Abstract
The analysis of Y-chromosome variation has provided valuable clues about the paternal history of domestic animal populations. The main goal of the current work was to characterize Y-chromosome diversity in 31 goat populations from Central Eastern (Switzerland and Romania) and Southern Europe (Spain and Italy) as well as in reference populations from Africa and the Near East. Towards this end, we have genotyped seven single nucleotide polymorphisms (SNPs), mapping to the SRY, ZFY, AMELY and DDX3Y Y-linked loci, in 275 bucks from 31 populations. We have observed a low level of variability in the goat Y-chromosome, with just five haplotypes segregating in the whole set of populations. We have also found that Swiss bucks carry exclusively Y1 haplotypes (Y1A: 24%, Y1B1: 15%, Y1B2: 43% and Y1C: 18%), while in Italian and Spanish bucks Y2A is the most abundant haplotype (77%). Interestingly, in Carpathian goats from Romania the Y2A haplotype is also frequent (42%). The high Y-chromosome differentiation between Swiss and Italian/Spanish breeds might be due to the post-domestication spread of two different Near Eastern genetic stocks through the Danubian and Mediterranean corridors. Historical gene flow between Southern European and Northern African goats might have also contributed to generate such pattern of genetic differentiation.
Introduction
Because of its male-limited transmission and lack of recombination1, Y-chromosome variation provides a simple and highly informative record of the paternal history of domestic species2–4. In general, Y-chromosome diversity is quite low due to small effective size (e.g. high variance in male reproductive success) and low mutation rate combined with the erosive effects of positive and purifying selection on diversity5. A first glimpse of caprine Y-chromosome variation was provided by Pidancier et al. (2006)6, who sequenced fragments of the amelogenin, Y-Linked (AMELY) and zinc finger protein, Y-Linked (ZFY) genes in wild and domestic goats. By doing so, they defined two common haplotypes C1 and C2 and a third rarer haplotype named C3. Further studies made possible to establish the existence of six Y-chromosome haplotypes i.e. Y1A, Y1B, Y1C, Y2A, Y2B, and Y2C7–10. Contrary to mitochondrial DNA, the analysis of caprine Y-chromosome variation evidenced the existence of a strong population structure in goats.
Waki et al.9 sequenced a fragment of the 3′UTR of the SRY gene in 210 Asian goats and demonstrated that the most frequent haplotype in Asia is Y1A (62%) followed by Y2B (30%). Asian breeds also showed marked differences with regard to Y-chromosome haplotype distribution9. Çinar-Kul et al.8 investigated Y-chromosome diversity in several Turkish goat breeds by partially sequencing the AMELY, ZFY and SRY genes and found that the most frequent haplotype was Y2A followed by Y1A, whilst a new Y2C minority haplotype was also identified. This study was of particular interest because Eastern Anatolia has been shown to be the unique primary centre of goat domestication11. Moreover, genotyping of four single nucleotide polymorphisms (SNPs) mapping to the sex determining region Y (SRY) gene in 46 Moroccan and 44 Portuguese bucks evidenced that haplotype frequencies are remarkably similar in both populations (Y2 is more frequent than Y1A and Y1B), suggesting the existence of gene flow between goats from the Iberian Peninsula and the Maghreb12.
Although Y-chromosome diversity has been reported in several European caprine breeds7,10,12, the number of sampled populations and individuals is still quite limited. Our goal was to overcome this drawback by genotyping seven Y-chromosome SNPs in 275 bucks from Southern Europe (Italy and Spain, N = 106), Central and East Europe (Switzerland and Romania, N = 113), Africa (Egypt, Burkina-Faso and Nigeria, N = 33) and the Near East (Iran and Oman, N = 23).
Methods
Sampling and sequencing of five Y-chromosome regions
Blood and hair were collected in 275 bucks belonging to 31 populations from nine different countries (Table 1). These samples came from four main geographic areas i.e. Central and East Europe, Southern Europe, Africa and Near East (Table 1). The isolation of genomic DNA was performed as described by Amills et al.4. Five regions of the AMELY, DEAD-box helicase 3, Y-linked (DDX3Y), ubiquitously transcribed tetratricopeptide repeat containing, Y-Linked (UTY) and ZFY genes were amplified by PCR and subsequently sequenced in a set of ten goats belonging to five different breeds (Saanen, Alpine, Murciano-Granadina, Palmera and Tinerfeña) to identify polymorphic sites. We used previously described ovine primers2 in four of the five selected regions (Supplementary Table S1 ). Polymerase chain reactions were carried out in a final volume of 30 μL containing 1.5 mm MgCl2, 200 μM dNTPs, 0.2 μM of each primer, 25 ng genomic DNA and 0.3 U Taq DNA polymerase (Biogen). The thermal profile was 94 °C for 5′ followed by 35 cycles of 94 °C during 30″, 55 °C for 1′ 30″ and 72 °C during 1′ 30″, ending with an extension step of 5′ at 72 °C. Amplicons were purified with the ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific) and sequenced with the BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific) and the corresponding amplification primers. Sequencing reactions were electrophoresed in an ABI310 Genetic Analyzer equipment (Applied Biosystems). Sequences were visualized with the SeqAnalysis software (Thermo Fisher Scientific) and aligned with the Multalin program13.
Table 1.
Distribution of Y-chromosome haplotypes in 31 goat populations.
| Area | BREED | Country | Y1A | Y1B1 | Y1B2 | Y1C | Y2 |
|---|---|---|---|---|---|---|---|
| Central and East Europe | Alpine | Switzerland | 8 | 2 | |||
| Appenzell | 8 | ||||||
| Camosciata delle Alpi | 1 | ||||||
| Chamois Coloured goat | 5 | ||||||
| Grisons Striped | 9 | ||||||
| Peacock Goat | 9 | ||||||
| Saanen | 3 | 15 | |||||
| St Gallen Booted goat | 9 | ||||||
| Toggenburg | 8 | ||||||
| Valais Blackneck | 9 | ||||||
| Verzasca goat | 8 | ||||||
| Carpathian | Romania | 10 | 1 | 8 | |||
| TOTAL (N = 113) | 33 | 14 | 41 | 17 | 8 | ||
| Southern Europe | Bermeya | Spain | 9 | ||||
| Blanca Andaluza | 1 | 1 | |||||
| Guadarrama | 10 | ||||||
| Malagueña | 4 | ||||||
| Murciano Granadina | 23 | ||||||
| Mallorquina | 10 | ||||||
| Pitiüsa | 15 | ||||||
| Blanca de Rasquera | 5 | ||||||
| Garganica | Italy | 1 | 1 | 1 | |||
| Sarda | 16 | 1 | 5 | ||||
| Maltese | 3 | ||||||
| TOTAL (N = 106) | 20 | 1 | 2 | 1 | 82 | ||
| Africa | Djallonke | Burkina Faso | 5 | ||||
| Sahel | 9 | ||||||
| African Dwarf Goat | Nigeria | 2 | |||||
| Nigerian | 2 | ||||||
| Zaraibi | Egypt | 15 | |||||
| TOTAL (N = 33) | 0 | 0 | 0 | 0 | 33 | ||
| Near East | Lori-Bakhtyari Goat | Iran | 8 | 2 | |||
| Esfahan Goat | 12 | ||||||
| Oman | Oman | 1 | |||||
| TOTAL (N = 23) | 20 | 0 | 0 | 0 | 3 | ||
Generation and analysis of genotypic data
Seven Y-chromosome polymorphisms, two discovered in this study and five reported in the literature6,7,10, were selected to be genotyped in a multiplex assay (Table 2). A total of 275 bucks were typed in the Veterinary Service of Molecular Genetics (http://svgm.es/ca/Home) of the Universitat Autònoma de Barcelona. Genomic DNA samples were distributed in 384-well sample plates together with the PCR mix. These reactions were transferred onto QuantStudio 12 K Flex OpenArray plates with the QuantStudio 12 K Flex AccuFill System (Thermo Fisher Scientific). Genotyping assays were run in a QuantStudio 12 K Flex real-time PCR instrument (Thermo Fisher Scientific) in standalone mode. Primers and probes are indicated in Supplementary Table S2. Nucleotide and haplotype diversities were calculated with the DnaSP software v514. The Network software v5 (www.fluxus-engineering.com) was used to construct a median-joining network with default parameters15.
Table 2.
Y-chromosome polymorphisms and haplotypes analyzed in the current work.
| SRY 2971 | SRY 3098 | SRY 1876 | AMELY 42 | ZFY 527 | ZFY 46 | DDX3Y 56 | Haplotype |
|---|---|---|---|---|---|---|---|
| T | G | A | C | A | C | G | Y1A |
| A | G | A | C | A | C | G | Y1B1 |
| A | G | A | C | A | T | G | Y1B2 |
| A | G | C | C | A | C | G | Y1C |
| T | A | A | C | G | C | C | Y2 |
Results and Discussion
Sequence alignment made possible to identify four unreported polymorphisms in the non-coding regions of the genes AMELY (g.310_313delATAT in Genbank MF448227-MF448228), DDX3Y (g.56 C > G in MF448229-MF448230 and g.390 T > C in MF448231-MF448232) and ZFY (g.46 C > T in MF448233-MF448234). The deletion detected in the gene AMELY and the polymorphism in the intron 7 of the DDX3Y gene were discarded from the genotyping panel due to technical reasons. We therefore aimed to genotype five Y-chromosome SNPs (SRY-2971T > A, SRY-3098G > A, SRY-1876A > C –GenBank D82963-, AMELY-42C > T –GenBank AY082491.1-, and ZFY-527A > G –GenBank AY082500) that had been reported in previous studies7–10 plus two SNPs discovered by us (DDX3Y g.56 C > G and ZFY g.46 C > T). Altogether, these mutations defined five haplotypes i.e. Y1A, Y1B1, Y1B2, Y1C and Y2A (Table 2) whose frequencies were estimated in 275 bucks from 31 populations (Table 1). In general, the diversity of the caprine Y-chromosome was quite low (2.1 polymorphisms/kb, Table 3), a result that matches previous data reported in sheep16. These findings might be explained by the low effective size of the Y-chromosome mainly as a consequence of the high variance in reproductive success of bucks and rams. Another important factor that decreases Y-chromosome variation is the removal of deleterious mutations by purifying selection17. Due to the non-recombining nature of the Y-chromosome, this type of selection can erase variability even at linked neutral sites located far away from the purged mutation17. Interestingly, the analysis of Y-chromosome variation in pigs has uncovered the existence of a remarkable level of polymorphism18. This discrepancy between goats and pigs regarding the amount of paternal variation could be due to the fact that pigs were independently domesticated at two distinct sites and on the basis of two gene pools (Asia and West) that diverged around 1 Ma ago. This resulted in the emergence of two main Y-chromosome haplogroups that are highly differentiated18. In strong contrast, all modern goats descend from bezoars domesticated at Eastern Anatolia11 and this is probably the main reason why all Y-chromosome haplotypes are remarkably similar. The median joining network displayed in Fig. 1 shows that the distribution of Y-chromosome diversity is tightly linked to geography. In goats from Switzerland, the Y1A and Y1B2 haplotypes are clearly predominant (67%), while Y1B1 (15%) and Y1C (18%) are less frequent and Y2A cannot be found. In stark contrast, the most frequent haplotype in Italian and Spanish goats is Y2A (77%) followed by Y1A (18%), whilst frequencies of Y1B and Y1C are almost negligible. Interestingly, the Y2A haplotype is the only one segregating in African breeds. In a previous study, Pereira et al.12 analyzed the Y-chromosome diversity of goats from Morocco and Portugal and also found that the most abundant haplotype in both populations is Y2, though Y1A and Y1B haplotypes could be also detected. In goats from Iran, we have found that Y1A is the most frequent haplotype (90%). In contrast, Çinar-Kul et al.8 demonstrated that in Turkey the Y2A haplotype is predominant with the exception of a single breed, Kilis, where Y1A segregates at high frequencies. The coexistence of several differentiated Y-chromosome haplotypes in this area is consistent with the high diversity that is usually found in primary domestication centers.
Table 3.
Caprine Y-chromosome diversity in four geographic areas.
| Geographic areas | Number of haplotypes | Haplotype diversity | Sample size |
|---|---|---|---|
| Central and East Europe | 5 | 0.747 | 113 |
| Southern Europe | 5 | 0.369 | 106 |
| Near East | 2 | 0.273 | 23 |
| Africa | 1 | 0.000 | 33 |
| TOTAL | 5 | 0.690 | 275 |
Figure 1.
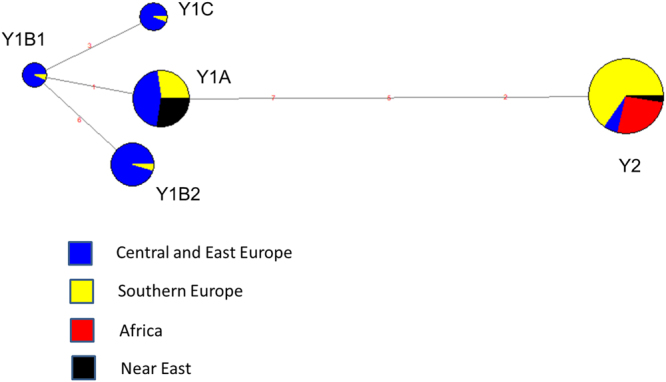
Median joining network based on the Y-chromosome genotypes of 275 goats from Southern European (Bermeya, Blanca Andaluza, Garganica, Guadarrama, Malagueña, Maltese, Murciano Granadina, Mallorquina, Pitiüsa, Blanca de Rasquera and Sarda breeds), Central and East European (Alpine, Appenzell, Camosciata delle Alpi, Chamois Coloured, Grisons Striped, Peacock Goat, Carpathian, Saanen, St Gallen Booted goat, Toggenburg, Valais Blackneck and Verzasca breeds), African (Djallonke, African Dwarf, Nigeria, Sahel and Zaraibi breeds) and Near Eastern (Iran and Oman) countries.
Central European breeds analysed in our study come exclusively from Switzerland. The analysis of autosomal variation in 10 Swiss breeds has shown that several of them (Appenzell, Toggenburg, Valais and Booted goat) display low levels of diversity but in general long runs of homozygosity (>15 Mb) are rare indicating the absence of recent inbreeding19. At this point, we do not know if Y-chromosome data obtained by us in Swiss populations can be extrapolated to other breeds from Central or North Europe. However, it is worth to emphasize that the patterns observed by Lenstra7 in North Central European breeds (i.e. Y1 haplotypes are abundant and Y2 is scarce) closely match those observed by us in Swiss populations. Significant genetic differences between Central and Southern European breeds have been also observed at the autosomal level. In this way, Cañón et al.20 genotyped a panel of microsatellites in 1,426 goats from 45 European and Near Eastern breeds and showed that Mediterranean and Central European breeds are clearly differentiated. This marked genetic differentiation could be due to several reasons. The dispersal of goats from the domestication center at Eastern Anatolia into Europe followed two main routes i.e. the Mediterranean and the Danubian corridors21. The Danubian route involved the transportation of goats through the continental heartland of Europe towards the Danube Valley and the central and northern plains of Europe21. Pastoralist communities were established in Greece and Bulgary 6,500 YBP and they subsequently moved north and eastwards until arriving to Scandinavia and the British Isles 4,000 YBP22. In stark contrast, the dissemination of domestic animals and plants through the Mediterranean corridor was essentially maritime23, with Neolithic seafarers reaching the Iberian Peninsula 7,700-7,300 YBP and Libya and Algeria 7,000 YBP. Thus, it is possible that the two Eastern Anatolian goat gene pools that followed the Danubian and Mediterranean corridors were subjected to serial founder effects resulting in the genetic differentiation of Swiss and Italian/Spanish European goat breeds. Indeed, the results of Çinar Kul et al.8 indicate that, in Turkish goat breeds, the Y1A haplotype is almost fixed in Kilis bucks, while Y2A nearly reached fixation in Abaza, Gurcu, Angora and Norduz bucks. This genetic heterogeneity of Turkish breeds would fit a scenario based on the post-domestication spread of different gene pools along the Danubian and Mediterranean corridors.
An alternative, but not excluding, explanation for this pattern of differentiation would be the existence of gene flow between Southern European and North African goat populations. Pereira et al.12 hypothesized that the similar frequencies of Y-haplotypes that they observed in Portuguese and Moroccan goats might be due to the occurrence of ancient genetic exchanges. Martínez et al.24 shed light on this issue by demonstrating, with the aid of coalescent genealogy samplers, the existence of a significant and bidirectional migration between northwest African and Iberian goat populations. In a subsequent study, Manunza et al.25 analysed the variation of Spanish and African breeds on the basis of 52,000 SNP genotypes and found evidences of admixture between Andalusian (Murciano-Granadina and Malagueña) and Tunisian goats. In a recent study, Decker et al.26 showed that Spanish cattle breeds had a 7.5–20% of African introgression into their genomes, and an indicine introgression was also observed in Italian breeds. Taken together, these results suggest that admixture events may have contributed to the genetic differentiation of Central and Southern European goats.
Particularly intriguing is the case of Carpathian bucks from Romania which displayed high frequencies (42%) of the Y2A haplotype. This might likely be the consequence of the historical commercial relationships between this region and Turkey, or even the arrival and establishment, during the 8–12th centuries, of Turkic nomad peoples (e.g. pechenegs and cumans) who migrated from the Central Asian steppes to Romania, very likely carrying their own livestock. Lack of arable land, drought and harsh and cold winters explain why nomadic pastoralism has been so prevalent in the Eurasian steppe since prehistoric times. Indeed, the analysis of sheep mitogenomes has revealed the existence of remarkable levels of genetic diversity in the Mongolian Plateau, suggesting that this region constituted one of the main centers of sheep dispersal across Asia27. Even nowadays, pastoral nomadism is the main form of land use in Mongolia, with one third of the population living as nomads from livestock breeding28.
Data presented by us and others7–9 indicate that Y-chromosome markers recapitulate much better the population structure of goat breeds than mitochondrial polymorphisms29. However, this does not mean that the maternal and paternal histories of goats are substantially different. The larger physical size of the Y-chromosome, if compared with the mitochondrial genome, facilitates the detection of more ancient drift signals30. Previous data based on microsatellite24 and SNP25 markers have revealed that Spanish goats are more closely related to their Central European counterparts than to the African ones, an outcome that does not fully match our analysis of Y-chromosome markers. The large scale study of genome-wide data from caprine breeds with a worldwide distribution will probably shed light on this and other issues that are essential to understand the history of goat pastoralism.
Ethics statement
Blood and hair root samples were collected from goats by trained veterinarians in the context of sanitation campaigns and parentage controls not directly related with our research project. For this reason, permission from the Universitat Autònoma de Barcelona Committee of Ethics in Animal Experimentation was not required. In all instances, veterinarians followed standard procedures and relevant national guidelines to ensure appropriate animal care.
Electronic supplementary material
Acknowledgements
This research was funded by the Spanish National Institute for Agriculture and Food Research and Technology (INIA, grants RZ2007-0005-C02-02, RZ2011-00015-C03-03 and RZ2011-00015-C03-01) and the Spanish Ministry of Economy and Competitiveness (grant AGL2016-76108-R). We also acknowledge financial support from the Spanish Ministry of Economy and Competitiveness, through the “Severo Ochoa Programme for Centres of Excellence in R&D” 2016-2019 (SEV‐2015‐0533). This research was also supported by the CERCA Programme of the Generalitat de Catalunya.
Author Contributions
Study design: O.V., M.A., C.D., J.J., V.A.B., J.V.D., F.G., F.P., J.C., A.P., Performed the molecular analysis: O.V., Performed the population genetic analyses:, M.A., B.B., Provision of the data: G.O.R., I.R., A.M., S.E., V.L., A.T., M.P., G.M.V., M.D.A., I.A., A.S., O.V. and M.A. wrote the manuscript and all authors read, corrected and approved its content.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15593-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 2.Meadows JRS, Hawken RJ, Kijas JW. Nucleotide diversity on the ovine Y chromosome. Anim. Genet. 2004;35:379–85. doi: 10.1111/j.1365-2052.2004.01180.x. [DOI] [PubMed] [Google Scholar]
- 3.Kantanen J, et al. Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus) Heredity (Edinb). 2009;103:404–15. doi: 10.1038/hdy.2009.68. [DOI] [PubMed] [Google Scholar]
- 4.Amills M, et al. Mitochondrial DNA diversity and origins of South and Central American goats. Anim. Genet. 2009;40:315–322. doi: 10.1111/j.1365-2052.2008.01837.x. [DOI] [PubMed] [Google Scholar]
- 5.Hellborg L, Ellegren H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol. Biol. Evol. 2004;21:158–163. doi: 10.1093/molbev/msh008. [DOI] [PubMed] [Google Scholar]
- 6.Pidancier N, Jordan S, Luikart G, Taberlet P. Evolutionary history of the genus Capra (Mammalia, Artiodactyla): discordance between mitochondrial DNA and Y-chromosome phylogenies. Mol. Phylogenet. Evol. 2006;40:739–49. doi: 10.1016/j.ympev.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Lenstra, J. A. Evolutionary and demographic history of sheep and goats suggested by nuclear mtDNA and Y-chromosome markers in Proceedings of the International Workshop ‘The role of biotechnology for the characterization of crop, forestry, animal and fishery genetic resources’ 97–100 (2005).
- 8.Cinar Kul B, et al. Y-chromosomal variation of local goat breeds of Turkey close to the domestication centre. J. Anim. Breed. Genet. 2015;132:449–453. doi: 10.1111/jbg.12154. [DOI] [PubMed] [Google Scholar]
- 9.Waki A, Sasazaki S, Kobayashi E, Mannen H. Paternal phylogeography and genetic diversity of East Asian goats. Anim. Genet. 2015;46:337–339. doi: 10.1111/age.12293. [DOI] [PubMed] [Google Scholar]
- 10.Pereira F, et al. A multiplex primer extension assay for the rapid identification of paternal lineages in domestic goat (Capra hircus): Laying the foundations for a detailed caprine Y chromosome phylogeny. Mol. Phylogenet. Evol. 2008;49:663–8. doi: 10.1016/j.ympev.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Naderi S, et al. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc. Natl. Acad. Sci. 2008;105:17659–17664. doi: 10.1073/pnas.0804782105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira F, et al. Tracing the history of goat pastoralism: new clues from mitochondrial and Y chromosome DNA in North Africa. Mol. Biol. Evol. 2009;26:2765–73. doi: 10.1093/molbev/msp200. [DOI] [PubMed] [Google Scholar]
- 13.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 15.Bandelt HJ, Forster P, Rohl A. median_joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, et al. Y chromosome haplotype diversity of domestic sheep (Ovis aries) in northern Eurasia. Anim Genet. 2014;45:903–907. doi: 10.1111/age.12214. [DOI] [PubMed] [Google Scholar]
- 17.Wilson Sayres MA, Lohmueller KE, Nielsen R. Natural selection reduced diversity on human Y chromosomes. PLoS Genet. 2014;10:e1004064. doi: 10.1371/journal.pgen.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramírez O, et al. Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds. Mol. Biol. Evol. 2009;26:2061–2072. doi: 10.1093/molbev/msp118. [DOI] [PubMed] [Google Scholar]
- 19.Burren A, et al. Genetic diversity analyses reveal first insights into breed-specific selection signatures within Swiss goat breeds. Anim. Genet. 2016;47:727–739. doi: 10.1111/age.12476. [DOI] [PubMed] [Google Scholar]
- 20.Cañón J, et al. Geographical partitioning of goat diversity in Europe and the Middle East. Anim. Genet. 2006;37:327–334. doi: 10.1111/j.1365-2052.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 21.Pereira F, Amorim A. Origin and spread of goat pastoralism in Encyclopedia of Life Sciences (John Wiley & Sons, Ltd. 2010 [Google Scholar]
- 22.Porter, V., Alderson, L., Hall, S. J. G. & Sponenberg D. P. Goats in Mason’s World Encyclopedia of Livestock Breeds and Breeding 350–352 (CABI, Oxfordshire, United Kingdom, 2016).
- 23.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez A, et al. Detecting the existence of gene flow between Spanish and North African goats through a coalescent approach. Sci. Rep. 2016;6:38935. doi: 10.1038/srep38935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manunza, A. et al. A genome-wide perspective about the diversity and demographic history of seven Spanish goat breeds. Genet. Sel. Evol. 48–52, 10.1186/s12711-016-0229-6 (2016). [DOI] [PMC free article] [PubMed]
- 26.Decker JE, et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 2014;10:e1004254. doi: 10.1371/journal.pgen.1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv FH, et al. Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 2015;32:2515–2533. doi: 10.1093/molbev/msv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lkhagvadorj D, Hauck M, Dulamsuren C, Tsogtbaatar J. Pastoral nomadism in the forest-steppe ecotone of the Mongolian Altai under a changing economy and a warming climate. Journal of Arid Environments. 2013;88:82–89. doi: 10.1016/j.jaridenv.2012.07.019. [DOI] [Google Scholar]
- 29.Luikart G, et al. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc. Natl. Acad. Sci. 2001;98:5927–5932. doi: 10.1073/pnas.091591198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


