Abstract
Xia-Gibbs syndrome is an autosomal dominant multisystem developmental disorder characterized by global developmental delay, hypotonia, obstructive sleep apnea, seizures, retrocerebellar cysts, delayed myelination, micrognathia, and mild dysmorphic features. Using whole-exome sequencing, we identified a de novo AHDC1 frameshift mutation c.2030_2030delG (p.G677Afs*52) in a Colombian patient, which was absent in both parents. Furthermore, we summarized the phenotypes of patients reported in the literature.
Keywords: AHDC1 mutation, Whole-exome sequencing, Xia-Gibbs syndrome
Many patients living with rare genetic diseases have gone through several rounds of genetic testing (e.g., karyotyping, candidate gene sequencing, aCGH, next-generation sequencing), frequently not receiving an accurate diagnosis or proper information regarding the prognosis of the disease. This may lead to treatment delay and a risk of morbidity/mortality [Gahl et al., 2012].
aCGH offers a higher diagnostic rate (15–20%) for genetic testing of individuals with unexplained developmental delay (DD) and/or intellectual disability (ID) compared to karyotyping, primarily because of its higher sensitivity for submicroscopic deletions and duplications [Miller et al., 2010]. However, whole-exome sequencing (WES) in DD/ID revealed de novo mutations in protein coding genes in 60% of individuals, having a higher performance [Hochstenbach et al., 2011].
Next-generation sequencing is a powerful tool for the diagnosis of mendelian disorders with variable phenotypes without a family history, leading to the discovery of new genes associated with diseases or a new disease associated with specific loci [Yang et al., 2013; Xue et al., 2015; Retterer et al., 2016]. The clinical yield of this test will continue to increase over time, allowing providers to efficiently arrive at a diagnosis [Lee et al., 2014; Xue et al., 2015]. According to Yang et al. [2014], this method identifies underlying genetic defects in 25% of patients referred for a possible genetic condition.
According to the American College of Medical Genetics (ACMG), indications for WES [ACMG, 2012] are when (1) clinical diagnostic assessment of a phenotypically affected individual with history data strongly implicate a genetic etiology, but the phenotype does not correspond with a specific disorder for a specific gene, (2) the phenotype demonstrates a high degree of genetic heterogeneity without a specific test available, and (3) specific genetics tests available for the phenotype have failed.
Xia-Gibbs syndrome (OMIM 615829) was mentioned for the first time by Xia et al. [2014] after analysis of parent-offspring trios of 4 probands with DD, hypotonia, mild dysmorphic features, sleep apnea, and other symptoms, finding 3 new allelic variants in the ADHC1 (AT hook DNA-binding motif containing 1) gene likely to be pathogenic; it was the first report in genome databases. Yang et al. [2015] reported 6 frameshifts or nonsense deleterious de novo variants and 1 recurrent variant in the same gene, associated with expressive language delay, hypotonia, and sleep apnea. Recently, Bosch et al. [2016] identified a de novo variant in an older patient with a history of DD and speech delay as well as characteristic facies. Previously, a de novo missense variant in a case of schizophrenia [Guipponi et al., 2014] and a de novo balanced translocation with a breakpoint in the AHDC1 intron 1 in a boy with bicuspid aortic valve, aortic coarctation, patent ductus arteriosus, and DD/ID have been reported in this gene [Quintero-Rivera et al., 2015]. Several large CNV deletions including the AHDC1 gene have been described in pediatric patients with DD/ID [Itsara et al., 2009; Cooper et al., 2011; Coe et al., 2014]. Although due to the large size of the deletions, it is uncertain whether AHDC1 is the only gene contributing to the neurodevelopmental phenotype. Xia et al. [2014] proposed that AHDC1 -associated ID is due to a dominant-negative mechanism, given the autosomal-dominant inheritance and the single coding exon of this gene.
The AHDC1 gene located in chromosome 1p36.11 encodes a protein of 1,603 amino acids, consisting of 7 exons with only 1 coding exon (exon 6), containing 2 AT-hooks, which likely function in DNA binding. AT-hook motifs are known as auxiliary protein motifs that cooperate with other DNA-binding activities and facilitate DNA structure changes [Aravind and Landsman, 1998]. The AHDC1 gene is part of the CBX family of proteins associated with human chromodomain-containing Polycomb proteins [Vandamme et al., 2011]. In vivo assays have demonstrated that AHDC1 interacts with several nuclear proteins involved in epigenetic regulation during development [Vandamme et al., 2011], mainly in neural sites and neuron proteins for transport [Uhlén et al., 2015]. In mice, Ahdc1 is expressed at embryonic day E11.5 and E16.5 in the developing brain [Quintero-Rivera et al., 2015], suggesting that AHDC1 may be involved in early brain development.
In this study, we described the first Colombian patient with Xia-Gibbs syndrome, with a new mutation in the AHDC1 gene identified by WES and compared the 12 cases reported in the literature to better understand the clinical phenotype and the association with the AHDC1 gene.
Patient and Methods
Case Report
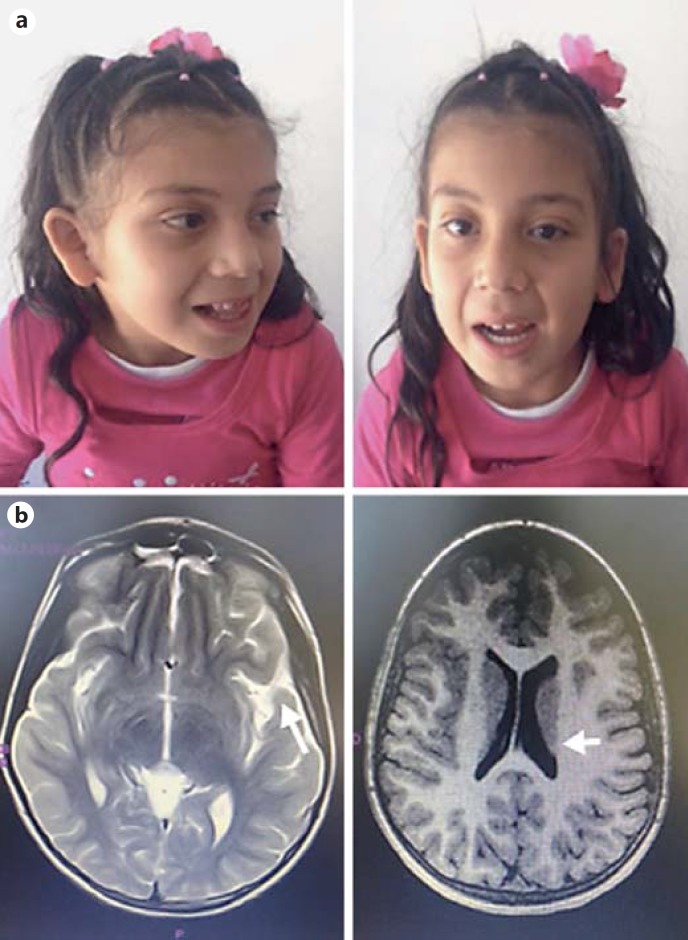
The 8-year-old Colombian girl is the first child born at term to 27-year-old nonconsanguineous parents after prolonged labor and perinatal hypoxia, with an Apgar score of 6 at 10 min. The child was admitted to the neonatal intensive care unit requiring mechanical ventilation for 27 days. Clinical follow-up showed hypotonia and DD (head control at 7 months, sitting at 12 months, standing at 24 months, walking at 28 months, and currently speaking only 2 words) with dysmorphic features including midfacial hypoplasia, hypertelorism, micrognathia, epicanthic fold, prominent teeth, upslanting palpebral fissures (Fig. 1), and laryngomalacia requiring 8 surgical interventions due to glottal stenosis. Her family history was unremarkable
Fig. 1.
a Patient at the age of 5 years. Facial features showing midfacial hypoplasia, hypertelorism, micrognathia, epicanthic fold, prominent teeth, and upslanting palpebral fissures. b Brain MRI showing frontal and temporal cortical atrophy, with loss of posterior ventricular white matter (arrows).
Brain MRI revealed frontal and temporal cortical atrophy with loss of posterior ventricular white matter (Fig. 1). The girl's karyotype, microarray, and metabolic tests were normal.
Molecular Analysis
WES was performed using Illumina HiSeq platform (overall performance of 90% exome coverage at 40× depth) identifying a new mutation in AHDC1 (c.2030_2030delG; p.Gly677AlafsX52) verified by Sanger sequencing and absent in both maternal and paternal DNA. It was therefore interpreted as a de novo event that results in a frameshift of the AHDC1 open reading frame and is predicted to cause a premature termination codon after 52 amino acids. Predicted truncating mutations in AHDC1 are absent in the genome mutation databases dbSNP and ExAC. The pathogenic effect of the described mutation in AHDC1 was predicted by the bioinformatic tools PolyPhen-2 (prediction “probably damaging” – score of 0.958) [Adzhubei et al., 2010], MutationTaster (prediction “disease causing”) [Schwarz et al., 2014], and SNPs&GO (RI = 8 Effect = neutral) [Calabrese et al., 2009]. Furthermore, glycine is a conserved amino acid between species; therefore, any change in this amino acid can generate alterations of the protein structure.
Discussion
Here, we report a proband with DD and dysmorphic facial features due to a frameshift mutation in AHDC1, which was discovered by WES. To date, 12 individuals with AHDC1 mutations have been reported in the literature. Developmental histories of patients evidenced that all had delayed speech and psychomotor delay; most individuals had mild dysmorphic facial features that could be seen at a young age and had a history of sleep disturbance because of anomalies of the upper airway structure.
The independent occurrence of 12 de novo mutational events at this locus in unrelated individuals with similar phenotypes is highly unlikely, suggesting that mutations in AHDC1 cause this complex disorder, which has not been previously reported in the literature since Xia et al. [2014]. It is remarkable that all cases were detected by WES after several genetic tests. Clinical features of the reported cases of Xia et al. [2014], Yang et al. [2015], and Bosch et al. [2016] compared with the clinical presentation of our case are summarized in Table 1.
Table 1.
Clinical featurs of patients with Xia-Gibbs syndrome
| Reference | Case | Gender | Age | Variant in AHDC1 | Developmental delay | Cognitive delay | Speech delay | Dysmorphic facial features | Sleep disturbance | Brain abnormality |
|---|---|---|---|---|---|---|---|---|---|---|
| Xia F et al. [2014] | 1 | F | 18 mo | c.2373_2374 delTG (p.Cys791Trpfs*57) | no sitting at 18 mo | unknown | no words at 18 mo | low-set ears, exotropia, upslanting palpebral fissures, micrognathia, flat nasal bridge | obstructive sleep apnea | retrocerebellar cyst |
| 2 | F | 4 yrs | c.2898delC (p.Tyr967Thrfs*175) | sitting at 19 mo, walking at 2 yrs | moderate | 2 words at 4 yrs | protuberant ears, upslanting palpebral fissures, flat nasal bridge | obstructive sleep apnea | thinning of the corpus callosum | |
| 3 | M | 8 yrs | c.2373_2374 delTG (p.Cys791Trpfs*57) | sitting at 9 mo, walking at 18 mo | mild | first words after 1 y, persistent speech therapy | protuberant low-set ears, small earlobes, hypertelorism, downslanting palpebral fissures, mild ptosis, micrognathia | obstructive sleep apnea | thinning of the corpus callosum | |
| 4 | M | 11 yrs | c.2547delC (p.Ser850 Profs*82) | sitting at 15 mo, no independent ambulation | moderate to severe | no words, noncommunicating autism | upturned earlobes, hypertelorism, exotropia, flat nasal bridge | snoring | thinning of the corpus callosum | |
| Yang et al. [2015] | 5 | M | 2 yrs | c.1945delG (p.A649Pfs*83) | no independent walking | unknown | no verbal speech | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | sleep apnea | normal |
| 6 | F | 7 yrs | c.2529_2545dell7 (p.D845Rfs*40) | walking at 3 yrs 10 mo | yes | current 2–3-word sentences | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | restless sleeper | periventricular leukomalacia, thin CC, subependymal cysts | |
| 7 | M | 5 yrs | c.1881delG (p.Q627Hfs*105) | unknown | moderate | unknown | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | unknown | unknown | |
| 8 | F | 4 yrs | c.1122dupC (p.G375Rfs* 3) | walking at 20 mo | yes | current single words | normal | unknown | small CC, loss of posterior ventricular white matter | |
| 9 | M | 9 yrs | c.3809delA (p.Q1270Rfs*75) | walking at 4 yrs, 9 mo | yes | no verbal speech | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | unknown | hypomyelination, thin CC, small brain stem | |
| 10 | M | 5 yrs | c.2373_2374delTG (p.C791Wfs*57) | walking at 2 yrs | yes | current 2-word sentences | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | unknown | prominent CSF, small arachnoid cyst vs. abnormal cisterna magna | |
| 11 | F | 16 yrs | c.1480 A>T (p.K494X) | unknown | yes | no verbal speech | bitemporal hollowing, brachycephaly, plagiocephaly, short nose, narrow mouth, high-arched palate, tented upper lip, and high, prominent forehead with prominent glabella | unknown | demyelination, involving subcortical U fibers, retrcerebellar cyst | |
| Bosch et al., [2016] | 12 F | 22 yrs | c.1402dupT p.Cys468Leufs*49 | delayed | unknown | first words at 4 yrs | long asymmetric face, proptosis, broad nose, full lips, broad mouth, wide-spaced teeth, small high palate, simple left ear, overfolding of the right superior helices, hyperlordosis, soft skin, 5th finger clinodactyly, fetal finger pads, pes equines, many moles | snoring | asymmetric wide ventricles, thin corpus callosum | |
| Present case | F | 8 yrs | c.2030_2030del (p.G677Afs*55) | sitting at 12 mo, walking at 28 mo | severe | currently speaking single words | midfacial hypoplasia, hypertelorism, micrognathia, epicanthic fold, prominent teeth, upslanting palpebral fissures | sleep apnea | frontal and temporal cortical atrophy, loss of posterior ventricular white matter | |
CC, corpus callosum; CSF, cerebrospinal fluid; mo, months; N/A, not applicable; yrs, years.
Xia et al. [2014] proposed that AHDC1 -associated ID is due to a dominant-negative mechanism, given the autosomal-dominant inheritance and the single coding exon of this gene [Xia et al., 2014]. This results in an insufficient dosage of encoded protein which would sever the link between DNA damage response and proper brain development [Quintero-Rivera et al., 2015]. This is based on a model in which the protein encoded by the gene with the gene dosage imbalance or mutation affects the bindings to other protein(s) to form a functional complex which may lead to the formation of inactive complexes [Poot et al., 2011].
Although there are no functional or computational studies of these proteins, we speculate that AHDC1 variants may interrupt protein translation, possibly disrupting the interaction with other proteins important for brain development and function.
WES has proven to be an important tool to improve our ability to diagnose patients with heterogeneous diseases; especially, the expanded use of WES has led us to find new genes causing diseases and new diseases caused by genes. Variants in AHDC1 should be validated with in vivo and in vitro assays to understand the mechanisms of the genotype/phenotype relationship.
Statement of Ethics
Written informed consent was obtained from our patient's legal guardian for publication of this case report. The study was approved by the ethics committee of the Roosevelt Pediatric Orthopedic Institute, Bogotá, Colombia.
Disclosure Statement
The authors have no conflicts of interest to disclose.
Acknowledgments
We thank the family for their cooperation and interest to participate in this study.
References
- ACMG Board of Directors Points to consider in the clinical application of genomic sequencing. Genet Med. 2012;14:759–761. doi: 10.1038/gim.2012.74. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet. 2016;24:660–665. doi: 10.1038/ejhg.2015.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli P, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl WA, Markello TC, Toro C, Fajardo KF, Sincan M, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med. 2012;14:51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Santoni FA, Setola V, Gehrig C, Rotharmel M, et al. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9:e112745. doi: 10.1371/journal.pone.0112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach R, Buizer-Voskamp JE, Vorstman JA, Ophoff RA. Genome arrays for the detection of copy number variations in idiopathic mental retardation, idiopathic generalized epilepsy and neuropsychiatric disorders: lessons for diagnostic workflow and research. Cytogenet Genome Res. 2011;135:174–202. doi: 10.1159/000332928. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, van der Smagt J, Brilstra E, Bourgeron T. Disentangling the myriad genomics of complex disorders, specifically focusing on autism, epilepsy, and schizophrenia. Cytogenet Genome Res. 2011;135:228–240. doi: 10.1159/000334064. [DOI] [PubMed] [Google Scholar]
- Quintero-Rivera F, Xi QJ, Keppler-Noreuil KM, Lee JH, Higgins AW, et al. MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus. Hum Mol Genet. 2015;24:2375–2389. doi: 10.1093/hmg/ddv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, et al. Proteomics: tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vandamme J, Völkel P, Rosnoblet C, Le Faou P, Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteomics. 2011;10:M110.002642. doi: 10.1074/mcp.M110.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F, Bainbridge MN, Tan TY, Wangler MF, Scheuerle AE, et al. De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am J Hum Genet. 2014;94:784–789. doi: 10.1016/j.ajhg.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–451. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, et al. De novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Cold Spring Harb Mol Case Stud. 2015;1:1–9. doi: 10.1101/mcs.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]