Abstract
Exemestane (EXE) is an aromatase inhibitor used for the prevention and treatment of breast cancer. The major metabolic pathway for EXE is reduction to form the active 17β-dihydro-EXE (17β-DHE) and subsequent glucuronidation to 17β-hydroxy-EXE-17-O-β-D-glucuronide (17β-DHE-Gluc) by UGT2B17. The aim of the present study was to determine the effects of UGT2B17 copy number variation on the levels of urinary and plasma 17β-DHE-Gluc and 17β-DHE in patients taking EXE. Ninety-six post-menopausal Caucasian breast cancer patients with ER+ breast tumors taking 25 mg EXE daily were recruited into this study. UGT2B17 copy number was determined by a real-time PCR copy number variant assay and the levels of EXE, 17β-DHE and 17β-DHE-Gluc were quantified by UPLC/MS in patients’ urine and plasma. A 39-fold decrease (P<0.0001) in the levels of creatinine-adjusted urinary 17β-DHE-Gluc was observed among UGT2B17 (*2/*2) subjects vs. subjects with the UGT2B17 (*1/*1) genotype. The plasma levels of 17β-DHE-Gluc was decreased 29-fold (P<0.0001) in subjects with the UGT2B17 (*2/*2) genotype vs. subjects with UGT2B17 (*1/*1) genotype. The levels of plasma EXE-adjusted 17β-DHE was 28% higher (P=0.04) in subjects with the UGT2B17 (*2/*2) genotype vs. subjects with the UGT2B17 (*1/*1) genotype. These data indicate that UGT2B17 is the major enzyme responsible for 17β-DHE-Gluc formation in vivo and that the UGT2B17 copy number variant may play a role in inter-individual variability in 17β-DHE levels in vivo.
Introduction
Breast cancer is the most frequently diagnosed cancer (excluding cancers of the skin) and second leading cause of cancer death in women, with a lifetime risk for breast cancer of 12.4% (1). An estimated 246,660 new cases of invasive breast cancer are expected to be diagnosed among women in the USA in 2016 (2). Approximately 75% of all breast cancers express estrogen receptors (ERs)(3). Treatment for early-stage ER+ breast cancer in post-menopausal women has primarily been focused on elimination of growth stimulation caused by estrogens. Aromatase inhibitors (AIs) act to abolish aromatase activity, blocking estrone/estradiol biosynthesis and preventing the growth stimulation that estrogens induce (4–6). Clinical trials have shown that the use of AIs increases disease-free survival and decreases the occurrence of contralateral breast cancer as compared to the use of selective estrogen receptor modulators like tamoxifen alone (7–10). Although AIs represent an improvement in breast cancer treatment, considerable inter-individual variability exists in patient response and in the manifestation of toxicity to these drugs (11–14). The causes of this variability have not been elucidated.
Exemestane (EXE), a steroidal substrate analog, is a 3rd generation AI that acts through a mechanism of suicide inhibition to inhibit the target aromatase enzyme, and is widely used for the adjuvant treatment and prevention of breast cancer in postmenopausal women as well as in the metastatic setting (15–18). After absorption, EXE is distributed extensively into tissues with 90% bound to plasma proteins (19). It is known that EXE is extensively metabolized by reduction of the 17-keto group by aldo-keto reductases and cytochrome P450s to form an active metabolite, 17-dihydroexemestane (17β-DHE), and subsequent addition of glucuronic acid (Gluc) to 17β-DHE to form 17β-DHE-glucuronide (17β-DHE-Gluc) (20–26). EXE is excreted roughly equally in the urine and feces, with the unchanged drug in urine less than 1% of the dose (19).
The UDP glucuronosyltransferase (UGT) superfamily of enzymes catalyze the glucuronidation of various compounds, including endogenous compounds as well as xenobiotics. Previous in vitro study have strongly implicated UGT2B17 as the major enzyme responsible for the glucuronidation of 17β-DHE (24). A polymorphic whole-gene deletion of the UGT2B17 gene has been identified with an allelic prevalence of ~30% in Caucasians (27–30), and this copy number variant (CNV) was associated with decreased formation of 17β-DHE-Gluc in human liver microsomes (HLMs) in vitro (24).
The goal of the present study was to examine the effect of the UGT2B17 CNV on 17β-DHE-Gluc formation and 17β-DHE levels in vivo, by examining the levels of EXE, 17β-DHE, and 17β-DHE-Gluc in 96 postmenopausal women with breast cancer taking EXE.
Materials and Methods
Chemicals and materials
Exemestane (EXE) and creatinine were purchased from Sigma-Aldrich (St Louis, Missouri, USA). 17β-hydroxy (OH)-EXE-17-O-β-D-Glucuronide (17β-DHE-Gluc), 17β-OH-EXE-D3-17-O-β-D-Glucuronide (D3-17β-DHE-Gluc), 17β-OH-EXE-d3 (D3-17β-DHE), EXE-19-D3 (D3-EXE), and D3-creatinine were purchased from Toronto Research Chemicals (North York, ON, Canada). 17β-DHE was synthesized as previously described to a purity of greater than 99% (25). Ammonium formate was obtained from Sigma-Aldrich (St Louis, Missouri, USA) while ammonium acetate and formic acid were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Acetonitrile was purchased from Merck (Kenilworth, NJ, USA). Solvent and buffer modifiers for liquid chromatography-mass spectrometry (LC/MS) analysis including acetonitrile, ammonium formate, ammonium acetate and formic acid were all LC/MS grade. Milli-Q water was used for making any solutions when needed. PureLink ® Genomic DNA Kits were purchased from Thermo Fisher Scientific (Waltham, MA, USA), and the TaqMan® Copy Number Reference Assay (RNase P, Human) and the UGT2B17 TaqMan® Copy Number Assay were purchased from Life Technologies (Carlsbad, CA, USA).
Subjects and samples
Ninety-six post-menopausal Caucasian breast cancer patients (age range: 35 to 89 y) with ER+ breast tumors taking 25 mg EXE daily (orally) and one healthy volunteer not taking EXE (used as a control) were recruited from the breast oncology clinic at the Penn State Hershey Cancer Institute into this study. All recruited subjects provided blood (10 cc) and urine (up to 50 mL). Patients were excluded from the study if they had been given EXE concurrently with adjuvant chemotherapy or if they were taking other adjuvant endocrine therapies, or were on chronic corticosteroid or megestrol acetate therapies. Approval was obtained from the Institutional Review Board at Penn State University with informed consent obtained from all subjects and with all specimens being de-identified. Specimens were obtained 4~6 hours after last pill ingestion by a trained nurse coordinator after patients had been taking EXE for at least 28 days. Pretreatment medical histories including a comprehensive list of current medications and results of physical and laboratory examinations were also collected for each subject. Blood was separated by differential centrifugation and buffy coat was used to extract genomic DNA. Aliquoted urine samples and plasma fractions of blood samples were stored at −80°C until analysis.
Sample preparation
Genomic DNA was purified from blood samples using PureLink ® Genomic DNA Kits. DNA quantity and purity were determined photometrically at 260 nm and 280 nm using the Thermo Scientific Nanodrop 2000 spectrophotometer (Waltham, MA, USA).
For EXE metabolite analysis, a 50-μL aliquot of each urine sample was first spiked with 10 μL of a mixture of deuterium-labeled internal standards in methanol, including D3-EXE (0.17 μM), D3-17β-DHE (1.7 μM) and D3-17β-DHE-Gluc (1.1 μM). Ninety μL of methanol was then added to extract EXE and its metabolites. After vortexing and subsequent centrifugation at 16,100 × g for 10 min at 4°C, an aliquot of 50 μL of supernatant was transferred to a sample vial for analysis by ultra-pressure liquid chromatography (UPLC)/mass spectrometry (MS).
For analysis of plasma, 10 μL of each plasma sample was first mixed with 10 μL of a mixture of deuterium-labeled internal standards as described above. Eighty μL of methanol was then added to precipitate proteins. After vortexing and subsequent centrifugation at 16,100 × g for 10 min at 4°C, an aliquot of 50 μL of supernatant was transferred to a sample vial for analysis by UPLC/MS.
UPLC/MS conditions
For the simultaneous analysis of EXE, 17β-DHE and 17β-DHE-Gluc in urine and plasma, samples prepared as described above were analyzed using a UPLC/MS system (Waters), consisting of an Acquity UPLC pump, an Acquity sample manager-FTN, an ACQUITY UPLC BEH column C18 (2.1×100 mm, 1.7 μm particle size), and a XEVO G2-S QTOF mass spectrometer. UPLC was performed at a flow rate of 0.4 mL/min with solvent A (5 mM ammonium formate and 0.01% formic acid in water) and solvent B (100% acetonitrile) using the following conditions for both urine and plasma specimens: 1 min at 35% solvent B, a linear gradient to 52% solvent B for 0.5 min, 2.5 min at 52% solvent B, a liner gradient to 100% solvent B for 0.5 min, followed by re-equilibrium with 35% solvent B for 1.5 min. The injection volume of each prepared urine and plasma sample was 2 μL. The column temperature was 35°C. The retention times were 2.96 min for EXE, 2.66 min for 17β-DHE and 0.99 min for 17β-DHE-Gluc (Supplement Figure 1). Corresponding peaks from the urine and plasma matched well with the internal standards. No EXE, 17β-DHE or 17β-DHE-Gluc peaks were observed in the urine or plasma specimens from the subject not treated with EXE.
The Waters XEVO G2-S QTOF MS was equipped with an electrospray ionization (ESI) probe operated in the positive-ion mode, with capillary voltage at 0.6 kV. Nitrogen was used as both the cone and desolvation gases with flow rates maintained at 50 and 800 L/h, respectively. Ultra-pure argon was used as the collision gas with a flow rate of 0.1 L/h. The source and desolvation gas temperatures were 120 and 500°C, respectively, with the mass spectrometer was operated in the MS/MS mode. The dwell time for each ion was 100 ms. The ion-related parameters for the 6 transitions monitored are listed in Table 1. The detection limits at which the signal to noise ratio was ≥ 3:1 for EXE, DHE and DHE-Gluc were 1 nM, 0.6 nM and 0.5 nM, respectively, in plasma, and 0.3 nM, 1 nM and 5 nM, respectively, in urine. The limit of quantification at which the signal to noise ratio was ≥ 10:1 for EXE, DHE and DHE-Gluc were 3 nM, 1.8 nM and 1.5 nM, respectively, in plasma, and 0.9 nM, 3 nM and 15 nM, respectively, in urine.
Table 1.
MS/MS transitions and ion optic parameters for EXE and EXE metabolites.
| ES+ MS/MS transition (m/z) | cone voltage (V) | collision energy (eV) | |
|---|---|---|---|
| 17β-DHE-Gluc | 475.23>281.19 | 20 | 15 |
| D3-17β-DHE-Gluc | 478.25>284.21 | 20 | 15 |
| 17β-DHE | 299.20>299.20 | 25 | 10 |
| D3-17β-DHE | 302.22>302.22 | 25 | 10 |
| EXE | 297.19>297.19 | 25 | 10 |
| D3-EXE | 300.20>300.20 | 25 | 10 |
Standard curves were constructed by plotting the ratio of analyte peak area to peak area of the corresponding internal standard (described above) versus concentration of analyte standard. The concentrations of stock standards were 1000 ppm, which were 3.4 mM, 3.4 mM and 2.1 mM in methanol for EXE, 17β-DHE and 17β-DHE-Gluc, respectively. A serial dilution of standards at concentrations ranging from 0.4 nM to 3.4 μM, 0.4 nM to 3.4 μM, and 0.25 nM to 2.1 μM were used to establish standard curves for EXE, 17β-DHE and 17β-DHE-Gluc, respectively. Analyte concentrations were determined by measuring the peak area ratios of analyte to internal standard and then calculating analyte concentration from the appropriate standard curve using Waters’ TargetLynx software.
Determination of urinary creatinine
A 10 μL aliquot of each urine sample was first spiked with 10 μL of internal standard (100 ppm D3-creatinine) and then diluted 1,000-fold with water before injecting 1 μL onto an ACQUITY UPLC HSS T3 column (2.1×100 mm, 1.8 μm particle size). UPLC was performed at a flow rate of 0.4 mL/min using the following conditions: 100% solvent A (5 mM ammonium acetate) for 1 min, a liner gradient to 100% solvent B (100% acetonitrile) for 2 min, 100% B for 1 min, followed by re-equilibrium for 1 min with 100% solvent A. The mass spectrometer was operated in positive MS scan mode with cone voltage at 20 V, other MS operation condition were the same as described above for EXE and its metabolites. Standards at concentrations ranging from 0.002 to 2 ppm were used to establish standard curves for quantification of urinary creatinine levels.
Genotyping of UGT2B17
The UGT2B17 CNV was determined by real-time PCR using the UGT2B17 TaqMan® Copy Number Assay and the TaqMan® Copy Number Reference Assay (RNase P, human). All real-time PCR analyses were performed in the Washington State University-Spokane Genomics Core Facility using a Bio-Rad CFX384 real-time PCR machine.
Statistical analysis
Statistical analyses were performed using Prism (version 6, Graph Pad Software, San Diego, CA). The Pearson correlation test was used to test the association between the levels of EXE and its metabolites in both urine and plasma. The Student’s t test (or the Mann-Whitney rank test when data were not normally distributed) and the trend test were used to compare levels of EXE, 17β-DHE and 17β-DHE-Gluc in patients stratified by UGT2B17 copy number genotypes. Hardy-Weinberg equilibrium was used to test the genotype distribution.
Results
EXE and its two major metabolites, 17β-DHE and 17β-DHE-Gluc, were separated by UPLC-MS as described in the Materials and Methods. The levels of EXE and its two major metabolites, 17β-DHE and 17β-DHE-Gluc, were determined simultaneously in individual urine and plasma specimens from 96 subjects taking EXE, and also in one control subject who did not take EXE. At least one of these metabolites were detected in all 96 plasma specimens and in 92 of the urine specimens (EXE and its metabolites were not detected in 4 urine specimens; these specimens were not incorporated into the genotype:phenotype analysis described below). Among the subjects taking EXE, the mean concentrations of EXE, 17β-DHE and 17β-DHE-Gluc were 23 nM, 6.0 nM and 46 nM, respectively, in plasma (n=96 specimens), and 161 nM, 9.1 nM and 1048 nM, respectively, in urine (92 specimens; Table 2).
Table 2.
Summary of EXE, 17β-DHE and 17β-DHE-Gluc concentrations in 96 plasma and 92 urine specimens from EXE-treated subjects.
| plasma (nM) | urine (nM) | urine, creatinine-adjusted (nmol/mg creatinine) | ||||
|---|---|---|---|---|---|---|
| mean (95% CI) | range | mean (95% CI) | range | mean (95% CI) | range | |
| EXE | 23 (17–29) | 0–157 | 161 (101–221) | 0.24–1709 | 0.41 (0.24–0.57) | 0.001–4.0 |
| 17β-DHE | 6.0 (5.1–6.9) | 0.03–30 | 9.1 (6.1–12) | 0–86 | 0.022 (0.014–0.029) | 0–0.21 |
| 17β-DHE-Gluc | 46 (31–60) | 0–461 | 1048 (525–1572) | 0.48–22562 | 1.8 (1.0–2.7) | 0.001–34 |
UGT2B17 genotypes were obtained for all 96 EXE-treated patients tested. In addition to the 35 subjects (37%) who exhibited the UGT2B17 (*1/*1) genotype (two wild-type UGT2B17 alleles), one subject (1.0%) exhibited three copies of the UGT2B17 gene, suggesting that this individual contained one wild-type UGT2B17 allele and one allele (termed the *3 allele) that contained two copies of the UGT2B17 gene, exhibiting a (*1/*3) genotype. Forty-four subjects (46%) were heterozygous with the UGT2B17 (*1/*2) genotype while 16 subjects (17%) exhibited the UGT2B17 null (*2/*2) genotype. This genotype distribution was consistent with Hardy-Weinberg equilibrium (P = 0.683) and the UGT2B17 deletion allelic frequency was 40% among the 96 subjects.
Among the 91 EXE-treated subjects for whom detectable levels of EXE or its two metabolites were observed in urine, a 2.1- (P=0.12) and 39- (P<0.0001) fold decrease in the urinary levels of creatinine-adjusted 17β-DHE-Gluc was observed among subjects with the UGT2B17 (*1/*2) and (*2/*2) genotypes, respectively, as compared to subjects with the UGT2B17 (*1/*1) genotype (Figure 1, panel A). A significant (P<0.0001) 18-fold decrease in the urinary levels of creatinine-adjusted 17β-DHE-Gluc was also observed among UGT2B17 (*2/*2) genotype subjects vs. subjects with the UGT2B17 (*1/*2) genotype. A significant trend (Ptrend=0.018) towards decreasing levels of creatinine-adjusted 17β-DHE-Gluc was observed with decreasing numbers of the UGT2B17*1 allele. No difference in the urinary levels of creatinine-adjusted 17β-DHE (Figure 1, panel B) or creatinine-adjusted EXE (Figure 1, panel C) were observed after stratifying by UGT2B17 genotype. Similar results were observed when all 96 urine specimens were included in the association analysis (result not shown).
Figure 1. Association between the UGT2B17 CNV and urinary creatinine-adjusted 17β-DHE-Gluc, 17β-DHE and EXE levels in EXE-treated subjects.
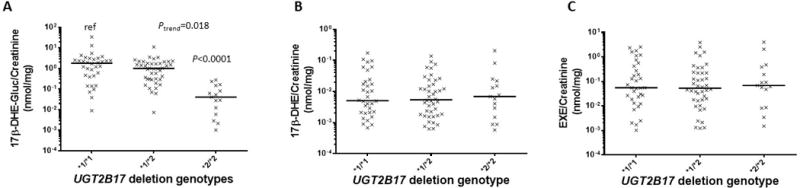
Subjects were stratified by UGT2B17 CNV genotype, with the UGT2B17*1 allele corresponding to the wild-type single-gene copy number and the UGT2B17*2 allele corresponding to the UGT2B17 gene deletion variant. Panel A, UGT2B17 CNV genotypes versus creatinine-adjusted 17β-DHE-Gluc (nmol/mg creatinine); Panel B, UGT2B17 CNV genotypes versus creatinine-adjusted 17β-DHE; and Panel C, UGT2B17 CNV genotypes versus creatinine-adjusted EXE. All values are expressed as the median for 91 subjects.
Similar to the patterns observed in urine, the levels of plasma 17β-DHE-Gluc were decreased by 1.9- (P=0.067) and 29-fold (P<0.0001) in subjects with the UGT2B17 (*1/*2) and (*2/*2) genotypes, respectively, as compared to the subjects with the UGT2B17 (*1/*1) genotype (Figure 2, panel A). A 15-fold decrease (P<0.0001) in the levels of plasma 17β-DHE-Gluc was also observed among UGT2B17 (*2/*2) genotype subjects vs. subjects with the UGT2B17 (*1/*2) genotype. A significant trend (Ptrend=0.0011) towards decreasing levels of plasma 17β-DHE-Gluc was observed with decreasing numbers of the UGT2B17*1 allele.
Figure 2. Association between the UGT2B17 CNV and plasma 17β-DHE-Gluc, 17β-DHE and EXE levels in EXE-treated subjects.
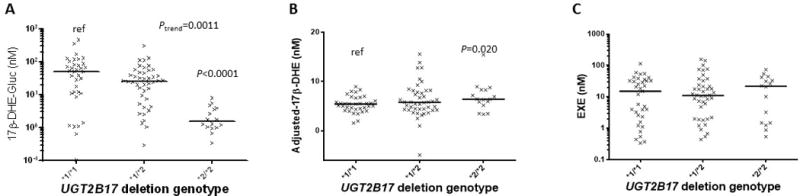
Subjects were stratified by UGT2B17 CNV genotype, with the UGT2B17*1 allele corresponding to the wild-type single-gene copy number and the UGT2B17*2 allele corresponding to the UGT2B17 gene deletion variant. Panel A, UGT2B17 CNV genotypes versus 17β-DHE-Gluc; Panel B, UGT2B17 CNV genotypes versus 17β-DHE; and Panel C, UGT2B17 CNV genotypes versus EXE. All values are expressed as the median for 95 subjects.
A strong association between the levels of 17β-DHE and EXE was observed in plasma (R2=0.56, P<0.0001). When the levels of plasma 17β-DHE were adjusted by EXE using linear regression, the mean levels of plasma EXE-adjusted-17β-DHE was 28% higher (P=0.02) in subjects with the UGT2B17 (*2/*2) genotype vs. subjects with the UGT2B17 (*1/*1) genotype (Figure 2, panel B). No difference in the levels of plasma EXE (Figure 2, panel C) were observed after stratifying by UGT2B17 genotype in the 95 subjects.
The creatinine-adjusted urinary levels of EXE, 17β-DHE and 17β-DHE-Gluc (1.7 nmol/mg, 0.043 nmol/mg, and 8.7 nmol/mg, respectively) in the single subject exhibiting the UGT2B17 (*1/*3) genotype were higher than the mean levels (0.41 nmol/mg, 0.024 nmol/mg and 3.0 nmol/mg, respectively), but lower than the maximum levels (2.5 nmol/mg, 0.17 nmol/mg and 34 nmol/mg, respectively), observed for these compounds in the thirty-five subjects with the UGT2B17 (*1/*1) genotype. The plasma levels of EXE and 17β-DHE-Gluc (50 nM and 158 nM, respectively) in this subject was also higher than the mean (21 nM and 72 nM, respectively) but lower than the maximum levels (114 nmol/mg and 461 nmol/mg, respectively) observed for these compounds in the UGT2B17 (*1/*1) genotype subjects. The plasma levels of 17β-DHE (7.3 nM) in the UGT2B17 (*1/*3) genotype subject was similar to the mean levels (5.4 nM) observed for these compounds in the UGT2B17 (*1/*1) genotype subjects. The significant pattern of decreased levels of 17β-DHE-Gluc in both urine and plasma and increased levels of 17β-DHE observed in plasma observed in subjects with the UGT2B17 (*2/*2) genotype vs subjects with the UGT2B17 (*1/*1) genotype was also observed when the subject with the UGT2B17 (*1/*3) genotype was combined with the UGT2B17 (*1/*1) genotype group in the comparative analysis (results not shown).
Discussion
Previous studies have demonstrated that 17β-DHE is the primary metabolite of EXE and that it exhibits an in vitro anti-aromatase activity similar to that observed for EXE (20–24). The major mode of detoxification of 17β-DHE is by glucuronidation, with UGT2B17 catalyzing greater than 95% of total 17β-DHE formation in HLM (24). In the present study, it was demonstrated that the common UGT2B17 gene deletion polymorphism is associated with large decreases in both urinary and plasma levels of 17β-DHE-Gluc, with the levels of 17β-DHE-Gluc reduced by 39- and 29-fold in urine and plasma, respectively, in subjects exhibiting the UGT2B17 (*2/*2) homozygous deletion genotype vs. subjects exhibiting the (*1/*1) genotype. This is consistent with that observed previously in HLM in vitro (24). It is also consistent with that observed in a recent small pharmacokinetic study of healthy volunteeers which demonstrated an ~8-fold difference in the AUC and Cmax of conjugated 17β-DHE between subjects with the UGT2B17 (*1/*1) vs. (*2/*2) genotypes (31). Together, these studies are consistent with an important role for UGT2B17 in the glucuronidation of this active EXE metabolite (21).
The UGT2B17 deletion polymorphism was associated with a 28% increase in plasma 17β-DHE levels in the present study. This contrasts with that observed in a recent study where no difference in 17β-DHE levels were observed in non-β-glucuronidase-treated plasma (31). The difference in plasma 17β-DHE levels vs. UGT2B17 genotype observed between studies is likely due to sample size or study design differences. The previous study sampled multiple time points from 14 healthy individuals after a single dose of EXE, while the current study examined samples from single time points from 96 breast cancer patients who daily doses of EXE for at least 4 weeks. In addition, the major toxicity associated with AIs including EXE is joint pain, bone loss, the propensity for bone fractures, and increased risk for osteoporosis (10, 32, 33). Other toxicities reported for AIs include a variety of gynecological events including vaginal bleeding, vaginal dryness or discharge and hot flashes (34, 35). Variability in changes of lipid profiles, manifestation of osteoporosis, and time of recurrence, were observed in many patients in several AI clinical trials (7). EXE-induced toxicities are not universal, with only a fraction of patients reporting any one of them, and the UGT2B17 deletion polymorphism could be contributing to this inter-individual variability. Also, the UGT2B17*2 deletion allele frequency varies between different racial groups, with subjects exhibiting the homozygous (*2/*2) deletion genotype most common in the Asian population (e.g. Korean, 67%; Chinese, 77%) as compared to that observed in Caucasians (e.g., Swedish, 9%) (36, 37). Therefore, 17β-DHE glucuronidation may not be a major pathway for EXE elimination in Asians and this could potentially affect EXE clinical efficacies between racial groups.
While increased plasma 17β-DHE levels were associated with increasing numbers of the UGT2B17 deletion allele, the magnitude of this increase was less than the corresponding decrease in plasma 17β-DHE-Gluc levels [e.g., 29-fold decrease for 17β-DHE-Gluc vs. 1.3-fold for 17β-DHE, comparing UGT2B17 (*2/*2) subjects with UGT2B17 (*1/*1) subjects]. This may be due to other excretion pathways also playing a role in EXE metabloism. Previous studies have shown that there was similar excretion of EXE in the urine vs bile, but it is unknown whether this was studied in subjects with wild-type UGT2B17 (19). It is possible that EXE excretion patterns (urine vs bile) also differ between UGT2B17 (1/*1) vs (*2/*2) subjects. It is also possible that other 17β-DHE metabolites accumulate in UGT2B17-deleted subjects, including the inactive 17β-hydroxy-6-hydroxymethylandrosta-1,4,6-trien-3-one and its secondary metabolite, 6α/β, 17β-dihydroxy-6α/β-hydroxymethylandrosta-1,4-dien-3-one, both of which were implicated as important metabolites of 17β-DHE (38). Studies investigating these possibilities are on-going.
The detection limits observed in the present study were between 0.5 and 1 nM for EXE, 17β-DHE and 17β-DHE-Gluc in plasma, 0.3 and 5 nM in urine. In addition, the quantification limits for EXE, DHE and DHE-Gluc were 3 nM, 1.8 nM and 1.5 nM, respectively, in plasma, and 0.9 nM, 3 nM and 15 nM, respectively, in urine. These limits are comparable to those observed in previous studies, where the lower limits of quantification were 0.2 ng/mL and 0.1 ng/mL for EXE and 17β-DHE, respectively, in plasma and the lower limits of detection were 1 ng/mL for both in urine (39, 40). Of the ninety-six subjects analyzed in the present study, all exhibited detectable EXE, 17β-DHE or 17β-DHE-Gluc in plasma with only 4 exhibiting levels that were below the detection limit for EXE, 17β-DHE and 17β-DHE-Gluc in urine. Together, these data suggest that the LC/MS method developed in the present study for the detection of EXE, 17β-DHE and 17β-DHE-Gluc is highly sensitive.
While UGT2B17 is the major enzyme responsible for DHE glucuronidation, many other enzymes are involved EXE metabolism. Cytosolic keto steroid reductases AKR1Cs and CBR1 are highly active in EXE reduction to DHE in vitro and several common variants in the cytosolic keto steroid reductases were associated with altered enzymatic activity in vitro (25). Cytochrome P450s 4A11 and 1A1/2 were also found to be responsible for formation of DHE, while CYP3A was active in EXE oxidation to form 6-hydroxymethylexemestane (26). Therefore, in addition to the UGT2B17 deletion polymorphism, functional polymorphisms in these aldo-keto reductases and CYP450s may also play a role in the therapeutic efficacy or toxicity of EXE in vivo.
A limitation of the present study was that the urine and blood specimens were obtained from subjects taking EXE near the Tmax. Since previous pharmacokinetic studies demonstrated a Cmax for EXE at 1~2 h post dose (41, 42), it is possible that metabolite concentrations may differ at the 4~6 hour time point that specimens were collected post-pill ingestion in the present study, Another potential limitation was that spot urine collections were performed in the present study. However, while 24-h urine collections are considered to be the most reliable type of urine sample (43), previous studies have also reported that a group average of many individual spot urine samples (as performed in the present study) also provides a good estimate of exposure (44).
In conclusion, the present study demonstrates that UGT2B17 is the major enzyme responsible for 17β-DHE-Gluc formation in vivo. It also showed that UGT2B17 genotypes are associated with altered 17β-DHE levels in the plasma of women taking EXE, and may therefore play a role in variability in EXE-induced toxicity as well as therapeutic efficacy between patients. Clinical trials examining this important possibility will be necessary to examine this directly.
Supplementary Material
Acknowledgments
We thank the Mass Spectrometry Core facility and the Genomic Core facility at Washington State University-Spokane for their help with UPLC-MS and genotyping, respectively. We are also very grateful to Zuping Xia in the Department of Pharmaceutical Sciences at Washington State University for providing 17β-DHE as a standard for these studies. This work was supported by a grant (RO1-CA164366) from the National Cancer Institute at the National Institutes of Health to P. Lazarus and a China Scholarship Council grant (File No. 201406600026) for Shaman Luo.
Footnotes
Conflicts
There are no competing financial interests or conflicts of interest for any of the authors of this manuscript.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review. 1975–2013 Available from: http://seer.cancer.gov/csr/1975_2013/
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos SM. Aromatase inhibitors for breast cancer in postmenopausal women. Oncologist. 2004;9(2):126–36. doi: 10.1634/theoncologist.9-2-126. [DOI] [PubMed] [Google Scholar]
- 5.Miller WR. Biology of aromatase inhibitors: pharmacology/endocrinology within the breast. Endocr Relat Cancer. 1999;6(2):187–95. doi: 10.1677/erc.0.0060187. [DOI] [PubMed] [Google Scholar]
- 6.Santen RJ, Brodie H, Simpson ER, Siiteri PK, Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr Rev. 2009;30(4):343–75. doi: 10.1210/er.2008-0016. [DOI] [PubMed] [Google Scholar]
- 7.Eisen A, Trudeau M, Shelley W, Messersmith H, Pritchard KI. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: a systematic review. Cancer Treat Rev. 2008;34(2):157–74. doi: 10.1016/j.ctrv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Ferretti G, Bria E, Giannarelli D, Felici A, Papaldo P, Fabi A, et al. Second- and third-generation aromatase inhibitors as first-line endocrine therapy in postmenopausal metastatic breast cancer patients: a pooled analysis of the randomised trials. Br J Cancer. 2006;94(12):1789–96. doi: 10.1038/sj.bjc.6603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arimidex TAoiCTG. Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9(1):45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 10.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–2. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 11.Campos SM, Guastalla JP, Subar M, Abreu P, Winer EP, Cameron DA. A comparative study of exemestane versus anastrozole in patients with postmenopausal breast cancer with visceral metastases. Clin Breast Cancer. 2009;9(1):39–44. doi: 10.3816/CBC.2009.n.007. [DOI] [PubMed] [Google Scholar]
- 12.Gluck S. Exemestane as first-line therapy in postmenopausal women with recurrent or metastatic breast cancer. Am J Clin Oncol. 2010;33(3):314–9. doi: 10.1097/COC.0b013e31819fdf9b. [DOI] [PubMed] [Google Scholar]
- 13.Paridaens R, Dirix L, Lohrisch C, Beex L, Nooij M, Cameron D, et al. Mature results of a randomized phase II multicenter study of exemestane versus tamoxifen as first-line hormone therapy for postmenopausal women with metastatic breast cancer. Ann Oncol. 2003;14(9):1391–8. doi: 10.1093/annonc/mdg362. [DOI] [PubMed] [Google Scholar]
- 14.Chia S, Gradishar W, Mauriac L, Bines J, Amant F, Federico M, et al. Double-blind, randomized placebo controlled trial of fulvestrant compared with exemestane after prior nonsteroidal aromatase inhibitor therapy in postmenopausal women with hormone receptor-positive, advanced breast cancer: results from EFECT. J Clin Oncol. 2008;26(10):1664–70. doi: 10.1200/JCO.2007.13.5822. [DOI] [PubMed] [Google Scholar]
- 15.Hong Y, Yu B, Sherman M, Yuan YC, Zhou D, Chen S. Molecular basis for the aromatization reaction and exemestane-mediated irreversible inhibition of human aromatase. Mol Endocrinol. 2007;21(2):401–14. doi: 10.1210/me.2006-0281. [DOI] [PubMed] [Google Scholar]
- 16.Petkov PI, Temelkov S, Villeneuve DL, Ankley GT, Mekenyan OG. Mechanism-based categorization of aromatase inhibitors: a potential discovery and screening tool. SAR QSAR Environ Res. 2009;20(7):657–78. doi: 10.1080/10629360903438347. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Chen S. Aromatase destabilizer: novel action of exemestane, a food and drug administration-approved aromatase inhibitor. Cancer Res. 2006;66(21):10281–6. doi: 10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- 18.Deeks ED, Scott LJ. Exemestane: a review of its use in postmenopausal women with breast cancer. Drugs. 2009;69(7):889–918. doi: 10.2165/00003495-200969070-00007. [DOI] [PubMed] [Google Scholar]
- 19.Pfizer. Aromasin Exemestane Tablets 2016. Available from: http://www.pfizer.com/files/products/uspi_aromasin.pdf.
- 20.Lonning PE. Pharmacological profiles of exemestane and formestane, steroidal aromatase inhibitors used for treatment of postmenopausal breast cancer. Breast Cancer Res Treat. 1998;49(Suppl 1):S45–52. doi: 10.1023/a:1006048722559. discussion S73–7. [DOI] [PubMed] [Google Scholar]
- 21.Evans TR, Di Salle E, Ornati G, Lassus M, Benedetti MS, Pianezzola E, et al. Phase I and endocrine study of exemestane (FCE 24304), a new aromatase inhibitor, in postmenopausal women. Cancer Res. 1992;52(21):5933–9. [PubMed] [Google Scholar]
- 22.Mareck U, Geyer H, Guddat S, Haenelt N, Koch A, Kohler M, et al. Identification of the aromatase inhibitors anastrozole and exemestane in human urine using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(12):1954–62. doi: 10.1002/rcm.2545. [DOI] [PubMed] [Google Scholar]
- 23.Traina TA, Poggesi I, Robson M, Asnis A, Duncan BA, Heerdt A, et al. Pharmacokinetics and tolerability of exemestane in combination with raloxifene in postmenopausal women with a history of breast cancer. Breast Cancer Res Treat. 2008;111(2):377–88. doi: 10.1007/s10549-007-9787-1. [DOI] [PubMed] [Google Scholar]
- 24.Sun D, Chen G, Dellinger RW, Sharma AK, Lazarus P. Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics. 2010;20(10):575–85. doi: 10.1097/FPC.0b013e32833b04af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platt A, Xia Z, Liu Y, Chen G, Lazarus P. Impact of nonsynonymous single nucleotide polymorphisms on in-vitro metabolism of exemestane by hepatic cytosolic reductases. Pharmacogenet Genomics. 2016 doi: 10.1097/FPC.0000000000000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamdem LK, Flockhart DA, Desta Z. In vitro cytochrome P450-mediated metabolism of exemestane. Drug Metab Dispos. 2011;39(1):98–105. doi: 10.1124/dmd.110.032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson W, 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84(4):707–14. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15(11):769–78. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 29.Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(4):823–8. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, Giambrone NE, Jr, Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res. 2010;70(19):7543–52. doi: 10.1158/0008-5472.CAN-09-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SM, Atchley DH, Murphy MA, Gurley BJ, Kamdem LK. Impact of UGT2B17 Gene Deletion on the Pharmacokinetics of 17-Hydroexemestane in Healthy Volunteers. J Clin Pharmacol. 2016;56(7):875–84. doi: 10.1002/jcph.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coates AS, Keshaviah A, Thurlimann B, Mouridsen H, Mauriac L, Forbes JF, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1–98. J Clin Oncol. 2007;25(5):486–92. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- 33.Coleman RE, Banks LM, Girgis SI, Kilburn LS, Vrdoljak E, Fox J, et al. Skeletal effects of exemestane on bone-mineral density, bone biomarkers, and fracture incidence in postmenopausal women with early breast cancer participating in the Intergroup Exemestane Study (IES): a randomised controlled study. Lancet Oncol. 2007;8(2):119–27. doi: 10.1016/S1470-2045(07)70003-7. [DOI] [PubMed] [Google Scholar]
- 34.Karaer O, Oruc S, Koyuncu FM. Aromatase inhibitors: possible future applications. Acta Obstet Gynecol Scand. 2004;83(8):699–706. doi: 10.1111/j.0001-6349.2004.00562.x. [DOI] [PubMed] [Google Scholar]
- 35.Grana G. Adjuvant aromatase inhibitor therapy for early breast cancer: A review of the most recent data. J Surg Oncol. 2006;93(7):585–92. doi: 10.1002/jso.20564. [DOI] [PubMed] [Google Scholar]
- 36.Jakobsson J, Ekstrom L, Inotsume N, Garle M, Lorentzon M, Ohlsson C, et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J Clin Endocrinol Metab. 2006;91(2):687–93. doi: 10.1210/jc.2005-1643. [DOI] [PubMed] [Google Scholar]
- 37.Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83(6):663–74. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzzetti F, Di Salle E, Longo A, Briatico G. Synthesis and aromatase inhibition by potential metabolites of exemestane (6-methylenandrosta-1,4-diene-3,17-dione) Steroids. 1993;58(11):527–32. doi: 10.1016/0039-128x(93)90029-m. [DOI] [PubMed] [Google Scholar]
- 39.Corona G, Elia C, Casetta B, Diana C, Rosalen S, Bari M, et al. A liquid chromatography-tandem mass spectrometry method for the simultaneous determination of exemestane and its metabolite 17-dihydroexemestane in human plasma. J Mass Spectrom. 2009;44(6):920–8. doi: 10.1002/jms.1566. [DOI] [PubMed] [Google Scholar]
- 40.Semenistaya EN, Dikunets MA, Viryus ED, Rodchenkov GM. Determination of exemestane and 17-hydroxyexemestane by high-performance liquid chromatography coupled with tandem mass spectrometry and high-resolution mass spectrometry. J Anal Chem+ 2010;65(5):498–506. [Google Scholar]
- 41.Wang LZ, Goh SH, Wong AL, Thuya WL, Lau JY, Wan SC, et al. Validation of a rapid and sensitive LC-MS/MS method for determination of exemestane and its metabolites, 17beta-hydroxyexemestane and 17beta-hydroxyexemestane-17-O-beta-D-glucuronide: application to human pharmacokinetics study. PLoS One. 2015;10(3):e0118553. doi: 10.1371/journal.pone.0118553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corona G, Elia C, Casetta B, Diana C, Rosalen S, Bari M, et al. A liquid chromatography-tandem mass spectrometry method for the simultaneous determination of exemestane and its metabolite 17-dihydroexemestane in human plasma. Journal of Mass Spectrometry. 2009;44(6):920–8. doi: 10.1002/jms.1566. [DOI] [PubMed] [Google Scholar]
- 43.Cornelis R, Heinzow B, Herber RF, Christensen JM, Poulsen OM, Sabbioni E, et al. Sample collection guidelines for trace elements in blood and urine. IUPAC Commission of Toxicology. J Trace Elem Med Biol. 1996;10(2):103–27. doi: 10.1016/s0946-672x(96)80018-6. [DOI] [PubMed] [Google Scholar]
- 44.Smith AH, Steinmaus CM. Arsenic in urine and drinking water. Environ Health Perspect. 2000;108(11):A494–5. doi: 10.1289/ehp.108-a494c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


