Abstract
STIL (SCL/TAL1 interrupting locus) is a core component of the centriole duplication process. STIL mutations have been associated with both autosomal recessive primary microcephaly (MCPH) and holoprosencephaly. In this report, we describe a family with multiple miscarriages and 2 terminations of pregnancy due to marked fetal microcephaly, delayed cortical gyrification, and dysgenesis of the corpus callosum. Whole exome sequencing allowed us to identify novel compound heterozygous mutations in STIL. The mutations lie, respectively, in the CPAP/CENPJ and the hsSAS6 interacting domains of STIL. M-phase synchronized amniocytes from both affected fetuses did not display an aberrant number of centrioles, as shown previously for either STIL-depleted or overexpressing cells. However, we observed an elongation of at least 1 centriole for each duplicated centrosome. These preliminary results may point to a novel mechanism causing MCPH and embryonic lethality in humans.
Keywords: Centriole length, Fetal microcephaly, STIL mutation
Autosomal recessive primary microcephaly (MCPH) is defined as an occipito-frontal head circumference (OFC) at birth reduced to at least 2–3 SDs below the mean compared to age, sex, and ethnicity matched controls, characterized by prenatal onset and a slower than average growth in OFC after birth [Mahmood et al., 2011; Morris-Rosendahl and Kaindl, 2015]. Some MCPH patients might display a simplified cortical gyral pattern, but the brain architecture is generally unaffected. Variable features observed in MCPH patients include cognitive impairment – ranging from mild to severe – reduced stature, and craniosynostosis [Mahmood et al., 2011; Barbelanne and Tsang, 2014; Verloes et al., 2017]. Currently (accessed April 2017), OMIM lists 17 genes that have been associated with MCPH. It is not surprising that several of these genes encode for proteins that localize to the centrosome or mitotic spindle pole (CPAP/CENPJ, STIL, CEP135, CEP152, and SASS6), underscoring the importance of the centrosome for proper cell division, especially during early neurogenesis, when timing and symmetry of neuroprogenitor cell division are tightly regulated [Paridaen and Huttner, 2014; Hardwick et al., 2015]. In general, MCPH-causing genes are involved in the regulation of centriole duplication, spindle positioning, and kinetochore attachment for proper chromosome segregation in mitosis but also in the maintenance of genome stability via the DNA damage repair response pathways [Alcantara and O'Driscoll, 2014; Morris-Rosendahl and Kaindl, 2015]. Morever, in recent years, it has become clear that MCPH has both a genetic and clinical overlap with other neurodevelopmental disorders, such as Seckel syndrome, Meier-Gorlin syndrome, and microcephalic primordial dwarfism. For instance, biallelic defects in the genes CPAP/CENPJ and CEP152 have been associated to both MCPH and Seckel syndrome [Bond et al., 2005; Al-Dosari et al., 2010; Guernsey et al., 2010; Kalay et al., 2011], CENPE mutations to MCPH and microcephalic primordial dwarfism [Mirzaa et al., 2014], while STIL mutations have been found both in patients with MCPH and holoprosencephaly (HPE) [Kumar et al., 2009; Kakar et al., 2015; Mouden et al., 2015], leading to the conclusion that this group of disorders may be considered as a clinical continuum rather than individual entities [Barbelanne and Tsang, 2014; Morris-Rosendahl and Kaindl, 2015].
In the last years, the implementation of massively parallel sequencing in both research and medical practice contributed significantly to the acceleration in the identification of disease-associated genes, especially for rare and clinically heterogeneous mendelian disorders [Gilissen et al., 2011; Dixon-Salazar et al., 2012; Need et al., 2012; Rabbani et al., 2012, 2014; Yang et al., 2013, 2014; Wang et al., 2014]. Although the interpretation of the clinical significance and pathogenicity of variants identified by whole exome (WES)/genome sequencing is often challenging [Richards et al., 2015; Amendola et al., 2016], these technologies allowed researchers to determine the underlying developmental and molecular processes disrupted in many neurodevelopmental disorders, improving the clinical management as well as genetic counseling [Green and Guyer, 2011; Soden et al., 2014; Morris-Rosendahl and Kaindl, 2015].
Here, we report a family with multiple miscarriages and 2 terminations of pregnancy because of profound fetal microcephaly (MC) associated with delayed gyrification and dysgenesis of the corpus callosum. WES allowed the identification of novel compound heterozygous mutations in STIL.
Since overexpression and depletion of STIL result, respectively, in the excess of formation and loss of centrioles [Kitagawa et al., 2011; Tang et al., 2011; Vulprecht et al., 2012; Arquint and Nigg, 2014], we looked for similar abnormalities in amniocytes of the affected fetuses. Surprisingly, upon functional testing, no centriolar depletion or amplification was observed. However, we detected a significant elongation of at least 1 centriole in each centrosome in the patients' cells. Since the variants are located in the CPAP/CENPJ and the hsSAS6 interacting domains, respectively, these results might point to a novel disease-causing mechanism, namely centriole elongation.
Case Report
This healthy nonconsanguineous couple was referred to our tertiary center at 21 weeks of gestation because of suspicion of MC. After 5 first-trimester miscarriages, the couple had a healthy daughter and a healthy son (Fig. 1). The 5 spontaneous miscarriages occurred between the 7th and 11th gestational week. Chromosomal causes of recurrent miscarriages were excluded in both parents. The 8th pregnancy resulted after ovulation induction with clomiphene citrate. Fetal ultrasound biometry measurements were within the range for gestational age (21 weeks post menstrual age), but the OFC and biparietal diameter were well below the 3rd percentile [157.6 mm (normal: 176 ± 20 mm) and 42.8 mm (normal: 49 ± 4 mm), respectively]. In the sagittal plane, the sloping forehead confirmed MC (Fig. 2A). Enhanced fetal neurosonography revealed a normally shaped but small cerebellum (3rd percentile) and a normal vermis (7.7 mm). The cavum septi pellucidi was present but appeared very small. The gyri were poorly developed, and sulci were absent (Fig. 2B). The corpus callosum appeared short and thick (11 mm), suggesting the diagnosis of a partial agenesis of the corpus callosum with an abnormal vascular pattern of the pericallosal artery (Fig. 2C). An amniocentesis was performed and revealed a normal fetal molecular karyotype. Mutation analysis of ARX was also normal. After multidisciplinary counseling, the couple opted to terminate the pregnancy at 25 weeks of gestation. Pathological examination of the male fetus did not reveal any additional external or internal macroscopic or microscopic anomalies.
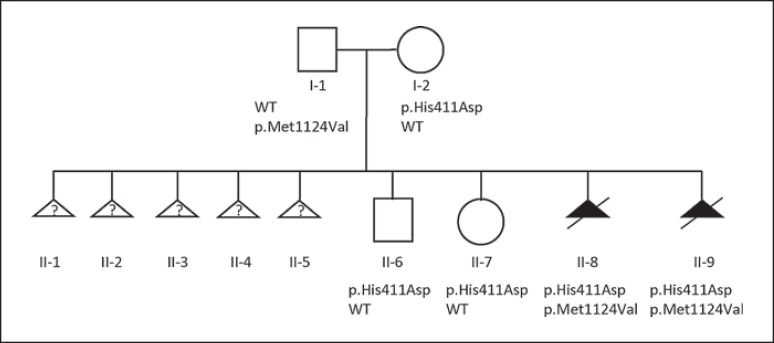
Fig. 1.
Pedigree of the family. WES was performed for individuals I-1, I-2, II-8 and II-9. Sanger sequencing demonstrated that II-6 and II-7 inherited the maternal p.His411Asp variant. WT, wild type.
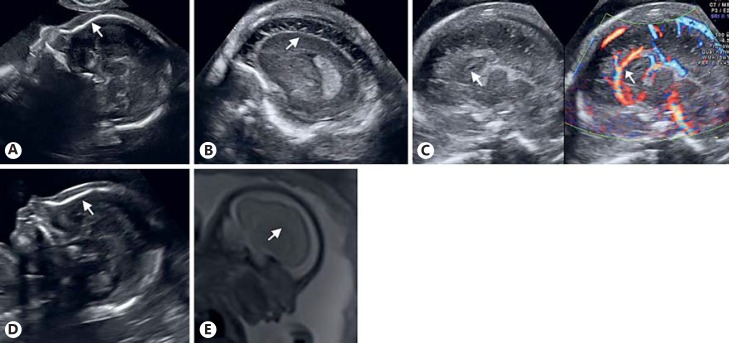
Fig. 2.
Prenatal imaging of the brain of both affected fetuses. A-C Second trimester ultrasound scans of the first affected fetus (II-8). A Sagittal plane showing the sloping forehead indicating microcephaly (arrow). B Enhanced neurosonogram showing poorly developed gyri and absent sulci (arrow). C Sagittal plane revealing a short corpus callosum with absent splenium and an aberrant course of the pericallosal artery, indicated by arrows. D, E Second trimester scan of the second affected fetus (II-9). D Sagittal plane showing the sloping forehead, indicating microcephaly (arrow). E Fetal MRI confirming the delayed gyration for gestational age. Dysgenesis of the corpus callosum is shown in the midsagittal view (arrow).
Within a year, the patient became pregnant again, without additional treatment. The second-trimester ultrasound scan (20 weeks post menstrual age) revealed an MC with a sloping fetal forehead (Fig. 2D). The biparietal diameter was 44.2 mm (normal: 47 ± 3.5 mm) and OFC was 164.9 mm (normal: 170 ± 20 mm), both below the 3rd percentile, but all other measurements were in line with gestational age. The small cavum septi pellucidi was associated with a short corpus callosum (<3rd percentile) because of an absent splenium [Malinger et al., 2012]. Further evaluation confirmed the abnormal development of the cortex, with flattening of the insula, absence of the lateral median and the lateral parieto-occipital sulci as well as pachygyria. Fetal MRI confirmed the ultrasound findings (Fig. 2E). Again, the couple chose to terminate the pregnancy. Due to autolysis, pathological examination of the fetal brain was not possible.
Materials and Methods
Library Preparation, Exome Enrichment and Massively Parallel Sequencing
First, 200 ng of genomic DNA per each individual were quantified using the Qubit® dsDNA Broad Range Assay kit on a Qubit Fluorometer (Thermo Fisher Scientific Inc.). Genomic DNA was fragmented using a Covaris M220 ultrasonicator (Covaris, Woburn). Sequencing libraries were prepared following the SureSelectXT Target Enrichment System for Illumina Paired-End Sequencing version B1 December 2014 (Agilent Technologies). The OneSeq Constitutional Research Panel was then used for exome enrichment according to the manufacturer's specification (Agilent Technologies). Sheared DNA, full-genomic and exome-enriched libraries quality was assessed with either Agilent DNA 1000 or DNA High Sensitivity chips run on an Agilent 2100 Bioanalyzer instrument. The samples were then sequenced on an Illumina HiSeq2500 machine in rapid mode using a paired-end 2×100 bp protocol. We obtained on average an 88-fold enrichment of the target region, a mean 30× coverage of 93% and 10× coverage of 97%. Sequence reads were aligned to the human genome reference sequence (Genome Assembly GRCh37/hg19) with the Burrows-Wheeler Aligner (BWA version 0.7.8). SAMtools (version 0.1.19) were used for SAM to BAM files conversion, sorting and indexing alignments. Picard tools (version 1.118) were used to compute quality metrics and mark PCR-generated duplicates. The Genome Analysis Toolkit (GATK version 3.2.2) software package was used to perform local realignment around indels and base quality score recalibration. SNPs and small indels were called using GATK HaplotypeCaller (version 3.2.2). Variants annotation was performed with ANNOVAR (version 11-0882013), including data sets from dbSNP137, the NHLBI 6500 Exome (version October 2012), and 1000 Genomes Projects (version April 2012) for variant frequencies, amino acid change, functional predictions from SIFT, Polyphen2, LRT, MutationTaster, PhyloP and GERP++ conservation scores.
Variants Filtering and Sanger Sequencing Validation
Trio-based filtering of all annotated variants collected on an Excel file was conducted manually according to several inheritance patterns using genotype predictions generated by the GATK UnifiedGenotyper tool (homozygous recessive, compound heterozygous, hemizygous, and de novo shared by the affected fetuses). “knownGene”-based annotations were used for further filtering: only exonic, splicing, and nonsynonymous variants were retained. Only variants completely absent from or with a minor allele frequency (MAF) <1% in 1000 Genomes Project, ESP6500si_ALL and dbSNP137 were retained. Candidate variants were further manually explored in the ExAC database (http://exac.broadinstitute.org/), Kaviar (http://db.systemsbiology.net/kaviar/cgi-pub/Kaviar.pl), in the updated versions of the aforementioned databases (1000 Genomes Phase 3, ESP6500SI-V2 and dbSNP147), and by the in-house developed NGS-Logistic software (https://ngsl.esat.kuleuven.be/), which collects anonymized exome sequencing data generated in 5 Belgian Human Genetics Centers [Ardeshirdavani et al., 2014]. Segregation analysis was performed in all living family members – including 2 unaffected siblings – by standard capillary electrophoresis. Sequencing primers were designed with the Primer3 web application (http://primer3.ut.ee/): for the p.His411Asp variant primers FWD 5′-AGTTGTGTGTCTTGGGTAGGTA-3′ and REV 5′-AGCTTCCCTGGTCAACTAACA-3′; for the p.Met1124Val variant primers FWD 5′-AAGGCTCTTTCTGACCACCA-3′, and REV 5′-AGTGTGTTGGCATGCATCAG-3′. PCR products were purified using the ExoSAP-IT® PCR Product Cleanup (Affymetrix) and Sanger sequencing reactions performed with the BigDye® Terminator v3.1 chemistry (Thermo Fisher Scientific Inc.), following bidirectional DNA sequencing with an ABI 3500 Series Genetic Analyzer (Applied Biosystems®). GenBank accession Nos. NM_001048166.1 and NP_001041631.1 were used everywhere in the paper for human STIL. The variants reported here were submitted to the LOVD 3.0 shared installation (individual 00089033, variants 0000147090 and 0000147091).
Immunofluorescence and Microscopy, Centriole Count and Length Measurement
Fetal amniocytes from the affected fetus and from a healthy control (invasive testing performed for advanced maternal age and anxiety, prenatal array result, arr(1–22)×2,(XY)×1), were grown on coverslips in AmnioMAX™ C-100 Basal Medium + Supplement (Gibco™) at 37°C and 5% CO2. At 70% confluency, cells were pre-synchronized for 24 h in a 2 mM thymidine-containing medium which blocks DNA synthesis arresting cells in G1/S phase. After thymidine washout, STLC-containing medium (7.5 µM) was added to arrest cells in prometaphase and cells incubated overnight. Before fixation, cells were released in normal medium for 2 h. Cells were then fixed in ice-cold methanol at −20°C for 10 min, followed by 3× 1% PBS wash, permeabilization in 0.2% PBST (0.2% Tween-20 in 1% PBS) for 5 min, 3× 1% PBS wash, 30-min incubation in blocking buffer (5% BSA/PBS + 0.1% Tween-20) at room temperature, 1-h incubation at room temperature in blocking buffer containing 1:500 of primary rabbit anti-Centrin-2 antibodies (sc-27793-R, Santa Cruz Biotechnology). After 3× 1% PBS wash, cells were incubated in blocking buffer containing a 1:1,000 dilution of secondary antibodies (A-11034 goat anti-rabbit Alexa Fluor® 488, Invitrogen) for 1 h at room temperature, followed by 3× 1% PBS wash and a 15-min incubation in DAPI-containing 1% PBS (1:10,000). Stained coverslips where then mounted with Mowiol® 4–88 (Sigma-Aldrich). Confocal images were acquired with a Leica TCS SPE laser-scanning confocal system mounted on a Leica DMI 4000B microscope and equipped with a Zeiss Plan Apochromat 63x 1.40NA oil DIC objective. Optical sections were acquired every 0.17 µm. Images were processed with the ImageJ software, using the 3D-project feature without interpolation and slice spacing of 0.1 µm to rotate images and determine CETN2-positive foci length.
Statistical Methods
XLSTAT tools package (Addinsoft) was used to calculate one-way ANOVA, and Tuckey and Dunnett tests for parametric multiple comparisons of CETN2-positive foci average lengths; k-proportions and Monte Carlo tests were used to determine differences in the proportions of the number of centrioles observed in mitotic amniocytes. For all tests, default settings were used.
Results
Compound Heterozygous STIL Mutations Are the Likely Cause of Fetal Microcephaly
We performed WES on genomic DNA of both affected fetuses and the parents (Fig. 1; individuals I-1, I-2, II-8, and II-9). Since the couple experienced several miscarriages, we hypothesized a homozygous or a combined heterozygous recessive inheritance pattern. Since both fetuses were male, we equally looked for X-linked maternally inherited variants, but did not identify any possible pathogenic/damaging candidates (online suppl. Table 1; see www.karger.com/doi/10.1159/000479666 for all suppl. material). Also putative de novo variants shared by the affected fetuses were absent, excluding parental germinal mosaicism as a possible mechanism [Poirier et al., 2013; Zillhardt et al., 2016]. After filtering, no homozygous rare coding variants were detected. Subsequently, we filtered for the presence of rare compound heterozygous mutations, leading to 4 potential candidate genes (ALDH4A1, STIL, TTN, and VWF; online suppl. Table 2). The missense variants identified in STIL (c.C1231G; p.His411Asp, maternally inherited and the c.A3370G; p.Met1124Val, paternally inherited) are predicted to be damaging by the most used prediction softwares and involve highly constrained amino acids as indicated by GERP++ scores (5.05 and 4.99, respectively). The C>G transversion leading to p.His411Asp is absent in all interrogated population genetics databases (dbSNP147, 1000 Genomes, Exome Variant Server, ExAC, and Kaviar), while the C>A transversion at the same position leading to p.His411Asn (rs746778024) is present in dbSNP147 and ExAC with a MAF of 0.00002 (2/121,298 alleles). The p.Met1124Val variant is found in dbSNP147 (rs776799930) and has been identified in the UK10K project with a MAF of 0.000013. Sanger sequencing confirmed the presence of both variants in the fetuses as well as the heterozygous state in both parents and showed that the 2 unaffected siblings carried the maternal p.His411Asp variant.
STIL is a centrosomal protein involved in the maintenance of centrosome integrity, mitotic spindle organization, and positioning [Pfaff et al., 2007; Castiel et al., 2011; Kitagawa et al., 2011; Vulprecht et al., 2012]. Most importantly, it represents a key factor required for proper centriole duplication [Stevens et al., 2010; Arquint et al., 2012]. Mutations in STIL are associated with MCPH7 in humans (OMIM 612703) [Kumar et al., 2009], while ablation of Stil (formerly Sil) – the mouse orthologue of STIL – causes mid-gestation lethality with marked growth retardation, prominent midline neural tube defects, and randomized axial asymmetry due to an impaired response to Shh signaling [Izraeli et al., 1999, 2001; David et al., 2014]. The zebrafish stil (formerly sil) loss-of-function homozygous mutant cassiopeia (csp) also displays embryonic lethal defects, with an increased number of mitotic cells displaying disorganized mitotic spindles often lacking one or both centrosomes [Pfaff et al., 2007]. We conclude that the compound heterozygous STIL mutations found by WES in the affected fetuses cause the severe neurodevelopmental defects observed by fetal ultrasound and MRI, possibly also causing the recurrent miscarriages for the couple.
STIL Mutations Cause Centriole Elongation
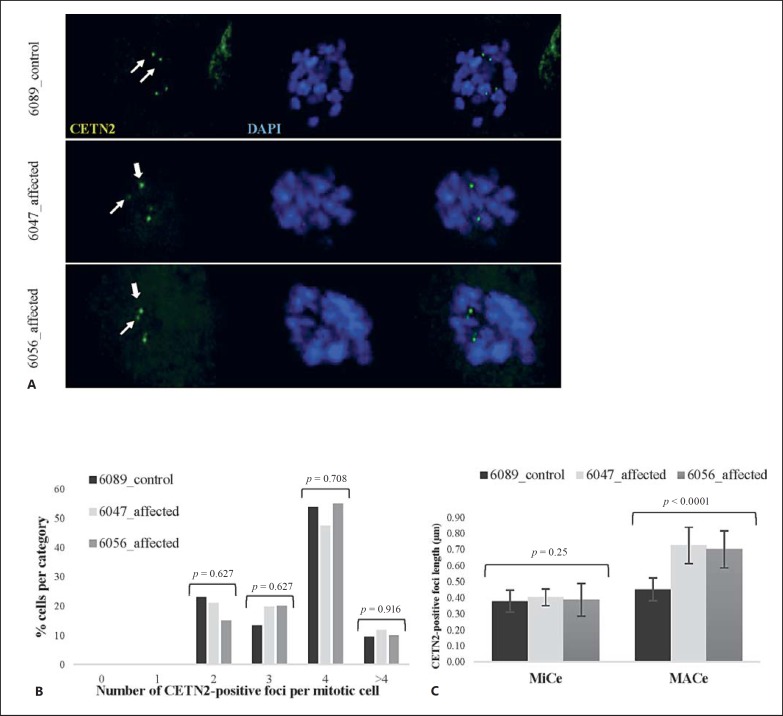
In light of the severe phenotype detected in the affected fetuses, we hypothesized that the compound heterozygous STIL mutations would be loss-of-function mutations and would recapitulate the depletion of STIL which blocks centriole duplication [Kitagawa et al., 2011; Tang et al., 2011; Arquint et al., 2012; Vulprecht et al., 2012], possibly causing embryonic lethality in this case. To test this hypothesis, we performed immunofluorescence experiments with antibodies against Centrin-2 (CETN2) to mark the centriolar structures in fetal amniocytes synchronized in prometaphase with the reversible KIF11 inhibitor STLC (S-trityl-L-cysteine). STLC blocks the separation of duplicated centrosomes and bipolar spindle formation without impairing centriole duplication itself [Skoufias et al., 2006]. Control and affected mitotic amniocytes (6089_control, n = 58; 6047_affected [II-8], n = 80; 6056_affected [II-9], n = 20) were evaluated in an immunofluorescence experiment run in duplicate (Fig. 3A). Surprisingly, no aberrant number of centrioles was observed in the affected fetal amniocytes compared to the controls (Fig. 3B), implying that the mutations detected in the fetuses did not have the hypothesized null effect nor mimic the multiple centriole formation observed in earlier overexpression experiments [Tang et al., 2011; Arquint et al., 2012]. However, when looking at the 3D projection, we noted that at least one centriole per duplicated centrosome appeared longer in affected versus control fetal amniocytes. To quantify this observation, the distance between the first and last fluorescent CETN2-positive stack per centriole was measured and used as an indicator of centriole length. The average length of the longest centriole of each centrosome (designated as “major centriole” or MACe) was compared to the average of the shortest ones (defined as “minor centriole” or MiCe). The average MiCe lengths in affected and control mitotic amniocytes were statistically equal (one-way ANOVA test p = 0.25; pairwise comparison tests, nonsignificant; online suppl. Table 3A). The control MACe average length is significantly different from both affected and control MiCe values (one-way ANOVA test p <0.0001, significance confirmed by pairwise comparison tests; online suppl. Table 3C). Since previous electron microscopy studies report that daughter centrioles are usually about 80% the length of the mother [Chrétien et al., 1997; Azimzadeh and Bornens, 2007; Uzbekov et al., 2012], we can assume that at least for the control amniocytes MiCe and MACe represent daughter and mother centrioles, respectively. On the other hand, average affected MACe lengths were significantly higher than in the control cells (one-way ANOVA test p <0.0001, confirmed by pairwise comparison tests, Fig. 3C; online suppl. Table 3B). The average MACe lengths of the amniocytes of both affected fetuses lie far outside the range of measurement uncertainty, making these values significantly different from all MiCes and control MACe.
Fig. 3.
Affected fetal amniocytes do not display an aberrant number of CETN2-positive foci in mitosis, but at least one elongated centriole per centrosome. A Confocal images of control and affected mitotic amniocytes stained with anti-CETN2 antibodies (green). Thin arrows represent minor centrioles (MiCes) and thick arrows represent major centrioles (MACes).B Number of CETN2-positive foci in affected versus unaffected mitotic amniocytes. K-proportions test p values are displayed, and nonsignificance was confirmed by the Monte Carlo method for low values (5,000 simulations, p values not shown). C Comparison of average MACe and MiCe lengths in affected versus control amniocytes (MiCe 6089_control = 0.38 ± 0.07 µm; 6047_affected = 0.40 ± 0.05 µm; 6056_affected = 0.39 ± 0.1 µm; MACe 6089_control = 0.45 ± 0.07 µm; 6047_affected = 0.72 ± 0.11 µm; 6056_affected = 0.70 ± 0.12 µm). One-way ANOVA p values are displayed (see online suppl. Table 3 for pairwise comparison tests between control and affected MiCes/MACes singularly).
Discussion
The STIL gene was originally identified through a fusion cDNA in a T-cell leukemia line resulting from an interstitial deletion between STIL and the adjacent 5′ UTR of SCL, a putative hematopoietic transcription factor [Aplan et al., 1990]. The gene contains 17 exons coding for a 1,288-amino acid protein. The protein is recruited at the proximal end of nascent daughter centrioles at the onset of centriole duplication together with the centriole duplication factor hsSAS-6 and the centriole length regulator CPAP/CENPJ [Tang et al., 2011]. This process is driven by PLK4 phosphorylation activity [Holland et al., 2010; Ohta et al., 2014; Kratz et al., 2015; Moyer et al., 2015]. When the centriole duplication has occurred, Cdk1 triggers STIL translocation from the centrosome to the cytoplasm where it is degraded via the anaphase-promoting complex/cyclosome-proteasome pathway [Arquint and Nigg, 2014]. STIL levels are tightly regulated during the cell cycle: the protein level is very low in G1 phase, increases gradually during and after the G1-S transition, and finally starts declining at the time of metaphase-anaphase transition [Arquint et al., 2012]. While depletion of STIL in human cells blocks centriole duplication, overexpression of the protein results in the formation of multiple daughter centrioles around a single mother centriole [Kitagawa et al., 2011; Tang et al., 2011; Arquint et al., 2012; Arquint and Nigg, 2014], a phenotype reminiscent of those observed for both hsSAS-6 and PLK4 overexpression [Habedanck et al., 2005; Leidel et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007; Rodrigues-Martins et al., 2007; Strnad et al., 2007; Cunha-Ferreira et al., 2009; Rogers et al., 2009; Marthiens et al., 2013; Coelho et al., 2015]. Interestingly, mutations in PLK4 have been associated with primordial dwarfism and retinopathy/chorioretinopathy, thus extending the clinical spectrum correlated to centriole defects [Martin et al., 2014; Shaheen et al., 2014; Tsutsumi et al., 2016].
For the first time, Kumar et al. [2009] reported the association between homozygous mutations in STIL and autosomal recessive primary MC. In that study, 3 homozygous truncating mutations in STIL were identified in members of 4 Indian families affected by MCPH derived from consanguineous marriages. Subsequently, several other families have been described (overview in Fig. 4; Table 1), and besides MC, other brain abnormalities including a simplified gyral pattern, reduction of the white matter, abnormal corpus callosum, and lobar HPE were often seen [Kumar et al., 2009; Bennett et al., 2014; Kakar et al., 2015; Mouden et al., 2015]. Fetal CNS developmental abnormalities caused by compound heterozygous STIL mutations have been described in a single previous study in which fetal MC was observed at the 20th gestational week by ultrasound imaging [Bennett et al., 2014]. In this case, MRI imaging performed at the 31st week of gestation revealed severe MC and anomalies consistent with HPE. However, postnatal imaging of the same case showed large symmetric bilateral schizencephaly or hydranencephaly with underlying migrational abnormalities rather than HPE. The 5 miscarriages occurring in our family may suggest a causal relationship between the STIL mutations and embryonic lethality in humans. Unfortunately, no material was present to perform genetic testing retrospectively. Despite this, the phenotype variability observed between individuals with the same STIL mutations [e.g., Kakar et al., 2015; Mouden et al., 2015] (Table 1) allows us to hypothesize that more severe brain malformations may have happened in the previous pregnancies.
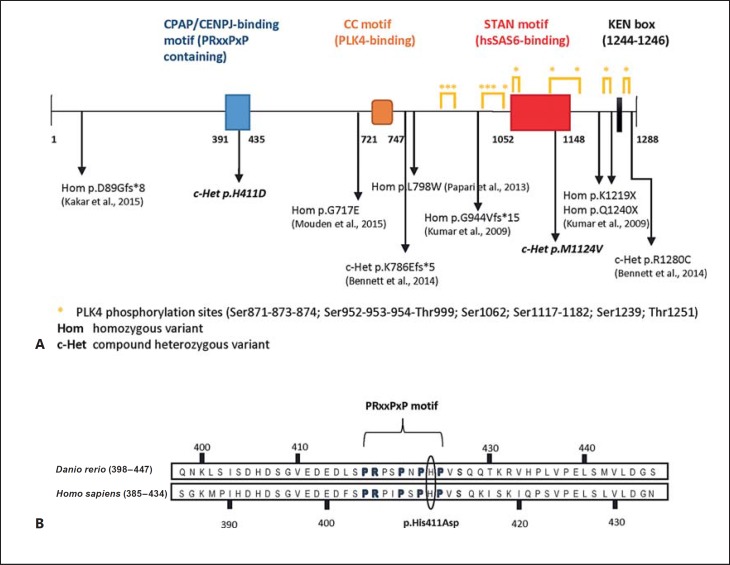
Fig. 4.
Schematic of STIL mutations reported so far, protein domains and PLK4-dependent phosphorylated sites. A The schematic depicts the CPAP/CENPJ-binding (amino acids 391–435), PLK4-binding (CC-motif, amino acids 721–747) and hsSAS6-binding (STAN motif, amino acids 1,052–1,148) domains. The KEN-box (amino acids 1,244–1,246) at the C-terminal end of the protein drives APC/C-mediated degradation of STIL [Arquint et al., 2012]. Below is an overview of the mutations described to date with their references as well as the novel mutations described in this report (in italics). B Sequence alignment of Danio rerio and Homo sapiens CPAP-binding domain of STIL. The p.His411Asp mutation lies inside the conserved PRxxPxP motif responsible for the interaction between STIL and the TCP domain of CPAP [Cottee et al., 2013].
Table 1.
Summary of reported families with mutations in STIL
| Kumar et al., 2009 | Darvish et al., 2010 | Papari et al., 2013 | Bennett et al., 2014 | Kakar et al., 2015 | Mouden et al., 2015 | This study | |
|---|---|---|---|---|---|---|---|
| Families reported, n | 4, consanguineous | 2, consanguineous | 1, consanguineous | 1, nonconsanguineous | 1, consanguineous | 1, consanguineous | 1, nonconsanguineous |
| Mutation(s) identified | hom p.Gln1240* (family IIS-17) hom p.Leu1219* (family IIS-28 and family IIS-45) hom p.Gly944Valfs*15 (family IIS-3) | unidentified | hom p.Leu798Trp* | c-Het p.Lys786Glufs*5 (paternal) p.Arg1280Cys (maternal) |
hom p.Asp89Glyfs*8 | hom p.Gly717Glu | c-Het p.His411Asp (maternal) p.Met1124Val (paternal) |
| Type of mutation | protein truncating | unidentified | missense | protein truncating; missense | protein truncating | missense | missense |
| Ethnicity | Indian | Iranian | Iranian | not reported | Pakistani | Turkish | Caucasian |
| Prenatal features | not reported | not reported | not reported | MC (OFC <−2 SD, 20th week ultrasonography), agyria and absent sulci, abnormal brain tissue consistent with holoprosencephaly and schizencephaly (31st week MRI) | not reported | not reported | II-8: MC, small cerebellum (P3), small CSP, poorly developed gyri, absent sulci, partial ACC (21st week ultrasonography) II-9: MC, small CSP, short corpus callosum, absence of lateral median and parieto-occipital sulci, pachygyria (20th week ultrasonography, fetal MRI) |
| OFC at birth | not reported | not reported | not reported | 26 cm (40 weeks) | not reported | not reported | / |
| Postnatal phenotype | primary MC (OFC <−4/−10 SD), mild to severe ID, developmental delay | primary MC, short stature, strabismus, ataxia, seizures | primary MC, mild ID | MC (OFC 33 cm at 6 months), hypsarrhythmia, questionable social smile and attention, mild hypotonia | severe MC (OFC ≤–9 SD), severe ID | II-3: severe MC (–7 SD), short stature (–3 SD), severe ID II-5: severe MC (–8 SD), short stature (–2 SD), self-aggression episodes, sleep disorders, developmental delay, tonic/clonic seizures | / |
| Postnatal brain MRI | not reported | not reported | not reported | agyria of frontal and temporal lobes with thickened cortex, interhemispheral cyst, ACC, hypoplastic pons, cerebellar dysgenesis (large symmetric bilateral schizencephaly or hydranencephaly with underlying migrational abnormalities) | severe foreshortened frontal lobe and small frontal horns, diffuse simplified gyral pattern, short dysmorphic corpus callosum, mildly small brainstem and cerebellum, lobar HPE | II-3: lobar HPE, absence of ventricular frontal horns, partial ACC II-5: semilobar HPE, atrophy of vermis, partial ACC, absence of occipital lobe | / |
ACC, agenesis of corpus callosum; c-Het, combined heterozygous; CSP, cavum septum pellucidum; hom, homozygous; HPE, holoprosencephaly; ID, intellectual disability; MC, microcephaly; OFC, occipitofrontal head circumference.
The STIL p.His411Asp mutation identified here is located in the proline-rich CPAP/CENPJ-interacting domain of the protein (Fig. 4), which has also been demonstrated to be responsible for the majority of the binding activity with CPAP/CENPJ [Cottee et al., 2013]. Yeast 2-hybrid screening suggested that the CP3 region of the human CPAP/CENPJ protein (residues 895–1,338) interacts with the STIL region between amino acids 231–619 [Tang et al., 2011], an interval further narrowed down by Cottee et al. [2013] to STIL residues 391–435. Interestingly, the homozygous missense CPAP/CENPJ mutation p.Glu1235Val found in individuals with MCPH6 [Bond et al., 2005] was shown to interfere significantly with the binding to STIL, strengthening the hypothesis that the aberrant interaction between STIL and CPAP/CENPJ could be a possible mechanism for MCPH in humans [Tang et al., 2011; Hatzopoulos et al., 2013]. Moreover, a systematic computational analysis of the structural consequences of several STIL missense polymorphisms identified rs147744459 (p.Arg242Cys) as a highly deleterious disease-associated nonsynonymous SNP. Although lying outside the CPAP/CENPJ-interacting domain identified by Cottee et al. [2013], molecular dynamic simulation showed a drastic alteration of the STILR242C protein structure, which may be responsible for the disruption of the centrosomal location of CPAP/CENPJ [Kumar et al., 2012]. Interestingly, in cooperation with CEP120 and SPICE1, CPAP/CENPJ controls procentriole elongation through its tubulin-dimer binding activity which promotes the polymerization of centriolar microtubules [Comartin et al., 2013; Lin et al., 2013]. A depletion of any of these proteins results in shorter daughter centrioles [Comartin et al., 2013]. Conversely, overexpression of either CPAP/CENPJ or CEP120 induces the assembly of overly long centrioles with atypical supernumerary centrioles branching out from them [Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009; Comartin et al., 2013; Lin et al., 2013]. The same results have been observed for depletion of the end-capping protein CP110, which normally localizes to the distal tips of both parental and daughter centrioles [Kleylein-Sohn et al., 2007; Schmidt et al., 2009]. Recently, more insight into the regulation of centriole length was gained with the identification of the N-terminal LID element of CPAP/CENPJ, which acts as a cap that sterically blocks tubulin addition to the centriole distal end, thus controlling this organelle size and ensuring the exceptionally slow growth rate of centriolar microtubules compared to their cytoplasmic counterparts [Sharma et al., 2016].
The second heterozygous mutation lies within the C-terminal STAN (STil/ANa2) domain of the protein (Fig. 4). This conserved motif (residues 1,052–1,148) is responsible for the interaction between STIL and the centriolar protein hsSAS6, an event facilitated by the PLK4-phosphorylation of 7 Ser-Thr localized in and around the same domain. This event is required for the recruitment and centriolar targeting of hsSAS6 and the initiation of procentriole assembly [Stevens et al., 2010; Dzhindzhev et al., 2014; Ohta et al., 2014]. Besides phosphorylating STIL, PLK4 binds directly to its CC-domain (residues 721–747), thus allowing its recruitment to the centrioles at the beginning of centriole duplication [Ohta et al., 2014]. The p.Met1124Val change found in this study results in the maintenance of a hydrophobic side chain, but this substitution could either prevent the PLK4-dependent phosphorylation of the surrounding residues or sterically impede the interaction between STIL and hsSAS6, in both cases resulting in the incorrect recruitment of the latter at the site of procentriole assembly. Alternatively, since hsSAS6 possesses an intrinsic microtubule assembly-promoting activity mediated by its C-terminal domain [Gupta et al., 2015], the p.Met1124Val mutation could promote hsSAS6 stabilization resulting in procentriolar microtubules over-elongation.
It is difficult to speculate how elongated centrioles can lead to MC. Centriole elongation has also been observed in OFD1 syndrome [oro-facial-digital syndrome 1 (OFDS1; OMIM 311200)], an X-linked ciliopathy characterized by malformations of the face, oral cavity, and digits [Ferrante et al., 2001; Thauvin-Robinet et al., 2005]. In addition, this disorder can be accompanied by variable CNS malformations and MC [Gurrieri et al., 2007]. OFD1 associates with the distal ends of centriolar microtubules, constrains mother and daughter centriole elongation, and is required for distal appendage and cilium formation. A number of disease-associated OFD1 mutations have been shown to reduce the ability of the protein to restrain the elongation of the distal portion of centrioles, thus resulting in abnormally long mother and daughter centrioles [Singla et al., 2010]. These defects adversely impact upon the duration of the different phases of the cell cycle which is particularly short in rapidly dividing neural progenitors, contributing to the pathogenesis of cortical abnormalities and MC [Alcantara and O'Driscoll, 2014]. We hypothesize that in our case an impairment of the normal regulation of centriole lengthening could have a similar effect. Further functional investigations on how missense STIL mutations located in binding domains of the protein affect the regulation of centriole elongation and the interaction with other centriolar proteins are definitely required in order to fully understand the molecular mechanisms linking centriole elongation to the pathogenesis of complex brain malformation and MC.
Statement of Ethics
The WES protocol was approved by the Institutional Review Board of the University Hospitals of Leuven, and informed consent was obtained from the parents before the procedure.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgments
We wish to thank the family members involved in this study. Alexa Fluor® secondary antibodies were a gift from Prof. Mathieu Bollen (Laboratory of Biosignaling and Therapeutics, Department of Cellular and Molecular Medicine, KUL, Belgium). We would also like to thank Sofie De Munter (Biosignaling and Therapeutics Lab) for helping with immunofluorescence optimization and confocal microscope settings. F.C. is a PhD aspirant and H.V.E. is a Clinical Investigator of the Research Foundation-Flanders (Fonds Wetenschappelijk Onderzoek, FWO, Belgium). This work was supported by grants from the KUL PFV/10/016 SymBioSys to J.R.V. and GOA/12/015 to J.R.V. and H.V.E., and the Belgian Science Policy Office Interuniversity Attraction Poles (BELSPO-IAP) program (project IAP P7/43-BeMGI).
References
- Alcantara D, O'Driscoll M. Congenital microcephaly. Am J Med Genet Part C Semin Med Genet. 2014;166:124–139. doi: 10.1002/ajmg.c.31397. [DOI] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. J Med Genet. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- Ardeshirdavani A, Souche E, Dehaspe L, Van Houdt J, Vermeesch JR, Moreau Y. NGS-logistics: federated analysis of NGS sequence variants across multiple locations. Genome Med. 2014;6:71. doi: 10.1186/s13073-014-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr Biol. 2014;24:351–360. doi: 10.1016/j.cub.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Barbelanne M, Tsang WY. Molecular and cellular basis of autosomal recessive primary microcephaly. Biomed Res Int. 2014;2014:547986. doi: 10.1155/2014/547986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett H, Presti A, Adams D, Rios J, Benito C, Cohen D. A prenatal presentation of severe microcephaly and brain anomalies in a patient with novel compound heterozygous mutations in the STIL gene found postnatally with exome analysis. Pediatr Neurol. 2014;51:434–436. doi: 10.1016/j.pediatrneurol.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Castiel A, Danieli MM, David A, Moshkovitz S, Aplan PD, et al. The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J Cell Sci. 2011;124:532–539. doi: 10.1242/jcs.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- Coelho PA, Bury L, Shahbazi MN, Liakath-Ali K, Tate PH, et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015;5:150209. doi: 10.1098/rsob.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D, Gupta GD, Fussner E, Coyaud É, Hasegan M, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, et al. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Darvish H, Esmaeeli-Nieh S, Monajemi G, Mohseni M, Ghasemi-Firouzabadi S, et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J Med Genet. 2010;47:823–828. doi: 10.1136/jmg.2009.076398. [DOI] [PubMed] [Google Scholar]
- David A, Liu F, Tibelius A, Vulprecht J, Wald D, et al. Lack of centrioles and primary cilia in STIL−/− mouse embryos. Cell Cycle. 2014;13:2859–2868. doi: 10.4161/15384101.2014.946830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar T, Silhavy J, Udpa N, Schroth J, Bielas S, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, et al. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr Biol. 2014;24:2526–2532. doi: 10.1016/j.cub.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Guyer MS, National Human Genome Research Institute Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta H, Badarudeen B, George A, Thomas GE, Gireesh KK, Manna TK. Human SAS-6 C-terminus nucleates and promotes microtubule assembly in vitro by binding to microtubules. Biochemistry. 2015;54:6413–6422. doi: 10.1021/acs.biochem.5b00978. [DOI] [PubMed] [Google Scholar]
- Gurrieri F, Franco B, Toriello H, Neri G. Oral-facial-digital syndromes: review and diagnostic guidelines. Am J Med Genet A. 2007;143A:3314–3323. doi: 10.1002/ajmg.a.32032. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hardwick LJ, Ali FR, Azzarelli R, Philpott A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 2015;359:187–200. doi: 10.1007/s00441-014-1895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, et al. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure. 2013;21:2069–2077. doi: 10.1016/j.str.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Good D, Dorward DW, et al. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature. 1999;399:691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Campaner S, Hahn H, et al. Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis. 2001;31:72–77. doi: 10.1002/gene.10004. [DOI] [PubMed] [Google Scholar]
- Kakar N, Ahmad J, Morris-Rosendahl DJ, Altmüller J, Friedrich K, et al. STIL mutation causes autosomal recessive microcephalic lobar holoprosencephaly. Hum Genet. 2015;134:45–51. doi: 10.1007/s00439-014-1487-4. [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, et al. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci. 2011;124:3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Lončarek J, Meng X, McEwen BF, Mogensen MM, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz AS, Bärenz F, Richter KT, Hoffmann I. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol Open. 2015;4:370–377. doi: 10.1242/bio.201411023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Rajendran V, Sethumadhavan R, Purohit R. In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ) FEBS Open Bio. 2012;2:285–293. doi: 10.1016/j.fob.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJC, et al. CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol. 2013;202:211–219. doi: 10.1083/jcb.201212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S, Ahmad W, Hassan MJ. Autosomal recessive primary microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J Rare Dis. 2011;6:39. doi: 10.1186/1750-1172-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinger G, Lev D, Oren M, Lerman-Sagie T. Non-visualization of the cavum septi pellucidi is not synonymous with agenesis of the corpus callosum. Ultrasound Obstet Gynecol. 2012;40:165–170. doi: 10.1002/uog.11206. [DOI] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- Martin CA, Ahmad I, Klingseisen A, Hussain MS, Bicknell LS, et al. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet. 2014;46:1283–1292. doi: 10.1038/ng.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Vitre B, Carpenter G, Abramowicz I, Gleeson JG, et al. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet. 2014;133:1023–1039. doi: 10.1007/s00439-014-1443-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Rosendahl DJ, Kaindl AM. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH) Mol Cell Probes. 2015;29:271–281. doi: 10.1016/j.mcp.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Mouden C, de Tayrac M, Dubourg C, Rose S, Carré W, et al. Homozygous STIL mutation causes holoprosencephaly and microcephaly in two siblings. PLoS One. 2015;10:e0117418. doi: 10.1371/journal.pone.0117418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol. 2015;209:863–878. doi: 10.1083/jcb.201502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, et al. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun. 2014;5:5267. doi: 10.1038/ncomms6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papari E, Bastami M, Farhadi A, Abedini SS, Hosseini M, et al. Investigation of primary microcephaly in Bushehr province of Iran: novel STIL and ASPM mutations. Clin Genet. 2013;83:488–490. doi: 10.1111/j.1399-0004.2012.01949.x. [DOI] [PubMed] [Google Scholar]
- Paridaen JT, Huttner WB. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15:351–364. doi: 10.1002/embr.201438447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol. 2007;27:5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, et al. Mutations in TUBG1DYNC1H1KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani B, Mahdieh N, Hosomichi K, Nakaoka H, Inoue I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet. 2012;57:621–632. doi: 10.1038/jhg.2012.91. [DOI] [PubMed] [Google Scholar]
- Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, et al. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Shaheen R, Al Tala S, Almoisheer A, Alkuraya FS. Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J Med Genet. 2014;51:814–816. doi: 10.1136/jmedgenet-2014-102790. [DOI] [PubMed] [Google Scholar]
- Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, et al. Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev Cell. 2016;37:362–376. doi: 10.1016/j.devcel.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, et al. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem. 2006;281:17559–17569. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Fu R, Wu K, Hsu W, Tang T. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- Tang C, Lin S, Hsu W, Lin Y, Wu C, et al. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauvin-Robinet C, Cossée M, Cormier-Daire V, Van Maldergem L, Toutain A, et al. Clinical, molecular, and genotype-phenotype correlation studies from 25 cases of oral-facial-digital syndrome type 1: a French and Belgian collaborative study. J Med Genet. 2005;43:54–61. doi: 10.1136/jmg.2004.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi M, Yokoi S, Miya F, Miyata M, Kato M, et al. Novel compound heterozygous variants in PLK4 identified in a patient with autosomal recessive microcephaly and chorioretinopathy. Eur J Hum Genet. 2016;24:1702–1706. doi: 10.1038/ejhg.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, Maurel D, Aveline P, Pallu S, Benhamou C, Rochefort G. Microscopy microanalysis centrosome fine ultrastructure of the osteocyte mechanosensitive primary cilium. Microsc Microanal. 2012;18:1430–1441. doi: 10.1017/S1431927612013281. [DOI] [PubMed] [Google Scholar]
- Verloes A, Drunat S, Gressens P, Passemard S. Primary autosomal recessive microcephalies and Seckel syndrome spectrum disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, et al., editors. University of Washington, Seattle: GeneReviews® [Internet]; 1993–2017. Initial posting: 2009 Sep 1; last update: 2013 Oct 31. [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012;125:1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci. 2014;127:4111–4122. doi: 10.1242/jcs.151753. [DOI] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillhardt JL, Poirier K, Broix L, Lebrun N, Elmorjani A, et al. Mosaic parental germline mutations causing recurrent forms of malformations of cortical development. Eur J Hum Genet. 2016;24:611–614. doi: 10.1038/ejhg.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data