of total variance in the data accounted for by the specific random effect in the model (Goldstein et al., 2002), in this case, vendor, site and participant.
Secondary multilevel analyses were also performed where fixed effects (predictors) were tested to account for variance attributed to acquisition- and participant-related effects. In this study, the effects of linewidth, NAA SNR, , age and sex on GABA measurement outcome were tested. Such a conditional model with a single predictor is formulated as:
| [5] |
This model includes an explanatory variable (x1ijk) with a grand mean slope (β1) and by-vendor and by-site random intercepts (v0k, s0jk) and random slopes (v1k, s1jk). At the vendor level, the random effects v0k and v1k are assumed to follow a bivariate normal distribution with zero means, variances and and covariance σv01. The covariance denotes the correlation between the predictor slopes and intercepts. The same definitions apply to the site-level parameters s0jk, s1jk, , and σs01. In this model, both the by-vendor and by-site intercepts and slopes of the explanatory variable are allowed to vary across each level. This “maximal” approach has been shown to reduce Type I error rates in linear mixed-effects models (Barr et al., 2013).
Linear mixed-effects models were fit in R (version 3.3.3; R Core Team, 2017) using the lme4 package (Bates et al., 2015) and maximum likelihood for model estimation. The outcome and continuous explanatory variables were standardized (by z-transformation) to aid model convergence and interpretability of model parameter estimates (Schielzeth, 2010). Goodness-of-fit was calculated as a log-likelihood statistic (−2logL). To test for significant random or fixed effects, chi-square likelihood ratio tests were performed by comparing the log-likelihood statistic of one model to that of a reduced model (i.e., a model excluding the random or fixed effect of interest). Likelihood ratio tests were bootstrapped 2,000 times using a parametric bootstrap method (Halekoh and Højsgaard, 2014). If an effect was significant, it was retained in the next assessed model; if not, it was removed. Specifically, the effects of vendor and site were tested first, the effects of acquisition-related variables (linewidth, NAA SNR, ) were tested second and the effects of participant-related variables (age, sex) were tested last.
A Pearson correlation coefficient was calculated to test the relationship between participants’ GABA+/Cr and MM-suppressed GABA/Cr values. This was done by using the residuals of the respective linear mixed-effects model that included only the effects that accounted for a significant amount of variance in either dataset. To illustrate the importance of accounting for systematic effects in the data, a correlational test was also conducted on the raw GABA+/Cr and MM-suppressed GABA/Cr values. The correlations were bootstrapped 10,000 times to produce 95% confidence intervals (CIs) using the bias-corrected and accelerated nonparametric bootstrap method (DiCiccio and Efron, 1996). For all inferential statistical tests, a p-value less than 0.05 was considered significant. Corrections for multiple comparisons were not applied.
3. Results
GABA-edited MRS data were successfully acquired at all 24 sites. Following quality control analysis, seven GABA+ and 19 MM-suppressed GABA datasets (3% and 7% of the total collected data for either acquisition, respectively) were removed from further analysis. All MM- suppressed GABA data from site G3 were excluded as consistent, excessive center frequency offsets (approximately −0.1 ppm on average) resulted in extremely small or absent GABA signals. Fig. 2 shows the mean ± 1 SD GABA+ and MM-suppressed GABA DIFF spectra for each vendor. Examples of the GABA+Glx signal fitting on individual acquisitions are provided in Fig. S1. Distinctive edited GABA peak lineshapes were seen for each vendor, likely a consequence of the different implementations of the MEGA-PRESS sequences between each vendor (Near et al., 2013b). GABA+/Cr and MM-suppressed GABA/Cr values, broken down by site and by vendor, are shown in Fig. 3. Mean ± 1 SD GABA+/Cr values were 0.123 ± 0.014 for GE, 0.111 ± 0.013 for Philips and 0.116 ± 0.012 for Siemens. Across all sites and vendors, GABA+/Cr was 0.116 ± 0.014. Coefficients of variation (CVs) were 11.5%, 11.6%, and 10.7% for GE, Philips and Siemens, and 12.0% across all vendors. The mean within-site CV was 9.5%. Mean MM-suppressed GABA/Cr values (and CVs) were 0.043 ± 0.013 (29.6%) for GE, 0.044 ± 0.014 (30.7%) for Philips and 0.041 ± 0.007 (17.3%) for Siemens, and 0.043 ± 0.012 (27.6%) across all sites and vendors. The mean within-site CV was 18.8%. The average ratio between MM-suppressed GABA/Cr and GABA+/Cr was 0.38 ± 0.11. Site-level GABA+/Cr and MM-suppressed GABA/Cr values are listed in Table 3.
Fig. 2.
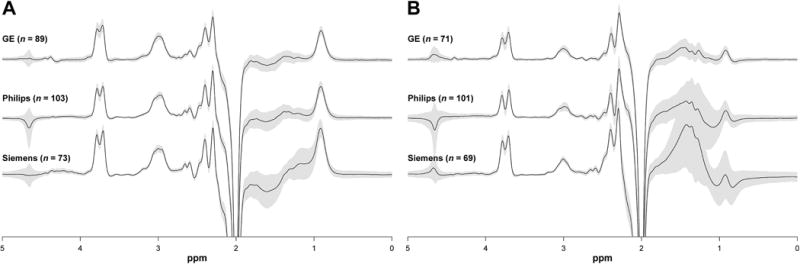
Vendor-mean GABA-edited DIFF spectra acquired by (A) GABA+ editing and (B) MM-suppressed GABA editing. The grey patches represent ± 1 SD. The associated sample sizes are shown in parentheses. Each individual DIFF spectrum was normalized to the amplitude of an unsuppressed water signal prior to averaging. The larger SD of the residual water signal (4.68 ppm) is in part a result of inconsistent water suppression (both between individual acquisitions and shot-to-shot) during the MEGA-PRESS experiment. The use of MOIST water suppression by some Philips sites also contributed to the larger SD in the mean Philips spectra.
Fig. 3.
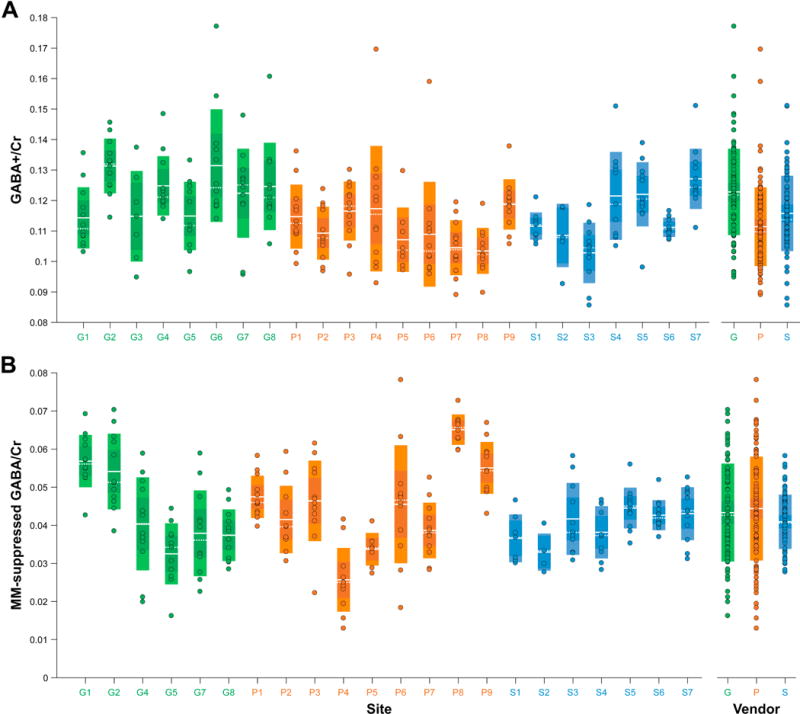
(A) GABA+/Cr and (B) MM-suppressed GABA/Cr measurements, displayed by site and by vendor. The boxes shaded with lighter colors represent ±1 SD and the darker boxes represent the 95% CI. The solid white lines denote the mean, while the dashed white lines denote the median. Sites are colored by vendor (GE sites in green, Philips sites in orange, Siemens sites in blue).
Table 3.
Quantification and data quality metrics for the GABA+ and MM-suppressed GABA data, displayed by site and by vendor (shown as mean ± SD).
| Site ID | GABA/Cr | Fit error (%) | Linewidth (Hz) | NAA SNR | GABA SNR | (ppm) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| GABA+ | MM-s GABA | GABA+ | MM-s GABA | GABA+ | MM-s GABA | GABA+ | MM-s GABA | GABA+ | MM-s GABA | MM-s GABA | MM-s GABA | |
|
|
||||||||||||
| G1 | 0.11 ± 0.01 | 0.06 ± 0.01 | 4.59 ± 0.63 | 6.16 ± 1.21 | 8.68 ± 0.66 | 8.70 ± 0.70 | 350 ± 58 | 343 ± 49 | 21 ± 3 | 10 ± 2 | 0.015 ± 0.012 | 0.014 ± 0.011 |
| G2 | 0.13 ± 0.01 | 0.05 ± 0.01 | 6.16 ± 1.99 | 8.02 ± 3.05 | 8.38 ± 0.63 | 8.55 ± 0.81 | 418 ± 76 | 412 ± 63 | 27 ± 5 | 12 ± 3 | −0.010 ± 0.012 | −0.014 ± 0.013 |
| G3 | 0.11 ± 0.01 | DE | 6.87 ± 1.39 | DE | 9.07 ± 1.54 | DE | 380 ± 29 | DE | 23 ± 3 | DE | −0.054 ± 0.027 | DE |
| G4 | 0.12 ± 0.01 | 0.04 ± 0.01 | 5.32 ± 1.36 | 10.32 ± 3.49 | 8.93 ± 0.74 | 9.07 ± 1.10 | 285 ± 41 | 279 ± 49 | 18 ± 3 | 7 ± 2 | −0.017 ± 0.011 | −0.016 ± 0.015 |
| G5 | 0.11 ± 0.01 | 0.03 ± 0.01 | 6.26 ± 2.40 | 10.11 ± 3.20 | 8.62 ± 0.65 | 8.43 ± 0.72 | 503 ± 90 | 445 ± 73 | 26 ± 3 | 8 ± 2 | 0.003 ± 0.021 | 0.004 ± 0.021 |
| G6 | 0.13 ± 0.02 | DNA | 6.58 ± 1.24 | DNA | 7.94 ± 0.47 | DNA | 427 ± 88 | DNA | 23 ± 4 | DNA | −0.005 ± 0.009 | DNA |
| G7 | 0.12 ± 0.01 | 0.04 ± 0.01 | 7.06 ± 1.84 | 10.78 ± 2.78 | 8.64 ± 0.67 | 8.67 ± 0.65 | 390 ± 70 | 345 ± 72 | 23 ± 5 | 9 ± 2 | −0.023 ± 0.014 | −0.026 ± 0.015 |
| G8 | 0.12 ± 0.01 | 0.04 ± 0.01 | 7.40 ± 2.71 | 11.25 ± 3.24 | 8.17 ± 0.60 | 8.10 ± 0.51 | 322 ± 67 | 330 ± 58 | 19 ± 3 | 8 ± 2 | −0.02 ± 0.012 | −0.020 ± 0.01 |
| All GE | 0.12 ± 0.01 | 0.04 ± 0.01 | 6.24 ± 1.95 | 9.43 ± 3.34 | 8.53 ± 0.79 | 8.59 ± 0.80 | 384 ± 93 | 358 ± 80 | 23 ± 5 | 9 ± 3 | −0.012 ± 0.022 | −0.010 ± 0.02 |
| P1 | 0.11 ± 0.01 | 0.05 ± 0.01 | 4.91 ± 0.49 | 6.42 ± 1.13 | 7.53 ± 0.39 | 7.41 ± 0.35 | 457 ± 76 | 495 ± 83 | 25 ± 5 | 11 ± 2 | −0.006 ± 0.005 | −0.004 ± 0.005 |
| P2 | 0.11 ± 0.01 | 0.04 ± 0.01 | 4.83 ± 0.73 | 8.48 ± 1.99 | 7.43 ± 0.39 | 7.55 ± 0.35 | 448 ± 73 | 421 ± 75 | 22 ± 4 | 9 ± 2 | 0.002 ± 0.003 | 0.0001 ± 0.003 |
| P3 | 0.12 ± 0.01 | 0.05 ± 0.01 | 5.40 ± 0.76 | 8.70 ± 1.69 | 7.88 ± 0.49 | 7.96 ± 0.46 | 351 ± 47 | 342 ± 64 | 19 ± 3 | 8 ± 2 | −0.009 ± 0.007 | −0.009 ± 0.009 |
| P4 | 0.12 ± 0.02 | 0.03 ± 0.01 | 6.40 ± 1.69 | 18.10 ± 9.76 | 7.41 ± 0.33 | 7.44 ± 0.37 | 466 ± 56 | 481 ± 88 | 26 ± 5 | 6 ± 1 | 0.004 ± 0.003 | 0.005 ± 0.004 |
| P5 | 0.11 ± 0.01 | 0.03 ± 0.00 | 4.60 ± 0.76 | 8.86 ± 2.04 | 7.74 ± 0.39 | 7.69 ± 0.38 | 473 ± 107 | 484 ± 76 | 26 ± 3 | 9 ± 1 | −0.009 ± 0.004 | −0.004 ± 0.004 |
| P6 | 0.11 ± 0.02 | 0.05 ± 0.02 | 6.17 ± 0.89 | 11.95 ± 4.59 | 7.73 ± 0.47 | 7.65 ± 0.37 | 340 ± 48 | 356 ± 47 | 19 ± 5 | 8 ± 2 | −0.005 ± 0.005 | −0.001 ± 0.005 |
| P7 | 0.10 ± 0.01 | 0.04 ± 0.01 | 5.37 ± 0.69 | 8.06 ± 2.56 | 9.08 ± 0.66 | 8.92 ± 0.59 | 418 ± 60 | 448 ± 80 | 24 ± 3 | 9 ± 2 | 0.010 ± 0.005 | 0.010 ± 0.006 |
| P8 | 0.10 ± 0.01 | 0.07 ± 0.00 | 4.72 ± 0.50 | 4.11 ± 0.51 | 7.35 ± 0.44 | 7.43 ± 0.42 | 659 ± 101 | 646 ± 139 | 34 ± 5 | 22 ± 3 | 0.018 ± 0.028 | 0.022 ± 0.027 |
| P9 | 0.12 ± 0.01 | 0.06 ± 0.01 | 4.88 ± 0.70 | 5.11 ± 0.98 | 7.41 ± 0.23 | 7.35 ± 0.26 | 458 ± 68 | 479 ± 65 | 26 ± 3 | 13 ± 3 | −0.004 ± 0.004 | −0.0004 ± 0.005 |
| All Philips | 0.11 ± 0.01 | 0.04 ± 0.01 | 5.28 ± 1.04 | 8.96 ± 5.56 | 7.73 ± 0.67 | 7.72 ± 0.62 | 449 ± 110 | 457 ± 114 | 25 ± 6 | 11 ± 5 | 0.0003 ± 0.013 | 0.002 ± 0.013 |
| S1 | 0.11 ± 0.00 | 0.04 ± 0.01 | 5.63 ± 1.00 | 5.39 ± 0.77 | 8.65 ± 1.33 | 8.47 ± 1.56 | 556 ± 103 | 562 ± 58 | 27 ± 4 | 14 ± 3 | −0.022 ± 0.011 | −0.031 ± 0.012 |
| S2 | 0.11 ± 0.01 | 0.03 ± 0.00 | 4.73 ± 0.46 | 7.17 ± 1.06 | 8.79 ± 0.43 | 8.89 ± 0.38 | 480 ± 99 | 473 ± 55 | 25 ± 4 | 10 ± 1 | 0.007 ± 0.017 | 0.010 ± 0.011 |
| S3 | 0.10 ± 0.01 | 0.04 ± 0.01 | 6.79 ± 0.82 | 8.81 ± 3.05 | 7.73 ± 0.44 | 7.74 ± 0.46 | 379 ± 65 | 377 ± 97 | 16 ± 4 | 11 ± 3 | 0.007 ± 0.015 | 0.004 ± 0.023 |
| S4 | 0.12 ± 0.01 | 0.04 ± 0.01 | 5.63 ± 0.50 | 7.47 ± 1.26 | 7.63 ± 0.24 | 7.49 ± 0.22 | 565 ± 104 | 489 ± 80 | 25 ± 4 | 12 ± 3 | −0.015 ± 0.007 | −0.024 ± 0.009 |
| S5 | 0.12 ± 0.01 | 0.04 ± 0.01 | 5.98 ± 0.76 | 6.64 ± 1.35 | 8.43 ± 1.14 | 8.40 ± 1.27 | 373 ± 56 | 347 ± 40 | 18 ± 1 | 10 ± 1 | −0.003 ± 0.013 | −0.009 ± 0.017 |
| S6 | 0.11 ± 0.00 | 0.04 ± 0.00 | 5.22 ± 0.73 | 5.00 ± 1.19 | 7.94 ± 0.47 | 8.06 ± 0.61 | 585 ± 90 | 568 ± 166 | 28 ± 3 | 15 ± 2 | −0.009 ± 0.012 | −0.011 ± 0.015 |
| S7 | 0.13 ± 0.01 | 0.04 ± 0.01 | 5.08 ± 0.76 | 5.92 ± 1.97 | 7.97 ± 0.49 | 7.97 ± 0.48 | 689 ± 151 | 653 ± 108 | 47 ± 10 | 25 ± 9 | −0.004 ± 0.013 | −0.016 ± 0.021 |
| All Siemens | 0.12 ± 0.01 | 0.04 ± 0.01 | 5.64 ± 0.94 | 6.61 ± 2.11 | 8.10 ± 0.83 | 8.07 ± 0.88 | 522 ± 148 | 495 ± 146 | 27 ± 11 | 14 ± 7 | −0.006 ± 0.015 | −0.012 ± 0.020 |
| Overall | 0.116 ± 0.014 | 0.043 ± 0.012 | 5.70 ± 1.45 | 8.43 ± 4.33 | 8.10 ± 0.83 | 8.07 ± 0.83 | 447 ± 128 | 439 ± 128 | 25 ± 8 | 11 ± 5 | −0.006 ± 0.018 | −0.005 ± 0.018 |
DE, data excluded; DNA, data not acquired; MM-s, MM-suppressed
Fig. 4 shows the distribution of data quality metrics, by site and by vendor, with numerical values also included in Table 3. Mean vendor fit error ranged from 5–6% for GABA+ editing and 7–9% for MM-suppressed GABA editing (Fig. 4A). NAA linewidth was within acceptable ranges for 3 T MRS, and approximately equal between the two edited acquisitions (overall: 8.10 Hz [GABA+] vs. 8.07 Hz [MM-suppressed GABA]) (Fig. 4B). The Philips data, however, showed lower linewidths on average over both acquisitions (7.73 Hz) compared to the GE (8.56 Hz; pairwise comparison: p < 0.001) and Siemens (8.09 Hz; pairwise comparison: p < 0.01) data. NAA SNR estimates were also consistent across acquisition type (overall: 447 [GABA+] vs. 439 [MM-suppressed GABA]), though some sites’ data exhibited relatively higher SNR values (Fig. 4C). This was most likely driven by differences in RF coil hardware. GABA SNR estimates were mostly consistent within acquisition type (Fig. 4D), with site-to-site variability tending to match the site-to-site variability in NAA SNR estimates. Average frequency offset varied to a degree across sites, with all Philips sites except P8 having relatively low offset due to the employment of frequency correction during data acquisition (Fig. 4E). As can be seen in Figs. 5A and S2A, the pattern of center frequency offset during acquisition was dominated by random effects and linear drift. In the case of Philips sites, there were additional regular corrections due to real-time center frequency updates. Occasional step-changes or spikes were observed due to participant motion, but these were relatively minor features. The median within-participant standard deviation of estimated phase offsets (averaged across acquisition type) was 2.74 degrees (GE), 1.09 degrees (Philips) and 5.93 degrees (Siemens).
Fig. 4.
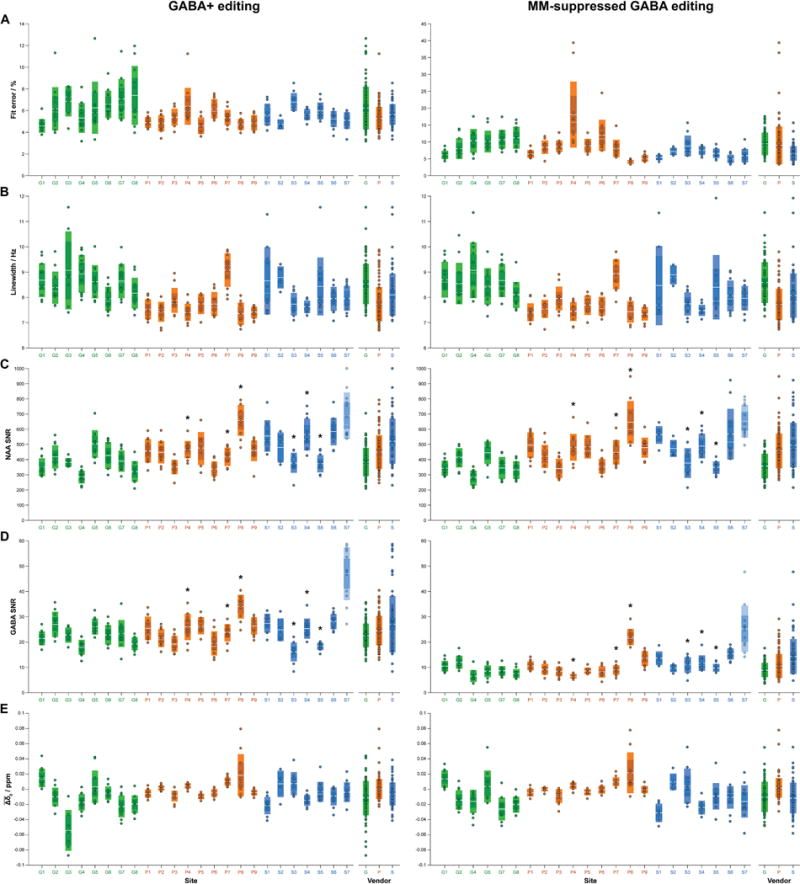
Quantitative quality metrics for the GABA+ (left column) and MM-suppressed GABA (right column) data, displayed by site and by vendor. Metrics are: (A) fit error; (B) NAA linewidth; (C) NAA SNR; (D) GABA SNR; and (E) average frequency offset . The boxes shaded with lighter colors represent ±1 SD and the darker boxes represent the 95% CI. The solid white lines denote the mean, while the dashed white lines denote the median. Sites are colored by vendor (GE sites in green, Philips sites in orange, Siemens sites blue). The asterisks in C and D denote sites with “unusual” transmit/receive RF hardware for the given vendor: sites P4, P7 and P8 had fully digital broadband RF hardware; sites S3, S4 and S5 used 20–64- and 12-channel head coils, respectively. Note that site S7’s NAA and GABA SNR estimates in C and D are transparent to highlight that the estimation of noise signal in these data was unreliable. For the Siemens data, the noise in the up- and downfield frequency ends of the spectrum was attenuated. Since site S7 acquired data with a spectral width shorter than the other Siemens sites (−3.5–13 ppm), the attenuated noise led to upward-biased SNR values.
Fig. 5.
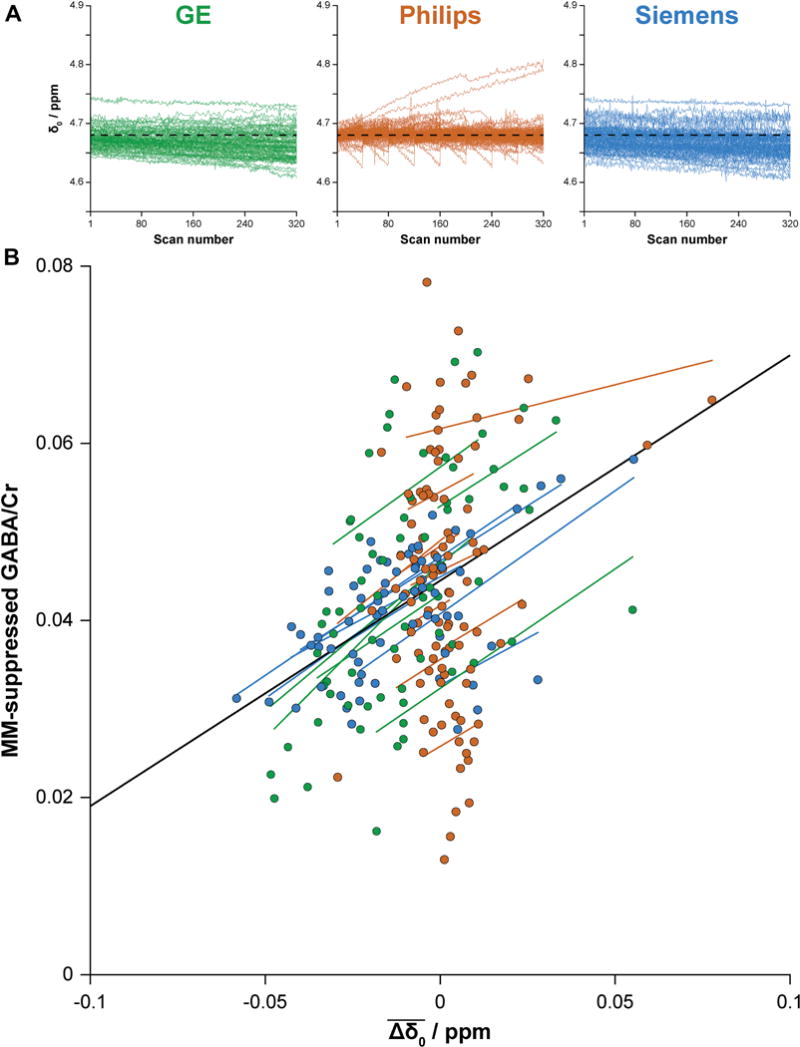
(A) B0 field changes during the MM-suppressed GABA editing experiment. The observed frequency of the residual water signal in each subspectrum is plotted against the scan number over the course of the acquisition (320 averages, ~10 min). Data from all participants are overlaid (separated by vendor). The dashed black lines represent the nominal water frequency (4.68 ppm). (B) Scatterplot illustrating the relationship between average frequency offset and MM-suppressed GABA/Cr as determined by the linear mixed-effects model. Individual measurements are color-coded by vendor (GE in green, Philips in orange, Siemens in blue). The black regression line shows the relationship between and MM-suppressed GABA/Cr over the entire dataset. Additional color-coded regression lines are shown for each site.
3.1 Multilevel analyses
Summaries of the linear mixed-effects models for the GABA+ and MM-suppressed GABA data are given in Tables S1 and S2. The initial unconditional multilevel analysis revealed significant effects of vendor [χ2(1) = 2.95, pboot = 0.02] and site [χ2(1) = 27.93, pboot < 0.001] on GABA+/Cr measurements. For the MM-suppressed GABA data, site effects were significant [χ2(1) = 111.49, pboot = 0.001] but vendor effects were not [χ2(1) < 0.1, pboot = 0.60]. The nonsignificant effect of vendor can be better understood by noticing that there was a strong overlap of the vendor-level distributions of MM-suppressed GABA/Cr as shown in Fig. 3B. Consequently, the vendor-level random effect was removed from subsequent models with the MM-suppressed data to simplify model fitting. The variance partition coefficients (VPCs) for the unconditional model of the GABA+ dataset showed that out of the total variance, 8.2% was attributed to vendor-level differences, 19.7% was attributed to site-level differences and 72.1% was attributed to participant-level differences. In the MM-suppressed GABA data, 50.4% of the total variance was attributed to site-level differences and 49.6% was attributed to participant-level differences.
Results of the secondary multilevel analyses showed no significant effects of linewidth or NAA SNR on GABA+/Cr [χ2(5) = 3.30, pboot = 0.31 and χ2(5) = 0.25, pboot = 0.95, respectively] or on MM-suppressed GABA/Cr [χ2(3) = 0.08, pboot = 0.98 and χ2(3) = 5.32, pboot = 0.10, respectively]. Average frequency offset was, however, significantly associated with both GABA+/Cr [χ2(5) = 11.72, pboot = 0.005] and MM-suppressed GABA/Cr [χ2(3) = 44.31, pboot <0.001] measurements. Of the variance remaining after accounting for site and vendor effects, accounted for 4.0% of variance in the GABA+ data and 21.0% of variance in the MM-suppressed GABA data. The association between and MM-suppressed GABA/Cr is shown in Fig. 5. By-site regression lines are consistent across sites and vendor, indicating a robust relationship. The same plot for GABA+/Cr is shown in Fig. S2.
Finally, the effects of age and sex on GABA measurement outcome were examined, after adjusting for , but no significant effects on either GABA+/Cr [age: χ2(7) = 3.52, pboot = 0.31; sex: χ2(7) = 0.37, pboot = 0.95] or MM-suppressed GABA/Cr [age: χ2(4) = 3.21, pboot = 0.33; sex: χ2(4) = 3.87, pboot = 0.24] were observed.
3.2 Correlational analysis
A correlational analysis of the residuals of the linear mixed-models including as a predictor showed that GABA+/Cr and MM-suppressed GABA/Cr were significantly correlated (r = 0.25, 95% CI: [0.15, 0.35], p < 0.001) (Fig. 6). Specifically, the shared variance between the two measurements, after adjusting for site, vendor and frequency offset effects, amounted to 6.3%.
Fig. 6.
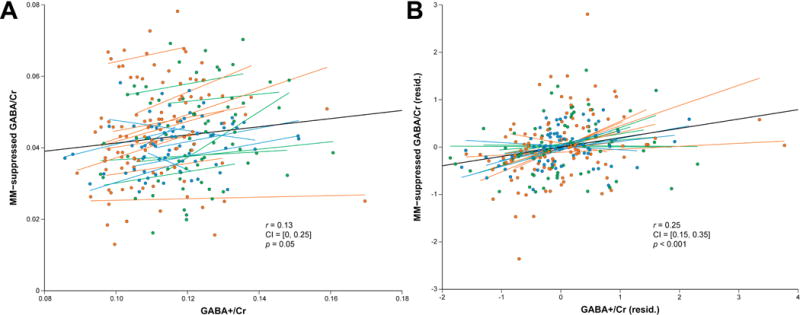
Scatterplots illustrating the relationship between GABA+/Cr and MM-suppressed GABA/Cr using (A) raw values and (B) residuals of the linear mixed-effects models after adjusting for systematic effects of vendor, site and . The Pearson correlation coefficients and p-values are shown, as are the 95% CIs of the coefficients.
4. Discussion
This is the largest multi-site study to date applying GABA-edited MRS in the human brain. The aims at the outset were to establish the extent to which GABA-edited measurements are influenced by site-, sequence- and vendor-specific differences, and to investigate sources of observed variance. Overall, the major findings can be summarized as follows:
The agreement between GABA+ values was surprisingly good, with whole-dataset CV (12%) not much higher than the mean within-site CV (10%), although site and vendor both contributed significantly to total variance.
Agreement between MM-suppressed GABA values was less good than GABA+, with much higher whole-dataset (28%) and mean within-site (19%) CVs. The amount of absolute variance in the MM-suppressed GABA data was, however, similar to the GABA+ data.
Average center frequency offset was a significant factor in both experiments, explaining a greater percentage of variance in the MM-suppressed experiment (21%) than in the GABA+ experiment (4%) after accounting for variance attributed to site and vendor effects.
The level of agreement between GABA+ measurement outcomes was better than anticipated. The whole-dataset CV reported in this study falls well within the range of inter-individual CVs observed for edited GABA+ measurements in the literature: 6–24% (Bogner et al., 2010; Evans et al., 2010; Geramita et al., 2011; Long et al., 2015; Mikkelsen et al., 2016a; O’Gorman et al., 2011). That a majority of the total variance in the data was participant-level variance indicates that initial steps taken to standardize acquisition parameters across vendors (most notably TR, TE and editing pulse bandwidth) were largely successful. The dominant proportion of variance attributed to within-site (i.e., between-participant) variability may in large part reflect a greater level of experience with the GABA+-edited acquisition across all platforms and greater success in standardizing the acquisitions (as well as a greater inherent robustness of this sequence to minor differences such as B0 field offsets).
The protocols used in this study may be considered as a standard, with the currently published data serving as a benchmark for sites applying GABA-edited MRS. Although the majority of sites within-vendor used the same pulse sequence, there were differences. One GE site (G1) used a different MEGA-PRESS implementation to the others, and had the lowest average GABA+ and highest average MM-suppressed GABA values. One Philips site did not use prospective frequency correction (P8), and gave the lowest average GABA+ values and highest average MM-suppressed GABA values. Two Siemens sites had locally modified sequences (compared to the rest), and one of these (S7) had the highest average GABA+ values. Thus, even small differences in sequence implementation seem to be enough to differentiate sites from the group. Further efforts to standardize sequence timings and editing pulse shapes within and between vendors would be expected to reduce vendor- and site-level variance. At this stage, both GE and Siemens have vendor-distributed research sequences in place, using proprietary RF pulse shapes, so this further standardization is a challenge to be taken up by the edited MRS community.
At this stage, it is clear that the MM suppression methodology is less consistent than the GABA+ method, with higher rates of data rejection (19 MM-suppressed GABA datasets vs. seven GABA+ datasets) and greater relative variance. One major contributor of variance that has been identified is frequency offset, with the data reproducing the approximately linear relationship observed by Edden et al. (2016). The ratio between MM-suppressed GABA and GABA+ measurements (0.38) is lower than expected. Typically, it is assumed that ~50% of the GABA+ signal is GABA (Harris et al., 2015a; Mikkelsen et al., 2016a; Shungu et al., 2016). This is largely explained by differential T2 relaxation between GABA signal at TE = 68 ms and TE = 80 ms (13% edited signal loss based on a T2 of 88 ms (Edden et al., 2012b)) and artificially reduced “MM-suppressed GABA” values due to negative MM co-editing (~5% edited signal loss due to mean of −0.005 ppm (see Edden et al., 2016)). The fraction of GABA+ signal that is MM will depend on the bandwidth of the editing pulse used, as will GABA signal losses in the MM-suppressed experiment. While differences in TE between vendors in the MM-suppressed acquisition added a level of methodological heterogeneity, the multilevel analysis did not consider vendor-level effects in the MM-suppressed data to be of statistical importance, in line with previous findings of a minimal effect of TE on the edited GABA signal between 68 and 80 ms (Edden et al., 2012c; Mikkelsen et al., 2016a). These data provide further evidence to support the recommendation of prospective frequency correction for MM-suppressed GABA-edited acquisitions (Edden et al., 2016). For most applications, it is more important that MM suppression removes MM-related variance, rather than MM signal per se. The greater variance in the MM-suppressed GABA results may also explain the weak correlation between GABA+/Cr and MM-suppressed GABA/Cr to some degree (although the statistical modeling approach used, which removes, e.g., site-level variance in the measures, is relatively conservative and will remove some real biologically driven variance).
One important strength of this dataset, in support of edited MRS of GABA, is the fact that, even over so large a dataset as this, there was no significant relationship between GABA measurements and independent metrics of data quality, such as NAA SNR and linewidth. Thus, even though the data quality metrics did vary site-to-site to some degree, tolerable levels (in the sense of not impacting GABA measurements) were achieved at all sites. However, it is acknowledged that these data were homogeneously acquired from a large voxel in a brain region where relatively favorable linewidth and SNR can be achieved. In contrast, associations between metabolite measurements, or their uncertainty, and SNR and/or linewidth are widely observed in investigations of linear combination modeling of unedited spectra (Bartha et al., 2007; Kanowski et al., 2004; Near et al., 2013a). With spectral editing, the goal is to attain an unambiguously resolved signal that allows for simple peak fitting and integration (Bogner et al., 2016; Harris et al., 2017), but with (short-TE) unedited spectra quantification is based on linear-combination fitting, the outcome of which depends on the degree of orthogonality of the basis-set, which itself depends on data quality (Graveron-Demilly, 2014). Although edited MRS of lower-concentration metabolites typically necessitates comparatively longer scan durations or larger voxels to achieve reasonable SNR, the advent of multiplexed editing (Chan et al., 2016, 2017a, 2017b; Oeltzschner et al., 2017; Saleh et al., 2016) and development of edited MRSI (Bogner et al., 2014; Hnilicová et al., 2016; Zhu et al., 2011) continues to improve the efficiency of spectral editing approaches.
A number of multi-site MRS studies have been conducted in the past, each with a specific focus. These focuses have included: unedited, short-TE MRS (Deelchand et al., 2015); low-field MRS (Träber et al., 2006); ultra-high field MRS (van de Bank, 2015); absolute quantification (Bovée et al., 1998; De Beer et al., 1998; Keevil et al., 1998; Soher et al., 1996); MRSI (Sabati et al., 2015; Wijnen et al., 2010); body MRS (Bolan et al., 2016; Scheenen et al., 2011); brain tumor classification (García-Gómez et al., 2009; Julià-Sapé et al., 2006; Tate et al., 2003; Vicente et al., 2013); and HIV-associated dementia (Chang et al., 2004; Lee et al., 2003; Sacktor et al., 2005). Even for short-TE methods, the degree of agreement between sites and scanners is highly dependent on the degree of acquisition homogeneity.
Edited MRS of GABA has a number of limitations, which are not directly addressed in this paper. The fact that MM-suppressed GABA measurements are so susceptible to B0 field changes resulting from scanner drift and participant head motion means that GABA+ is still the most widely used edited GABA measure, in spite of the ~50% MM contribution. However, measures of GABA that effectively remove the MM contamination would have clearer biochemical significance than GABA+ measurements, and this paper establishes the importance of future research dedicated to obtaining MM-suppressed GABA measures with less sensitivity to B0 field offsets. The application of MM suppression is strongly motivated by the desire to remove MM-related variance, and further development to improve the robustness of MM suppression remains important. Even without this MM contamination, the interpretation of MRS measures of total GABA concentration is complex – and the extent to which it is an index of GABAergic neurotransmission (beyond simply being a marker of GABAergic interneuron cell density) is the subject of ongoing debate (Myers et al., 2016; Rae, 2014; Stagg et al., 2011). This paper also does not explore the complexities of GABA quantification by tissue water-referencing, a popular alternative to Cr-referencing. Additional aspects of water-referenced quantification (such as site-to-site segmentation differences) will contribute to the variability of water-referenced GABA measurements across vendors, research sites and individuals (e.g., see Gasparovic et al., 2006; Harris et al., 2015b; Mikkelsen et al., 2016b).
In conclusion, an international consortium collected a large dataset of GABA-edited MRS measurements, the first study of this size for in vivo MRS of GABA. These data support the use of GABA-edited MRS for multi-site, multi-vendor studies, with site and vendor contributing a surprisingly small amount of total variance to GABA+ measurements.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 EB016089, R01 EB023963 and P41 EB015909. Data collection was supported by the Shandong Provincial Key Research and Development Plan of China (2016ZDJS07A16) and the National Natural Science Foundation of China for Young Scholars (no. 81601479). IDW thanks Mrs. J. Bigley of the University of Sheffield MRI Unit for her assistance with data acquisition. JJP was supported by NIAAA grant K23 AA020842. MPS was supported by NIH grant F32 EY025121. NAJP receives salary support from NIH grant K99 MH107719. The authors acknowledge implementation contributions from a number of employees of Siemens Medical Solutions, including Dr. Keith Heberlein and Dr. Sinyeob Ahn, to the Siemens WIP sequences, which are shared with several research sites under sequence-specific agreements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barr DJ, Levy R, Scheepers C, Tily HJ. Random effects structure for confirmatory hypothesis testing: Keep it maximal. J Mem Lang. 2013;68:255–278. doi: 10.1016/j.jml.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha R. Effect of signal-to-noise ratio and spectral linewidth on metabolite quantification at 4 T. NMR Biomed. 2007;20:512–521. doi: 10.1002/nbm.1122. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67 doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Bhagwagar Z, Wylezinska M, Jezzard P, Evans J, Boorman E, M Matthews P, J Cowen P. Low GABA concentrations in occipital cortex and anterior cingulate cortex in medication-free, recovered depressed patients. Int J Neuropsychopharmacol. 2008;11:255–260. doi: 10.1017/S1461145707007924. [DOI] [PubMed] [Google Scholar]
- Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJW, Strasser B, Marjańska M, Trattnig S, Grant E, Rosen B, Andronesi OC. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. doi: 10.1016/j.neuroimage.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner W, Gruber S, Doelken M, Stadlbauer A, Ganslandt O, Boettcher U, Trattnig S, Doerfler A, Stefan H, Hammen T. In vivo quantification of intracerebral GABA by single-voxel 1H-MRS—How reproducible are the results? Eur J Radiol. 2010;73:526–531. doi: 10.1016/j.ejrad.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Bogner W, Hangel G, Esmaeili M, Andronesi OC. 1D-spectral editing and 2D multispectral in vivo 1H-MRS and 1H-MRSI - Methods and applications. Anal Biochem. 2016 doi: 10.1016/j.ab.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Bolan PJ, Kim E, Herman BA, Newstead GM, Rosen MA, Schnall MD, Pisano ED, Weatherall PT, Morris EA, Lehman CD, Garwood M, Nelson MT, Yee D, Polin SM, Esserman LJ, Gatsonis CA, Metzger GJ, Newitt DC, Partridge SC, Hylton NM. MR spectroscopy of breast cancer for assessing early treatment response: Results from the ACRIN 6657 MRS trial. J Magn Reson Imaging. 2016 doi: 10.1002/jmri.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann S, Ghisleni C, Poil S, Martin E, Ball J, Eich-Höchli D, Edden RAE, Klaver P, Michels L, Brandeis D, O’Gorman RL. Developmental changes in gamma-aminobutyric acid levels in attention-deficit/hyperactivity disorder. Transl Psychiatry. 2015;5:e589. doi: 10.1038/tp.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- Bovée W, Canese R, Decorps M, Forssell-Aronsson E, Le Fur Y, Howe F, Karlsen O, Knijn A, Kontaxis G, Kg̋el H, McLean M, Podo F, Slotboom J, Vikhoff B, Ziegler A. Absolute metabolite quantification by in vivo NMR spectroscopy: IV. Multicentre trial on MRSI localisation tests. Magn Reson Imaging. 1998;16:1113–1125. doi: 10.1016/S0730-725X(98)00120-9. [DOI] [PubMed] [Google Scholar]
- Boy F, Evans CJ, Edden RAE, Lawrence AD, Singh KD, Husain M, Sumner P. Dorsolateral prefrontal γ-aminobutyric acid in men predicts individual differences in rash impulsivity. Biol Psychiatry. 2011;70:866–872. doi: 10.1016/j.biopsych.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix MK, Ersland L, Hugdahl K, Dwyer GE, Grüner R, Noeske R, Beyer MK, Craven AR. Within- and between-session reproducibility of GABA measurements with MR spectroscopy. J Magn Reson Imaging. 2017 doi: 10.1002/jmri.25588. [DOI] [PubMed] [Google Scholar]
- Chan KL, Oeltzschner G, Schär M, Barker PB, Edden RAE. Spatial Hadamard encoding of J-edited spectroscopy using slice-selective editing pulses. NMR Biomed. 2017a;30:e3688. doi: 10.1002/nbm.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Puts NAJ, Schär M, Barker PB, Edden RAE. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016;76:11–19. doi: 10.1002/mrm.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Saleh MG, Oeltzschner G, Barker PB, Edden RAE. Simultaneous measurement of Aspartate, NAA, and NAAG using HERMES spectral editing at 3 Tesla. Neuroimage. 2017b doi: 10.1016/j.neuroimage.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Lee PL, Yiannoutsos CT, Ernst T, Marra CM, Richards T, Kolson D, Schifitto G, Jarvik JG, Miller EN, Lenkinski R, Gonzalez G, Navia BA. A multicenter in vivo proton-MRS study of HIV-associated dementia and its relationship to age. Neuroimage. 2004;23:1336–1347. doi: 10.1016/j.neuroimage.2004.07.067. [DOI] [PubMed] [Google Scholar]
- De Beer R, Barbiroli B, Gobbi G, Knijn A, Kg̋el H, Langenberger K, Tkac I, Topp S. Absolute metabolite quantification by in vivo NMR spectroscopy: III. Multicentre 1H MRS of the human brain addressed by one and the same data-analysis protocol. Magn Reson Imaging. 1998;16:1107–1111. doi: 10.1016/S0730-725X(98)00119-2. [DOI] [PubMed] [Google Scholar]
- Deelchand DK, Adanyeguh IM, Emir UE, Nguyen TM, Valabregue R, Henry PG, Mochel F, Öz G. Two-site reproducibility of cerebellar and brainstem neurochemical profiles with short-echo, single-voxel MRS at 3T. Magn Reson Med. 2015;73:1718–1725. doi: 10.1002/mrm.25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCiccio TJ, Efron B. Bootstrap confidence intervals. Stat Sci. 1996;11:189–228. doi: 10.1214/ss/1032280214. [DOI] [Google Scholar]
- Donahue MJ, Near J, Blicher JU, Jezzard P. Baseline GABA concentration and fMRI response. Neuroimage. 2010;53:392–398. doi: 10.1016/j.neuroimage.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Drenthen GS, Barendse EM, Aldenkamp AP, van Veenendaal TM, Puts NAJ, Edden RAE, Zinger S, Thoonen G, Hendriks MPH, Kessels RPC, Jansen JFA. Altered neurotransmitter metabolism in adolescents with high-functioning autism. Psychiatry Res Neuroimaging. 2016;256:44–49. doi: 10.1016/j.pscychresns.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Barker PB. Spatial effects in the detection of γ-aminobutyric acid: Improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58:1276–1282. doi: 10.1002/mrm.21383. [DOI] [PubMed] [Google Scholar]
- Edden RAE, Crocetti D, Zhu H, Gilbert DL, Mostofsky SH. Reduced GABA concentration in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2012a;69:750–753. doi: 10.1001/archgenpsychiatry.2011.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T2 in vivo with J-difference editing: Application to GABA at 3 Tesla. J Magn Reson Imaging. 2012b;35:229–234. doi: 10.1002/jmri.22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Oeltzschner G, Harris AD, Puts NAJ, Chan KL, Boer VO, Schär M, Barker PB. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J Magn Reson Imaging. 2016;44:1474–1482. doi: 10.1002/jmri.25304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Puts NAJ, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012c;68:657–661. doi: 10.1002/mrm.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40:1445–1452. doi: 10.1002/jmri.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emir UE, Tuite PJ, Öz G. Elevated pontine and putamenal GABA levels in mild-moderate Parkinson disease detected by 7 Tesla proton MRS. PLoS One. 2012;7:e30918. doi: 10.1371/journal.pone.0030918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, McGonigle DJ, Edden RAE. Diurnal stability of γ-aminobutyric acid concentration in visual and sensorimotor cortex. J Magn Reson Imaging. 2010;31:204–209. doi: 10.1002/jmri.21996. [DOI] [PubMed] [Google Scholar]
- Foerster BR, Callaghan BC, Petrou M, Edden RAE, Chenevert TL, Feldman EL. Decreased motor cortex γ-aminobutyric acid in amyotrophic lateral sclerosis. Neurology. 2012;78:1596–1600. doi: 10.1212/WNL.0b013e3182563b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster BR, Pomper MG, Callaghan BC, Petrou M, Edden RAE, Mohamed MA, Welsh RC, Carlos RC, Barker PB, Feldman EL. An imbalance between excitatory and inhibitory neurotransmitters in amyotrophic lateral sclerosis revealed by use of 3-T proton magnetic resonance spectroscopy. JAMA Neurol. 2013;70:1009–1016. doi: 10.1001/jamaneurol.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm J, Merboldt KD, Hänicke W. Localized proton spectroscopy using stimulated echoes. J Magn Reson. 1987;72:502–508. doi: 10.1016/0022-2364(87)90154-5. [DOI] [PubMed] [Google Scholar]
- Fujihara K, Narita K, Suzuki Y, Takei Y, Suda M, Tagawa M, Ujita K, Sakai Y, Narumoto J, Near J, Fukuda M. Relationship of γ-aminobutyric acid and glutamate+glutamine concentrations in the perigenual anterior cingulate cortex with performance of Cambridge Gambling Task. Neuroimage. 2015;109:102–108. doi: 10.1016/j.neuroimage.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Gaetz W, Bloy L, Wang DJ, Port RG, Blaskey L, Levy SE, Roberts TPL. GABA estimation in the brains of children on the autism spectrum: Measurement precision and regional cortical variation. Neuroimage. 2014;86:1–9. doi: 10.1016/j.neuroimage.2013.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez JM, Luts J, Julià-Sapé M, Krooshof P, Tortajada S, Robledo JV, Melssen W, Fuster-García E, Olier I, Postma G, Monleón D, Moreno-Torres À, Pujol J, Candiota AP, Martínez-Bisbal MC, Suykens J, Buydens L, Celda B, Van Huffel S, Arús C, Robles M. Multiproject–multicenter evaluation of automatic brain tumor classification by magnetic resonance spectroscopy. Magn Reson Mater Physics, Biol Med. 2009;22:5–18. doi: 10.1007/s10334-008-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, Jung RE, Morrison LA. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn Reson Med. 2006;55:1219–1226. doi: 10.1002/mrm.20901. [DOI] [PubMed] [Google Scholar]
- Geramita M, van der Veen JW, Barnett AS, Savostyanova AA, Shen J, Weinberger DR, Marenco S. Reproducibility of prefrontal γ-aminobutyric acid measurements with J-edited spectroscopy. NMR Biomed. 2011;24:1089–1098. doi: 10.1002/nbm.1662. [DOI] [PubMed] [Google Scholar]
- Goldstein H, Browne W, Rasbash J. Partitioning variation in multilevel models. Underst Stat. 2002;1:223–231. doi: 10.1207/S15328031US0104_02. [DOI] [Google Scholar]
- Graveron-Demilly D. Quantification in magnetic resonance spectroscopy based on semi-parametric approaches. Magn Reson Mater Physics, Biol Med. 2014;27:113–130. doi: 10.1007/s10334-013-0393-4. [DOI] [PubMed] [Google Scholar]
- Greenhouse I, King M, Noah S, Maddock RJ, Ivry RB. individual differences in resting corticospinal excitability are correlated with reaction time and GABA content in motor cortex. J Neurosci. 2017;37:2686–2696. doi: 10.1523/JNEUROSCI.3129-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halekoh U, Højsgaard S. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models - The R package pbkrtest. J Stat Softw. 2014;59:1–32. doi: 10.18637/jss.v059.i09. [DOI] [Google Scholar]
- Harris AD, Glaubitz B, Near J, John Evans C, Puts NAJ, Schmidt-Wilcke T, Tegenthoff M, Barker PB, Edden RAE. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72:941–8. doi: 10.1002/mrm.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Puts NAJ, Barker PB, Edden RAE. Spectral-editing measurements of GABA in the human brain with and without macromolecule suppression. Magn Reson Med. 2015a;74:1523–1529. doi: 10.1002/mrm.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Puts NAJ, Edden RAE. Tissue correction for GABA-edited MRS: Considerations of voxel composition, tissue segmentation, and tissue relaxations. J Magn Reson Imaging. 2015b;42:1431–1440. doi: 10.1002/jmri.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AD, Saleh MG, Edden RAE. Edited 1H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn Reson Med. 2017;77:1377–1389. doi: 10.1002/mrm.26619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler G, van der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced prefrontal glutamate/glutamine and γ-aminobutyric acid levels in major depression determined using proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2007;64:193. doi: 10.1001/archpsyc.64.2.193. [DOI] [PubMed] [Google Scholar]
- Hayes AF. A primer on multilevel modeling. Hum Commun Res. 2006;32:385–410. doi: 10.1111/j.1468-2958.2006.00281.x. [DOI] [Google Scholar]
- Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45:517–520. doi: 10.1002/1522-2594(200103)45:3<517::AID-MRM1068>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hnilicová P, Považan M, Strasser B, Andronesi OC, Gajdošík M, Dydak U, Ukropec J, Dobrota D, Trattnig S, Bogner W. Spatial variability and reproducibility of GABA-edited MEGA-LASER 3D-MRSI in the brain at 3 T. NMR Biomed. 2016;29:1656–1665. doi: 10.1002/nbm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen X, Gu H, Yang Y. Resting-state glutamate and GABA concentrations predict task-induced deactivation in the default mode network. J Neurosci. 2013;33:18566–18573. doi: 10.1523/JNEUROSCI.1973-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julià-Sapé M, Acosta D, Mier M, Arùs C, Watson D. A multi-centre, web-accessible and quality control-checked database of in vivo MR spectra of brain tumour patients. Magn Reson Mater Physics, Biol Med. 2006;19:22–33. doi: 10.1007/s10334-005-0023-x. [DOI] [PubMed] [Google Scholar]
- Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4T. NMR Biomed. 2008;21:22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- Kanowski M, Kaufmann J, Braun J, Bernarding J, Tempelmann C. Quantitation of simulated short echo time 1H human brain spectra by LCModel and AMARES. Magn Reson Med. 2004;51:904–912. doi: 10.1002/mrm.20063. [DOI] [PubMed] [Google Scholar]
- Kapogiannis D, Reiter DA, Willette AA, Mattson MP. Posteromedial cortex glutamate and GABA predict intrinsic functional connectivity of the default mode network. Neuroimage. 2013;64:112–119. doi: 10.1016/j.neuroimage.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keevil S, Barbiroli B, Brooks JC, Cady E, Canese R, Carlier P, Collins D, Gilligan P, Gobbi G, Hennig J, Kügel H, Leach M, Metzler D, Mlynárik V, Moser E, Newbold M, Payne G, Ring P, Roberts J, Rowland I, Thiel T, Tkác I, Topp S, Wittsack H, Wylezinska M, Zaniol P, Henriksen O, Podo F. Absolute metabolite quantification by in vivo NMR spectroscopy: II. A multicentre trial of protocols for in vivo localised proton studies of human brain Magn Reson Imaging. 1998;16:1093–1106. doi: 10.1016/S0730-725X(98)00118-0. [DOI] [PubMed] [Google Scholar]
- Kegeles LS, Mao X, Stanford AD, Girgis R, Ojeil N, Xu X, Gil R, Slifstein M, Abi-Dargham A, Lisanby SH, Shungu DC. Elevated prefrontal cortex γ-aminobutyric acid and glutamate-glutamine levels in schizophrenia measured in vivo with proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2012;69:449–59. doi: 10.1001/archgenpsychiatry.2011.1519. [DOI] [PubMed] [Google Scholar]
- Lange T, Ko CW, Lai PH, Dacko M, Tsai SY, Buechert M. Simultaneous detection of valine and lactate using MEGA-PRESS editing in pyogenic brain abscess. NMR Biomed. 2016;29:1739–1747. doi: 10.1002/nbm.3660. [DOI] [PubMed] [Google Scholar]
- Lee PL, Yiannoutsos CT, Ernst T, Chang L, Marra CM, Jarvik JG, Richards TL, Kwok EW, Kolson DL, Simpson D, Tang CY, Schifitto G, Ketonen LM, Meyerhoff DJ, Lenkinski RE, Gonzalez RG, Navia BA. A multi-center 1H MRS study of the AIDS dementia complex: Validation and preliminary analysis. J Magn Reson Imaging. 2003;17:625–633. doi: 10.1002/jmri.10295. [DOI] [PubMed] [Google Scholar]
- Long Z, Dyke JP, Ma R, Huang CC, Louis ED, Dydak U. Reproducibility and effect of tissue composition on cerebellar γ-aminobutyric acid (GABA) MRS in an elderly population. NMR Biomed. 2015;28:1315–1323. doi: 10.1002/nbm.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick DA. GABA as an inhibitory neurotransmitter in human cerebral cortex. J Neurophysiol. 1989;62:1018–1027. doi: 10.1152/jn.1989.62.5.1018. [DOI] [PubMed] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(SICI)1099-1492(199810)11:6<266::AID-NBM530>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M, Singh KD, Sumner P, Evans CJ. Comparison of the repeatability of GABA-edited magnetic resonance spectroscopy with and without macromolecule suppression. Magn Reson Med. 2016a;75:946–953. doi: 10.1002/mrm.25699. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M, Singh KD, Brealy JA, Linden DEJ, Evans CJ. Quantification of γ-aminobutyric acid (GABA) in 1H MRS volumes composed heterogeneously of grey and white matter. NMR Biomed. 2016b;29:1644–1655. doi: 10.1002/nbm.3622. [DOI] [PubMed] [Google Scholar]
- Mullins PG, McGonigle DJ, O’Gorman RL, Puts NAJ, Vidyasagar R, Evans CJ, Cardiff Symposium on MRS of GABA. Edden RAE. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. doi: 10.1016/j.neuroimage.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaraswamy SD, Edden RAE, Jones DK, Swettenham JB, Singh KD. Resting GABA concentration predicts peak gamma frequency and fMRI amplitude in response to visual stimulation in humans. Proc Natl Acad Sci U S A. 2009;106:8356–8361. doi: 10.1073/pnas.0900728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JF, Nutt DJ, Lingford-ughes AR. γ-aminobutyric acid as a metabolite: Interpreting magnetic resonance spectroscopy experiments. J Psychopharmacol. 2016;30:422–427. doi: 10.1177/0269881116639298. [DOI] [PubMed] [Google Scholar]
- Near J, Andersson J, Maron E, Mekle R, Gruetter R, Cowen P, Jezzard P. Unedited in vivo detection and quantification of γ-aminobutyric acid in the occipital cortex using short-TE MRS at 3 T. NMR Biomed. 2013a;26:1353–1362. doi: 10.1002/nbm.2960. [DOI] [PubMed] [Google Scholar]
- Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. 2015;73:44–50. doi: 10.1002/mrm.25094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Evans CJ, Puts NAJ, Barker PB, Edden RAE. J-difference editing of gamma-aminobutyric acid (GABA): Simulated and experimental multiplet patterns. Magn Reson Med. 2013b;70:1183–1191. doi: 10.1002/mrm.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Ho YCL, Sandberg K, Kumaragamage C, Blicher JU. Long-term reproducibility of GABA magnetic resonance spectroscopy. Neuroimage. 2014;99:191–196. doi: 10.1016/j.neuroimage.2014.05.059. [DOI] [PubMed] [Google Scholar]
- O’Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: Reproducibility and gender effects. J Magn Reson Imaging. 2011;33:1262–1267. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G, Puts NAJ, Chan KL, Boer VO, Barker PB, Edden RAE. Dual-volume excitation and parallel reconstruction for J-difference-edited MR spectroscopy. Magn Reson Med. 2017;77:16–22. doi: 10.1002/mrm.26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öngür D, Prescot AP, McCarthy J, Cohen BM, Renshaw PF. Elevated gamma-aminobutyric acid levels in chronic schizophrenia. Biol Psychiatry. 2010;68:667–670. doi: 10.1016/j.biopsych.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrou M, Pop-Busui R, Foerster BR, Edden RA, Callaghan BC, Harte SE, Harris RE, Clauw DJ, Feldman EL. Altered excitation-inhibition balance in the brain of patients with diabetic neuropathy. Acad Radiol. 2012;19:607–612. doi: 10.1016/j.acra.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peugh JL. A practical guide to multilevel modeling. J Sch Psychol. 2010;48:85–112. doi: 10.1016/j.jsp.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Puts NAJ, Edden RAE, Evans CJ, McGlone F, McGonigle DJ. Regionally specific human GABA concentration correlates with tactile discrimination thresholds. J Neurosci. 2011;31:16556–16560. doi: 10.1523/JNEUROSCI.4489-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts NAJ, Wodka EL, Harris AD, Crocetti D, Tommerdahl M, Mostofsky SH, Edden RAE. Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Res. 2016:1–12. doi: 10.1002/aur.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RB, Shungu DC, Mao X, Nestadt P, Kelly C, Collins KA, Murrough JW, Charney DS, Mathew SJ. Amino acid neurotransmitters assessed by proton magnetic resonance spectroscopy: Relationship to treatment resistance in major depressive disorder. Biol Psychiatry. 2009;65:792–800. doi: 10.1016/j.biopsych.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. https://www.R-project.org. [Google Scholar]
- Rae CD. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem Res. 2014;39:1–36. doi: 10.1007/s11064-013-1199-5. [DOI] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of γ-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90:5662–6. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LM, Krause BW, Wijtenburg SA, McMahon RP, Chiappelli J, Nugent KL, Nisonger SJ, Korenic SA, Kochunov P, Hong LE. Medial frontal GABA is lower in older schizophrenia: a MEGA-PRESS with macromolecule suppression study. Mol Psychiatry. 2016;21:198–204. doi: 10.1038/mp.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabati M, Sheriff S, Gu M, Wei J, Zhu H, Barker PB, Spielman DM, Alger JR, Maudsley AA. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med. 2015;74:1209–1220. doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor N, Skolasky RL, Ernst T, Mao X, Selnes O, Pomper MG, Chang L, Zhong K, Shungu DC, Marder K, Shibata D, Schifitto G, Bobo L, Barker PB. A multicenter study of two magnetic resonance spectroscopy techniques in individuals with HIV dementia. J Magn Reson Imaging. 2005;21:325–333. doi: 10.1002/jmri.20272. [DOI] [PubMed] [Google Scholar]
- Saleh MG, Oeltzschner G, Chan KL, Puts NAJ, Mikkelsen M, Schär M, Harris AD, Edden RAE. Simultaneous edited MRS of GABA and glutathione. Neuroimage. 2016;142:576–582. doi: 10.1016/j.neuroimage.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheenen TWJ, Fütterer J, Weiland E, van Hecke P, Lemort M, Zechmann C, Schlemmer HP, Broome D, Villeirs G, Lu J, Barentsz J, Roell S, Heerschap A. Discriminating cancer from noncancer tissue in the prostate by 3-dimensional proton magnetic resonance spectroscopic imaging: A prospective multicenter validation study. Invest Radiol. 2011;46:25–33. doi: 10.1097/RLI.0b013e3181f54081. [DOI] [PubMed] [Google Scholar]
- Schielzeth H. Simple means to improve the interpretability of regression coefficients. Methods Ecol Evol. 2010;1:103–113. doi: 10.1111/j.2041-210X.2010.00012.x. [DOI] [Google Scholar]
- Shibata K, Sasaki Y, Bang JW, Walsh EG, Machizawa MG, Tamaki M, Chang LH, Watanabe T. Overlearning hyperstabilizes a skill by rapidly making neurochemical processing inhibitory-dominant. Nat Neurosci. 2017;20:470–475. doi: 10.1038/nn.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungu DC, Mao X, Gonzales R, Soones TN, Dyke JP, van der Veen JW, Kegeles LS. Brain γ-aminobutyric acid (GABA) detection in vivo with the J-editing 1H MRS technique: a comprehensive methodological evaluation of sensitivity enhancement, macromolecule contamination and test-retest reliability. NMR Biomed. 2016;29:932–942. doi: 10.1002/nbm.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveri MM, Sneider JT, Crowley DJ, Covell MJ, Acharya D, Rosso IM, Jensen JE. rontal lobe γ-aminobutyric acid levels during adolescence: Associations with impulsivity and response inhibition. Biol Psychiatry. 2013;74:296–304. doi: 10.1016/j.biopsych.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders TAB, Bosker RJ. Multilevel Analysis: An Introduction to Basic and Advanced Multilevel Modeling. SAGE Publications; London: 2012. [Google Scholar]
- Soher BJ, Hurd RE, Sailasuta N, Barker PB. Quantitation of automated single-voxel proton MRS using cerebral water as an internal reference. Magn Reson Med. 1996;36:335–339. doi: 10.1002/mrm.1910360302. [DOI] [PubMed] [Google Scholar]
- Stagg CJ, Bachtiar V, Johansen-Berg H. What are we measuring with GABA magnetic resonance spectroscopy? Commun Integr Biol. 2011;4:573–575. doi: 10.4161/cib.16213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING) Magn Reson Med. 1997;38:311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- Tate AR, Majós C, Moreno A, Howe FA, Griffiths JR, Arús C. Automated classification of short echo time in in vivo 1H brain tumor spectra: A multicenter study. Magn Reson Med. 2003;49:29–36. doi: 10.1002/mrm.10315. [DOI] [PubMed] [Google Scholar]
- Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47:1009–1012. doi: 10.1002/mrm.10146. [DOI] [PubMed] [Google Scholar]
- Träber F, Block W, Freymann N, Gür O, Kucinski T, Hammen T, Ende G, Pilatus U, Hampel H, Schild HH, Heun R, Jessen F. A multicenter reproducibility study of single-voxel 1H-MRS of the medial temporal lobe. Eur Radiol. 2006;16:1096–1103. doi: 10.1007/s00330-005-0108-y. [DOI] [PubMed] [Google Scholar]
- van de Bank BL, Emir UE, Boer VO, van Asten JJA, Maas MC, Wijnen JP, Kan HE, Oz G, Klomp DWJ, Scheenen TWJ. Multi-center reproducibility of neurochemical profiles in the human brain at 7T. NMR Biomed. 2015;28:306–316. doi: 10.1002/nbm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J, Fuster-Garcia E, Tortajada S, García-Gómez JM, Davies N, Natarajan K, Wilson M, Grundy RG, Wesseling P, Monleón D, Celda B, Robles M, Peet AC. Accurate classification of childhood brain tumours by in vivo 1H MRS - a multicentre study. Eur J Cancer. 2013;49:658–67. doi: 10.1016/j.ejca.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. 2007;25:1032–1038. doi: 10.1016/j.mri.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen JP, van Asten JJA, Klomp DWJ, Sjobakk TE, Gribbestad IS, Scheenen TWJ, Heerschap A. Short echo time 1H MRSI of the human brain at 3T with adiabatic slice-selective refocusing pulses; reproducibility and variance in a dual center setting. J Magn Reson Imaging. 2010;31:61–70. doi: 10.1002/jmri.21999. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Grandelis A, Maddock RJ. Dorsolateral prefrontal cortex GABA concentration in humans predicts working memory load processing capacity. J Neurosci. 2016;36:11788–11794. doi: 10.1523/JNEUROSCI.1970-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Maddock RJ, Rokem A, Silver MA, Minzenberg MJ, Ragland JD, Carter CS. GABA concentration is reduced in visual cortex in schizophrenia and correlates with orientation-specific surround suppression. J Neurosci. 2010;30:3777–3781. doi: 10.1523/JNEUROSCI.6158-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Edden RAE, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med. 2011;65:603–609. doi: 10.1002/mrm.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


