Abstract
Given the cognitive and behavioral effects following in utero Δ9-tetrahydrocannabinol (THC) exposure that have been reported in humans and rodents, it is critical to understand the precise consequences of THC on developing human neurons. Here, we utilize excitatory neurons derived from human-induced pluripotent stem cells (hiPSCs), and report that in vitro THC exposure reduced expression of glutamate receptor subunit genes (GRIA1, GRIA2, GRIN2A, and GRIN2B). By expanding these studies across hiPSC-derived neurons from individuals with a variety of genotypes, we believe that a hiPSC-based model will facilitate studies of the interaction of THC exposure and the genetic risk factors underlying neuropsychiatric disease vulnerability.
Keywords: Cannabinoid, Pluripotent stem cell, Gene expression
Introduction
Cannabis is the most prevalent illicit drug of abuse [1]. Although prenatal exposure is associated with negative effects on fetal brain development in humans [2, 3, 4, 5, 6, 7, 8, 9], cannabis use is self-reported by over 25% of pregnant women [10]. The major psychoactive ingredient in cannabis is Δ9-tetrahydrocannabinol (THC), which acts primarily via the G-protein coupled cannabinoid receptor 1 (CB1R) [11], signaling via the activation of ERK kinases and/or the inhibition of adenyl cyclase (reviewed in [12, 13]).
CNR1 (CB1R) expression is detected in the human fetal cortex in the first trimester, increasing during development [14, 15] and ultimately found in both excitatory and inhibitory neurons and in glia cells in the adult brain (reviewed in [16]). Evidence from neuron subtype-specific deletions suggests that CB1R may have different functions and pharmacological properties in different cell types (reviewed in [17]) and that its subcellular localization may mediate distinct signaling [18, 19]. Endocannabinoids play critical roles during fetal brain development, being involved in neuronal differentiation, survival and the regulation of neurotransmitter systems [20, 21, 22].
Exposure to THC has been shown to alter neuronal excitability [21, 23, 24] via changes in expression of synaptic components, such as glutamate receptor subunits (GRIA1, GRIA2, GRIN1, GRIN2A, and GRIN2B) [21, 24, 25] as well as CNR1 [26, 27] and COX2 [21, 28], two important mediators of intracellular cannabinoid signaling.
Resolving the consequences of prenatal THC on human fetal brain development is complicated by variable THC exposures (dose, timing, and duration) between pregnancies, confounding exposures to multiple illicit drugs [29], and genotype-dependent effects [30]. Because neural cells differentiated from human-induced pluripotent stem cells (hiPSCs) most resemble fetal brain tissue [31, 32, 33, 34], they provide an unprecedented platform for studying the molecular, cellular, and functional results of fetal THC exposure to neural cells, across a variety of genetic backgrounds. In this study, we demonstrate that THC treatment of human excitatory neurons (whether generated by NGN2 induction from hiPSCs (NGN2 – hiPSC neurons) [35] or hiPSC-derived neural progenitor cells (NGN2 -NPC neurons) [36], or via directed differentiation from NPCs (forebrain neurons) [37, 40] recapitulated several known molecular consequences of THC exposure, such as changes in glutamate receptor subunit expression, at least partially in a CB1R-dependent manner.
Methods
hiPSC Reprogramming and NPC Differentiation
For cohort 1, human fibroblasts were obtained in collaboration with Judith Rapoport, MD (NIMH) as previously described [38] (NSB553, NSB2607, NSB690). Derivation of hiPSCs was done by Sendai virus reprogramming and then cultured on mEF plates using HUES media ((DMEM/F12 [Life Technologies], 20% KO-Serum Replacement [Life Technologies], 1× Glutamax [Life Technologies], 1× NEAA [Life Technologies], 55 μM β-mercaptoethanol [Sigma], and 20 ng/mL FGF2 [Invitrogen]). hiPSCs were passaged approximately 1:3 every 5–7 days with 1 mg/mL Collagenase (Invitrogen) in DMEM (Life Technologies) and fed every day. Passage-matched hiPSCs were used for all experiments. Cohort 1 NPCs were derived from hiPSCs using dual-SMAD inhibition as previously described [39, 40]. NPCs were cultured on Matrigel (BD)-coated plates in NPC media (DMEM/F12, 1× N2 [Life Technologies], 1× B27-RA [Life Technologies], and 20 ng/mL FGF2), maintained at high density and passaged with Accutase (Millipore).
For cohort 2, human fibroblasts were obtained from ATCC (CRL-2522) and Coriell (GM03440, GM03651, GM04506, and AG09319). hiPSCs were reprogrammed using tetracycline-inducible lentiviral vectors and differentiated to NPCs as previously described [40].
Passage-matched NPCs were used for all experiments. All hiPSC and NPCs used were mycoplasma-free (Table 1).
Table 1.
Case and control hiPSC and NPC lines
| Cohort | Individual | Source | Sex | hiPSC/NPC line ID |
|---|---|---|---|---|
| 1 | C1 | NIH | male | NSB553 hiPSC#2 / NSB553 hiPSC#S1 NPC#1 |
| 1 | C2 | NIH | male | NSB2607 hiPSC#4 / hiPSC#4 NPC#1 |
| 1 | C3 | NIH | male | NSB690 hiPSC#3 / NSB690 hiPSC#2 NPC#1 |
| 1 | C4 | NIH | female | NSB3183 hiPSC#1 |
| 1 | C5 | NIH | female | NSB3121 hiPSC#1 |
| 2 | Ca | ATCC | male | BJ hiPSC#2 NPC#A |
| 2 | Cb | Coriell | male | GM03440 hiPSC#5 NPC#1 |
| 2 | Cc | Coriell | female | GM03651 hiPSC#A NPC#A |
| 2 | Cd | Coriell | female | GM04506 hiPSC#B NPC#A |
| 2 | Ce | Coriell | female | AG09319 hiPSC#2 NPC#A |
| 2 | S1 | Coriell | male | GM01792 hiPSC#1 NPC#A, GM01792 hiPSC#1 NPC#E |
| 2 | S2 | Coriell | male | GM02038 hiPSC#1 NPC#A, GM02038 hiPSC#1 NPC#B |
| 2 | S3 | Coriell | female | GM01835 hiPSC#1 NPC#5 |
| 2 | S4 | Coriell | female | GM02497 hiPSC#1 NPC#C |
Neural Induction/Differentiation
NGN2 -hiPSC neurons: hiPSCs were washed with PBS, dissociated with Accutase (Millipore), and plated on Matrigel (BD)-coated plates in MEF-conditioned HUES (DMEM/F12, 20% Knockout Serum Replacement, 1× NEAA, 1× Glutamax, 1× β-mercaptoethanol, 20 ng/mL FGF2) media with ROCK inhibitor (10 μM Y27631). Tetracyline-inducible lentivirus expressing NGN2 was constituted in MEF-conditioned HUES media and spinfected into dispersed hiPSCs (1,000 g, 1 h) after 24 h recovery (day −1). Media was changed at day 0 to neural differentiation media (DMEM/F12, 1× B27, 1x N2, 20 ng/mL BDNF, 20 ng/mL GDNF, 1 mM d-cAMP, 200 nM ascorbic acid, 1 μg/μL laminin, and 1 μg/mL doxycycline) to commence neural induction. Cells were selected with 0.2 μg/mL puromycin from days 2 to 4. Media was replaced at day 2; subsequently, only 50% of the media was changed every other day.
NGN2 -NPC neurons: NPCs were dissociated with Accutase, plated on Matrigel, spinfected with doxycycline-inducible NGN2 lentivirus, and selected with 0.2 μg/mL puromycin, as previously described [36]. Neurons were fed neural differentiation media every other day for 3 weeks after induction.
hiPSC forebrain neurons: NPCs were dissociated with Accutase, plated on polyornithine/laminin- (Fig. 3b, c, d) or Matrigel- (Fig. 3e, f, g, Fig. 4) coated plates for 6 weeks as previously described [40]. Neurons were fed neural differentiation 1–2 times per week for 6 weeks.
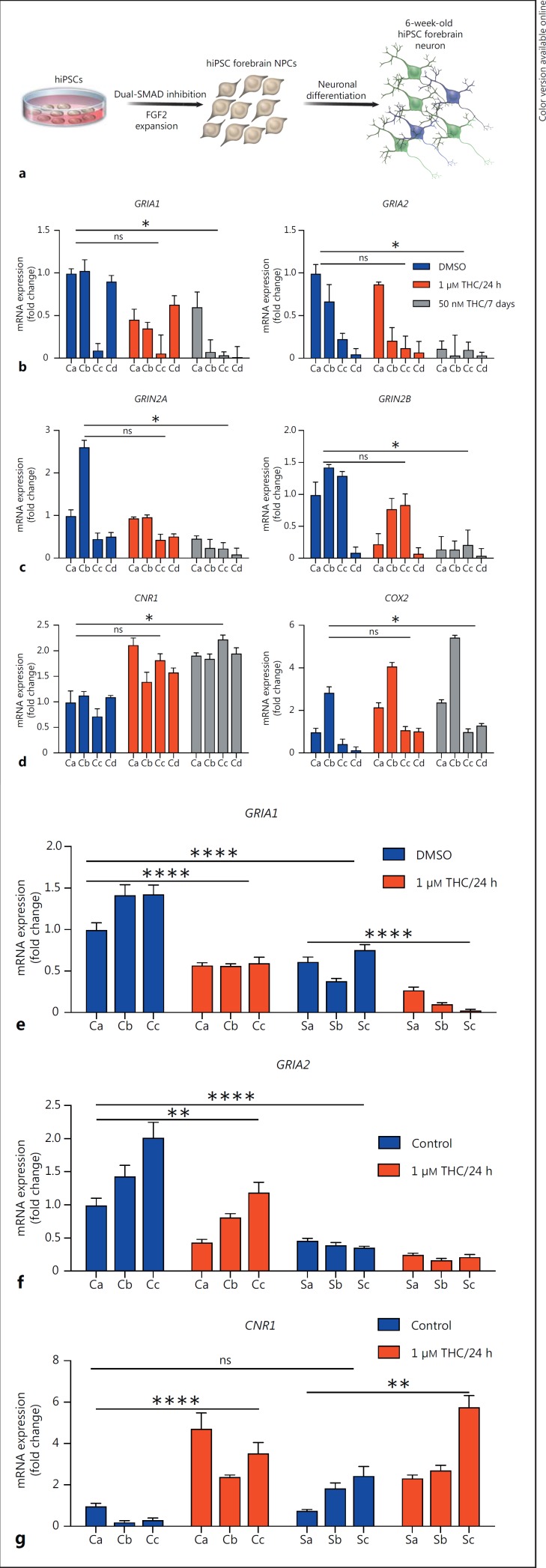
Fig. 3.
THC-induced alteration in gene expression in hiPSC forebrain neurons from cases with schizophrenia and healthy controls. a Schematic of directed differentiation from hiPSCs. b–d Real-time PCR analysis of GRIA1, GRIA2, GRIN2A, GRIN2B, CNR1, and COX2 expression in hiPSC forebrain neurons treated with DMSO (control), 1 μM THC for 24 h, and 50 nM THC for 7 days. Ca–Cd indicate neurons induced from 4 different individuals (controls a–d). Values are expressed as mean ± SEM, relative to DMSO-treated levels for Ca. e–g Real-time PCR analysis of GRIA1, GRIA2, and CNR1 expression in hiPSC forebrain neurons treated with DMSO (control) or 1 μM THC for 24 h. Ca–Cc indicate neurons induced from 3 controls and Sa-Sc from 3 patients with schizophrenia (controls and schizophrenia patients a–c). Values are expressed as mean ± SEM, relative to DMSO-treated levels for Ca. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. p SZ-CNR1 acute vs. chronic = 0.0008. USA). Data are presented as the mean ± SEM and were analyzed by two-way ANOVA with the Sidak multiple comparison test or Friedman test with Dunn's multiple comparisons test. Values are expressed as mean ± SEM. p > 0.05 (ns); * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001.
Fig. 4.
THC-induced alteration in gene expression in hiPSC forebrain neurons from cases with schizophrenia and healthy controls. a–c Real-time PCR analysis of GRIA1, GRIA2, and CNR1 expression in hiPSC forebrain neurons treated with DMSO (control) or THC (5 nM) for 7 days. Ca-Cd indicate neurons induced from 4 controls and Sa-Sd from 4 patients with schizophrenia (controls and schizophrenia patients a–d). Values are expressed as mean ± SEM, relative to DMSO-treated levels for Ca. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
THC was dissolved in DMSO to 1 mg/mL and prepared as previously described [38]; in all experiments, an equivalent volume of DMSO was used as a vehicle control. THC treatment of NGN2 -hiPSC and NGN2 -NPC neurons occurred with each media change (every other day) for the final 7 days of maturation (5 nM every 48 h for 7 days); induced neurons were harvested for experiments on the 21st day. For forebrain neurons, cells were treated with either 5 nM THC every 48 h for 7 days or with acute THC exposure (1 μM THC for 24 h) and chronic THC treatment (50 μM THC daily for 7 days) immediately prior to harvest at 6 weeks. The CB1R antagonist SR141716A (RIM, Tocris) was dissolved in DMSO and used at a final concentration of 20 nM; an equivalent volume of DMSO was used as a vehicle control.
Lentivirus Generation
Lentivirus production was as previously described [36]: 12.2 μg lentiviral DNA, 8.1 μg MDL-gagpol, 3.1 μg Rev-RSV, and 4 μg CMV-VSVG were mixed together with 500 μL of Opti-MEM (Life Technologies) and 1 μg/μL polyethylenimine (Polysciences) 25kD linear and added per 15 cm plate of HEK 293T cells. Medium was changed 5 h later. Virus was harvested from media supernatant 48 h after transfection and again 48 h later. Virus was concentrated by ultracentrifugation at 13,000 g for 2 h, resuspended in DMEM, aliquoted, and stored at −80°C.
qRT-PCR
Total RNA was extracted from cells using Trizol (Life Technologies). One-step qRT-PCR was performed from 500 ng RNA, and the results reported include at least 3 independent RNA preparations (qRT-PCR from cohort 2 for Fig. 3 was from an independent RNA preparation and required, following Trizol RNA purification, clean up with chloroform followed by ethanol precipitation with glycogen and conversion to cDNA using the high-capacity RNA-to-cDNA kit [Thermofisher]). Primers spanned intron/exon junctions (except for ACTIN and MAP2); primer specificity and the absence of genomic RNA contamination was confirmed by the melting curve of the products. The PCR cycling parameters were 94°C for 2 min, 40 cycles at 94°C for 15 s, 60°C for 20 s, and 72°C for 40 s. The comparative threshold cycle value (Ct) method was used for data analysis. mRNA values were normalized to both GAPDH and ACTIN by dividing the expression level (per technical replicate) by the average of the 2 housekeeping Ct values (per technical replicate). Primer sequences are listed in online supplementary Table 1.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 7.0b for Mac OS X (GraphPad Software, San Diego, CA, USA). Data are presented as the mean ± SEM and were analyzed by two-way ANOVA with the Sidak multiple comparison test or Friedman test with Dunn's multiple comparisons test. Values are expressed as mean ± SEM. p > 0.05 (ns); * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001.
Results
THC Exposure Alters Gene Expression of Glutamate Receptor Subunits
We and others have previously demonstrated that NGN2 -hiPSC neurons [35], NGN2 -NPC neurons [36], and directed differentiation forebrain neurons [37, 40] have neuronal morphology (online suppl. Fig. 1, 2; see www.karger.com/doi/10.1159/000477762 for all online suppl. material), are positive for neuronal markers such as βIII-TUBULIN, the dendritic marker MAP2AB (online suppl. Fig. 2), the excitatory synaptic marker VGLUT1, and other synaptic proteins. Gene expression studies indicate that a variety of neuronal enzymes and synaptic proteins are expressed and that these cells are most similar to fetal forebrain tissue [31, 42]. All three populations of neurons undergo action potentials and show evidence of spontaneous neuronal activity [35, 36, 37].
In mice, THC is typically used to induce dose- and time-dependent alterations in gene expression in vivo at concentrations from 5 to 10 mg/kg body weight for 4 h to 7 days; it has been used on primary mouse neurons in vitro at doses ranging from 25 nM to 30 μM for 12–20 h (Table 2). Critically, because THC induced cell death in two human neuroblastoma cell lines (SK-N-SH and NUB-6) in treatments as low as 20 nM THC [43], we first treated hiPSC-derived neurons with a minimal dose of THC (5 nM for 7 days), increasing in later experiments (50 nM THC for 7 days and 1 μM THC for 24 h).
Table 2.
Summary of the effect of THC on gene expression
| Gene | Evidence | Species/brain region | Dose | Method | Direction | NGN2-hiPSC neurons | NGN2-NPC neurons | Forebrain neurons |
|---|---|---|---|---|---|---|---|---|
| GRIA1 | Fan et al. [24] | mouse hippocampus | in vivo: 10 mg/kg/day, i.p., for 7 days | qPCR and western blot | decrease | decrease | decrease | decrease |
| Suarez et al. [52] | rat cerebellum | in vivo: 5 mg/kg, orally from gestational day 5 to postnatal day 20 | IHC | decrease | ||||
| GRIA2 | Fan et al. [24] | mouse hippocampus | in vivo: 10 mg/kg/day, i.p., for 7 days | qPCR and western blot | decrease | decrease | decrease | decrease |
| Suarez et al. [52] | rat cerebellum | in vivo: 5 mg/kg, orally from gestational day 5 to postnatal day 20 | IHC | decrease | ||||
| GRIN2A | Fan et al. [24] | mouse hippocampus | in vivo: 10 mg/kg/day, i.p., for 7 days | qPCR and western blot | decrease | decrease | decrease | decrease |
| GRIN2B | Fan et al. [24] | mouse hippocampus | in vivo: 10 mg/kg/day, i.p., for 7 days | qPCR and western blot | decrease | decrease | decrease | decrease |
| CNR1 | Zhuang et al. [27] | mouse hippocampus, cerebellum and striatum | in vivo: 10 mg/kg/day, i.p. from 6 h to 21 days | qPCR | increase in cerebellum and hippocampus; decrease in striatum | increase | increase | increase |
| COX2 | Chen et al. [21] | NG108-15 cells and hippocampal cell culture | in vitro: 30 µM for 12 h in vivo: 10 mg/kg/day, from 4 h to 7 days | qPCR and western blot | increase | increase | increase | increase |
| Mestre et al. [28] | murine brain-derived endothelial cells | in vitro: 25 nM, 100 nM, and 1 µM for 20 h | WIN55; western blot | increase | ||||
We tested the effect of 7-day 5-nM THC treatment, relative to DMSO-treated vehicle controls, on glutamate receptor gene expression across 3-week-old NGN2 – hiPSC neurons from 5 independent control hiPSC lines (cohort 1, controls 1–5) as well as 3-week-old NGN2 -NPC neurons from 3 independent control NPC lines (cohort 1, controls 1–3), in order to determine if THC effects are consistent in excitatory neurons derived from multiple control individuals via differing methodologies. Across 3 independent experimental replicates (triplicate biological samples within each experimental replicate) for both NGN2 -hiPSC and NGN2 -NPC neurons, THC treatment reduced GRIA1, GRIA2, GRIN2A, and GRIN2B expression (NGN2 -hiPSC neurons: p GRIA1 = 0.0345, p GRIA2 = 0.0064, p GRIN1 = 0.0518, p GRIN2A < 0.0001, p GRIN2B < 0.0001, p CNR1 < 0.0001, p COX2 < 0.0001; NGN2 -NPC neurons: p GRIA1 = 0.0004, p GRIA2 = 0.0367, p GRIN1 = 0.6205, p GRIN2A < 0.0001, p GRIN2B < 0.0001, p CNR1 = 0.0054, p COX2 < 0.0001; two-way ANOVA with Holm-Sidak test for multiple comparisons). Changes in GRIA1 were confirmed across 2 independent experimental replicates of western blot, from 3 controls each (cohort 1, controls 1–3), which demonstrated that GLUA1 (p = 0.0367) (online suppl. Fig. 3), but not MAP2 (p = 0.6342) (online suppl. Fig. 4), protein levels were decreased following 5-nM THC treatment of NGN2 -NPC neurons. THC also increased CNR1 and COX2 expression in NGN2 -hiPSC and NGN2 -NPC neurons (Fig. 1, 2; Table 2).
Fig. 1.
THC-induced alteration in gene expression in NGN2 -hiPSC neurons. a Schematic of NGN2 -hiPSC neuron induction from hiPSCs. b–d Real-time PCR analysis of GRIA1, GRIA2, GRIN1, GRIN2A, GRIN2B, CNR1, and COX2 expression in NGN2 -hiPSC neurons treated with DMSO (control) or THC (5 nM) for 7 days. C1-C5 indicate neurons induced from 5 different individuals (controls 1–5). Values are expressed as mean ± SEM, relative to DMSO-treated levels for C1. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Fig. 2.
THC-induced alteration in gene expression in NGN2 -NPC neurons. a Schematic of NGN2 -NPC neuron induction from hiPSC forebrain NPCs. b–d Real-time PCR analysis of GRIA1, GRIA2, GRIN1, GRIN2A, GRIN2B, CNR1, and COX2 expression in NGN2 -NPC neurons treated with DMSO (control) or THC (5 nM) for 7 days. C1–C3 indicate neurons induced from 3 different individuals (controls 1–3). Values are expressed as mean ± SEM, relative to DMSO-treated levels for C1. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
To validate these findings, we further tested the effect of acute THC exposure (1 μM THC for 24 h), relative to DMSO-treated vehicle controls, of 6-week-old forebrain neurons generated via directed differentiation from 4 independent control NPC lines (cohort 2, controls a–d) (triplicate biological samples within 1 experimental replicate). Again, we observed that THC treatment reduced GRIA1, GRIA2, GRIN2A, and GRIN2B expression and increased CNR1 and COX2 expression (p GRIA1/acute = 0.1542, p GRIA1/chronic = 0.0267, p GRIA2/acute = 0.9590, p GRIA2/chronic = 0.0417, p GRIN2A/acute = 0.9701, p GRIN2A/chronic = 0.02709, p GRIN2B/acute = 0.9636, p GRIN2B/chronic = 0.0106, p CNR1/acute = 0.0512, p CNR1/chronic = 0.012, p COX2/acute = 0.1542, p COX2/chronic = 0.0267, nonparametric Friedman ANOVA with Dunn's multiple comparison test) (Fig. 3b, c, d). Similar to the changes observed in controls, when this same THC treatment paradigm (1 μM THC treatment for 24 h) was applied to 6-week-old forebrain neurons derived from 3 cases with schizophrenia, we again observed reduced GRIA1 and GRIA2 and increased CNR1 expression in acute and chronic THC-treated neurons (p GRIA1 control vs. acute < 0.0001; p GRIA1 control vs. SZ-GRIA1 control < 0.0001; p SZ-GRIA1 control vs. acute < 0.0001; p GRIA2 control vs. acute = 0.0017; p GRIA2 control vs. SZ-GRIA2 control < 0.0001; p SZ-GRIA2 control vs. acute = 0.5479; p CNR 1 control vs. acute < 0.0001; p CNR 1 control vs. SZ-CNR1-control = 0.8261; p SZ-CNR1 control vs. acute = 0.028) (Fig. 3e, f, g).
Finally, when using a THC treatment paradigm consistent with that used for the NGN2 neurons (5 nM THC treatment for 7 days on 6-week-old forebrain neurons), we also observed reduced GRIA1 and GRIA2 and increased CNR1 (p GRIA1 < 0.0001, p SZ-GRIA1 < 0.0001, p GRIA2 < 0.0001, p SZ-GRIA2 < 0.0001, p CNR1 < 0.0001, p SZ-CNR1 < 0.0003 (Fig. 4).
Although these data suggest that neurons derived from cases with schizophrenia and controls respond similarly to THC, given the size of this small cohort, we caution that we are likely underpowered to conclude that there are (or are not) meaningful diagnosis-dependent differences in THC response.
Altered Gene Expression by THC Involves a CB1-Dependent Mechanism
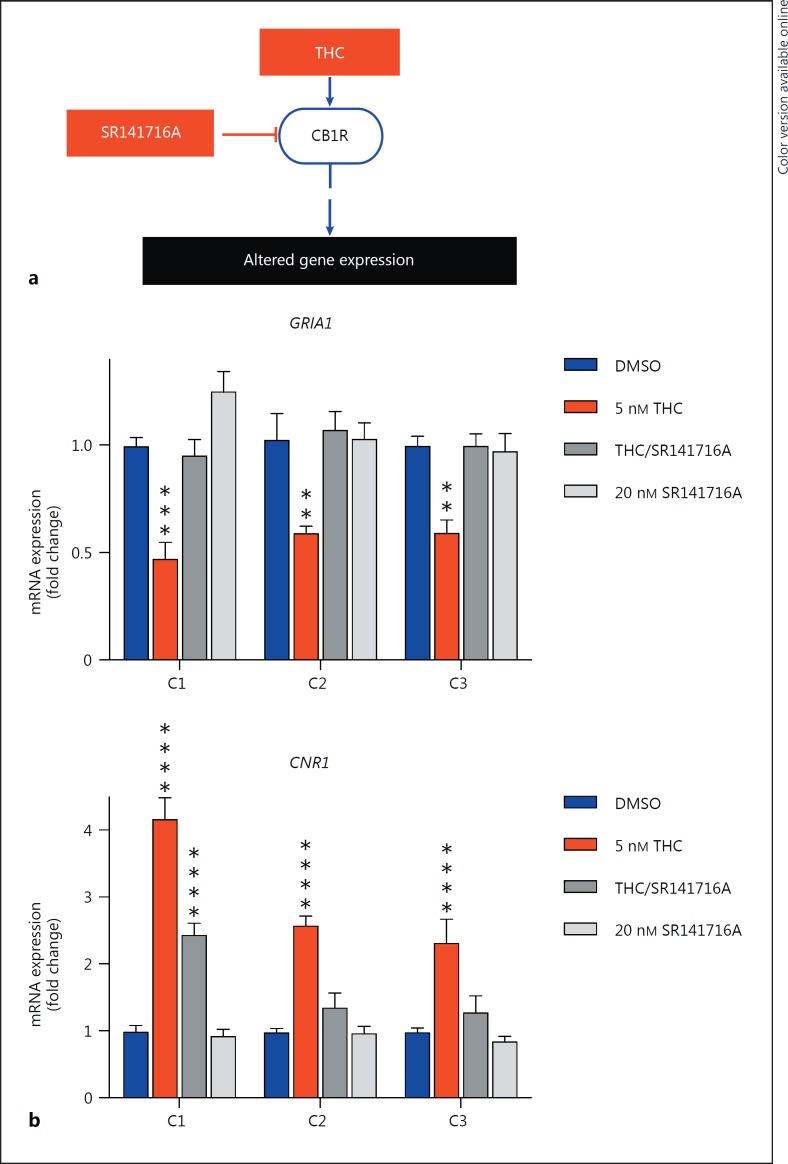
Because CB1R is the predominant cannabinoid receptor expressed in the central nervous system, we tested whether THC-mediated gene expression changes were dependent on CB1R activity, by treating with SR141716A, a selective CB1R antagonist [44]. THC-induced changes in GRIA1 and CNR1 mRNA expression in 3-week-old NGN2 -NPC neurons were blocked by concurrent 20-nM SR141716A treatment (triplicate biological samples within 2 experimental replicates), suggesting that THC-mediated effects on GRIA1 and CNR1 may be dependent on CB1R activity (GRIA1: p THC = 0.0312, p THC/SR141715A = 0.9767, p SR141715A = 0.9048; CNR1: p THC = 0.0045, p THC/SR141715A = 0.7052, p SR141715A = 0.9963, ordinary one-way ANOVA followed by Dunnett's test for multiple comparisons) (Fig. 5).
Fig. 5.
THC-induced alteration in gene expression in NGN2 -NPC neurons is mediated by CB1 receptor. a Model of the effect of THC on gene expression via the CB1 receptor. b Real-time PCR analysis of GRIA1 and CNR1 after treatment with THC (5 nM) or SR141716A (20 nM) for 7 days.
Discussion
Cannabinoids result in decreased glutamate receptor subunit protein levels in mice and a subsequent functional impairment in glutamatergic transmission [21, 24]. Consistent with this, we demonstrate that THC induced similar effects on NGN2 -hiPSC neurons, NGN2 -NPC neurons, and forebrain neurons from control individuals, indicating that hiPSC-derived neurons may serve as a human cell-based platform for studying the molecular and cellular effects of THC on developing human neural cells across a variety of genetic backgrounds. Overall, our results are consistent with results from animal studies of THC effects (Table 2). Of course, functional studies, including an examination of synaptic density, size, and activity, will be necessary to confirm the synaptic effects predicted by our gene expression analyses.
Notably, we observed variation in expression levels of a number of the glutamatergic genes between control individuals, a finding that is consistent with postmortem analyses [45] and likely reflects both the genetic variation between individuals as well as technical differences that arise from both the reprogramming and neuronal differentiation/induction processes. For this reason, THC-treated neurons were compared to their isogenic vehicle-treated controls.
The established role of endogenous cannabinoids and CB1R in fine-tuning brain development during sensitive developmental periods suggests that overstimulation of cannabinoid signaling by THC may perturb critical physiological processes at their most vulnerable periods and result in deficits in cortical circuits. Indeed, prenatal cannabinoid exposure is associated with lower IQs and delinquent behaviors in children [2, 3]. Moreover, adolescent cannabis use is associated with an increased risk of developing schizophrenia and/or accelerated onset of symptoms [5, 46, 47, 48, 49]. Emerging evidence suggests that cannabinoid exposure may have complex interactions with genetic factors associated with schizophrenia and other neuropsychiatric disorders (reviewed in [50, 51]), such as the link between CNR1 gene polymorphisms and cannabinoid exposure on both brain structure and clinical outcome in schizophrenia patients [30]. By expanding this work across a variety of hiPSC-derived neurons from individuals with different schizophrenia risk alleles, we hope that our hiPSC-based model will facilitate studies of the interaction of THC exposure and genetic risk factors underlying schizophrenia vulnerability.
Conclusions
In light of widespread cannabis use, our understanding of the molecular and cellular effects of THC on human neural cells must improve. Our study suggests that cannabinoid exposure results in changes in glutamate signaling in developing human neurons.
Statement of Ethics
The participants provided written informed consent.
Disclosure Statement
The authors declare that no conflicts of interest exist.
Author Contributions
I.V.O. contributed to the experimental design as well as to the qPCR experiments and analysis. H.M. and K.S. also contributed to qPCR experiments and analysis. E.K.F. contributed confocal imaging. I.V.O. and K.J.B. designed the experiments and wrote the manuscript.
Acknowledgements
Kristen J. Brennand is a New York Stem Cell Foundation – Robertson Investigator. The Brennand Laboratory is supported by the Brain and Behavior Research Foundation, Brain Research Foundation, NIH grants R01 MH101454 and R01 MH106056, and the New York Stem Cell Foundation. Ifeanyi V. Obiorah was supported by the National Institute of Drug Abuse grant T32DA007135.
References
- 1.Russo EB. History of cannabis and its preparations in saga, science, and sobriquet. Chem Biodivers. 2007;4:1614–1648. doi: 10.1002/cbdv.200790144. [DOI] [PubMed] [Google Scholar]
- 2.Day NL, Leech SL, Goldschmidt L. The effects of prenatal marijuana exposure on delinquent behaviors are mediated by measures of neurocognitive functioning. Neurotoxicol Teratol. 2011;33:129–136. doi: 10.1016/j.ntt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldschmidt L, Richardson GA, Willford JA, Day NL. Prenatal marijuana exposure and intelligence test performance at age six. J Am Acad Child Adolesc Psychiatry. 2008;47:254–263. doi: 10.1097/CHI.0b013e318160b3f0. [DOI] [PubMed] [Google Scholar]
- 4.Warner TD, Roussos-Ross D, Behnke M. It's not your mother's marijuana: effects on maternal-fetal health and the developing child. Clin Perinatol. 2014;41:877–894. doi: 10.1016/j.clp.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fried PA, Watkinson B, Gray R. Differential effects on cognitive functioning in 13- to 16-year-olds prenatally exposed to cigarettes and marihuana. Neurotoxicol Teratol. 2003;24:427–436. doi: 10.1016/s0892-0362(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 6.Jutras-Aswad D, Dinieri JA, Harkany T, Hurd YL. Neurobiological consequences of maternal cannabis on human fetal development and its neuropsychiatric outcome. Eur Arch Psychiatry Clin Neurosci. 2009;259:395–412. doi: 10.1007/s00406-009-0027-z. [DOI] [PubMed] [Google Scholar]
- 7.Richardson GA, Day NL, Goldschmidt L. Prenatal alcohol, marijuana and tobacco use: infant mental and motor development. Neurotoxicol Teratol. 1995;17:479–487. doi: 10.1016/0892-0362(95)00006-d. [DOI] [PubMed] [Google Scholar]
- 8.Schneider M. Cannabis use in pregnancy and early life and its consequences: animal models. Eur Arch Psychiatry Clin Neurosci. 2009;259:383–393. doi: 10.1007/s00406-009-0026-0. [DOI] [PubMed] [Google Scholar]
- 9.Morris CV, DiNieri JA, Szutorisz H, Hurd YL. Molecular mechanisms of maternal cannabis and cigarette use on human development. Eur J Neurosci. 2011;34:1574–1583. doi: 10.1111/j.1460-9568.2011.07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Americal College of Obstetricians and Gynecologists Committee on Obstetric Practice Committee Opinion No. 637: Marijuana use during pregnancy and lactation. Obstet Gynecol. 2015;126:234–238. doi: 10.1097/01.AOG.0000467192.89321.a6. [DOI] [PubMed] [Google Scholar]
- 11.Howlett AC, Fleming RM. Cannabinoid inhibition of adenylate cyclase: pharmacology of the response in neuroblastoma cell membranes. Mol Pharmacol. 1984;26:532–538. [PubMed] [Google Scholar]
- 12.Diaz-Alonso J, Guzman M, Galve-Roperh I. Endocannabinoids via CB1 receptors act as neurogenic niche cues during cortical development. Philos Trans R Soc Lond B Biol Sci. 2012;367:3229–3241. doi: 10.1098/rstb.2011.0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkany T, Guzman M, Galve-Roperh I, Berghuis P, Devi LA, Mackie K. The emerging functions of endocannabinoid signaling during CNS development. Trends Pharmacol Sci. 2007;28:83–92. doi: 10.1016/j.tips.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Biegon A, Kerman IA. Autoradiographic study of pre- and postnatal distribution of cannabinoid receptors in human brain. Neuroimage. 2001;14:1463–1468. doi: 10.1006/nimg.2001.0939. [DOI] [PubMed] [Google Scholar]
- 15.Mato S, Del Olmo E, Pazos A. Ontogenetic development of cannabinoid receptor expression and signal transduction functionality in the human brain. Eur J Neurosci. 2003;17:1747–1754. doi: 10.1046/j.1460-9568.2003.02599.x. [DOI] [PubMed] [Google Scholar]
- 16.Busquets Garcia A, Soria-Gomez E, Bellocchio L, Marsicano G. Cannabinoid receptor type-1: breaking the dogmas. F1000 Research. 2016;5((F1000 Faculty Rev)):990. doi: 10.12688/f1000research.8245.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer A. Genetic manipulation of the endocannabinoid system. Handb Exp Pharmacol. 2015;231:129–183. doi: 10.1007/978-3-319-20825-1_5. [DOI] [PubMed] [Google Scholar]
- 18.Mendizabal-Zubiaga J, Melser S, Bénard G, Ramos A, Reguero L, Arrabal S, Elezgarai I, Gerrikagoitia I, Suarez J, Rodriguez De Fonseca F, Puente N, Marsicano G, Grandes P. Cannabinoid CB1 receptors are localized in striated muscle mitochondria and regulate mitochondrial respiration. Front Physiol. 2016;7:476. doi: 10.3389/fphys.2016.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan D, Drysdale AJ, Lafourcade C, Pertwee RG, Platt B. Cannabidiol targets mitochondria to regulate intracellular Ca2+ levels. J Neurosci. 2009;29:2053–2063. doi: 10.1523/JNEUROSCI.4212-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campolongo P, Trezza V, Ratano P, Palmery M, Cuomo V. Developmental consequences of perinatal cannabis exposure: behavioral and neuroendocrine effects in adult rodents. Psychopharmacology (Berl) 2011;214:5–15. doi: 10.1007/s00213-010-1892-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen R, Zhang J, Fan N, Teng ZQ, Wu Y, Yang H, Tang YP, Sun H, Song Y, Chen C. Δ(9)-THC-caused synaptic and memory impairments are mediated through COX-2 signaling. Cell. 2013;155:1154–1165. doi: 10.1016/j.cell.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathur BN, Tanahira C, Tamamaki N, Lovinger DM. Voltage drives diverse endocannabinoid signals to mediate striatal microcircuit-specific plasticity. Nat Neurosci. 2013;16:1275–1283. doi: 10.1038/nn.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltesz I, Alger BE, Kano M, Lee SH, Lovinger DM, Ohno-Shosaku T, Watanabe M. Weeding out bad waves: towards selective cannabinoid circuit control in epilepsy. Nat Rev Neurosci. 2015;16:264–277. doi: 10.1038/nrn3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan N, Yang H, Zhang J, Chen C. Reduced expression of glutamate receptors and phosphorylation of CREB are responsible for in vivo d9-THC exposure-impaired hippocampal synaptic plasticity. J Neurochem. 2010;112:691–702. doi: 10.1111/j.1471-4159.2009.06489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, Kesner P, Metna-Laurent M, Duan T, Xu L, Georges F, Koehl M, Abrous DN, Mendizabal-Zubiaga J, Grandes P, Liu Q, Bai G, Wang W, Xiong L, Ren W, Marsicano G, Zhang X. Acute cannabinoids impair working memory through astroglial CB1 receptor modulation of hippocampal LTD. Cell. 2012;148:1039–1050. doi: 10.1016/j.cell.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 26.Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, Ren Y, Miller ML, Blitzer RD, Hurd YL. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology. 2014;39:1315–1323. doi: 10.1038/npp.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang S, Kittler J, Grigorenko EV, Kirby MT, Sim LJ, Hampson RE, Childers SR, Deadwyler SA. Effects of long-term exposure to delta9-THC on expression of cannabinoid receptor (CB1) mRNA in different rat brain regions. Brain Res Mol Brain Res. 1998;62:141–149. doi: 10.1016/s0169-328x(98)00232-0. [DOI] [PubMed] [Google Scholar]
- 28.Mestre L, Correa F, Docagne F, Clemente D, Guaza C. The synthetic cannabinoid WIN 55,212-2 increases COX-2 expression and PGE2 release in murine brain-derived endothelial cells following theiler's virus infection. Biochem Pharmacol. 2006;72:869–880. doi: 10.1016/j.bcp.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 29.Singer LT, Nelson S, Short E, Min MO, Lewis B, Russ S, Minnes S. Prenatal cocaine exposure: drug and environmental effects at 9 years. J Pediatr. 2008;153:105–111. doi: 10.1016/j.jpeds.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho BC, Wassink TH, Ziebell S, Andreasen NC. Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr Res. 2011;128:66–75. doi: 10.1016/j.schres.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, Sasai Y, Alvarez-Buylla A, Rubenstein JL, Kriegstein AR. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O'Rourke NA, Nguyen KD, Smith SJ, Huguenard JR. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Sudhof TC. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho SM, Hartley BJ, Tcw J, Beaumont M, Stafford K, Slesinger PA, Brennand KJ. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods. 2016;101:113–124. doi: 10.1016/j.ymeth.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topol A, Hartley BJ, English J, Hauberg ME, Tran N, Rittenhouse CA, Simone A, Ruderfer DM, Johnson J, Readhead B, Hadas Y, Gochman PA, Wang YC, Shah H, Cagney G, Rapaport J, Gage FH, Dudley JT, Sklar P, Mattheisen M, Cotter D, Fang G, Brennand KJ. Dysregulation of miRNA-9 in a subset of schizophrenia patient-derived neural progenitor cells. Cell Rep. 2016;15:1024–1036. doi: 10.1016/j.celrep.2016.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topol A, Tran NN, Brennand KJ. A guide to generating and using hiPSC derived NPCs for the study of neurological diseases. J Vis Exp. 2015;96:e52495. doi: 10.3791/52495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennand KJ, Simone A, Jou J, Gelboin- Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szutorisz H, Egervari G, Sperry J, Carter JM, Hurd YL. Cross-generational THC exposure alters the developmental sensitivity of ventral and dorsal striatal gene expression. Neurotoxicol Teratol. 2016;58:107–114. doi: 10.1016/j.ntt.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Stadler M, Weiss R, Church GM. Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol. 2014;10:760. doi: 10.15252/msb.20145508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher T, Golan H, Schiby G, PriChen S, Smoum R, Moshe I, Peshes-Yaloz N, Castiel A, Waldman D, Gallily R, Mechoulam R, Toren A. In vitro and in vivo efficacy of non-psychoactive cannabidiol in neuroblastoma. Curr Oncol. 2016;23:S15–S22. doi: 10.3747/co.23.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinaldi-Carmona M, Barth F, Heaulme M, Shire D, Calandra B, Congy C, Martinez S, Maruani J, Neliat G, Caput D, Ferrara P, Soubrie P, Breliere JC, Le Fur G. SR141716A, a potent and selective antagonist of the brain cannabinoid receptor. FEBS Lett. 1994;350:240–244. doi: 10.1016/0014-5793(94)00773-x. [DOI] [PubMed] [Google Scholar]
- 45.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caspi A, Moffitt TE, Cannon M, McClay J, Murray R, Harrington H, Taylor A, Arseneault L, Williams B, Braithwaite A, Poulton R, Craig IW. Moderation of the effect of adolescent-onset cannabis use on adult psychosis by a functional polymorphism in the catechol-O-methyltransferase gene: longitudinal evidence of a gene X environment interaction. Biol Psychiatry. 2005;57:1117–1127. doi: 10.1016/j.biopsych.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 47.Arseneault L, Cannon M, Witton J, Murray RM. Causal association between cannabis and psychosis: examination of the evidence. Br J Psychiatry. 2004;184:110–117. doi: 10.1192/bjp.184.2.110. [DOI] [PubMed] [Google Scholar]
- 48.Rabner J, Gottlieb S, Lazdowsky L, LeBel A. Psychosis following traumatic brain injury and cannabis use in late adolescence. Am J Addict. 2016;25:91–93. doi: 10.1111/ajad.12338. [DOI] [PubMed] [Google Scholar]
- 49.Schneider M. Puberty as a highly vulnerable developmental period for the consequences of cannabis exposure. Addict Biol. 2008;13:253–263. doi: 10.1111/j.1369-1600.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- 50.Bortolato M, Bini V, Tambaro S. Vulnerability factors for the psychiatric and behavioral effects of cannabis. Pharmaceuticals (Basel) 2010;3:2799–2820. doi: 10.3390/ph3092799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone DT, Hill MN, Rubino T. Adolescent cannabis use and psychosis: epidemiology and neurodevelopmental models. Br J Pharmacol. 2010;160:511–522. doi: 10.1111/j.1476-5381.2010.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suarez I, Bodega G, Fernandez-Ruiz J, Ramos JA, Rubio M, Fernandez B. Down-regulation of the AMPA glutamate receptor subunits GluR1 and GluR2/3 in the rat cerebellum following pre- and perinatal delta9-tetrahydrocannabinol exposure. Cerebellum. 2004;3:66–74. doi: 10.1080/14734220310017230. [DOI] [PubMed] [Google Scholar]