Abstract
Beige adipocyte differentiation within white adipose tissue, referred to as browning, is seen as a possible mechanism for increasing energy expenditure. The molecular regulation underlying the thermogenic browning process has not been entirely elucidated. Here, we identify the zinc finger transcription factor EGR1 as a negative regulator of the beige fat program. Loss of Egr1 in mice promotes browning in the absence of external stimulation and leads to an increase of Ucp1 expression, which encodes the key thermogenic mitochondrial uncoupling protein-1. Moreover, EGR1 is recruited to the proximal region of the Ucp1 promoter in subcutaneous inguinal white adipose tissue. Transcriptomic analysis of subcutaneous inguinal white adipose tissue in the absence of Egr1 identifies the molecular signature of white adipocyte browning downstream of Egr1 deletion and highlights a concomitant increase of beige differentiation marker and a decrease in extracellular matrix gene expression. Conversely, Egr1 overexpression in mesenchymal stem cells decreases beige adipocyte differentiation, while increasing extracellular matrix production. These results reveal a role for Egr1 in blocking energy expenditure via direct Ucp1 transcription repression and highlight Egr1 as a therapeutic target for counteracting obesity.
Introduction
White fat browning is a mechanism that produces heat and limits weight gain. The understanding of the molecular regulation underlying white fat browning has sparked interest to counteract obesity.
The adipose tissue of humans and other mammals contains white adipose tissue (WAT) and brown adipose tissue (BAT). WAT and BAT are developmentally and functionally distinct and contain white and brown adipocytes, respectively1–3. More recently, a third type of adipocytes has been described within WAT, beige adipocytes. Morphological and molecular analyses showed that brown and beige adipocytes are remarkably similar and express the same thermogenic markers4. However beige adipocytes, in contrast to brown adipocytes, express thermogenic markers only after external stimulations, such as cold exposure, starvation, exercise or hormone treatment5. In the adult, beige adipocytes are produced by the trans-differentiation of mature white adipocytes4 or by de novo differentiation of progenitors6 in response to external stimulations. This process is referred to as “browning” or “beigeing”2,7.
Because the increase of WAT is observed in many metabolic diseases, WAT browning represents a promising therapeutic approach. Consequently, it is crucial to decipher the molecular aspects underlying the beige differentiation program. Adipogenesis is triggered by a core adipogenic network, starting with the expression of Cebpb (CCAAT/enhancer binding protein ß), which activates the expression of Pparg (Peroxisome proliferator-activated receptor γ) and Cebpa (CCAAT/enhancer binding protein α), which in turn activates Ppara (Peroxisome proliferator-activated receptor α) expression8. Consistent with its thermogenic function, brown/beige differentiated adipocytes express high levels of UCP1, a mitochondrial protein that uncouples oxidative phosphorylation from ATP synthesis9,10. The Krebs cycle enzymes, such as OGDH (oxoglutarate dehydrogenase), SUCLA2 (succinate-Coenzyme A ligase) and COX8B (Cytochrome C Oxidase Subunit VIIIb)11,12 are also involved in heat production in beige/brown adipose tissue. Consistent with their anti-fat function, brown/beige differentiated adipocytes express factors involved in lipolysis such as PLIN5 (Perilipin 513) and CIDEA (Cell Death-Inducing DFFA-Like Effector A12). Beige adipocyte differentiation relies on the expression of a set of transcriptional activators2,3. PRDM16 (PR domain containing 16) is considered as a master regulator of the brown/beige program via direct interaction with transcription factors, such as C/EBPβ, PPARα, PPARγ, and PGC-1α (Peroxisome proliferator-activated receptor Gamma Coactivator 1-alpha14–16). Of note, beige and white differentiation programs share transcriptional regulators, such as C/EBPβ, which has been shown to be sufficient for Ucp1 transcription via direct binding to the Ucp1 proximal promoter in vitro 17,18. Moreover, Cebpb mutant mice display defective thermoregulation19. In addition to transcriptional regulators, growth factors such as FGF21 (Fibroblast Growth Factor-21) and BMP4 (Bone morphogenetic Protein-4), adipokines such as leptin and hormones such as T3 (Triiodothyronin 3) have been identified as being able to induce the brown/beige fat phenotype2,20,21. The T4 to T3 converting enzyme Desiodase 2 (DIO2) is also involved in the browning process22.
The zinc finger transcription factor EGR1 (Early Growth Response-1) is involved in multiple processes including cell proliferation, differentiation, migration, apoptosis, and inflammation in many cell types23–27. Egr1 is expressed in adult adipose tissues28,29 where its overexpression has been linked to obesity and obesity-associated metabolic disorders in both humans and mouse models28,29. Conversely, Egr1-deficient mice are protected from diet-induced obesity29. Consistently, EGR1 inhibits lipolysis and promotes fat accumulation in cultured adipocytes by directly repressing the transcription of the adipose triglyceride lipase (ATGL) gene30. Surprisingly, Egr1 overexpression represses white adipocyte differentiation in the 3T3-L1 and C3H101/2 cell lines31,32.
To understand how Egr1 can both be linked with obesity and adverse metabolic outcomes while repressing differentiation of white adipocytes in culture, we investigated the role of Egr1 in white adipose tissue development during the postnatal period in female mice. We analysed the consequences of Egr1 inhibition for subcutaneous inguinal white adipose tissue (SC-WAT) formation during postnatal and adult periods, using a mouse model deficient for Egr1, with no external stimulation. We also assessed the consequences of Egr1 overexpression for beige differentiation in mesenchymal stem cells.
Results and Discussion
Egr1−/− mice display inguinal subcutaneous white adipose tissue browning with no external stimulation
The subcutaneous inguinal white adipose tissue (SC-WAT) expands during the post-natal period33 and is the largest white fat depot in mice10,34. Moreover, mouse SC-WAT is comparable in terms of location to the large gluteofemoral subcutaneous depot in humans that is linked to increased risk of developing obesity-related morbidities and mortality35. The size and weight of SC-WAT fat pads were similar in Egr1 +/+ and Egr1 −/− 4-month-old mice, although the total body weight was slightly reduced in Egr1 −/− mice compared to control mice (Fig. 1A–C). These observations suggest that Egr1 loss-of-function leads to a reduced body weight via an increased numbers of active beige adipocytes and potentially through unknown global metabolic processes. Egr1 expression in SC-WAT was detected in blood vessels (Fig. 1D, arrow a) as previously described36 and in adipocytes (Fig. 1D, arrows b,c).
Figure 1.
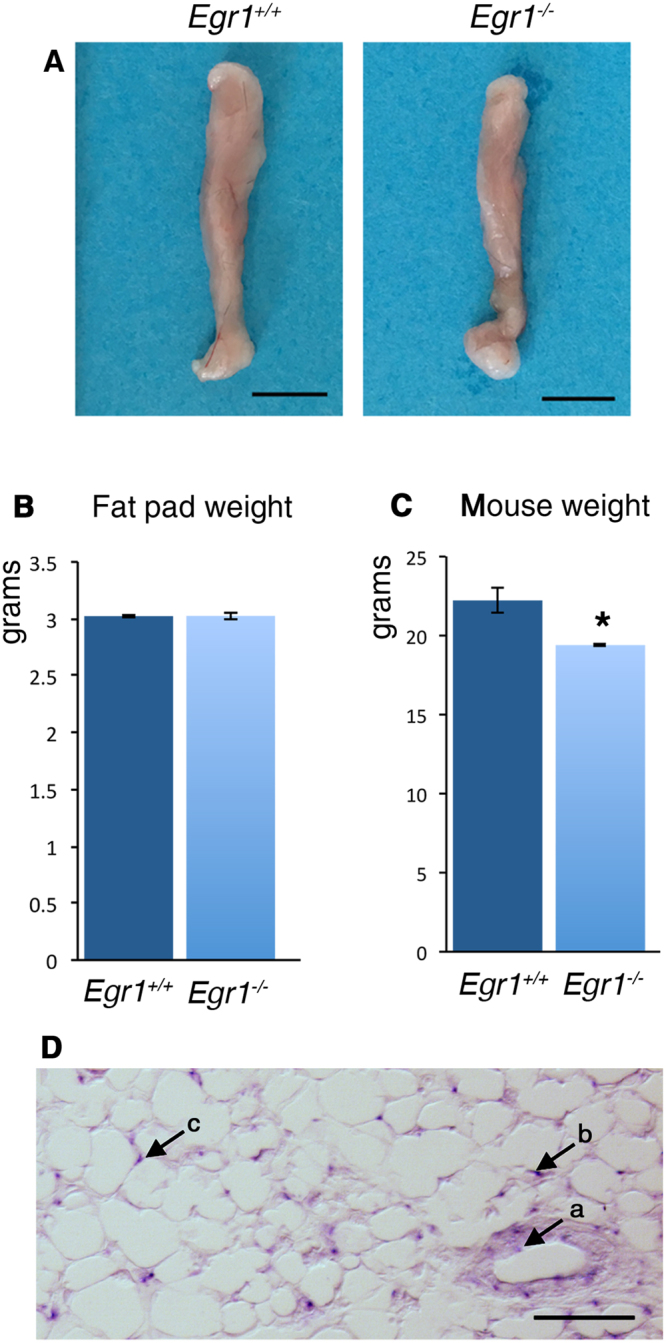
Phenotype of inguinal subcutaneous white adipose tissue in 4-month-old Egr1 −/− mice. (A) Pictures of fat pads (SC-WAT) from 4-month-old Egr1 +/+ and Egr1 −/− mice. Scale bars: 5 mm. (B) Weight in grams of SC-WAT of 4-month-old Egr1 +/+ and Egr1 −/− mice. The graph shows mean ± standard deviations of 6 Egr1 +/+ fat pads and 8 Egr1 −/− fat pads. (C) Weight in grams of 4-month-old wild-type and mutant mice. The graph shows means ± standard deviations of 4 Egr1 +/+ and 4 Egr1 −/− mice. The p-value was obtained using the Mann-Whitney test. Asterisk indicates the p-value *P < 0.05. (D) SC-WAT of 1-month-old mice was longitudinally sectioned. 6 µm sections were hybridized with the DIG-labeled antisense probe for Egr1 (blue). Arrow a points Egr1 expression in blood vessels. Arrows b and c indicate Egr1 expression in white adipocytes. Scale bars: 50 µm.
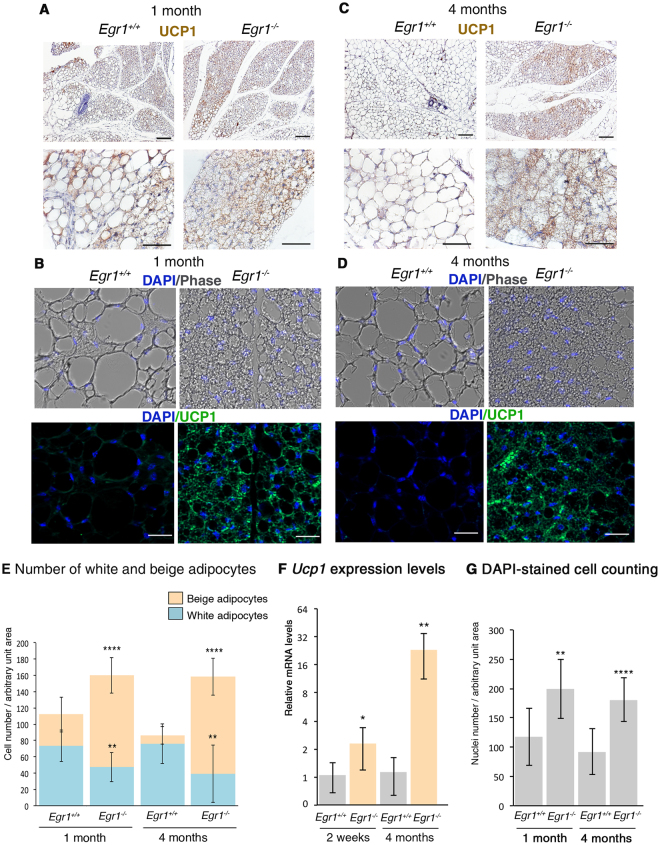
The beige adipocytes in SC-WAT of Egr1 +/+ and Egr1 −/− mice were identified by the expression of UCP1 detected both by immunodetection with DAB staining (Fig. 2A,C) or fluorescent staining (Fig. 2B,D) and by the multilocular aspect of the lipid droplets observed in phase contrast images. The white adipocytes were identified by a unilocular lipid droplet observed in phase contrast images (Fig. 2A–D). SC-WAT from 1-month-old (post-natal) and 4-month-old (adult) Egr1 −/− mice exhibited increased UCP1 staining compared to Egr1 +/+ mice (Fig. 2A–D). At birth and during post-natal period, the inguinal subcutaneous adipose tissue of wild-type mice contains beige adipocytes that are out-numbered by the development of increasing numbers of white adipocytes37 (Fig. 2E). In contrast, we observed a significant increase of the proportion of beige adipocytes and a reduction in the proportion of white adipocytes in SC-WAT of Egr1 −/− mice compared to Egr1 +/+ mice (Fig. 2E). Consistently, the Ucp1 mRNA expression levels were increased in Egr1-deficient SC-WATs compared to equivalent control SC-WATs (Fig. 2F). SC-WATs of Egr1 −/− mice exhibited a higher number of cells compared to Egr1 +/+ mice at 1 and 4 months of age (Fig. 2G), suggesting that EGR1 repressed adipocyte proliferation. However, we did not observe any difference in cell proliferation rates between 1-month-old and 4-month-old Egr1 −/− mice compared to respective controls (see Supplementary Fig. 1). These observations suggest that EGR1 affects cell proliferation of adipocyte progenitors at early stages of adipose tissue development, before 1 month of age. Contrary to our observations, EGR1 has recently been identified to promote self-renewal of adipose progenitors of chin and knee in physio-pathological conditions38. These opposite results highlight the heterogeneity of adipocyte progenitors and adipocytes depending on the fat pad location. Heterozygous Egr1 +/− mice exhibited an intermediate browning phenotype compared to Egr1 +/+ and Egr1 −/− 4-month-old mice (see Supplementary Fig. 2A). We observed an increase in the number of adipocytes and a higher proportion of beige adipocytes in SC-WAT of Egr1 +/− mice compared to Egr1 +/+ mice (see Supplementary Fig. 2B–C). However, the increase was smaller compared to Egr1 −/− mice (Fig. 1E,G).
Figure 2.
Egr1 leads to inguinal subcutaneous white adipose tissue browning in postnatal and 4 month-old mice. (A–D) Sections of SC-WAT of 1-month-old (A,B) and 4-month-old (C,D) Egr1 +/+ and Egr1 −/− mice were immuno-stained with UCP1 antibody. Nuclei were visualized with hematoxilin (A,C) or DAPI (B,D). (B,D) upper and lower panels are Dapi/Phase and UCP1/DAPI views of the same field. Scale bars: (A,C) lower magnification 100 µm, higher magnification 50 µm; (B,D) 25 µm. (E) White and beige adipocyte number was counted in arbitrary unit areas of transverse sections of SC-WAT of 1-month-old Egr1+/+ (N = 10) and Egr1 −/− (N = 11) mice and 4-month-old Egr1 +/+ (N = 13) and Egr1 −/− (N = 14) mice. Graphs show means of counts generated from 10 to 14 sections for each sample ± standard deviations. Asterisks indicate the p-values obtained using the Mann-Whitney test, comparing beige or white adipocyte number between mutant and control mice **P < 0.01, ****P < 0.0001. (F) RT-qPCR analysis of expression levels for beige adipocyte differentiation marker Ucp1 in SC-WAT of 2-week-old and 4-month-old Egr1 −/− mice compared to Egr1 +/+ mice. Graphs show means ± standard deviations of 5 samples from 2-week-old Egr1 +/+ and Egr1 −/− mice, 6 samples from 4-month-old wild-type mice and 5 samples from 4-month-old Egr1 −/− mice. The Ucp1 mRNA levels of control (Egr1 +/+) SC-WAT were normalized to 1. The relative mRNA levels were calculated using the 2−ΔΔCt method. The p-values were obtained using the Mann-Withney test. Asterisks indicate the p-values *p < 0.05, **p < 0.01. (G) Cell number in SC-WAT in Egr1 +/+ and Egr1 −/− mice. Number of nuclei (DAPI-positive cells) was counted in arbitrary unit areas of transverse sections of SC-WAT of 1 month-old Egr1 +/+ (N = 10) and Egr1 −/− (N = 11) mice and 4 month-old Egr1 +/+ (N = 13) and Egr1 −/− (N = 11) mice. Graphs show means of 10 to 13 sections for each sample ± standard deviations. Asterisks indicate the p-values obtained using the Mann-Whitney test, comparing beige or white adipocyte number between mutant and control mice **P < 0.01, **** P < 0.0001.
We conclude that the increase of Ucp1 transcript levels, of UCP1 protein and in the density of UCP1+ cells in SC-WAT of Egr1 −/− mice (Fig. 2) evidences an increase of WAT browning in Egr1 −/− mice with no external stimulation. This result is consistent with the UCP1 increase in visceral perigonadal white adipose previously observed in Egr1 −/− mice under high fat diet feeding29. We conclude that Egr1 deficiency promotes spontaneous SC-WAT browning without external stimulation. These results indicate that the presence of Egr1 in white adipocytes represses spontaneous WAT browning.
Molecular signature of inguinal subcutaneous white adipose tissue browning downstream of Egr1
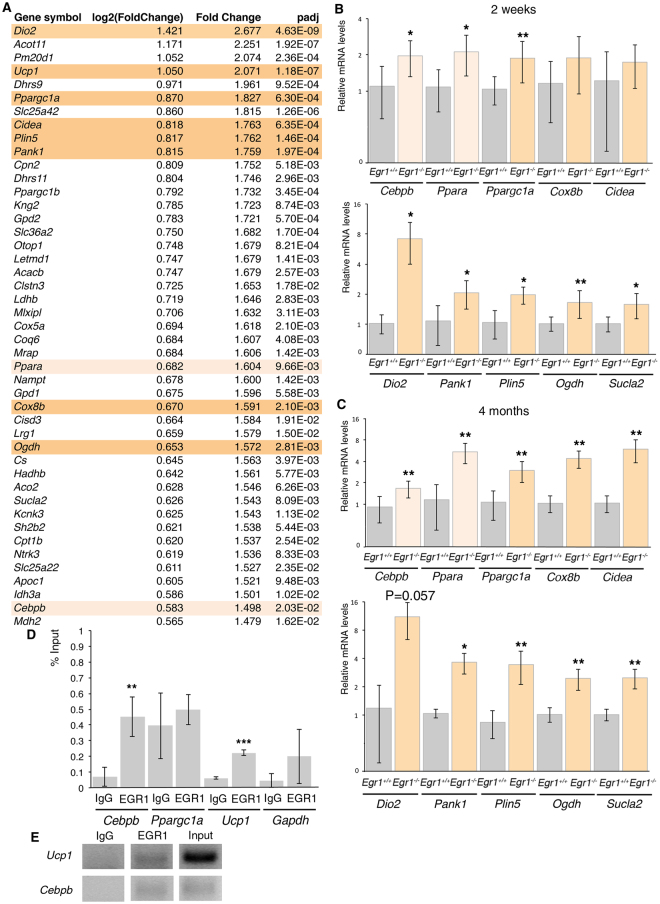
In order to define the molecular signature underlying WAT browning downstream of Egr1, we performed RNA-sequencing of SC-WAT of 2-week-old Egr1 +/+ and Egr1 −/− mice. 336 differentially expressed genes were significantly detected in Egr1-deficient SC-WAT compared to control SC-WAT. The 132 upregulated differentially expressed genes (Fig. 3A, see Supplementary Fig. 3) were subjected to functional annotation clustering according to their Gene Ontology (GO) classification, in the “Biological Process” category (see Supplementary Fig. 4). Among the 132 upregulated genes, the GO terms “NADH metabolic process”, “Tricarboxylic acid cycle”, “Brown fat cell differentiation” and “Fatty acid metabolic process” exhibited the highest enrichment scores (see Supplementary Fig. 4). Consistent with the beige phenotype (Fig. 2), the key beige adipocyte markers, Ppargc1a, Ucp1, Cox8b, Cidea 7 and other genes known to be involved in the beige differentiation program, Dio2, Pank1, Plin5, Ogdh and Sucla2 11,22,39,40 were identified as upregulated genes (Fig. 3A). The increased expression of these beige genes was confirmed by RT-qPCR at 2 weeks and 4 months of age (Fig. 3B,C). In addition, the generic adipogenesis regulators also known to be involved in beige differentiation, Cepbb 41 and Ppara 42 displayed an increased expression in Egr1-deficient SC-WAT (Fig. 3A–C). Interestingly, there was no modification of expression of signalling molecules controlling beige differentiation such as FGF21, BMP4 or Leptin. This indicates that the transcription factor EGR1 negatively regulates the transcription of beige differentiation markers. To test whether this regulation was direct, we performed Chromatin immunoprecipitation (ChIP) experiments from the SC-WAT of 2-week-old mice on key beige markers. EGR1 was recruited to the Ucp1 proximal promoter in SC-WAT (Fig. 3D,E), showing a direct transcriptional regulation by EGR1. EGR1 was also recruited to the Cebpb promoter (Fig. 3D,E) but not to that of Ppapgc1 gene (Fig. 3D), highlighting a direct and an indirect transcriptional regulation of these two genes by EGR1. These results show that EGR1 exerts its transcriptional repression of the beige program at two levels at least, through the direct recruitment of the main beige differentiation marker Ucp1 and also through the direct recruitment to the Cebpb gene, which is known to regulate Ucp1 transcription17.
Figure 3.
Transcriptomic analysis of subcutaneous inguinal adipose tissue of postnatal Egr1 −/− versus Egr1 +/+ mice shows upregulation of beige adipocyte markers. (A) List of the first 45 upregulated genes in 6 inguinal subcutaneous fat pads of 3 Egr1 −/− versus 3 Egr1 +/+ 2-week-old mice. (B,C) RT-qPCR analysis of the expression levels for generic adipocyte differentiation markers Cebpb, Ppara, beige adipocyte differentiation marker, Ppargc1a, Cox8b, Cidea, Dio2, Pank1, Plin5, Ogdh and Sucla2 in SC-WAT of 2-week-old (B) and 4-month-old (C) Egr1 −/− mice compared to Egr1 +/+ mice. For each gene, the mRNA levels of control (Egr1 +/+) SC-WAT were normalized to 1. Graphs show means ± standard deviations of 5 samples from 2-week-old Egr1 +/+ mice and Egr1 −/− mice, 6 samples from 4-month-old Egr1 +/+ mice and 5 samples from Egr1 −/− mice. The relative mRNA levels were calculated using the 2−ΔΔCt method. The p-values were obtained using the Mann-Withney test. Asterisks indicate the p-values *p < 0.05, **p < 0.01. (D) Chromatin Immunoprecipitation (ChIP) assays were performed from 60 fat pads of wild type 2-week-old mice with antibodies against EGR1 or IgG2 as irrelevant antibody in three independent biological experiments. ChIP products were analyzed by RT-q-PCR (N = 2). Primers targeting the proximal promoter regions of Cebpb and Ucp1 revealed the recruitment of EGR1 in the vicinity of these sequences, while primers targeting the proximal promoter regions of Ppargc1a and Gapdh (negative controls) did not show any immunoprecipitation with EGR1 antibody compared to IgG2 antibody. Results were represented as percentage of the input. Error bars showed standard deviations. The p-values were obtained using the Mann-Withney test. Asterisks indicate the p-values, **p < 0.01, ***p < 0.001. (E) ChIP-qPCR samples were loaded on agarose gel and confirmed a specific amplification of Cebpb and Ucp1 promoter regions after chromatin immunoprecipitation using EGR1 antibody. No DNA was immunoprecipitated by irrelevant IgG antibody. Input chromatin was diluted 2 times for Ucp1 qPCR and 4 times for Cebpb qPCR.
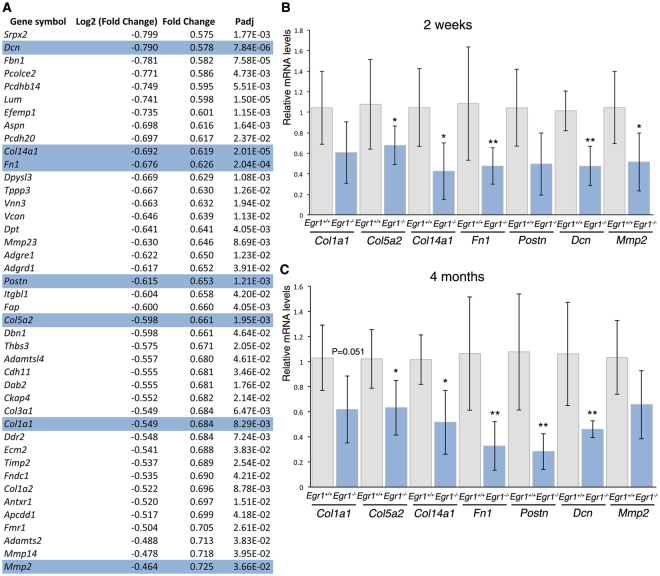
The 204 downregulated differentially expressed genes (Fig. 4A, see Supplementary Fig. 5) in SC-WAT of Egr1 −/− mice were enriched for the GO terms “Collagen fibril organization”, “Collagen catabolic process” and “Extracellular matrix organization” (see Supplementary Fig. 6). WAT produces extracellular matrix (ECM) whose composition and remodelling is crucial for adipocyte function43. Conversely, the expansion of adipose tissue during obesity leads to tissue remodelling and is associated with overexpression of Col1a1, Col5a2, Fn1, Dcn and the matrix metalloprotease Mmp2 genes44–48. In the transcriptome of Egr1-deficient SC-WAT, Col1a1, Col1a2, Col5a2, Col14a1, Fn1, Post, Dcn and Mmp2 were downregulated (Fig. 4A), which was confirmed by RT-qPCR in SC-WAT of 2 week- and 4 month-old mice (Fig. 4B,C). We conclude that Egr1-deficiency represses ECM genes associated with obesity. Our results indicate that WAT browning is associated with alteration of ECM composition. The inverse correlation between WAT browning and ECM is consistent with the suppression of brown adipogenesis in favour of fibrogenesis in mice49.
Figure 4.
Transcriptomic analysis of the subcutaneous inguinal adipose tissue of postnatal Egr1 −/− versus Egr1 +/+ mice reveals downregulation of extracellular matrix genes. (A) List of downregulated extracellular matrix genes in 6 inguinal subcutaneous fat pads of 3 Egr1 −/− versus 3 Egr1 +/+ 2-week-old mice. (B,C) RT-qPCR analysis of gene expression levels for extracellular matrix genes, Col1a1, Col5a2, Col14a1, Fn1, Postn, Dcn and Mmp2, in SC-WAT of 2-week-old (B) and 4-month-old (C) Egr1 +/+ and Egr1 −/− mice. For each gene, the mRNA levels of control (Egr1 +/+) SC-WAT were normalized to 1. Graphs show means ± standard deviations of 5 samples from 2-week-old Egr1 +/+ mice and Egr1 −/− mice, 6 samples from 4-month-old Egr1 +/+ mice and 5 samples from Egr1 −/− mice. The relative mRNA levels were calculated using the 2−ΔΔCt method. The p-values were obtained using the Mann-Withney test. Asterisks indicate the p-values *p < 0.05, **p < 0.01.
The concomitant upregulation of beige differentiation genes and downregulation of ECM genes is a signature of WAT browning downstream of Egr1 deletion without any external stimulation.
Mutual exclusive expression of Fibronectin and UCP1 in inguinal subcutaneous adipose tissue of Egr1−/− mice
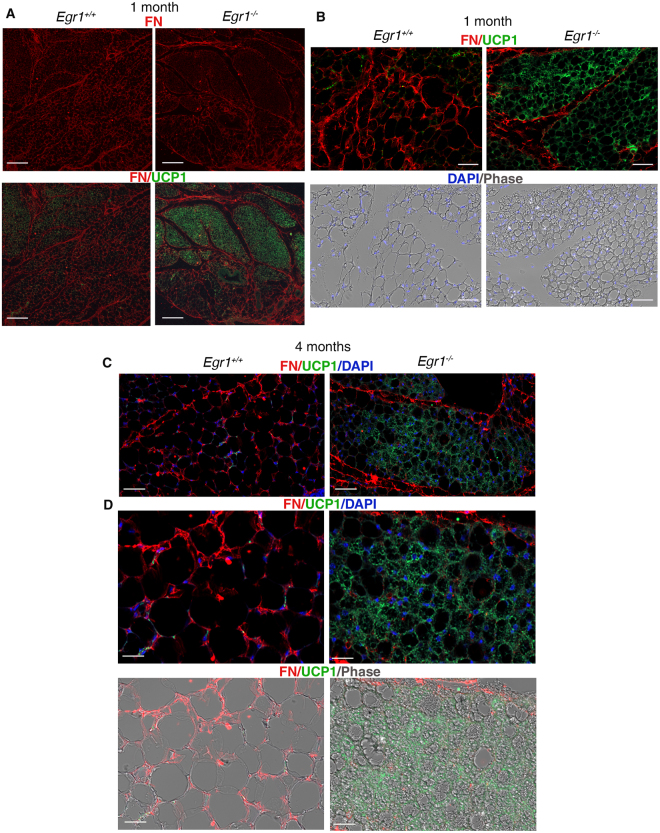
Adipocyte ECM is an important component of white adipogenesis in normal or pathological conditions43,44,48. However, little is known for ECM-adipocyte interaction during beige adipogenesis. The ECM protein Fibronectin (FN) has been linked with obesity and adipose tissue fibrosis50. EGR1 has been previously shown to directly regulate Fn1 transcription in human glioblastoma cells51. We analysed FN expression in SC-WAT in Egr1 deletion conditions (Fig. 5). Although immunohistochemistry on tissue section is not a quantitative technique, we observed a global decrease of FN expression in SC-WAT, concomitant with an increase of UCP1 expression in 1 month Egr1 −/− compared to Egr1 +/+ mice (Fig. 5A). The decrease of FN protein (Fig. 5A) was fully consistent with the down-regulation of Fn1 mRNA levels (Fig. 4B,C) in Egr1-deficient SC-WAT. FN expression was not observed around the UCP1 + beige adipocytes in SC-WAT of 1-month-old Egr1 −/− mice, while FN was produced by white adipocytes in 1-month-old Egr1 +/+ mice (Fig. 5B). It has to be noticed that FN was observed surrounding the beige adipocyte areas (Fig. 5B). Similar results were found at 4 months of age, where FN was not detected around UCP1+ beige adipocytes, while being expressed around white adipocytes in 4-month-old Egr1 +/+ mice and Egr1 +/+ mice, respectively (Fig. 5D). We conclude that WAT browning in Egr1 −/− mice is associated with an absence of FN expression by UCP1+ beige adipocytes. This result highlights an inverse correlation between the browning process and the expression of the ECM protein FN.
Figure 5.
Fibronectin localization in SC-WATs of Egr1 +/+ and Egr1 −/− mice. SC-WATs of 1-month-old (A,B) and 4-months-old (C,D) Egr1 +/+ and Egr1 −/− mice were sectioned transversely and immuno-stained with Fibronectin FN (red) and UCP1 (green) antibodies. Nuclei were visualized with DAPI (blue). Individual channel or merged channels are indicated in panels. (A) Low magnifications show that FN (red) is less expressed in UCP1-positive areas (green) compared to UCP1-negative areas in 1-month-old Egr1 +/+ and Egr1 −/− mice. Scale bars, 200 µm. (B) High magnifications show that FN is absent around UCP1 + beige adipoctytes in 1-month-old Egr1 −/− compared to Egr1 +/+ mice, while being present around white adipoctyes in Egr1 +/+ mice. Scale bars, 50 µm. (C,D) At 4 months of age, FN is also absent around UCP1 + beige adipoctyes of SC-WATs from Egr1 −/−, while being present around white adipocytes in Egr1 +/+ mice. Scale bars, (C) 50 µm (D) 25 µm. (B–D) FN surrounds the regions of beige adipocytes.
Forced Egr1 expression in mouse mesenchymal stem cells reduces beige marker expression and promotes extracellular matrix gene expression
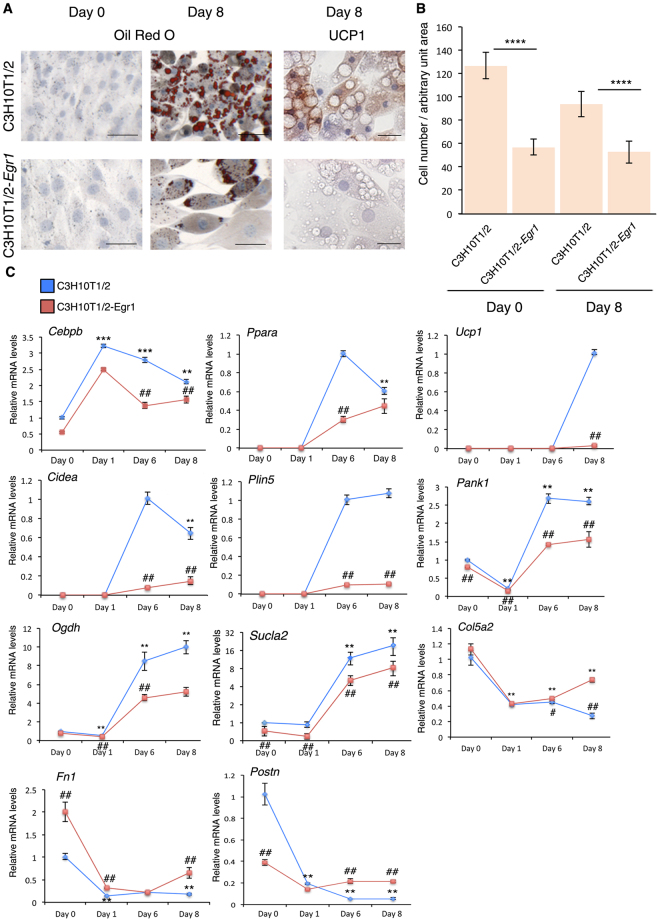
The spontaneous WAT browning in Egr1 −/− mice and the direct transcriptional regulation of Ucp1 gene by EGR1 in SC-WAT suggested that EGR1 repressed beige adipocyte differentiation. EGR1 gain-of-function experiments were performed in mouse mesenchymal stem cells, C3H10T1/2 cells, cultured under beige adipocyte differentiation conditions. Consistent with the increase in the number of adipocytes in SC-WAT of Egr1 −/− (Fig. 2G), we observed a decreased number of C3H10T1/2-Egr1 cells compared to C3H10T1/2 cells at day 0 and after 8 days of culture in the beige differentiation medium (Fig. 6A,B). Under beige stimulation, C3H10T1/2 cells acquired a beige phenotype, visualized by the appearance of numerous small lipid droplets and UCP1 expression within their cytoplasm (Fig. 6A). In contrast, C3H10T1/2-Egr1 cells did not express UCP1 under beige stimulation, showing that EGR1 repressed the expression of the key thermogenic beige marker (Fig. 6A). Consistent with the absence of UCP1 protein (Fig. 6A), Ucp1 mRNA levels were never increased in the context of Egr1 overexpression (Fig. 6C). This observation fits with EGR1 recruitment to Ucp1 promoter observed in SC-WAT (Fig. 3D,E). However, small lipid droplets were still observed in C3H10T1/2-Egr1 cells, indicating that EGR1 repressed part of the beige phenotype through the repression of UCP1, but did not fully abolish the formation of lipid droplets (Fig. 6A). The expression of Cebpb and Ppara genes was significantly reduced in C3H10T1/2-Egr1 cells compared to control cells as that of Cidea, Plin5, Pank1, Ogdh and Sucla2 genes (Fig. 6C). This showed that beige differentiation and the heat-producing ability of C3H10T1/2 cells were impaired upon Egr1 overexpression. Egr1 overexpression also blocked white adipocyte differentiation in C3H10T1/2 cells (see Supplementary Fig. 7), as previously observed32. The inhibition of both beige and white differentiation programs by EGR1 is to be related with the direct (Cebpb) and indirect transcriptional regulation of generic adipogenesis genes by EGR1 (Fig. 3B–D).
Figure 6.
Egr1 gain-of-function decreases beige adipocyte differentiation in mouse mesenchymal stem cells. (A) C3H10T1/2 and C3H10T1/2-Egr1 cells subjected to beige adipocyte differentiation for 8 days were then stained with Oil Red O and Hematoxilin/Eosin at Day 0 (confluence) and Day 8, or immuno-stained with UCP1 antibody and counterstained with Hematoxilin/Eosin at Day 8. Scale bars: Oil red O staining 50 µm, UCP1 immunostaining 25 µm. (B) C3H10T1/2 and C3H10T1/2-Egr1 cell density at day 0 and after 8 days in beige differentiation medium. Graphs show means ± standard deviations of cell number from 10 pictures in each condition. The p-values were obtained using the Mann-Whitney test. Asterisks indicate the p-value ****P < 0.0001. (C) RT-qPCR analysis of the expression levels for the adipocyte transcriptional activators Cebpb and Ppara, the beige markers, Ucp1, Cidea, Plin5, Pank1, Ogdh, Sucla2 and the extracellular components Col5a2, Fn1 and Postn in C3H10T1/2 and C3H10T1/2-Egr1 cells subjected to beige adipocyte differentiation. For each gene, the mRNA levels of the control C3H10T1/2 cells at Day 0 or from the first day of detection were normalised to 1. Cebpb, Pank1, Ogdh, Sucla2, Col5a2, Fn1 and Postn expression was detected from Day 0, Ppara, Cidea and Plin5 expression was detected from Day 6, Ucp1 expression was detected at day 8. The graphs show the relative levels of mRNAs in C3H10T1/2 and C3H10T1/2-Egr1 cells at different time points (Day 0, Day 1, Day 6, and Day 8) of beige adipocyte differentiation compared to C3H10T1/2 cells at Day 0 or to C3H10T1/2 cells from the first day of gene detection. For each time point, graphs show means ± standard deviations of 6 samples. The p values were calculated using the Mann-Withney test. The relative mRNA levels were calculated using the 2^−ΔΔCt method. Asterisks indicate the p-values of gene expression levels in C3H10T1/2-Egr1 cells or C3H10T1/2 cells compared to Day 0 (Ogdh and Col5a2) or from the first day of gene detection (Ppara, Cidea, Ucp1 and Plin5), **P < 0.01. #Indicate the p-values of gene expression levels in C3H10T1/2-Egr1 versus C3H10T1/2 cells, for each time point, #P < 0.05, ##P < 0.01.
In order to assess whether EGR1 promotes the expression of ECM genes in mesenchymal stem cells in the context of adipocyte differentiation, we analysed the expression of Col5a2, Fn1 and Postn in C3H10T1/2 and C3H10T1/2-Egr1 cells during beige (Fig. 6D) and white (see Supplementary Fig. 7) adipocyte differentiation. The expression of Col5a2, Fn1 and Postn genes was upregulated in Egr1 overexpressing cells, showing that EGR1 activated the expression of ECM genes during adipocyte differentiation. The positive regulation of ECM genes by EGR1 during adipocyte differentiation was consistent with similar regulation in the context of fibrosis, atherosclerosis and tendon repair32,52. We conclude that forced EGR1 expression in mouse mesenchymal stem cells reduces beige marker expression, while promoting ECM gene expression.
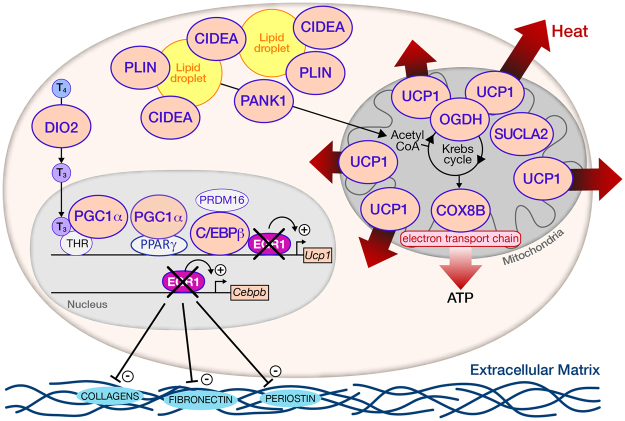
In summary, the deletion of Egr1 induces WAT browning by the release of the EGR1-mediated repression of the Cebpb and Ucp1 promoters (Fig. 7A,B). Egr1 loss-of-function causes the overexpression of the beige adipocyte differentiation genes Cebpb and Ppargc1, which both activate the expression of the thermogenic marker Ucp1 through a recruitment to its promoter18,53 (Fig. 7B). In addition, the beige adipocytes metabolic genes Dio2, Cidea, Plin, Pank1, Cox8b, Ogdh and Sucla2a are also upregulated (Fig. 7B) to induce the browning of SC-WAT in Egr1 −/− mice, without any cold stimulation or fasting. The expression of ECM genes is reduced in the context of Egr1 loss-of-function (Fig. 7B). Reciprocally, Egr1 gain-of-function represses the expression of Cebpb and Ucp1 presumably through the recruitment of EGR1 to their promoters (Fig. 7C). In addition, the beige adipocyte metabolic genes Dio2, Cidea, Plin, Pank1, Cox8b, Ogdh and Sucla2a are also downregulated (Fig. 7C), which prevents the differentiation of mesenchymal stem cells into beige adipocytes. The expression of ECM genes is enhanced in the context of Egr1 gain-of-function (Fig. 7C).
Figure 7.
EGR1 regulates beige adipocytes differentiation, metabolism and extracellular matrix formation. Egr1 loss-of-function upregulates the expression of genes encoding C/EBPß, PGC1α, UCP1, COX8B, SUCLA2, OGDH, CIDEA, PLIN5, PANK1 and DIO2, leading to a significant browning of SC-WAT in Egr1 −/− mice. Egr1 deletion downregulates the expression of genes encoding the ECM proteins Collagens, Fibronectin and Perisotin. Our study confirms the opposite correlation between adipose tissue browning and fibrogenesis.
The upregulated expression profile of beige differentiation markers and downregulated profile of ECM genes in Egr1-deficient WAT define a molecular signature of beige adipocyte differentiation program and constitute a protective signature against white adipocyte lipid accumulation. This study identifies Egr1 deficiency as a therapeutic approach to counteract obesity.
Methods
All experimental procedures using mice were conducted in accordance with the European guidelines (2010/63/UE) and were approved by the French National Ethic Committee for animal experimentation N°05 and are registered under the number 01789.02.
Mouse lines
The Egr1 gene was inactivated by homologous recombination with insertion of the LacZ coding sequence within the Egr1 5′ untranslated region in addition to a frameshift mutation upstream of the DNA-binding domain of Egr1 54. The line was maintained on a C57BL/6 J background (Janvier, France). All animals were kept under controlled photo-period (lights on 08:00–20:00 hours) and a diet of commercial rodent chow and tap water ad libitum.
Age-matched groups of 2-week-old, 1-month-old and 4-month-old Egr1 +/+ and Egr1 −/− female mice derived from heterozygous intercrosses of Egr1 +/− were used for the RNA-sequencing, RT-qPCR and immunostaining experiments.
For the RNA-sequencing experiments, we used 3 Egr1 +/+ and 3 Egr1 −/− female mice from 4 different litters. Among these animals, 1 Egr1 +/+ and 1 Egr1 −/− mice were littermates.
For the RT-qPCR experiments, we used 5 Egr1 +/+ and 5 Egr1 −/− 2-week-old female mice from 7 different litters. Among them, 1 Egr1 +/+ and 1 Egr1 −/− mice were littermates. We used 6 Egr1 +/+ and 5 Egr1 −/− 4-month-old female mice from 8 different litters.
For the immunostainings experiments, we used 3 Egr1 +/+ and 2 Egr1 −/− 1-month-old female mice. Among them, 2 Egr1 +/+ and 2 Egr1 −/− were littermates. We used 3 Egr1 +/+, 3 Egr1 −/− and 2 Egr1 +/− 4-month-old female mice. Among these mice, 1 Egr1 +/+, 1 Egr1 −/− and 1 Egr1 +/− were littermates.
In situ hybridization to adipose tissue sections
Inguinal subcutaneous fat pads were isolated from 1-month-old female mice, fixed in 4% paraformaldehyde overnight and processed for in situ hybridization to 6 mm wax tissue sections as previously described55. The digoxigenin-labeled mouse Egr1 probe was used as previously described54.
RNA isolation, sequencing and transcriptomic analysis
Fresh inguinal subcutaneous fat pads were removed from 2-week-old euthanized Egr1 +/+ (N = 3) and Egr1 −/− (N = 3) female mice and homogenized using a mechanical disruption device (Lysing Matrix A, Fast Prep MP1, 4 × 30 s, 6 m.s−1). Total RNA was isolated using the RNeasy mini kit (Qiagen) with 15 min of DNase I (Qiagen) treatment according to the manufacturer’s protocol. Preparation of cDNA libraries and sequencing was performed at the “Ecole Normale Supérieure” Genomic Platform (Paris, France). Ribosomal RNA depletion was performed with the Ribo-Zero kit (Epicentre), using 500 ng of total RNA. Libraries were prepared using the strand specific RNA-Seq library preparation ScriptSeq V2 kit (Epicentre). 51-bp paired-end reads were generated using a HiSeq. 1500 device (Illumina). A mean of 56.9 ± 6.3 million reads passing the Illumina quality filter were obtained for each of the 6 samples. Reads were mapped against the mus musculus reference genome (UCSC Dec. 2011, GRCm38/mm10) using TopHat v2.1.056, Bowtie (v2.2.5)57, and the Release M8 (GRCm38.p4) GTF annotations as a guide. Read counts were assigned to gene features using Feature Counts v1.4.6.p558 and differential expression analysis was performed with DESeq. 2 v1.6.359. Full details of the Galaxy workflow used in this study can be retrieved via the following link: https://mississippi.snv.jussieu.fr/u/emmanuellehavis/w/copy-of-grasostendon-differential-expression-2. Gene Ontology analysis on differentially expressed genes (Padj < 0.05) was performed with DAVID Bioinformatic Resources 6.860. Sequencing data was uploaded to the Gene Expression Omnibus (GEO) database under the accession number GSE91058.
RNA isolation, Reverse-Transcription and quantitative real time PCR
Fresh inguinal subcutaneous fat pads were removed from 2-week-old and 4-month-old euthanized Egr1 +/+ and Egr1 −/− female mice and homogenized using a mechanical disruption device (Lysing Matrix A, Fast Prep MP1, 4 × 30 s, 6 m.s−1). Total RNA was isolated using the RNeasy mini kit (Qiagen) with 15 min of DNase I (Qiagen) treatment according to the manufacturer’s protocol.
For RT-qPCR analyses, 500 ng RNA was Reverse-Transcribed using the High Capacity Retrotranscription kit (Applied Biosystems). Quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) using primers listed in Supplementary Table 1. We used Actb as housekeeping gene for the analysis of the SC-WAT from 2-week-old mice and Rplp0 for the analysis of the SC-WAT from 4-month-old mice and C3H10T1/2 and C3H10T1/2-Egr1 cells. The relative mRNA levels were calculated using the 2−ΔΔCt method61. The ΔCt values were obtained by calculating the differences: Ct(gene of interest) – Ct(housekeeping gene) in each sample. We obtained the ΔΔCt values by calculating the differences between ΔCt(Experiment) and the average of ΔCt(control) values. For mRNA level analysis in SC-WAT, Egr1 −/− values were considered as experimental and Egr1 +/+ as controls. The controls were normalized to 1.
5 to 6 independent RNA samples of 2-week-old and 4-month-old Egr1 +/+ and Egr1 −/− female mice were analysed in duplicate. For mRNA level analysis in cell cultures, C3H10T1/2-Egr1 values were considered as experimental and C3H10T1/2 as controls. 6 independent RNA samples were analysed in duplicate for each time point.
Chromatin Immunoprecipitation
ChIP assays were performed with previously reported protocol62 on the inguinal subcutaneous adipose tissue isolated from 60 2-week-old mice, homogenized using a mechanical disruption device (Lysing Matrix A, Fast Prep MP1, 3 × 30 sec). 8 µg of the rabbit polyclonal anti-Egr-1/Krox24 (C-19) antibody (Santa Cruz Biotechnology) or 8 µg of the goat anti-mouse IgG2b (Southern biotechnology) were used to immunoprecipitate 30 µg of sonicated chromatin. ChIP products were analyzed by quantitative PCR. 15 µg of chromatin was isolated before chromatin immunoprecipitation, to be used as positive control for the PCR experiments (Input). ChIP products and Inputs were analyzed by quantitative PCR to amplify the promoter regions upstream the Cebpb (−660 bp; −530 bp), Ppargc1a (−860 bp; −730 bp), Ucp1 (−170 bp; + 20 bp) and Gapdh (−2,9 Kb; −2,7 Kb; negative control) coding sequences. qPCR amplicons were loaded on a 1% agarose gel. The primer list is displayed in Supplementary Table 1.
Immunohistochemistry
Fresh inguinal subcutaneous fat pads were removed from 1-month-old and 4-month-old euthanized Egr1 +/+ and Egr1 −/− female mice, fixed in 4% formaldehyde overnight at 4 °C and processed for immunohistochemistry on 12 µm wax tissue sections, as previously described63. After wax removal, for UCP1 immunodetection, heat-induced epitope retrieval was performed by incubating sections 5 min at 95 °C in Glycine-HCl buffer (0.05 M Glycine, pH3.5). UCP1 protein was detected using rabbit polyclonal antibody (1:200, ab10983, Abcam), followed by secondary anti-rabbit HRP conjugate antibody (1:200, 170-6515, Biorad) and DiaminoBenzidine Tetra-Hydrochloride protocol (DAB) staining or followed by secondary anti-rabbit fluorescent antibody (1:200, Goat anti-Rabbit Alexa 488, A11008, Invitrogen) staining.
For fibronectin immunodetection, heat-induced epitope retrieval was performed by incubating sections 7 min at 95 °C in citrate buffer (10 mM, pH 6). Fibronectin protein was detected using mouse monoclonal antibody (1:200, F7387, Sigma), followed by secondary anti-mouse fluorescent antibody (1:200, Goat anti-Mouse Alexa 555, A21422, Invitrogen) staining.
Nuclei were visualised either by Hematoxylin & Eosin (H&E) histological staining using a standard protocol or by DAPI staining according to manufacturer’s instructions (DAPI, D9542, Sigma).
C3H10T1/2 and C3H10T1/2-Egr1 cells were cultured in beige or white adipocyte differentiation medium for 8 and 10 days, respectively, on cover slips. Cells were fixed with 4% Paraformaldehyde (Sigma) for 15 min. UCP1 protein was detected using rabbit polyclonal antibody (1:200, ab10983, Abcam), followed by secondary anti-rabbit HRP conjugate antibody (1:200, 170-6515, Biorad) and DiaminoBenzidine Tetra-Hydrochloride protocol (DAB) staining. Hematoxylin & Eosin (H & E) histological staining was performed using a standard protocol.
Cell number measurements
All cell number measurements were performed using the free software ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2012).
To quantify the number of white and beige adipocytes in SC-WAT of Egr1 +/++/+ and Egr1 −/− mice, cell number was counted per arbitrary unit area on sections originating from 1-month-old and 4-month-old female mice. The beige adipocytes were identified by the expression of UCP1 detected by immunodetection with DAB staining or fluorescent staining and by the multilocular aspect of the lipid droplets combined with positive Hematoxylin or DAPI nucleus staining. The white adipocytes were identified both by a unilocular lipid droplet and positive DAPI staining.
The number of beige and white cells in 1-month-old mice was counted from 10 sections originating from 6 fat pads of 2 Egr1 +/+ mice and from 11 sections originating from 4 fat pads of 2 Egr1 −/− mice. The number of beige and white cells in 4-month-old mice was counted from 13 sections originating from 6 fat pads of 3 Egr1 +/+ mice, from 14 sections originating from 6 fat pads of 3 Egr1 −/− mice and from 8 sections originating from 4 fat pads of 2 Egr1 +/− mice.
To quantify the total number of cells in SC-WAT of Egr1 +/+ and Egr1 −/− mice, the number of DAPI + cells was counted per arbitrary unit area on sections originating from 1-month-old and 4-month-old female mice. The number of DAPI + cells in 1-month-old mice was counted from 9 sections originating from 6 fat pads of 3 Egr1 +/+ mice and from 11 sections originating from 4 fat pads of 2 Egr1 −/− mice. The number of DAPI + cells in 4-month-old mice was counted from 11 sections originating from 6 fat pads of 3 Egr1 +/+ mice, from 12 sections originating from 6 fat pads of 3 Egr1 −/− mice and from 8 sections originating from 4 fat pads of 2 Egr1 +/− mice.
To quantify the number of C3H10T1/2 and C3H10T1/2-Egr1 cells at Day 0 and Day 8 of beige adipocyte differentiation conditions the number of Hematoxylin-positive cells was counted per unit area from 10 wells for each condition.
Cell cultures
Mouse mesenchymal stem cells, C3H10T1/264 and the stable Egr1 overexpressing counterparts, C3H10T1/2-Egr132 cells, were plated to 6-well plates at a density of 330,000 cells/well and grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen) supplemented with 10% foetal bovine serum (FBS, Sigma), 1% penicillin-streptomycin (Sigma), 1% Glutamin (Sigma), 800 μg/ml G418 Geneticin (Sigma) and incubated at 37 °C in humidified atmosphere with 5% CO2.
Confluent cells were cultured in beige differentiation induction medium for 2 days and in beige maturation medium for 6 days according to published protocols65. Day 0 corresponds to the addition of beige differentiation induction medium on confluent cells. Beige differentiation induction medium includes DMEM, 10% FBS, 1% penicillin-streptomycin, 10 µg/mL Insulin (Sigma), 0.25 µM Dexamethasone (Sigma), 0.5 mM 3-Isobutyl-1-methylxanthine (IBMX, Sigma), 50 nM 3.3′,5-Triiodo-L-thyronine sodium salt (T3, Sigma), 20 µM Curcumin (Sigma). The beige maturation medium comprises DMEM, 10% FBS, 1% penicillin-streptomycin, 10 µg/mL Insulin (Sigma), 50 nM 3,3′,5-Triiodo-L-thyronine sodium salt (T3, Sigma), 20 µM Curcumin (Sigma), 1 µM Rosiglitazone (Sigma). The maturation medium was changed every 2 days. Cells subjected to beige adipocyte differentiation medium were fixed for histological analysis or lysed for gene expression analysis at Day 0, Day 1, Day 6 and Day 8.
Confluent cells were cultured in white differentiation induction medium for 2 days and in white maturation medium for 8 days. Day 0 corresponds to the addition of white differentiation medium. White differentiation induction medium includes DMEM, 10% FBS, 1% penicillin-streptomycin, 10 µg/mL Insulin (Sigma), 0.25 µM Dexamethasone (Sigma), 0.5 mM 3-Isobutyl-1-methylxanthine (IBMX, Sigma), 30 nM 3.3′,5-Triiodo-L-thyronine sodium salt (T3, Sigma). The white maturation medium comprises DMEM, 10% FBS, 1% penicillin-streptomycin and 10 µg/mL Insulin (Sigma). The maturation medium was changed every 2 days. Cells subjected to white adipocyte differentiation medium were stopped at Day 0, Day 1, Day 4 and Day 10 for histological and gene expression analysis by RT-qPCR.
Oil Red O staining
C3H10T1/2 and C3H10T1/2-Egr1 cells were cultured in beige or white adipocyte differentiation medium for 8 and 10 days, respectively. Cells were fixed with 4% Paraformaldehyde (Sigma) for 15 min and washed twice with excess distilled H2O (Millipore). 60% Isopropanol was added for 5 min and replaced with an Oil Red O (Sigma) staining mixture, consisting of Oil Red O solution (0.5% Oil Red O dye in Isopropanol) and water in a 6:4 ratio, for 15 min. Cells were rinsed three times in distilled H2O, followed by a standard Hematoxylin & Eosin staining protocol.
Statistical analyses
Data was analysed using the non-parametric Mann-Withney test or ANOVA test with Graphpad Prism V6. Results are shown as means ± standard deviations. The p-values are indicated either with the value or with * or #.
Electronic supplementary material
Acknowledgements
We thank Kacey Marra, Peter Rubin and Erin Kershaw from the University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania, United States for comments on the manuscript and their expertise in adipose tissue biology. We thank Estelle Hirsinger from IBPS, Paris, France for comments on the manuscript. We thank Marie-Ange Bonnin from IBPS, Paris, France for technical support. We thank Sophie Lemoine and Stéphane Le Crom, from IBENS, Paris, France and Christophe Antoniewsky from ARTbio Bioinformatics Analysis Facility, Paris, France, for the bioinformatics analyses of the RNA-sequencing. We thank Sophie Gournet for illustrations. This work was supported by the Fondation pour la Recherche Médicale (FRM) DEQ. 20140329500 and FDT20150532272, Institut national de la santé et de la recherche Médicale (Inserm), Centre National de la Recherche Scientifique (CNRS), Université Pierre et Marie Curie (UPMC), Sorbonne Universités Emergence (SU-16-R-EMR-33) and the Agence Nationale de la Recherche (contracts ANR-10-BLAN-1219, ANR- 12-BSV1-0038). The École normale supérieure genomic platform was supported by the France Génomique national infrastructure, funded as part of the “Investissements d’Avenir” program managed by the Agence Nationale de la Recherche (contract ANR-10-INBS-09).
Author Contributions
C.M., acquisition, analysis and interpretation of data. M.B., acquisition, analysis and interpretation of data. K.A., contributed to unpublished essential data, analysis and interpretation of histology data, drafting the article. M.O., analysis and interpretation of bioinformatics data. F.C., acquisition of RNA-sequencing data. D.D., conception, design, analysis and interpretation of data, drafting the article, funding. E.H., conception, design, analysis and interpretation of data, drafting the article, funding.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Cécile Milet and Marianne Bléher contributed equally to this work.
Delphine Duprez and Emmanuelle Havis jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16543-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Delphine Duprez, Email: delphine.duprez@upmc.fr.
Emmanuelle Havis, Email: emmanuelle.havis@upmc.fr.
References
- 1.Berry DC, Stenesen D, Zeve D, Graff JM. The developmental origins of adipose tissue. Development. 2013;140:3939–49. doi: 10.1242/dev.080549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014;10:24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 3.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 2013;19:1252–63. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 4.Kajimura S, Spiegelman BM, Seale P. Brown and beige fat: Physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat. Cell Biol. 2013;15:659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 6.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia RA, Roemmich JN, Claycombe KJ. Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr. Metab. (Lond). 2016;13:24. doi: 10.1186/s12986-016-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peirce V, Carobbio S, Vidal-Puig A. The different shades of fat. Nature. 2014;510:76–83. doi: 10.1038/nature13477. [DOI] [PubMed] [Google Scholar]
- 9.Klaus S, Casteilla L, Bouillaud F. RD. The uncoupling protein UCP: a membraneous mitochondrial ion carrier exclusively expressed in brown adipose tissue. Int J Biochem. 1991;23:1773883. doi: 10.1016/0020-711X(91)90062-R. [DOI] [PubMed] [Google Scholar]
- 10.Shabalina I, et al. UCP1 in Brite/Beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013;5:1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 11.Forner F, et al. Proteome Differences between Brown and White Fat Mitochondria Reveal Specialized Metabolic Functions. Cell Metab. 2009;10:324–335. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallardo-Montejano VI, et al. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat. Commun. 2016;7:12723. doi: 10.1038/ncomms12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puigserver, P. et al. A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. 92, 829–839 (1998). [DOI] [PubMed]
- 15.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seale, P. et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. 121, 53–56 (2011). [DOI] [PMC free article] [PubMed]
- 17.Yubero P, et al. Dominant negative regulation by c-Jun of transcription of the uncoupling protein-1 gene through a proximal cAMP-regulatory element: a mechanism for repressing basal and norepinephrine-induced expression of the gene before brown adipocyte differentiation. Mol. Endocrinol. 1998;12:1023–37. doi: 10.1210/mend.12.7.0137. [DOI] [PubMed] [Google Scholar]
- 18.Villarroya F, Peyrou M, Giralt M. Transcriptional regulation of the uncoupling protein-1 gene. Biochimie. 2017;134:86–92. doi: 10.1016/j.biochi.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Carmona MC, et al. Defective thermoregulation, impaired lipid metabolism, but preserved adrenergic induction of gene expression in brown fat of mice lacking C/EBPβ. Biochem. J. 2005;389:47–56. doi: 10.1042/BJ20050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SH, Plutzky JB. Fat and Browning for the Treatment of Obesity and Related Metabolic Disorders. Diabetes Metab. J. 2016;40:12–21. doi: 10.4093/dmj.2016.40.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forest C, Joffin N, Jaubert A-M, Noirez P. What induces watts in WAT? Adipocyte. 2016;5:136–152. doi: 10.1080/21623945.2016.1187345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Jesus LA, et al. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 2001;108:1379–1385. doi: 10.1172/JCI200113803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckmann AM, Wilce P. Egr transcription factors in the nervous system. Neurochem. Int. 1997;31:477–51. doi: 10.1016/S0197-0186(96)00136-2. [DOI] [PubMed] [Google Scholar]
- 24.Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J. Biol. Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 25.Pagel J-I, Deindl E. Early growth response 1–a transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 2011;48:226–35. [PubMed] [Google Scholar]
- 26.Sakamoto KM, Fraser JK, Lee HJ, Lehman E, Gasson JC. Granulocyte-macrophage colony-stimulating factor and interleukin-3 signaling pathways converge on the CREB-binding site in the human egr-1 promoter. Mol. Cell. Biol. 1994;14:5975–85. doi: 10.1128/MCB.14.9.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai-Morris CH, Cao XM, Sukhatme VP. 5’ flanking sequence and genomic structure of Egr-1, a murine mitogen inducible zinc finger encoding gene. Nucleic Acids Res. 1988;16:8835–46. doi: 10.1093/nar/16.18.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, et al. Egr-1 decreases adipocyte insulin sensitivity by tilting PI3K/Akt and MAPK signal balance in mice. EMBO J. 2011;30:3754–3765. doi: 10.1038/emboj.2011.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. Dietary obesity-induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2. Sci. Rep. 2013;3:1476. doi: 10.1038/srep01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti P, et al. Insulin inhibits lipolysis in adipocytes via the evolutionarily conserved mTORC1-Egr1-ATGL-mediated pathway. Mol. Cell. Biol. 2013;33:3659–66. doi: 10.1128/MCB.01584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle, K. B. et al. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. 782–789, doi:10.1038/cdd.2009.11 (2009). [DOI] [PMC free article] [PubMed]
- 32.Guerquin, M.-J. et al. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 123 (2013). [DOI] [PMC free article] [PubMed]
- 33.Cereijo R, Giralt M, Villarroya F. Thermogenic brown and beige/brite adipogenesis in humans. Ann. Med. 2014;47:169–77. doi: 10.3109/07853890.2014.952328. [DOI] [PubMed] [Google Scholar]
- 34.Waldén, T. B., Hansen, I. R., Timmons, J. A., Cannon, B. & Nedergaard, J. Recruited vs. nonrecruited molecular signatures of brown, ‘ brite,’ and white adipose tissues. 1, 19–31 (2012). [DOI] [PubMed]
- 35.Chusyd, D. E., Wang, D., Huffman, D. M. & Nagy, T. R. Relationships between Rodent White Adipose Fat Pads and Human White Adipose Fat Depots. Front. Nutr. 3 (2016). [DOI] [PMC free article] [PubMed]
- 36.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 37.Lasar D, Julius A, Fromme T, Klingenspor M. Biochimica et Biophysica Acta Browning attenuates murine white adipose tissue expansion during postnatal development ✰. BBA - Mol. Cell Biol. Lipids. 2013;1831:960–968. doi: 10.1016/j.bbalip.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Ravaud C, Paré M, Azoulay S, Dani C, Ladoux A. Impairment of the activin A autocrine loop by lopinavir reduces self-renewal of distinct human adipose progenitors. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-02807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christian M. Transcriptional fingerprinting of ‘browning’ white fat identifies NRG4 as a novel adipokine. Adipocyte. 2014;4:50–54. doi: 10.4161/adip.29853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosell M, et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am. J. Physiol. Endocrinol. Metab. 2014;306:E945–64. doi: 10.1152/ajpendo.00473.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajimura S, et al. Initiation of myoblast/brown fat switch through a PRDM16-C/EBP-b transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barberá MJ, et al. Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 2001;276:1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- 43.Mariman ECM, Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010;67:1277–1292. doi: 10.1007/s00018-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Divoux A, Clement K. Architecture and the extracellular matrix: The still unappreciated components of the adipose tissue. Obes. Rev. 2011;12:494–503. doi: 10.1111/j.1467-789X.2010.00811.x. [DOI] [PubMed] [Google Scholar]
- 45.Berger E, et al. Pathways commonly dysregulated in mouse and human obese adipose tissue: FAT/CD36 modulates differentiation and lipogenesis. Adipocyte. 2015;4:161–80. doi: 10.4161/21623945.2014.987578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolton K, et al. Decorin is a secreted protein associated with obesity and type 2diabetes. Int. J. Obes. (Lond). 2008;32:1113–1121. doi: 10.1038/ijo.2008.41. [DOI] [PubMed] [Google Scholar]
- 47.Dubois SG, et al. Potential role of increased matrix metalloproteinase-2 (MMP2) transcription in impaired adipogenesis in type 2 diabetes mellitus. Biochem. Biophys. Res. Commun. 2008;367:725–728. doi: 10.1016/j.bbrc.2007.12.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henegar C, et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 2008;9:R14. doi: 10.1186/gb-2008-9-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, et al. AMPKα1 deficiency suppresses brown adipogenesis in favor of fibrogenesis during brown adipose tissue development. Biochem. Biophys. Res. Commun. 2017;491:508–514. doi: 10.1016/j.bbrc.2017.06.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chun T-H. Peri-adipocyte ECM remodeling in obesity and adipose tissue fibrosis. Adipocyte. 2012;1:89–95. doi: 10.4161/adip.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, Yao J, Mercola D, Adamson E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J. Biol. Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- 52.Buechler C, Krautbauer S, Eisinger K. Adipose tissue fibrosis. World J. Diabetes. 2015;6:548–53. doi: 10.4239/wjd.v6.i4.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Topilko P, et al. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol. Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 55.Bonnin MA, et al. Six1 is not involved in limb tendon development, but is expressed in limb connective tissue under Shh regulation. Mech. Dev. 2005;122:573–585. doi: 10.1016/j.mod.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Havis, E., Anselme, I. & Schneider-Maunoury, S. Whole embryo chromatin immunoprecipitation protocol for the in vivo study of zebrafish development. Biotechniques40 (2006). [DOI] [PubMed]
- 63.Wang H, et al. Stable, conditional, and muscle-fiber-specific expression of electroporated transgenes in chick limb muscle cells. Dev. Dyn. 2011;240:1223–1232. doi: 10.1002/dvdy.22498. [DOI] [PubMed] [Google Scholar]
- 64.Reznikoff, C. a, Brankow, D. W. & Heidelberger, C. Establishment and Characterization of a Cloned Line of C3H Mouse Embryo Cells Sensitive to Postconfluence Inhibition of Division Establishment and Characterization of a Cloned Line of C3H Mouse Embryo Cells Sensitive to Postconfluence Inhibition of. 3231–3238 (1973). [PubMed]
- 65.Lone, J., Choi, J. H., Kim, S. W. & Yun, J. W. ScienceDirect Curcumin induces brown fat-like phenotype in 3T3-L1 and primary white adipocytes. J. Nutr. Biochem. 1–10, doi:10.1016/j.jnutbio.2015.09.006 (2015). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.