Abstract
The majority of long noncoding RNAs (lncRNAs) is still poorly characterized with respect to function, interactions with protein-coding genes, and mechanisms that regulate their expression. As for protein-coding RNAs, epigenetic deregulation of lncRNA expression by alterations in DNA methylation might contribute to carcinogenesis. To provide genome-wide information on lncRNAs aberrantly methylated in breast cancer we profiled tumors of the C3(1) SV40TAg mouse model by MCIp-seq (Methylated CpG Immunoprecipitation followed by sequencing). This approach detected 69 lncRNAs differentially methylated between tumor tissue and normal mammary glands, with 26 located in antisense orientation of a protein-coding gene. One of the hypomethylated lncRNAs, 1810019D21Rik (now called Esrp2-antisense (as)) was identified in proximity to the epithelial splicing regulatory protein 2 (Esrp2) that is significantly elevated in C3(1) tumors. ESRPs were shown previously to have a dual role in carcinogenesis. Both gain and loss have been associated with poor prognosis in human cancers, but the mechanisms regulating expression are not known. In-depth analyses indicate that coordinate overexpression of Esrp2 and Esrp2-as inversely correlates with DNA methylation. Luciferase reporter gene assays support co-expression of Esrp2 and the major short Esrp2-as variant from a bidirectional promoter, and transcriptional regulation by methylation of a proximal enhancer. Ultimately, this enhancer-based regulatory mechanism provides a novel explanation for tissue-specific expression differences and upregulation of Esrp2 during carcinogenesis. Knockdown of Esrp2-as reduced Esrp2 protein levels without affecting mRNA expression and resulted in an altered transcriptional profile associated with extracellular matrix (ECM), cell motility and reduced proliferation, whereas overexpression enhanced proliferation. Our findings not only hold true for the murine tumor model, but led to the identification of an unannotated human homolog of Esrp2-as which is significantly upregulated in human breast cancer and associated with poor prognosis.
Introduction
The importance of long noncoding RNAs (lncRNAs) for the regulation of both developmental as well as tumorigenic processes is increasingly recognized. LncRNAs influence expression or stability of protein-coding RNAs, and act as hosting genes encoding for microRNAs or as microRNA decoys.1, 2, 3, 4 LncRNAs also affect translation and stability of proteins.2, 5 They can control expression of genes in a localized, gene-specific fashion6 or by targeting large chromosomal regions.1, 2, 3, 4 LncRNAs influence DNA methylation or the chromatin landscape by interacting with modifiers of epigenetic marks, thereby recruiting these modifiers to specific DNA loci, and subsequent gene silencing or activation.5, 7, 8
Identification and functional evaluation of lncRNAs has become an area of substantial scientific interest, for example by analyzing differential expression for de novo identification of lncRNAs.9, 10 In addition, information on chromatin marks for active transcription (H3K4me3 and H3K36me3) was combined with tiling microarray data to locate novel lncRNAs.11 RNA-seq allows the detection of lncRNAs at a genome-wide scale.9, 10 Elucidation of biological functions of lncRNAs is aided by bioinformatic strategies, for example, by analyzing the genomic context of the lncRNAs, or by placing them in a network of coexpressed genes.10 Experimentally, immunoprecipitation of RNAs is used to detect interaction partners.9 Despite these achievements in the discovery of novel lncRNAs, regulatory mechanisms of lncRNA expression are poorly understood, and especially genome-wide studies for epigenetic regulation of lncRNAs are still scarce.
In the present study, we hypothesized that epigenetic deregulation of lncRNA expression might contribute to carcinogenesis. We performed a genome-wide screen for differentially methylated lncRNA promoters in tumor samples of a mouse model for human breast cancer vs normal mammary tissue and identified a series of candidate regions in antisense orientation to protein-coding genes. One of the hypomethylated lncRNAs was 1810019D21RIK (termed Esrp2-antisense (as)) located in vicinity of the epithelial splicing regulatory protein 2 (Esrp2), which was upregulated in C3(1) tumors.
ESRP2 and its closely related isoform ESRP1 are crucial to maintain an epithelial-specific RNA splicing program. Loss of these splicing factors leads to epithelial-to-mesenchymal transition (EMT),12, 13, 14 which is an important process involved in development, tumor progression, malignant transformation, and metastasis formation.15, 16 In carcinogenesis, ESRPs appear to have a context-dependent dual function. Both isoforms have been found up- or downregulated in human cancers, and both high and low expression levels were associated with poor prognosis.17, 18, 19
We here report that coordinate expression of Esrp2 and Esrp2-as from a bidirectional promoter is regulated by differential methylation of a proximal enhancer. Knockdown and overexpression studies suggest that Esrp2-as is important to maintain Esrp2 protein expression and function. Our results are not limited to the mouse model, but led to the discovery of a novel human homolog of Esrp2-as with elevated levels in human breast cancer, associated with elevated risk of cancer recurrence.
Results
Genome-wide screen to identify differentially methylated lncRNAs
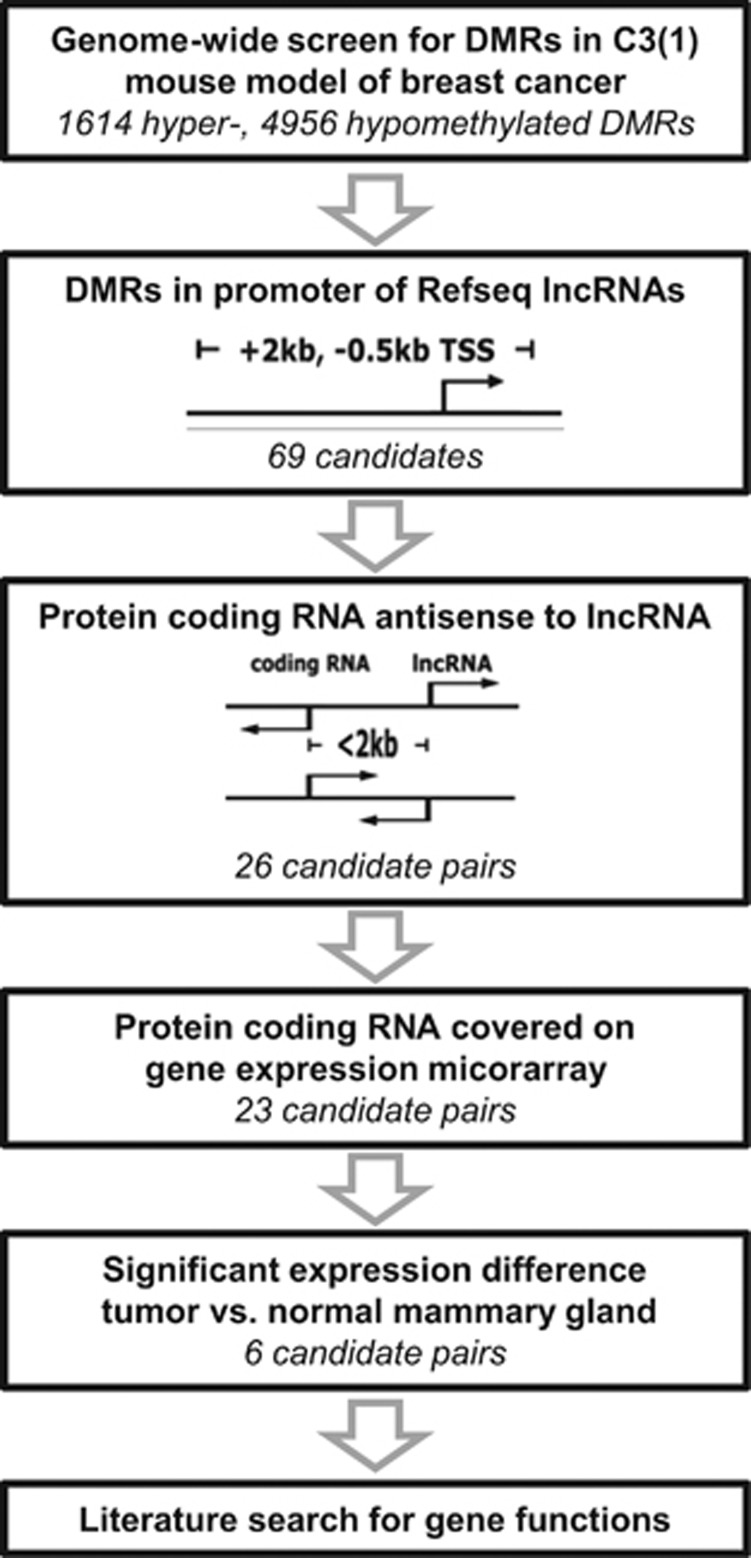
For the detection of lncRNAs with aberrant methylation during carcinogenesis, we made use of the transgenic C3(1) SV40TAg (C3(1)) transgenic mouse model of human breast cancer.20, 21 We performed a genome-wide screen by ‘Methylated CpG Immunoprecipitation’ to enrich for highly methylated DNA fragments,22 followed by next generation sequencing (MCIp-seq). Comparison of tumor samples with mammary glands of age-matched wildtype (WT) control animals identified 6570 differentially methylated regions (DMRs) (Figure 1). By overlapping DMRs with promoters of mouse Refseq annotated lncRNAs, we identified 37 hyper- and 32 hypomethylated lncRNA promoters (Table 1). RNA-seq analyses of M6 and M27H4 tumor cell lines derived from the C3(1) mouse model23 and 3T3-L1 murine adipocytes indicated that about half of the identified lncRNAs were expressed (Supplementary Table S1). We further focused on lncRNA candidates with neighboring mRNAs in antisense orientation and thus identified 26 pairs of protein-coding and noncoding RNAs with significant correlation of expression levels (Supplementary Figure S1A). Except for two lncRNA/mRNA pairs (Foxd2os/ Foxd2, F730043M19Rik/Atxn7l1), coding genes were up to 2800-fold higher expressed than their antisense lncRNAs (Supplementary Fig S1B). Using published microarray data for the C3(1) mouse model,24 we found Pcdh7, Gabrg3, and Hoxa11 significantly downregulated, and Otud7a, Lsr, and Esrp2 (Supplementary Figure S1C) upregulated in tumors vs normal mammary glands. We selected the Esrp2/Esrp2-as pair for further analysis of epigenetic gene regulation, owing to the functional role of ESRP2 in regulation of epithelial-to-mesenchymal transition.
Figure 1.
Schematic representation of the screening strategy.
Table 1. Candidate lncRNAs with differentially methylated promoters and neighboring protein coding genes.
| DMR |
lncRNA |
Protein coding gene |
Expression difference |
Referencea | |||
|---|---|---|---|---|---|---|---|
| Genomic position (mm10) | Gene symbol | ID | Gene symbol | ID | Tumor –Normalb | P-valuec | |
| Hypermethylation | |||||||
| chr1:180332417-180332819 | Gm5069 | NR_003623 | Itpkb | NM_001081175 | |||
| chr2:74762966-74763423 | Haglr | NR_110445 | Hoxd1 | NM_010467 | −0.10 | 0.55 | |
| chr3:37896886-37897206 | Gm20755 | NR_040559 | |||||
| chr3:82876021-82876531 | Rbm46os | NR_040382 | Rbm46 | NM_001277170 | |||
| chr4:114907033-114907726 | Foxd2os | NR_030721 | Foxd2 | NM_008593 | −0.34 | 0.09 | |
| chr4:145463820-145464350 | Smarca5-ps | NR_002888 | |||||
| chr4:152697098-152697448 | Gm833 | NR_033138 | |||||
| chr5:19226691-19226991 | 4921504A21Rik | NR_102341 | Magi2 | NM_001170746 | 0.64 | 0.23 | |
| chr5:57718135-57718436 | 4932441J04Rik | NR_015588 | Pcdh7 | NM_018764 | −1.26 | 0.0007*** | 25 |
| chr6:47943214-47943743 | Zfp783 | NR_027963 | |||||
| chr6:52315701-52316027 | 5730457N03Rik | NR_038163 | Evx1 | NM_007966 | −0.34 | 0.10 | |
| chr6:6864808-6865158 | Dlx6os2 | NR_002839 | |||||
| chr7:14623181-14623487 | Nlrp5-ps | NR_045119 | |||||
| chr7:40899025-40899517 | A230077H06Rik | NR_040329 | Vstm2b | NM_021387 | −0.49 | 0.13 | |
| chr7:51511083-51511470 | Ano5 | NR_073508 | |||||
| chr7:57386794-57387157 | Gm9962 | NR_033504 | Gabrg3 | NM_008074 | −0.45 | 0.04* | 26 |
| chr7:63444419-63444734 | 4930554H23Rik | NR_131089 | Otud7a | NM_130880 | 2.36 | 0.00009**** | 27 |
| chr7:79515301-79515977 | AI854517 | NR_040312 | |||||
| chr8:120230039-120230397 | A330074K22Rik | NR_110496 | |||||
| chr8:121084994-121085403 | Fendrr | NR_045471 | Foxf1 | NM_010426 | 0.30 | 0.11 | |
| chr8:84912426-84912992 | Gm38426 | NR_103491 | |||||
| chr8:99416005-99416420 | A330008L17Rik | NR_132435 | Cdh8 | NM_007667 | 0.01 | 0.97 | |
| chr9:71216962-71217270 | Gm3458 | NR_110518 | Aldh1a2 | NM_009022 | −0.39 | 0.13 | |
| chr10:93336092-93336432 | Gm17745 | NR_038014 | |||||
| chr11:95143377-95143691 | Dlx4os | NR_040279 | Dlx4 | NM_007867 | 0.05 | 0.90 | |
| chr12:20815469-20816034 | 1700030C10Rik | NR_015521 | |||||
| chr13:111868178-111868548 | Gm15326 | NR_130345 | |||||
| chr13:72816340-72816825 | D730050B12Rik | NR_046196 | |||||
| chr14:118234395-118234797 | LOC105245869 | NR_131969 | Sox21 | NM_177753 | −0.27 | 0.37 | |
| chr14:39472112-39472839 | LOC432842 | NR_131973 | Nrg3 | NM_008734 | −0.15 | 0.59 | |
| chr15:83366702-83367300 | 1700001L05Rik | NR_027980 | |||||
| chr16:98081806-98082497 | A630089N07Rik | NR_015491 | |||||
| chr17:34094655-34095214 | BC051537 | NR_046183 | |||||
| chr17:80373020-80373349 | Gm10190 | NR_028385 | |||||
| chr19:30539362-30539867 | Ppp1r2-ps3 | NR_003650 | |||||
| chrX:12159988-12160665 | 2900008C10Rik | NR_045434 | |||||
| chrX:12761797-12762195 | Gm14634 | NR_045852 | Med14 | NM_012005 | −0.31 | 0.13 | |
| Hypomethylation | |||||||
| chr1:133269974-133270300 | Gm19461 | NR_037984 | |||||
| chr1:136696173-136696502 | Platr22 | NR_037986 | |||||
| chr1:71888158-71888468 | Gm8883 | NR_027658 | |||||
| chr2:118112952-118113258 | Gm13986 | NR_126479 | Thbs1 | NM_011580 | |||
| chr3:116968252-116968565 | 4930455H04Rik | NR_040596 | Palmd | NM_023245 | −0.09 | 0.84 | |
| chr5:112206149-112206560 | 1700028D13Rik | NR_045377 | |||||
| chr5:112206149-112206560 | 1700028D13Rik | NR_045378 | |||||
| chr5:135000770-135001082 | Wbscr25 | NR_026907 | |||||
| chr5:143758048-143758457 | D130017N08Rik | NR_015486 | |||||
| chr6:52165042-52165362 | Hoxaas2 | NR_131182 | Hoxa2 | NM_010451 | −0.39 | 0.28 | |
| chr6:52243386-52243706 | Hoxa11os | NR_015348 | Hoxa11 | NM_010450 | −0.31 | 0.05* | 28, 29 |
| chr7:28496984-28497589 | 1700028B04Rik | NR_033605 | |||||
| chr7:30971903-30972229 | Fam187b | NR_038860 | Lsr | NM_017405 | 3.57 | 0.00004**** | 30 |
| chr8:106133504-106133810 | 1810019D21Rik (Esrp2-as) | NR_040344 | Esrp2 | NM_176838 | 1.55 | 0.0005*** | 12, 14 |
| chr8:11005553-11005971 | 9530052E02Rik | NR_046017 | Irs2 | NM_001081212 | |||
| chr8:70774591-70774966 | 2010320M18Rik | NR_029440 | Pik3r2 | NM_008841 | 0.47 | 0.11 | |
| chr9:40333291-40333624 | 1700110K17Rik | NR_040728 | |||||
| chr9:51875640-51875974 | Arhgap20os | NR_033560 | |||||
| chr11:102377699-102378035 | Bloodlinc | NR_131196 | |||||
| chr11:112711182-112711489 | BC006965 | NR_024085 | |||||
| chr12:11204247-11204597 | 9530020I12Rik | NR_131083 | |||||
| chr12:33149419-33149899 | F730043M19Rik | NR_015602 | Atxn7l1 | NM_028139 | −0.18 | 0.53 | |
| chr13:44216558-44216865 | A330076C08Rik | NR_045088 | |||||
| chr13:63296450-63296765 | Gm16907 | NR_045794 | |||||
| chr14:105589344-105589691 | 9330188P03Rik | NR_102319 | |||||
| chr14:118363552-118363938 | 1700044C05Rik | NR_045624 | |||||
| chr14:118922434-118923043 | Dzip1 | NR_130725 | |||||
| chr14:55071799-55072110 | Zfhx2os | NR_004444 | |||||
| chr15:103147888-103148190 | D930007P13Rik | NR_045743 | |||||
| chr16:95928970-95929274 | 1600002D24Rik | NR_040484 | |||||
| chr17:56554141-56554483 | Gm20219 | NR_130128 | |||||
| chr19:53076038-53076348 | 1700054A03Rik | NR_045320 | |||||
Abbreviation: lncRNA, long noncoding RNA.
Bold indicates Polycomb-associated RNAs.31
Ref.: Reference describing the function of significantly differentially expressed genes.
Expression data were taken from Herschkowitz et al.24 and include 8 TG tumor samples and 5 FVB/N WT mammary glands. Values are reported as the differences between tumor and normal samples in log2 array intensities relative to a pool of whole mouse embryo RNA.
P-values were calculated using two-sided Student’s t-test, *P< 0.05, ***P< 0.001, ****P< 0.0001.
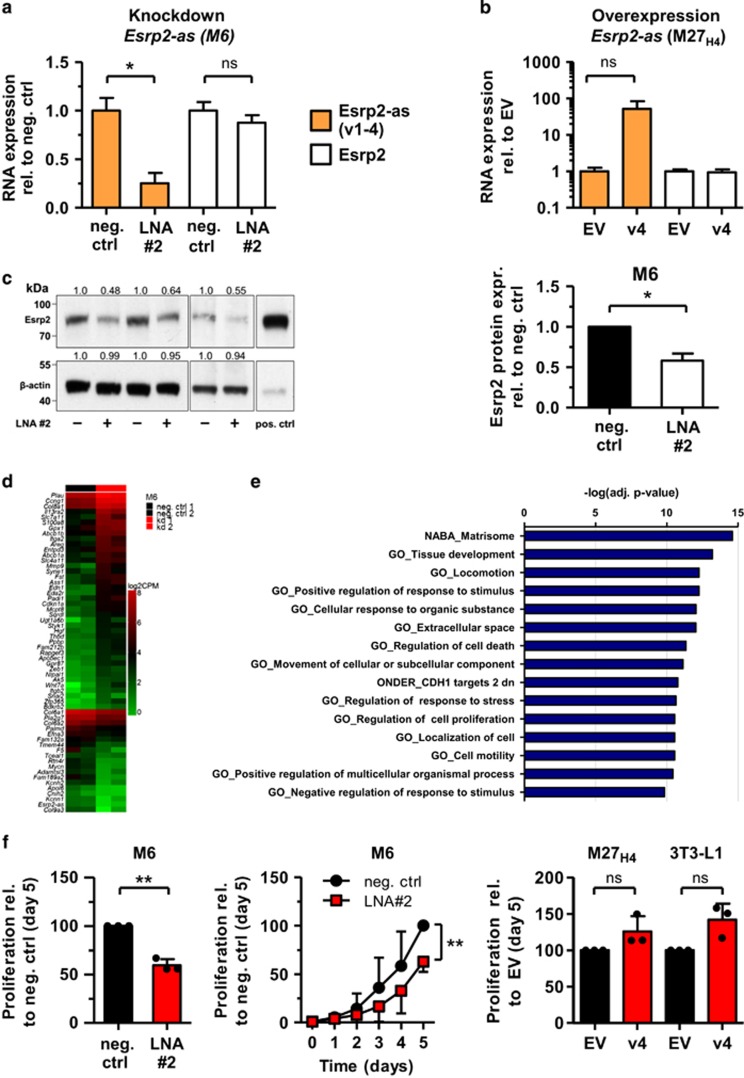
Esrp2 and Esrp2-as are coordinately overexpressed in C3(1) mammary gland tumors
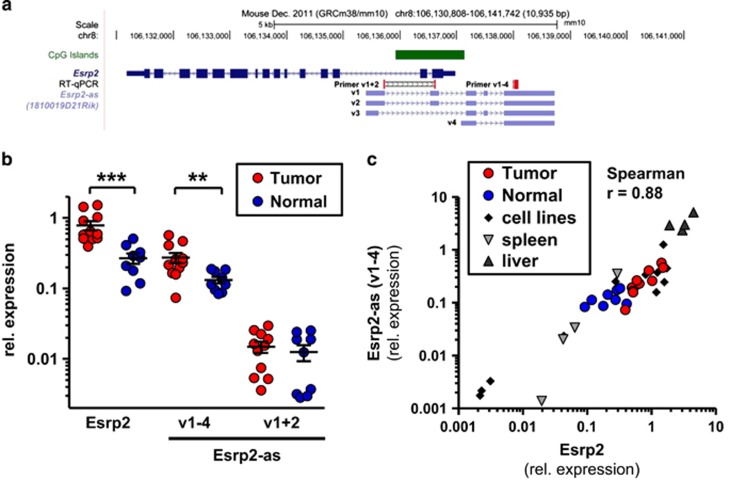
Esrp2-as is expressed as four annotated transcripts with a length between 1.2 and 1.6 kb (Figure 2a). Three long transcript variants (v1-3) share a common TSS approximately 1.6 kb downstream of the Esrp2 TSS. In contrast, the TSS for the short variant (v4) is about 100 bp upstream of the Esrp2 TSS. RT–qPCR analyses with primers detecting variants v1-4 or v1+2 revealed that all variants together were about 20-fold higher expressed than the long variants v1+2, suggesting that the short variant v4 represents the major transcript. Publicly accessible FANTOM5 CAGE-seq data32, 33 (Cap Analysis of Gene Expression) confirmed transcription initiation for both the short and the long variants of Esrp2-as (Supplementary Figure S2), underlining expression of all transcript variants in mouse mammary glands. In tumor tissue, Esrp2 and Esrp2-as v1-4 levels were significantly 3- and 2-fold elevated, whereas the long variants v1+2 were not differentially expressed (Figure 2b). Expression of Esrp2 and the antisense transcripts v1-4 were highly correlated in tumors and normal tissues from the mouse model and in various mouse cell lines (Figure 2c). Similarly, expression of Esrp2-as v1+2 and v1-4 were highly correlated (Supplementary Figure S3 and S4).
Figure 2.
Esrp2 and Esrp2-as are coordinately overexpressed in C3(1) mammary gland tumors. (a) Genomic organization of Esrp2 (dark blue) and Esrp2-as variants 1-4 (v1-4, light blue). Location of primers detecting Esrp2-as v1+2 or v1-4, respectively, is indicated in red. (b) Relative expression levels as determined by RT–qPCR are significantly different between tumor (n=11) and normal samples (n=9) for Esrp2 and Esrp2-as (v1-4), but not for Esrp2-as (v1+2). Samples were derived from animals aged 20-24 weeks, and expression levels were normalized to three reference genes (Hprt1, Tbp, β-Actin). Mann-Whitney U test, **P<0.01, ***P<0.001. (c) Esrp2 and Esrp2-as v1-4 expression assessed by RT–qPCR highly correlates in tumors, normal mammary gland, cell lines, spleen, and liver samples. Spearman’s rank correlation coefficient rho=0.88 (P <0.0001).
These results pointed either to a mutual regulation of expression between the coding and noncoding transcripts, or to a common control mechanism.
Esrp2 CpG island shores are differentially methylated in C3(1) tumors and cell lines
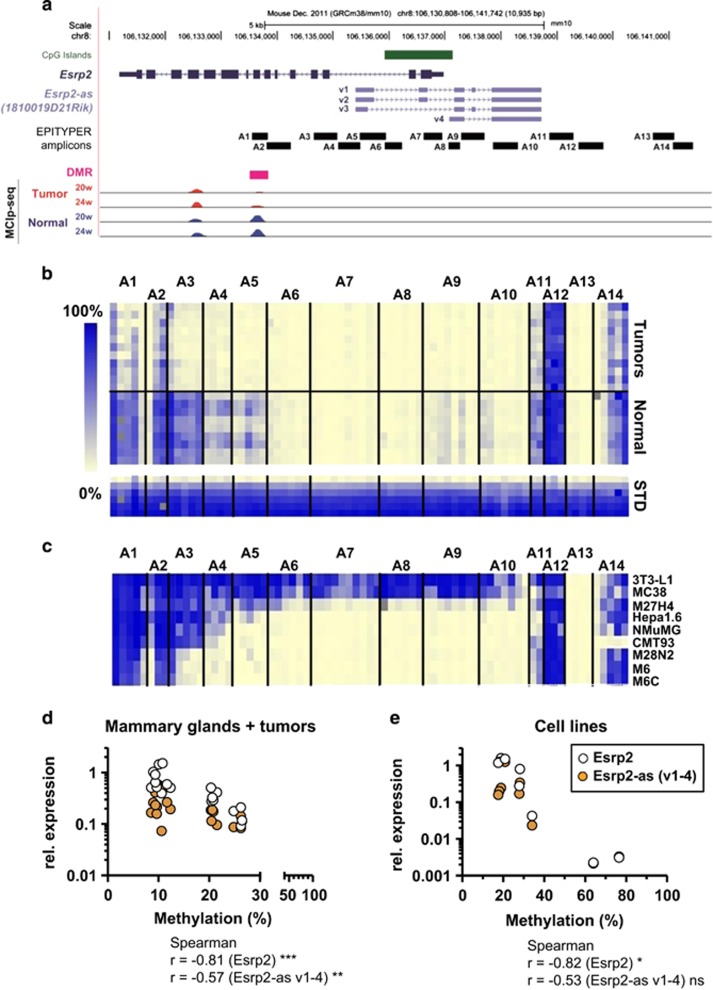
The CGI located in the Esrp2 promoter region was generally unmethylated in normal mouse tissues, whereas CGI shores (regions of 2 kb on either side of a CGI) displayed variable methylation between tissues that express Esrp2 and Esrp2-as (liver, kidney, stomach, lung) and those that do not (spleen and heart; Supplementary Figure S5). Thus, we designed EPITYPER amplicons covering the DMR (Figure 3a, Amplicon A1), the CGI (A6-A8), as well as the CGI shore regions (A2-A5, A9-A14) for quantitative methylation analyses. The central region spanning the CGI (A6-A8) was unmethylated in both tumor and normal tissue, whereas the DMR (A1) and individual CpG units in the shore regions covered by amplicons A3-A5 and A9-A11 were hypomethylated in tumor tissue (Figure 3b). Analysis of spleen and liver samples of the C3(1) model confirmed good concordance of EPITYPER data with published whole genome bisulfite sequencing (WGBS) data (Supplementary Figure S6). 3T3-L1 preadipocytes and MC38 colon carcinoma cells were 70–90% methylated in the analyzed region, M27H4 cells showed intermediate levels of methylation, and the M6 cell line was unmethylated, similar to C3(1) tumors (Figure 3c).
Figure 3.
DNA methylation levels inversely correlate with expression levels of Esrp2 and Esrp2-as. (a) Upper: genomic organization of Esrp2 (dark blue) and Esrp2-as variants 1-4 (v1-4, light blue) and the CGI overlapping the Esrp2 TSS (green). Positions of EPITYPER Amplicons A1-A14 are indicated by horizontal black bars, covering the DMR (pink), the CGI, and CGI shores on both sides. Lower: MCIp-seq detection of methylated DNA fragments in tumors (red) and normal WT mammary glands (blue) of animals at 20 and 24 weeks of age. Each lane represents average reads obtained for three individual samples. (b,c) Heatmap of DNA methylation levels in tumor samples (n=11) and normal mammary gland tissue (n=9) (b), or of various murine cell lines (c), with each row representing one individual sample and each column one CpG unit comprising of 1 to 4 individual CpG sites. Methylation levels are depicted by a color-coded gradient from 0% (light yellow) to 100% methylation (blue). Gray squares indicate failed measurements. (d,e) Correlation between average methylation of amplicons A1-A14 and Esrp2/Esrp2-as expression levels normalized to three reference genes, calculated by Spearman’s rank correlation for tumor/normal tissues (d) and cell lines (e). *P <0.05, **P<0.01, ***P<0.001.
Consistent with a gene silencing function of promoter methylation, 3T3-L1 and MC38 cell lines had lowest expression levels of Esrp2 and Esrp2-as (Supplementary Figure S4A). We observed strong negative correlation between sense/antisense transcript expression and methylation levels in tumor samples and normal tissue (Figure 3d) as well as in cell lines (Figure 3e). These results confirmed that the region around the Esrp2 TSS is differentially methylated, and methylation inversely correlated with expression of both the protein-coding and the noncoding RNA.
Demethylation induces reexpression of Esrp2 and Esrp2-as
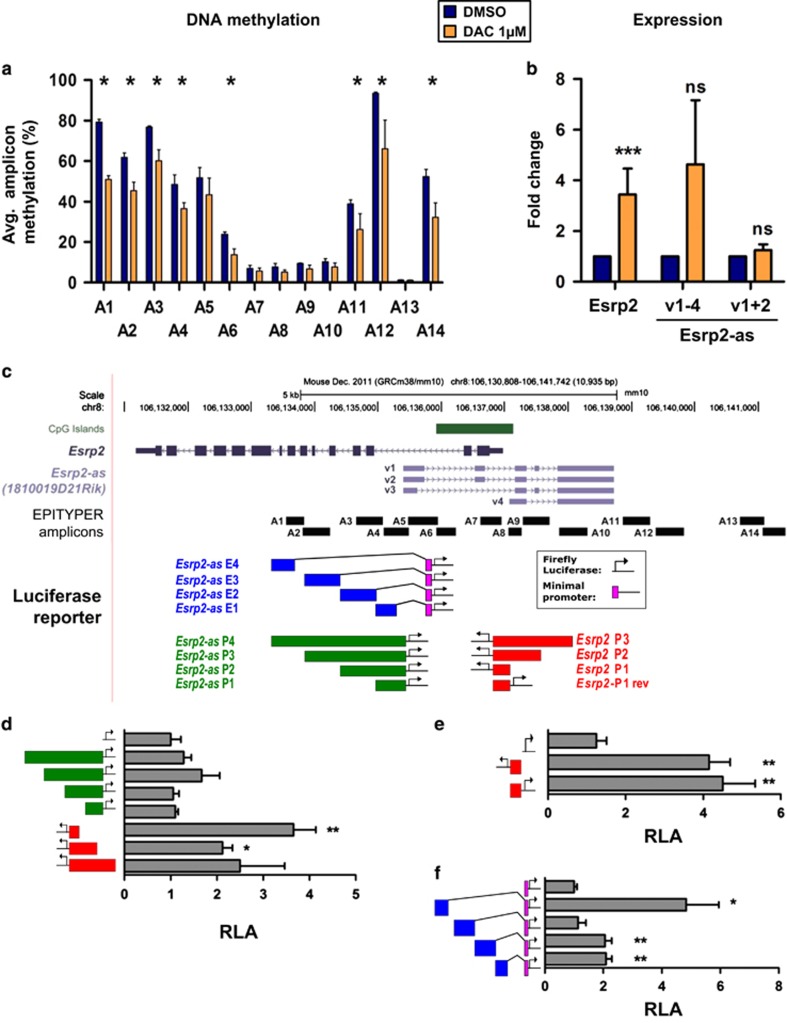
To functionally test the correlation of Esrp2 and Esrp2-as methylation and gene expression, we performed demethylation experiments in M27H4 cells by treatment with 1 μM Decitabine (DAC). DNA methylation levels decreased by 10-30% in amplicons that were >20% methylated in the DMSO solvent control (Figure 4a). Both Esrp2 and Esrp2-as v1-4 transcript levels increased by 3- and 4-fold, whereas the increase in Esrp2-as v1+2 levels was only marginal (Figure 4b). These results suggested concomitant regulation of Esrp2 and the short Esrp2-as variant v4 expression by methylation. The long Esrp2 transcript variants might be regulated independently.
Figure 4.
Esrp2 and Esrp2-as are coordinately expressed from a bidirectional promoter and regulated by methylation of a proximal enhancer. (a) Treatment of M27H4 cells with 1 μm DAC for 72 h with daily renewal decreases DNA methylation levels. Depicted is the mean methylation±SD for amplicons A1-A14 of three independent experiments. Mann-Whitney U test (one-sided), *P <0.05. (b) DAC treatment induces reexpression of Esrp2 and Esrp2-as v1-4 in M27H4 cells. Depicted is the mean expression±SD normalized to DMSO solvent control of three independent experiments. Unpaired Student’s t-test (one-sided), ***P <0.001, NS, not significant. (c) Schematic representation of luciferase reporter constructs to determine promoter and enhancer activity of fragments covering the Esrp2 region. Location of Esrp2 and Esrp2-as are depicted in gray and promoter constructs are marked in red and green. Enhancer reporters are displayed by blue boxes combined with a minimal promoter (purple). (d,e) Reporter plasmids with firefly luciferase for promoter (d) and bidirectional promoter (e) activity were transiently transfected into Hepa1.6 cells and luciferase activity was measured 48 h post transfection and normalized to a co-transfected CMV-Renilla luciferase construct. Transfections were conducted parallel in 8 technical replicates and reported is the mean±SEM of four independent experiments normalized to the pGL4.10 EV. (f) Reporter constructs for enhancer activity. Transfections and measurements were conducted and reported as in (d) and normalized to the pGL4.23 EV. Unpaired Student’s t-test (two-sided), asterisks represent comparisons against EV. *P <0.05, **P<0.01. RLA, relative luciferase activity.
Luciferase reporter assays confirm a bidirectional promoter and an enhancer region
To explore in more detail the genomic regions involved in regulation of Esrp2 and Esrp2-as expression we performed dual luciferase reporter assays using promoter constructs (P) of Esrp2 and the long variants of Esrp2-as v1-3 (Figure 4c). Reporter assays confirmed promoter activity for the Esrp2 P-fragments. The P1 fragment located closest to the Esrp2 TSS was associated with strong 4-fold induction of luciferase activity compared to EV. Analyses with the Esrp2-as P1-P4 constructs resulted in weak, insignificant induction of luciferase activity (Figure 4d). Luciferase assays with Esrp2 P1 in reverse orientation relative to the luciferase gene (sense to the Esrp2-as v4 transcript) revealed almost equally high luciferase activity as for the Esrp2 P1 fragment (Figure 4e), indicating that the Esrp2 P1 region has bidirectional promoter activity for expression of both Esrp2 and Esrp2-as.
ChIP-seq data available for murine liver and kidney34 demonstrated occupancy by enhancer marks H3K4me1 and H3K27ac in the region next to the TSS of the long Esrp2-as variants v1-3, suggesting that this region might have enhancer functions. We thus analyzed Esrp2-as E1–E4 regions in luciferase vectors to test for enhancer activity. The E4 region led to strongest activation of the reporter construct with a minimal promoter (Figure 4f), but not with the Esrp2 P1 promoter (Supplementary Fig S7).
These data explain coordinate expression of Esrp2 and Esrp2-as from a bidirectional promoter in cooperation with an enhancer region.
Knockdown of Esrp2-as reduces Esrp2 protein expression without affecting the mRNA level
LncRNAs have been reported to target gene activating or repressive functions to specific DNA loci.2, 5, 8 This prompted us to analyze whether Esrp2-as might epigenetically control expression of Esrp2 by a similar mechanism. Knockdown of Esrp2-as with two locked nucleic acid antisense Gapmers (LNAs) targeting the last exon of the Esrp2-as variants efficiently reduced antisense transcript levels by 60–80%, but did not influence Esrp2 mRNA levels (Figure 5a and Supplementary Figure S8). Equally, transient overexpression of Esrp2-as v4 in M27H4 cells did not affect Esrp2 transcript levels (Figure 5b). Additional knockdown or overexpression attempts in various other cell lines produced similar results (Supplementary Figure S8). To exclude the possibility that expression of the coding transcript regulates noncoding RNA levels, we also generated a M27H4 cell population that stably overexpressed Esrp2. Under these conditions, Esrp2-as levels were not affected. This was also true for an Esrp2-overexpressing 3T3-L1 preadipocyte population (Supplementary Figure S9). From these data we concluded that Esrp2 and Esrp2-as do not transcriptionally regulate expression of the transcript on the other strand.
Figure 5.
Knockdown of Esrp2-as reduces Esrp2 protein levels, globally alters expression of genes involved in cellular motility and inhibits cell proliferation. (a) Knockdown of Esrp2-as by Locked Nucleic Acid antisense Gapmers (LNAs) in the M6 cell line for 72 h. Expression levels were assessed by RNA-seq; CPM values are normalized to cells transfected with negative control LNA. Mean±SD of two independent experiments. Statistical significance was assessed using Student’s t-test with *P <0.05. (b) M27H4 cells were transiently transfected with constructs containing Esrp2-as v4. Expression levels 72 h post-transfection were assessed by RNA-seq; CPM values are normalized to cells transfected with the empty vector (EV). Mean±s.d. of three independent experiments. Statistical significance was assessed using Student’s t-test with Welch’s correction (P =0.116, NS). (c) Esrp2 protein expression in the M6 cell line 72 h after knockdown of Esrp2-as with LNA#2. M27H4 cells stably overexpressing Esrp2 were used as a positive control (pos. ctrl). Band intensities were normalized to β-actin levels and are semi-quantitatively evaluated relative to the negative control using TINA v2.09. Depicted is the mean±SD of three independent experiments. Statistical significance was assessed using a one-sample t-test with the neg. control set to 1.0, with *P <0.05. (d) Heatmap of top 40 up- and top 20 downregulated genes after Esrp2-as knockdown in M6 cells. Depicted are log2 CPM values of two biological replicates. (e) Overlap of differentially expressed genes after knockdown of Esrp2-as with gene sets in MSigDB using GSEA.35 (f) Cell proliferation was assessed by SRB staining on five consecutive days starting 24 h post transfection (day 0). Cell proliferation was adjusted for the number of seeded cells and is depicted in relation to the negative control or EV at day 5 (set as 100%). Shown is the mean±SD of three independent experiments (represented by black dots). Statistical significance was assessed using the one-sample t-test with **P<0.01, NS, not significant.
Besides influencing transcriptional regulation, lncRNAs have been reported to affect protein translation or stability.2, 3, 5 By western blotting experiments, we could demonstrate that knockdown of Esrp2-as led to a significant 40% reduction of Esrp2 protein levels in the M6 cell line (Figure 5c). Overexpression of the short variant v4 was however not sufficient to induce detectable Esrp2 protein levels in M27H4 and 3T3-L1 cells, in which the gene is silenced by promoter methylation (Supplementary Figure S10).
Knockdown of Esrp2-as induces extracellular matrix (ECM) and EMT regulators and reduces cell proliferation
To further investigate the function of Esrp2-as, we assessed genome-wide changes in gene expression by RNA-seq after knockdown and overexpression. Overexpression of the short Esrp2-as v4 had only minor influence on gene expression in M27H4 and 3T3-L1 cells, with significant transcriptional changes (FDR-adjusted P-value <0.05) of only 1 and 5 genes, respectively (Supplementary Table S2). This argues against regulatory potential in trans. Reducing the level of the noncoding RNA by about 80% in the M6 cell line by LNA#2 led to significant alterations in expression of 185 genes, with the majority of genes (76%) being upregulated (Figure 5d, Supplementary Table S2).
For further analyses we focused on the knockdown effects. Differentially expressed genes were significantly enriched in extracellular matrix (ECM) proteins, including collagens and glycoproteins, ECM regulators, and secreted factors (collectively defined as the matrisome36). Further enriched gene ontologies include tissue development, locomotion and motility, regulation of cell death and cell proliferation, and cellular response to stimuli and stress (Figure 5e and Supplementary Table S3). A strong influence of Esrp2-as knockdown on ECM and cell motility was confirmed by Ingenuity Pathway Analysis (IPA; www.qiagen.com/ingenuity) (Supplementary Table S4 and Supplementary Figure S11). The top #1 network represents many ECM proteins as well as the ECM receptor integrin B2 and Zeb1, a transcriptional repressor and inducer of EMT. Top #2 and #3 networks were associated with ‘Cell Morphology, Connective Tissue Development and Function’ and ‘Cell Cycle and Cancer’ with p53 central to the network (Supplementary Figure S11). We verified significant upregulation of several ECM and EMT-associated factors including Zeb1 and consequent downregulation of E-cadherin by RT–qPCR analyses. The pattern of expression changes induced by Esrp2-as knockdown recapitulated the expression differences observed in primary murine mammary epithelial cells (Mecs) versus mammary fibroblasts. mRNA levels of Esrp2, Esrp2-as and the epithelial marker Cdh1 were high in Mecs, but barely detectable in fibroblasts, which instead highly expressed MMP9, Plau, Prl2c3 and Zeb1 (Supplementary Figure S12A).
RNA-seq data revealed reduced expression of luminal cell differentiation markers after Esrp2-as knockdown in M6 cells, whereas expression of additional EMT transcription factors37 and cancer stem cell makers38 was increased (Supplementary Figure S12B). All of these results point to the activation of EMT after Esrp2-as knockdown and are consistent with the loss of Esrp2 protein expression.
As a functional consequence of the altered gene expression profiles, knockdown of Esrp2-as led to 40% reduced cell proliferation detectable from day 3 post transfection (Figure 5f). Concomitantly, we observed downregulation of cell proliferation markers Ki67 and Pcna, and significant upregulation of the cell cycle inhibitor p21 and several targets of p53 that are associated with tumor-suppressive or cell growth-inhibitory function (S11 Network #3 and Supplementary Figure S12B). Conversely, overexpression of the short Esrp2-as v4 resulted in slight (albeit not significant) induction of cell growth in both M27H4 and 3T3-L1 cell lines (Figure 5f).
Collectively, these data indicate that knockdown of Esrp2-as and the concomitant loss in Esrp2 protein expression lead to alterations in gene expression suggestive of a more mesenchymal/fibroblast-like phenotype with enhanced cell migratory potential and reduced cell proliferation capacity, whereas Esrp2-as overexpression promotes proliferation.
ESRP2 and ESRP2-AS are hypomethylated and overexpressed in human breast cancer, and indicate poor prognosis
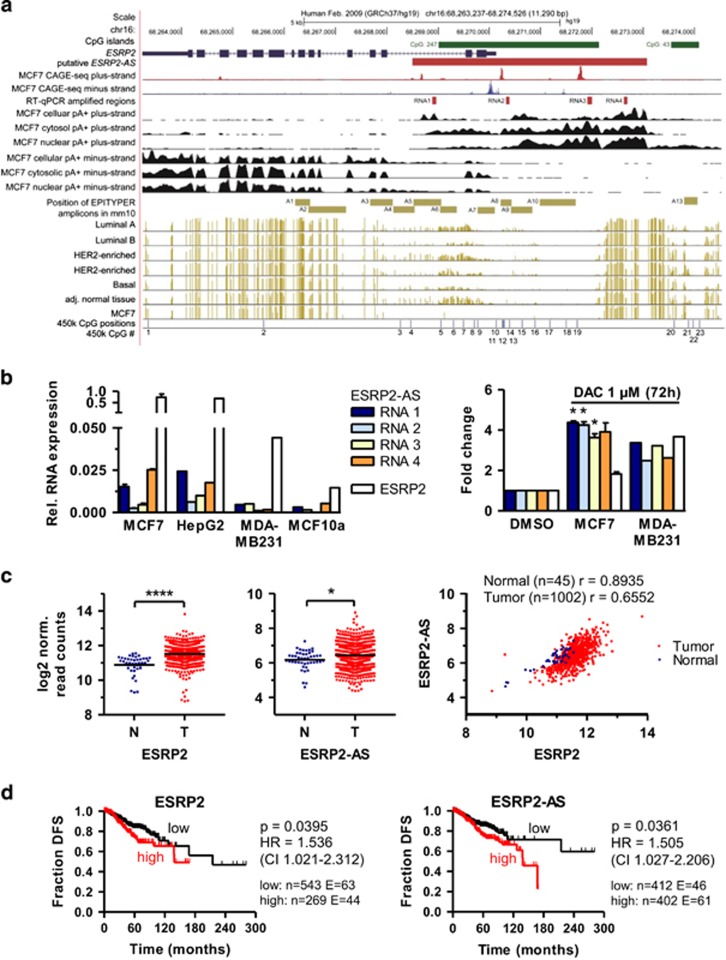
A human homolog of Esrp2-as has not been annotated yet, but various lines of evidence indicate its existence. (i) CAGE-seq data for MCF7 human breast cancer cells (FANTOM5,32) suggested transcription initiation at locations corresponding to the TSSs of mouse Esrp2-as (Figure 6a). (ii) Strand-specific RNA-seq datasets of polyA-enriched MCF7 RNA (provided by ENCODE39 in the UCSC genome browser40) indicated transcription of a human ESRP2-AS homolog (Figure 6a). (iii) Alignment analyses of the mouse Esrp2-as v1 and the putative human ESRP2-AS (red bar in Figure 6a) revealed 67.75% identity of both transcripts, especially at the 5′-end that showed a higher degree of conservation than the 3′-end (Supplementary Figure S13). (iv) We were able to detect ESRP2-AS expression in four human tumor cells lines by RT–qPCR (Figure 6b). We observed highest expression in estrogen receptor-positive MCF7 breast cancer cells and in HepG2 human hepatoma cells, and lower expression levels in the basal-like breast cancer cell lines MDA-MB231 and MCF10a. The pattern of expression using four different primer pairs was similar in all cell lines, supporting the notion that the human ESRP2 antisense transcript is expressed. (v) DAC treatment of MCF7 and MDA-MB231 cells led to a 1.8- to 4.4-fold increase in expression levels of both ESRP2 and ESRP2-AS transcripts, corroborating epigenetic regulation in human breast cancer cell lines (Figure 6b).
Figure 6.
A human homolog of Esrp2-as is overexpressed in human breast cancer and associated with poor prognosis. (a) UCSC Genome Browser40 scheme of the human ESPR2 locus on chr16 depicts the genomic location of the human ESRP2 gene (blue), CpG islands (green), genomic location of ESRP2-AS (red), MCF-7 CAGE-seq peaks for the positive (red) and negative strand (red), positions of RT–qPCR primers to quantify expression of ESRP2-AS (RNA1-4, red), strand-specific RNA-seq signals obtained with MCF7 whole cell lysates, cytosolic and nuclear fraction (black), location of mouse EPITYPER MassArray amplicons (using UCSC genome browser liftover tool, beige), DNA methylation levels for individual CpG sites (indicated by beige lines) assessed by WGBS (downloaded from TCGA), CpG sites covered on the Illumina 450 k methylation array (blue). (b) Left: Relative expression of ESRP2-AS (using four primer pairs designated as RNA1-4) and ESRP2 in MCF7 (n=2), HepG2 (n=1), MDA-MB231 (n=1) and MCF10a (n=1) human (cancer) cell lines, assessed by RT–qPCR. Right: Fold change of ESRP2 and ESRP2-AS levels in MCF7 (n=2) and MDA-MB231 cells (n=1) after DAC treatment, relative to DMSO solvent control set as 1. Statistical significance was assessed using the one-sample t-test with *P <0.05. (c) ESRP2 (left) and ESRP2-AS (middle) expression in human breast cancer samples from the TCGA BRCA breast invasive carcinoma dataset. RNA-seq data (log2 normalized read counts) for 1002 tumor samples were compared to 45 normal control samples by two-sided Student’s t-test with ****P <0.0001, *P<0.05. Right: Spearman correlation of ESPR2 and ESRP2-as levels (log2 norm. read counts) in normal breast tissue (blue) and tumor tissue (red) in the TCGA BRCA dataset. (d) Kaplan–Meier curves of disease-free survival are plotted for ESRP2 and ESRP2-AS expression. A total of 814 informative breast cancer cases from TCGA BRCA with an overall number of 107 events are separated by expression levels into a low (black) and high (red) expressing group. P-values for log-rank Mantel-Cox test; HR (hazard ratio) with 95% confidence interval indicated in parenthesis.
To compare expression of ESRP2 and ESPR2-AS in human breast tumors, we obtained RNA-seq read counts for 1002 tumor samples and 45 normal controls from ‘The Cancer Genome Atlas’ consortium (TCGA, http://cancergenome.nih.gov). ESRP2 was significantly overexpressed in tumor samples compared to normal breast tissue (Figure 6c). Since bioinformatic tools did not recognize ESPR2-AS as a novel transcript, we computed normalized read counts for the 4.6 kb region spanning the putative transcript in MCF7 cells (Figure 6a). Expression of ESPR2-AS was about 40-fold lower than that of the ESRP2 mRNA, but significantly elevated in tumor tissue vs normal breast tissue. Consistently, ESRP2 and ESRP2-AS expression levels highly correlated (Figure 6c).
WGBS data provided by TCGA demonstrated that the CGI covering the upstream promoter region of ESRP2 was lowly methylated, whereas the region overlapping with the mouse EPITYPER amplicons A4-A6 was hypomethylated in tumor samples compared to adjacent normal tissue (Figure 6a). The methylation patterns were very similar to those observed in the C3(1) mouse model (Figure 3b). We extracted 450 k DNA methylation data for 636 breast tumors and 35 normal controls from TCGA. Breast tumors were significantly hypomethylated compared to normal breast tissue in the region covered by CpG sites 3-7, downstream of the ESRP2 TSS (Supplementary Figure S14A). Methylation and expression levels of both transcripts inversely correlated, with highest anti-correlation in normal breast tissue, especially at CpG sites 3-6, 13 and 15. Inverse correlation in tumor tissue was weaker than in normal tissue and spread over the CGI from CpG sites 3-16 (Supplementary Figure S14B).
High expression of both ESRP2 and ESRP2-AS was significantly associated with lower disease-free survival in the TCGA breast cancer cohort, based on the analysis of 814 informative cancer cases with a total of 107 events and a follow-up time up to 281 months (Figure 6d). Using the Kaplan–Meier plotter survival analysis tool,41 we could confirm that high ESRP2 mRNA levels reduce the fraction of relapse-free survival, overall survival, and distant metastasis-free survival of breast cancer cases analyzed together or when cases were subdivided into breast cancer subtypes (Supplementary Figure S15).
Discussion
The present study aimed to identify lncRNAs epigenetically regulated during breast carcinogenesis in the C3(1) mouse model.20, 24 A recent study by Li et al. followed a similar approach in human breast cancer.42 The authors identified several hundred differentially expressed miRNAs and lncRNAs with aberrantly methylated promoters. These ncRNAs had high diagnostic potential and were involved in several pathways dysregulated in human breast cancer. EVX1-AS, MAGI2-AS, FOXD2-AS, FENDRR and HOXA11-AS were among the 69 differentially methylated lncRNAs identified in our study, emphasizing the relevance of the mouse model for human breast cancer.43, 44 Histological, transcriptomic and miRNA expression analyses have shown that this model best reflects the aggressive Luminal B and basal-like subtypes,24, 45, 46 which are associated with high mortality and poor prognosis.47, 48
Few of the differentially methylated lncRNAs have been functionally analyzed, with the exception of Haglr, Fendrr, and Hoxa11as that regulate expression of protein-coding genes.49, 50, 51, 52 Interestingly, RNA immunoprecipitation followed by sequencing identified that in embryonic stem cells, 17 of the 69 lncRNAs (indicated in Table 1) were associated with EZH2, an important member of the PRC2 Polycomb repressive complex.31 This observation suggests that these lncRNAs might be actively involved in the recruitment of the PRC2 complex. Genes near hypermethylated lncRNAs were enriched in members of the homeobox family of developmental regulators.53 Hypermethylation of homeobox genes has been identified as an early epigenetic event in human breast cancer,54, 55 acting in parallel with Polycomb repression to reduce the regulatory plasticity of these key regulatory genes.56 Accordingly, gene set enrichment analyses (GSEA)35, 57 revealed that eight of the 17 genes located close to hypermethylated lncRNAs were targets of the PRC2 or marked by the repressive histone modification H3K27me3 in differentiated or progenitor cells.58, 59, 60 Our study now adds epigenetic regulation of lncRNAs located close to these genes as an additional layer of regulation.
Nine of the hypomethylated lncRNAs were associated with protein-coding genes in their vicinity. These genes were amplified (Hoxa11, Hoxa2) or silenced by methylation (Irs2, Thbs1) in human breast cancer,61, 62 or downregulated in metastases from malignant melanoma compared to primary tumors (Esrp2, Palmd, Lsr).63 LSR levels were shown to correlate with ERα expression in human breast cancer, with lower expression in patient samples with lymph node invasion and distant metastases.30
We were interested in investigating whether epigenetic regulation of lncRNAs might influence expression of protein-coding genes in their vicinity, using the Esrp2/Esrp2-as pair as an example.
We verified hypomethylation and coordinate upregulation of Esrp2 and Esrp2-as in tumors of the C3(1) model compared to normal mammary glands. Demethylation experiments as well as luciferase reporter assays demonstrated that co-expression of Esrp2 and the short Esrp2-as variant v4 is regulated by methylation of a putative enhancer region proximal to a bidirectional promoter, which was generally lowly methylated. Reporter assays also confirmed that fragment E4 (corresponding to amplicon A1) had enhancer activity (Figure 4). These results are consistent with the observation that enhancer methylation often correlates even better with gene expression than promoter methylation.64, 65
To analyze whether the long Esrp2-as variants v1-3 are expressed from an independent promoter, we performed reporter assays using constructs covering the putative promoter region of the long variants. The results were inconclusive (Figure 4). This might be due to a lack of essential transcription factors in the Hepa1.6 cell line used for transfection, indicated by 400-fold lower expression levels of Esrp2-as v1+2 relative to v1-4 compared to 20-fold lower levels in most other cell lines (Supplementary Figure S4). Expression of Esrp2-as v1+2 highly correlated with v1-4 and Esrp2 in various tissues and cell lines (Supplementary Figure S3). However, the fact that different from v1-4, the long variants v1+2 were not overexpressed in C3(1) mouse tumors (Figure 2b) and also did not get re-expressed after DAC treatment (Figure 4b) suggests that additional factors contribute to regulation of expression of these long variants.
Comparison of WGBS and RNA-seq data for various murine tissues suggested that differential methylation in the putative promoter region of the long variants (covered by amplicons A3-A5) might contribute to cell type- and tissue-specific expression of Esrp2 and Esrp2-as (Supplementary Figure S5). Differential methylation was verified in spleen versus liver (Supplementary Figure S6). We also confirmed low expression of Esrp2 and Esrp2-as in murine mammary fibroblast versus Mecs. The long variants v1+2 were undetectable in fibroblasts, whereas expression levels of Esrp2 and Esrp2-as v1-4 were 13- and 10.5-fold higher in primary murine Mecs than in fibroblasts (Supplementary Figure S12). The preferential epithelial expression of Esrps was noted previously66 and led to their designation as epithelial splicing regulatory proteins.66 Earlier work also indicated that induction of EMT by TGFB1,(ref. 13) overexpression of EMT-inducing TFs or knockdown of E-cadherin completely abrogated Esrp expression.66
Knockdown or overexpression experiments did not support a role of Esrp2-as in transcriptional regulation of Esrp2 (Figure 5, Supplementary Figure S8). Rather, knockdown of Esrp2-as led to a significant reduction in Esrp2 protein levels. LncRNAs can alter translation of protein-coding genes without affecting mRNA levels, for example, by formation of a ribonucleoprotein complex with RNA-binding proteins, as shown for the regulation of E-cadherin by translational regulatory lncRNA (treRNA) with tumor- and metastasis-promoting properties.67 LncRNAs can also influence the association of mRNAs with active polysomes, such as lincRNA-p21 regulating β-catenin and JUNB levels,68 GAS5 interacting with MYC mRNA to reduce its translation,69 or lncRNA-Uchl1 controlling Ubiquitin carboxy terminal hydrolase-L1 (Uchl1) translation in mice.70 Alternatively, direct lncRNA-protein interaction can influence protein stability, as reported for the interaction of BMI1, a member of the PRC1 repressive complex, with oncogenic FAL1(ref. 6) and for lncRNA PVT1 interacting with MYC, thus preventing MYC phosphorylation and degradation.71, 72 Further analyses have to clarify how Esrp2-as contributes to enhancing Esrp2 translation or protein stability.
Knockdown of Esrp2-as in M6 cells resulted in significant differential expression of 185 genes, many of which code for ECM core proteins (collagens, Multimerin-2, Netrin-G2), ECM remodelers15 (MMP9, Plau, Serpins), or secreted factors regulating the ECM36, 73 (Supplementary Table S2 and Supplementary Table S3, Supplementary Figure S12). Alterations in the ECM facilitate cell migration, and consistently, we observed upregulation of several transcription factors controlling EMT,16 such as Zeb1, Zeb2, Snail2, and Twist2, resulting in reduced expression of E-cadherin. Knockdown of Esrp2-as also induced a more cancer stem cell-like transcriptional profile, with reduced expression of differentiation markers (Epcam, Gata3(ref. 74)) and upregulation of stem cell markers, such as Sca-1(ref. 75), Lgr6(ref. 76) and Procr.77
It is currently thought that ESRPs are repressed by EMT regulators such as Zeb1(ref. 13), and subsequent alternative splicing events contribute to EMT.17 Here we show that downregulation of Esrp2-as and loss of Esrp2 protein increased expression of the EMT regulators. These findings suggest more complex regulatory circuits and feedback loops between up- and downstream factors involved in regulation of EMT than currently anticipated. Lineage tracing78, 79 and conditional knockin and knockout mouse models might be a tool to delineate causes and consequences of Esrp2-as expression during carcinogenesis.80, 81
Knockdown of Esrp2-as in the M6 cell line resulted in reduced cell proliferation and reduced expression of proliferation markers Pcna and Ki67 concomitant with upregulation of genes with anti-proliferative or tumor-suppressive properties, including the cyclin-dependent kinase inhibitor p21, TP53Inp1(ref. 82), Ass1(ref. 83), Inka2(ref. 84), and Edar2r.85 Conversely, overexpression of Esrp2-as in M27H4 and 3T3-L1 cells weakly induced cell proliferation (Figure 5f). Since Esrp2-as acts at the translational level or directly targets Esrp2 protein, and these cell lines do not express detectable levels of Esrp2 protein, the effects of Esrp2-as overexpression might be underestimated.
Collectively, our data support the described context-dependent dual function of ESRP2 in cancer progression.19 Elevated levels maintain an epithelial phenotype and proliferative capacity, whereas low levels, as observed at the invasive front in oral squamous cell carcinoma86 and at the resection border in colon cancer87 facilitated EMT, migration, and invasion into the ECM. Both up- and downregulation of ESRP2 could therefore contribute to tumor progression. In addition, ESRP2 expression might be subject to intra-tumor heterogeneity, with higher levels in the center and reduced expression at the periphery in response to signals from the ECM,13 consistent with the plastic expression in oral cancers.86 In clear cell renal carcinoma, ESRP2 splicing function, but not mRNA levels, was an important indicator of prognosis, and splicing was facilitated by ubiquitination of ESRP2 by the ubiquitin ligase Arkadia.88 In this context, it will be interesting to investigate whether the regulatory role of Esrp2-as on Esrp2 protein might involve an influence on post-translational modifications, as described for the interaction of PVT1 and MYC.71
LncRNAs generally show less interspecies conservation than protein-coding genes.89, 90 Nonetheless, using an in silico approach we identified a novel, unannotated human homolog of Esrp2-as that shares 67.5% identity with the long Esrp2-as v1 (Supplementary Figure S13B). We confirmed expression and epigenetic regulation by RT–qPCR analyses and DAC treatment of human cancer cell lines. Using public data from TCGA we detected that ESRP2 and ESRP2-AS expression is upregulated in human breast tumors and that DNA methylation inversely correlated with expression. Upregulation of both transcripts is associated with a significantly elevated hazard ratio for tumor relapse. These findings support the value of mouse models for investigations of human diseases and in providing clinically relevant results.43, 45, 91
Taken together, we here present a resource of lncRNAs that are epigenetically regulated during mammary carcinogenesis in the C3(1) mouse model of breast cancer. Several of the transcripts have been identified as differentially methylated in human breast cancer. We provide novel evidence for the coordinated epigenetic regulation of the splicing regulator Esrp2 and its antisense transcript Esrp2-as by differential methylation of a proximal enhancer region. Esrp2-as affects Esrp2 translation or protein stability, and loss of function experiments indicate a role in the transcriptional regulation of EMT and cell proliferation. These findings are highly relevant for human breast cancer, as our analyses led to the identification of a novel human homolog of Esrp2-as, which is upregulated in human breast cancer and indicates poor prognosis.
Materials and methods
Mouse handling and sample collection
The FVB/N C3(1) SV40TAg mouse model of human breast cancer was previously described.20, 21, 23 Mice were housed and bred in the animal facility of the German Cancer Research Center. Genotyping of mice was conducted by PCR with the following primer pairs (C3(1): fwd: 5′-GGACAAACCACAACTAGAATGCAG-3′, rev: 5′-CAGAGCAGAATTGTGGAGTGG; WT: fwd 5′-GTCAGTCGAGTGCACAGTTT-3′ rev: 5′-CAAATGTTGCTTGTCTGGTG-3′). Animals were sacrificed by CO2 at 20 and 24 weeks of age, and tumors and mammary glands of age-matched WT mice were resected and immediately flash frozen in liquid nitrogen. The study was approved by the state Animal Care and Use Committee (Regierungspräsidium Karlsruhe) as regulated by German federal law for animal welfare under the registration number 35-9185.82/A-15/08.
Nucleic acid isolation
DNA and RNA from tissue samples and cell lines were isolated using the DNA & RNA allPrep Kit or the RNeasy Mini Kit (both Qiagen, Hilden, Germany) following the manufacturer’s instructions, including an on-column DNAse I digest for RNA isolation (RNase-free DNAse set, Qiagen). DNA and RNA were quality checked by Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and quantified by Nano Drop spectrophotometry and Qubit fluorimetry (Thermo Scientific, Wilmington, USA).
MCIp-seq
MCIp-seq92, 93 was used for genome-wide DNA methylation analysis as in Sonnet et al.92 with minor modifications. In brief, 5 μg DNA in 120 μl EB buffer was sheared to≈150 bp using a S2 sonicator (Covaris, Woburn, MA, USA) for 6 cycles (duty cycle 20%, intensity 5, burst per cycle 200, time: 60 s). Size distribution was confirmed on a DNA High sensitivity Chip (Agilent Bioanalyzer), before proceeding with MCIp reaction using a SX8G-V52 robot (Diagenode, Liège, Belgium) for automated processing. 60 μg of MBD2-Fc protein and 40 μl of protein A coated paramagnetic beads (Diagenode) were used for the reaction.
DNA eluted with the highest salt concentration was submitted to the DKFZ Genomics and Proteomics Core Facility for library preparation and next generation sequencing on an Ilumina HiSeq 2000 sequencer (single read 50 bp). We processed DNA from 3 tumors and 3 WT mammary glands each for the 20 and 24 week age groups for MCIp analysis.
Alignment of reads to the mouse reference genome mm10 was performed with Burrows Wheeler aligner (BWA)94 and duplicate as well as bad quality reads were removed with Picard (https://broadinstitue.github.io/picard) and Samtools.95 Saturation efficiency and CG-coverage by the R-MEDIPs package96 provided an additional control step. Data can be accessed under Gene Expression Omnibus (GEO) accession number GSE77096.
DMR calling and candidate selection
For calling of DMRs, reads for animals of the same age group and genotype were combined and analyzed with Homer (findPeaks: FDR<0.001, P-value<0.0001, size 150 bp, minDist 300 bp).97 Co-ocurrence of DMRs was calculated by the mergePeaks command (-d 300 –venn) and DMRs were overlapped with promoter regions of 3639 Refseq lncRNA (+2 kb, -0.5 kb TSS) (GRCm38/mm10, http://genome.ucsc.edu,98) by Bedtools intersectBed.99 We selected lncRNAs with a neighboring protein-coding RNA in antisense orientation, and the TSSs should be maximally 2 kb apart. We requested that gene expression in the C3(1) mouse model24 was significantly different (P<0.05, two-sided, unpaired Student’s t-test) between tumor samples and mammary glands for the protein-coding RNAs.
cDNA synthesis and RT–qPCR
Five hundred nanogram to 1 μg of RNA was reverse transcribed with Superscript II reverse transcriptase (Life Technologies, Darmstadt, Germany) using 200 ng random hexamers. Real-time qPCR analysis was performed using the Universal Probe Library system (Roche, Penzberg, Germany) using a program of 15 min at 95 °C followed by 45 cycles of 10 s at 95 °C, 20 s at 55 °C and 10 s at 72 °C on a Lightcycler480 Real-Time PCR System (Roche). Expression levels of target genes were normalized to three housekeeping genes (Hprt1, Tbp, β-Actin) according to the Livak method.100 Primers and respective probe numbers are listed in Supplementary Table S5.
Quantitative methylation analysis by EPITYPER
Five hundred nanogram to 1 μg genomic DNA was sodium bisulfite treated using the EZ methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. Quantitative DNA methylation analysis of single CpG units was performed using EPITYPER technology (Agena Bioscience, San Diego, CA, USA) as previously described.55, 101 Primers are listed in Supplementary Table S6.
Cell lines and cell culture
The M28N2, M27H4, M6 and M6C cell lines (kindly provided by Cheryl Jorcyk, Boise State University) were derived from the C3(1) model.23 3T3-L1 mouse preadipocytes,102 platinum-E (Plat-E) retroviral packaging cells103 and NMuMG mouse mammary gland epithelial cells104 were a kind gift from Daniel Mathow, and Hepa1.6 murine hepatoma cells105 were generously provided by Ursula Klingmüller (both DKFZ Heidelberg). The human breast cancer cell line MCF7 was provided by the cell line repository of the German Cancer Research Center. HepG2 liver and MDA-MB231 basal-like breast cancer cell lines were derived from American Type Culture Collection (Manassas, VA, USA). The MCF10a cell line was kindly provided by Doris Mayer (DKFZ, Heidelberg, Germany). Cell lines were cultivated in DMEM +10% FCS in a humidified atmosphere at 5% CO2 and 37 °C. For NMuMG cells, medium contained 10 μg/ml insulin, and for MCF10a cells, DMEM/Ham-F12 medium was supplemented with 5% horse serum, 0.5 μg/ml hydrocortisone, 0.02 μg/ml rHuEGF and 5 μg/ml insulin. Primary murine mammary epithelial cells and fibroblasts were isolated and cultured as described previously.106, 107 Identity of cell lines was verified by single tandem repeat typing. Cell lines were checked regularly for mycoplasma contamination. DNA and RNA from MC38 murine colon carcinoma cells and CMT93 murine rectum carcinoma cells were provided by Christoph Weigel (DKFZ Heidelberg, Germany).
Decitabine treatment
M27H4, MCF7 and MDA-MB231 cells were treated with 1 μm DAC (Decitabine, Sigma-Aldrich, Taufkirchen, Germany) dissolved in DMSO (0.5% final concentration) for 72 h starting at 24 h post seeding and renewing the medium every 24 h.
Cloning of overexpression and luciferase reporter constructs
Esrp2-as transcript variants v1 and v4 were PCR amplified from cDNA with Phusion high fidelity Polymerase (New England Biolabs, Frankfurt a.m., Germany) attaching KpnI and SalI sites. Fragments were subcloned into the pCR2.1 vector via the TOPO-TA cloning Kit (Life Technologies, Darmstadt, Germany). The fragments were excised by KpnI and SalI restriction digest and ligated into the respectively cut pCRII-cGFP-bGH vector (a kind gift of Sven Diederichs, DKFZ Heidelberg, Germany).
For dual luciferase reporter assays, sequences covering regions upstream of the Esrp2 and the Esrp2-as TSS were PCR amplified from genomic DNA attaching HindIII and/or NheI restriction sites. Fragments were inserted into the respectively cut pGL4.10 and pGL4.23 vectors (Promega Madison, WI, USA) for assessment of promoter or enhancer activity. Correct sequences of clones were ascertained by Sanger sequencing (GATC biotech, Konstanz, Germany). Primer sequences are listed in Supplementary Table S7.
Dual luciferase reporter assays
Hepa1.6 cells were reverse transfected with 40 ng of pGL4.10 or pGL4.23 firefly luciferase reporter constructs using 0.2 μl GenJet (Ver.II-LnCAP) (SignaGen Laboratories, Rockville, MD, USA) and 10 ng CMV-Renilla luciferase as a transfection normalization control. The plasmids were separately adjusted to 5 μl with plain DMEM and incubated for 5 min before mixing. The transfection reagent was diluted in 5 μl DMEM and after short mixing immediately added to the plasmids. After incubation for 15 min, the transfection mixture was added to the wells of a 384 well plate with≈6000 cells per well. Luciferase activity was measured after 48 h on a Spectramax microplate reader as previously described.108 Measurements were taken for 8 technical transfection replicates of 4 independent experiments and normalized to the respective pGL4.10 or pGL4.23 EV.
LNA antisense Gapmer-mediated knockdown
Custom designed antisense oligos (Exiqon, Vedbaek, Denmark) against Esrp2-as and the negative control oligo A were reverse transfected into cell lines M6, M28N2 and NMuMG with Dharmafect 1 solution (GE Healthcare, Buckinghamshire, United Kingdom). For transfections, LNAs at 20 nm and Dharmafect1 (2 μl per well for 12-plates) were mixed in serum-free DMEM according to manufacturer’s instructions. The transfection mix was added to the cells and incubated for 72 h (M6, NMuMG) or 96 h (M6, M28N2). In M6 cells, a long-term knockdown series was performed by repeatedly transfecting cells for four times in 96 h intervals. Esrp2-as and Esrp2 expression was analyzed by RT–qPCR and RNA-seq.
Overexpression of Esrp2-as and Esrp2
For overexpression of Esrp2-as in M27H4, Hepa 1.6, and 3T3-L1 cells, constructs (v1 or v4) were diluted in serum-free DMEM and mixed with respectively diluted TransIT-Lt1 transfection reagent (Mirus Bio LLC, Madison, WI, USA) at a ratio of 3:1 Transfection reagent to plasmid DNA. 20–30 min after mixing, the transfection mix was evenly distributed to cells, which were seeded 18–24 h before (≈1 × 105 cells per well in six-well plates). Cells were harvested after 72 h (M27H4, 3T3-L1) or 48 h (Hepa 1.6 and 3T3-L1) and analyzed by RT–qPCR or RNA-seq.
The retroviral vector pMXs-IRES-Blast-Esrp2-FF109 and the corresponding empty vector with GFP were a gift from Russ P Carstens (University of Pennsylvania). Retroviral infection of M27H4 and 3T3-L1 cells was performed as in,110 followed by selection with Blasticidin S (Sigma-Aldrich, Taufkirchen, Germany) at 5 μg/ml for stable incorporation.
Protein extraction and western blot analysis
Whole cell lysates were prepared by lysing washed cells in RIPA buffer (50 mm Tris-HCl pH7.5, 1% NP-40, 0.25% sodium deoxycholate, 150 mm sodium chloride, 2 mm magnesium chloride, 0.1% sodiumdodecylsulfate) on ice for 10 min with 2–3 steps of thorough vortexing. Protein content was determined using the Bicinchoninic acid assay (Thermo Fisher Scientific, Dreieich, Germany). Equal amounts of proteins were supplemented with 4x TruPage Lithium dodecyl sulfate (LDS) loading buffer (Sigma, Germany) and denatured by boiling at 95 °C for 20 min. Lysates were loaded onto precasted 4-20% Bis-Tris polyacrylamide TruPage gels (Sigma, Germany). Electrophoresis, blotting, detection and quantification were performed as described in Pappa et al.111 For detection of Esrp2, anti-Esrp2 monoclonal antibody (210-301-C32S, Rockland Immunochemicals, USA) was used at 1:250 dilution. Equal loading was ensured by detecting β-actin (sc-47778, 1:10 000, Santa Cruz Biotechnology Inc., Heidelberg, Germany).
RNA Ribodepletion and RNA-seq
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) from two to three independent replicates each of M6 cells after knockdown of Esrp2-as using LNA#2 or transfected with negative control LNA for 72 h, and M27H4/3T3-L1 cells after overexpression of Esrp2-as v4 or transfected with EV for 72 h. Ribosomal RNA was removed from the RNA samples using the Ribo-Zero Gold rRNA Removal Kit (MRZG12324, Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Ribodepleted RNA was purified using the RNAeasy MinElute Kit (Qiagen). Ribodepleted RNA (10 ng) was subjected to strand-specific library preparation with the SureSelect Strand-Specific Library Preparation Kit (G9691A, Agilent Technologies, Santa Clara, CA, USA). For high-throughput RNA sequencing, six samples were multiplexed on one lane and a 100 base-pair paired-end protocol was applied on the Illumina HiSeq4000 platform (Genomics and Proteomics Core Facility of the DKFZ, Heidelberg, Germany).
After quality control and adapter trimming with fastqc and cutadapt,112 the sequences were aligned using HISAT2(ref. 113) with the following parameters: —rna-strandness RF, —max-intronlen 20000, —no-unal, —dta. The quality of the alignment was assessed with qualimap.114 The transcripts were assembled using the Stringtie's115, 116 -eB option and the Ensembl mouse GRCm38 assembly (release 85) as a reference. The raw counts of the genes were calculated with the Stringtie's prepDE.py script. Statistical analysis was then performed with the edgeR117, 118 R package. During the analysis, technical replicates were averaged and used together as a biological replicate. Data can be accessed under Gene Expression Omnibus (GEO) accession number GSE96641.
Gene ontology enrichment of genes differentially expressed in M6 cells after knockdown of Esrp2-as was performed using GSEA MSigDB v5.1,.35 M6 expression data (limited to significantly up- and downregulated genes with FDR-adjusted P-value <0.05 and CPM (counts per million) >0.5) were analyzed and networks generated through the use of QIAGEN’s Ingenuit Pathway Analysis tool (IPA (www.qiagen.com/ingenuity), QIAGEN Redwood City).
Determination of cell proliferation by Sulforhodamine B (SRB) staining
M6, M27H4 and 3T3-L1 cells were collected 24 h after Esrp2-as knockdown and overexpression and reseeded at 1000 and 4000 cells/well in 96-well plates. Cells were fixed at day 1–6 post-transfection with 50 μl ice-cold 10% trichloroacetic acid. Further processing, staining with SRB and data analysis was performed as described previously.111
ESRP2/ESRP2-AS expression analysis
MCF-7 CAGE-seq and strand-specific MCF7 mRNA expression data was derived from ENCODE39 using the UCSC genome browser.40 TCGA BRCA breast cancer RNA-seq data processing was started with sliced bam files in the region chr16, 67557312-68844434 (hg38). The raw counts of ESRP2 and the ESRP2 antisense transcript were counted with HTSeq119 (parameters used: -m intersection-nonempty -i gene_id -r pos -s no) using gencode v22 as a reference. The ESRP2 antisense transcript region was added to the reference file with the coordinates chr16:68234577-68239177. Normalized read counts were analyzed for differential expression in 1002 breast tumor samples and 45 normal control samples. The association between expression and risk of relapse was represented by Kaplan–Meier plots and tested with log-rank Mantel-Cox test (GraphPad Prism).
Statistical analyses
Statistical tests were performed using GraphPad Prism version 6 (GraphPad Software Inc., San Diego, CA, USA), Microsoft Excel 2007, and R. Correlation analyses are based on Spearman’s rank analysis. Effects of knockdown and overexpression were expressed in relation to the negative or empty vector control set as 1 or 100%. Normal distribution of the data was checked. Single-sample or two-tailed Student’s t-tests were used for comparisons. Equal variance between groups was tested, and if applicable, t-test with Welch’s correction was used. Rank sum analysis by Mann-Whitney U test or Wilcoxon rank sum test was conducted as indicated. If not stated otherwise, n≥3 biological replicates were analyzed. All data are expressed as mean±standard deviation unless otherwise stated. P-values are given as *P <0.05, **P<0.01, ***P<0.001, ****P<0.0001 and NS, not significant.
Acknowledgments
KH was supported by a Ph.D. scholarship provided by the DKFZ Helmholtz International Graduate School for Cancer Research. We are grateful to Megumio Onishi Seebacher for providing barcoded oligonucleotide sequences, the Genome and Proteome core facility of the DKFZ for providing technical support and excellent MCIp-seq and RNA-seq services, and Lorena Salgueiro and Rocio Sotillo Roman for their instant support in isolating murine mammary epithelial cells and fibroblasts. We further want to thank Oliver Mücke and Monika Helf for excellent technical assistance, and Thomas Risch, Maïwen Caudron-Herger, Odilia Popanda and Dieter Weichenhan for helpful discussion. The results reported here are in part based upon data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer 2015; 1: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013; 14: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 2012; 19: 1068–1075. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013; 52: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013; 14: 880–893. [DOI] [PubMed] [Google Scholar]
- Hu X, Feng Y, Zhang D, Zhao Sihai D, Hu Z, Greshock J et al. A functional genomic approach identifies fal1 as an oncogenic long noncoding RNA that associates with BMI1 and Represses p21 Expression in Cancer. Cancer Cell 2014; 26: 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 2014; 55: 604–614. [DOI] [PubMed] [Google Scholar]
- Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci 2012; 2: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Xu C. Study strategies for long non-coding RNAs and their roles in regulating gene expression. Cell Mol Biol Lett 2015; 20: 323–349. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 2010; 29: 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S et al. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 2012; 31: 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011; 7: e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 2014; 15: 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110. [DOI] [PubMed] [Google Scholar]
- Gottgens EL, Span PN, Zegers MM. Roles and regulation of epithelial splicing regulatory proteins 1 and 2 in Epithelial-Mesenchymal Transition. Int Rev Cell Mol Biol 2016; 327: 163–194. [DOI] [PubMed] [Google Scholar]
- Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 2017; 16: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A, Saitoh M, Miyazawa K. Dual Roles for Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) in Cancer Progression. Adv Exp Med Biol 2016; 925: 33–40. [DOI] [PubMed] [Google Scholar]
- Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene 2000; 19: 1020–1027. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A 1994; 91: 11236–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res 2006; 66: 6118–6128. [DOI] [PubMed] [Google Scholar]
- Holzer RG, MacDougall C, Cortright G, Atwood K, Green JE, Jorcyk CL. Development and Characterization of a Progressive Series of Mammary Adenocarcinoma Cell Lines Derived from the C3(1)/SV40 Large T-antigen Transgenic Mouse Model. Breast Cancer Res Treat 2003; 77: 65–76. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AM, Tian AX, Zhang RX, Ge J, Sun X, Cao XC. Protocadherin-7 induces bone metastasis of breast cancer. Biochem Biophys Res Commun 2013; 436: 486–490. [DOI] [PubMed] [Google Scholar]
- Iwakiri M, Mizukami K, Ikonomovic MD, Ishikawa M, Abrahamson EE, DeKosky ST et al. An immunohistochemical study of GABA A receptor gamma subunits in Alzheimer's disease hippocampus: relationship to neurofibrillary tangle progression. Neuropathology 2009; 29: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Pei L, Wang L, Zhang F, Hu X, Gui Y. Snail1-dependent transcriptional repression of Cezanne2 in hepatocellular carcinoma. Oncogene 2014; 33: 2836–2845. [DOI] [PubMed] [Google Scholar]
- Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int 2014; 14: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JA, Lee BB, Kim Y, Park SE, Heo K, Hong SH et al. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget 2013; 4: 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves DK, Fagan-Solis KD, Dunphy K, Oliver SD, Scott DW, Fleming JM. The role of lipolysis stimulated lipoprotein receptor in breast cancer and directing breast cancer cell behavior. PLoS ONE 2014; 9: e91747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010; 40: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V et al. A promoter-level mammalian expression atlas. Nature 2014; 507: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015; 16: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 2007; 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the "omics" era. Matrix Biol 2016; 49: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir M, Damineni S, Mukherjee G, Kondaiah P. Activin-A signaling promotes epithelial–mesenchymal transition, invasion, and metastatic growth of breast cancer. Npj Breast Cancer 2015; 1: 15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci 2017; 74: 951–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res 2013; 41: D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Li S, Lu J, Chen J, Wang Y et al. Genome-wide DNA methylome analysis reveals epigenetically dysregulated non-coding RNAs in human breast cancer. Sci Rep 2015; 5: 8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CN, Green JE. Genomic analyses as a guide to target identification and preclinical testing of mouse models of breast cancer. Toxicol Pathol 2010; 38: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CN, Green JE. Unlocking the power of cross-species genomic analyses: identification of evolutionarily conserved breast cancer networks and validation of preclinical models. Breast Cancer Res 2008; 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C et al. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res 2007; 67: 8065–8080. [DOI] [PubMed] [Google Scholar]
- Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D et al. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol 2011; 12: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Birnbaum D. Basal breast cancer: a complex and deadly molecular subtype. Curr Mol Med 2012; 12: 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E et al. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol 2014; 32: 2794–2803. [DOI] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013; 24: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lindsay J, Feng ZP, Frankenberg S, Hu Y, Carone D et al. Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics 2012; 13: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau YM, Pando S, Taylor HS. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. J Clin Endocrinol Metab 2002; 87: 2674–2680. [DOI] [PubMed] [Google Scholar]
- Yarmishyn A, Batagov A, Tan J, Sundaram G, Sampath P, Kuznetsov V et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics 2014; 15: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer 2002; 2: 777–785. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res 2009; 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faryna M, Konermann C, Aulmann S, Bermejo JL, Brugger M, Diederichs S et al. Genome-wide methylation screen in low-grade breast cancer identifies novel epigenetically altered genes as potential biomarkers for tumor diagnosis. FASEB J 2012; 26: 4937–4950. [DOI] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA 2008; 105: 12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res 2008; 68: 9532–9540. [DOI] [PubMed] [Google Scholar]
- Wang W, Huper G, Guo Y, Murphy SK, Olson JA Jr, Marks JR. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene 2005; 24: 2705–2714. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Koczan D, Thiesen HJ, Ibrahim SM, Gross G, Spang R et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res 2007; 13: 806–815. [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 2012; 47: 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol 2013; 14: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol 2012; 22: 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J 2013; 32: 2672–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One 2014; 9: e107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012; 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Tseng Y-Y, Bagchi A. The PVT1-MYC duet in cancer. Mol Cell Oncol 2015; 2: e974467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014; 512: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Naba A. Overview of the matrisome—an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 2012; 4: a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asch-Kendrick R, Cimino-Mathews A. The role of GATA3 in breast carcinomas: a review. Hum Pathol 2016; 48: 37–47. [DOI] [PubMed] [Google Scholar]
- Batts TD, Machado HL, Zhang Y, Creighton CJ, Li Y, Rosen JM. Stem cell antigen-1 (sca-1) regulates mammary tumor development and cell migration. PLoS One 2011; 6: e27841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaas L, Pucci F, Messal HA, Andersson AB, Josue Ruiz E, Gerling M et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol 2016; 18: 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 2015; 517: 81–84. [DOI] [PubMed] [Google Scholar]
- Drake LE, Macleod KF. Tumour suppressor gene function in carcinoma-associated fibroblasts: from tumour cells via EMT and back again? J Pathol 2014; 232: 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 2014; 28: 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mouse models in cancer research. Adv Cancer Res 2010; 106: 113–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 2017; 9: 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma T, Filippone MG, Pierantoni GM, Fusco A, Soddu S, Zannini M. Pax8 has a critical role in epithelial cell survival and proliferation. Cell Death Dis 2013; 4: e729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Wu WR, Wang YH, Wang JW, Fang FM, Tsai JW et al. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin Cancer Res 2013; 19: 2861–2872. [DOI] [PubMed] [Google Scholar]
- Minden A. The pak4 protein kinase in breast cancer. ISRN Oncol 2012; 2012: 694201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punj V, Matta H, Chaudhary PM. X-linked ectodermal dysplasia receptor is downregulated in breast cancer via promoter methylation. Clin Cancer Res 2010; 16: 1140–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 2014; 289: 27386–27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloria AJ, Hoflmayer D, Kienzl P, Lopatecka J, Sampl S, Klimpfinger M et al. Epithelial splicing regulatory protein 1 and 2 paralogues correlate with splice signatures and favorable outcome in human colorectal cancer. Oncotarget 2016; 7: 73800–73816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani A, Koinuma D, Seimiya H, Miyazono K. The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma. Oncogene 2016; 35: 3514–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014; 1840: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet 2014; 30: 121–123. [DOI] [PubMed] [Google Scholar]
- Sun H, Chen J, Qian W, Kang J, Wang J, Jiang L et al. Integrated long non-coding RNA analyses identify novel regulators of epithelial-mesenchymal transition in the mouse model of pulmonary fibrosis. J Cell Mol Med 2016; 20: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnet M, Baer C, Rehli M, Weichenhan D, Plass C. Enrichment of methylated DNA by methyl-CpG immunoprecipitation. Methods Mol Biol 2013; 971: 201–212. [DOI] [PubMed] [Google Scholar]
- Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics 2007; 90: 314–323. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Li H, Gu Y, Davidson NE, Zhou Q. Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis 2010; 31: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014; 30: 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004; 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 2014; 47: 11 12 11–11 12 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 2005; 102: 15785–15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell 1974; 3: 127–133. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000; 7: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Deckers M, van Dinther M, Buijs J, Que I, Löwik C, van der Pluijm G et al. The tumor suppressor Smad4 is required for transforming growth factor β–induced epithelial to mesenchymal transition and bone metastasis of breast cancer Cells. Cancer Res 2006; 66: 2202–2209. [DOI] [PubMed] [Google Scholar]
- Darlington GJ. Liver cell lines. Methods Enzymol 1987; 151: 19–38. [DOI] [PubMed] [Google Scholar]
- Smalley MJ. Isolation, culture and analysis of mouse mammary epithelial cells. Methods Mol Biol 2010; 633: 139–170. [DOI] [PubMed] [Google Scholar]
- Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mammary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Genes Dev 2009; 23: 1677–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C, Oakes CC, Ruppert AS, Claus R, Kim-Wanner S-Z, Mertens D et al. Epigenetic silencing of miR-708 enhances NF-κB signaling in chronic lymphocytic leukemia. Int J Cancer 2015; 137: 1352–1361. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009; 33: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roring M, Herr R, Fiala GJ, Heilmann K, Braun S, Eisenhardt AE et al. Distinct requirement for an intact dimer interface in wild-type, V600E and kinase-dead B-Raf signalling. EMBO J 2012; 31: 2629–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa G, Strathmann J, Lowinger M, Bartsch H, Gerhauser C. Quantitative combination effects between sulforaphane and 3,3'-diindolylmethane on proliferation of human colon cancer cells in vitro. Carcinogenesis 2007; 28: 1471–1477. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011; 17: 10–12 Available at http://journal.embnet.org/index.php/embnetjournal/article/view/200 (accessed on 29 December 2016). [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016; 32: 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016; 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015; 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.