Abstract
Whole-genome and transcriptome sequencing were performed to identify potential therapeutic strategies in the absence of viable treatment options for a patient initially diagnosed with vulvar adenocarcinoma. Genomic events were prioritized by comparison against variant distributions in the TCGA pan-cancer data set and complemented with detailed transcriptome sequencing and copy-number analysis. These findings were considered against published scientific literature in order to evaluate the functional effects of potentially relevant genomic events. Analysis of the transcriptome against a background of 27 TCGA cancer types led to reclassification of the tumor as a primary HER2+ mammary-like adenocarcinoma of the vulva. This revised diagnosis was subsequently confirmed by follow-up immunohistochemistry for a mammary-like adenocarcinoma. The patient was treated with chemotherapy and targeted therapies for HER2+ breast cancer. The detailed pathology and genomic findings of this case are presented herein.
Keywords: neoplasm of the breast, neoplasm of the genitourinary tract
INTRODUCTION
Background on the Disease
Vulvar cancers represent 5% of gynecologic cancers and <1% of all cancers in women (PDQ Adult Treatment Editorial Board 2017). Approximately 90% of vulvar cancers are squamous cell carcinomas, the majority of which are associated with high-risk human papilloma virus (HPV) (Del Pino et al. 2013). Vulvar adenocarcinomas, which include primary tumors and metastases to the vulva (Neto et al. 2003), account for most of the remaining vulvar cancers and are not associated with HPV. The differential diagnosis of vulvar adenocarcinomas is complex, including primary adenocarcinomas (mammary-like adenocarcinoma, mucinous carcinoma, adenoid cystic carcinoma, Bartholin gland adenocarcinoma, and carcinoma arising from extramammary Paget disease [EMPD]) and metastatic disease. Metastatic disease forms 5%–8% of all vulvar cancers (Neto et al. 2003).
Mammary-like adenocarcinomas of the vulva (MLAV) are rare primary vulvar cancers that resemble primary breast carcinomas (Alligood-Percoco et al. 2015). They are locally aggressive tumors that frequently recur and have lymph node metastasis in ∼60% of cases (Abbott and Ahmed 2006). MLAV are thought to arise from mammary-like glands in the vulva. These glands were first reported in 1875 (Hartung 1875), and at the time were thought to be supernumerary breast tissue remnants located along the milk lines. Current understanding suggests they are modified vulvar eccrine glands that can give rise to a variety of tumors including vulvar adenocarcinomas (van der Putte 1994). Treatment guidelines are traditionally the same across vulvar carcinomas, but new evidence suggests the transposition of some of the preexisting diagnostic and treatment approaches for breast carcinomas to mammary-like adenocarcinomas of the vulva (McMaster et al. 2013); these include sentinel node biopsy and molecular subtyping combined with adjuvant therapy (Abbott and Ahmed 2006; Perrone et al. 2009; Kazakov et al. 2011; Butler et al. 2014; Tessier-Cloutier et al. 2017).
Here we present a case study of a poorly differentiated vulvar adenocarcinoma, which was subsequently reclassified as a HER2+ MLAV upon genomic analysis. We present the initial pathology, genomic findings, confirmatory immunohistochemistry (IHC), and clinical follow-up after treatments based on the revised genomics-based diagnosis. To our knowledge, this is the first case of an MLAV described with detailed whole-genome and transcriptome sequencing analysis. This case highlights the utility of genomics-guided tumor-type characterization in confirming differential diagnosis for vulvar adenocarcinoma with mammary-like origins and suggests the potential for using genomic characterization to guide the staging and diagnostic histopathology of poorly differentiated tumors that often have indeterminate histologic or immunohistochemical findings. To this end, this study provides the first integrative genomic analysis of a mammary-like adenocarcinoma arising in the vulva and provides insights into the mutational status of such tumors.
Clinical Background
A 60-yr-old woman presented with a bleeding vulvar mass. There was no family history of cancer malignancies. Multiple masses were noted on physical examination, specifically a 2.5 cm firm right labium maius mass, a 2.5 cm bleeding vaginal introital mass, and bilateral inguinal lymphadenopathy up to 3.0 cm. The entire vulva was “desquamated,” but no specific skin lesion associated with the right labium maius mass was noted nor any proximity to the Bartholin gland. The largest lymph node was a left external iliac lymph node measuring 3.6 cm. No breast masses were identified in the physical examination. Furthermore, the patient reported that a history of remote mammograms had shown no malignancy. Bilateral mammograms were also negative for breast malignancy. Chest X-ray showed no metastatic pulmonary disease. Positron emission tomography/computed tomography (PET/CT) scans showed a hypermetabolic mass in the medial aspect of the right labium maius (3.2 × 2.1 cm, maximal SUV 24.0) and an adjacent FDG-avid circumferential mass in the vaginal introitus (3.0 × 2.0 cm, maximal SUV 23.6). No other potential sites for a primary tumor were identified. The uterus was enlarged but showed physiologic uptake. The clinical and radiologic findings were consistent with stage IV vulvar cancer with metastatic disease to the bilateral inguinal, retrocaval, external iliac, and common iliac lymph nodes. An initial vulvar biopsy was taken at this point and pathology findings reported “poorly differentiated infiltrating carcinoma, favor [sic] poorly differentiated adenocarcinoma” (see Pathology analysis).
The patient was treated with four rounds of carboplatin and paclitaxel. Repeat imaging by PET/CT demonstrated response in all areas except for the inguinal lymph nodes. Subsequently, radiotherapy was delivered to the entire field of disease involvement at baseline, with radiotherapy boosts delivered to the still FDG-avid inguinal lymph nodes. At the 6-wk follow-up appointment following the completion of radiotherapy, a large left supraclavicular lymph node was palpated, and a fine needle aspirate (FNA) confirmed this to be a poorly differentiated adenocarcinoma consistent with metastatic vulvar adenocarcinoma.
In the absence of subsequent standard treatment options, the patient was enrolled in the Personalized OncoGenomics (POG) Project at the BC Cancer Agency (BCCA) to identify potentially actionable targets and to validate the pathology diagnosis of vulvar adenocarcinoma. No additional genetic testing was done, and no other treatment was received between the initial vulvar biopsy and presentation of metastasis in the left supraclavicular lymph node or during the genomic analysis. The sample from the metastatic mass in the left supraclavicular lymph node (subsequently referred to as the recurrence biopsy) was submitted to POG for sequencing and analysis. A detailed clinical time line is provided in Supplemental Table 1.
Pathology Analysis
Pathology of Initial Vulvar Biopsy
The tumor was composed of nests and cords of large pleomorphic epithelioid cells with hyperchromatic nuclei and showed no definite gland formation, papillary structures, or intraluminal vacuoles. However, there were occasional cells showing possible signet-ring features and intraluminal vacuoles. Mitotic figures were easily identified. No overlying epidermis or normal mammary-like glands were present. IHC performed at the receiving hospital revealed that the tumor was positive for CK7 and Ber-EP4 and negative for CEA, CK5, CK20, MART-1, and S100. PAS-diastase was negative for definite intraluminal murin. Summarily, in the initial assessment of the vulvar biopsy, the poorly differentiated morphology and the nonspecific immunoprofile resulted in a broad differential diagnosis, including poorly differentiated vulvar squamous cell carcinoma, poorly differentiated vulvar adenocarcinoma (Bartholin gland adenocarcinoma, MLAV, or adenocarcinoma arising from extramammary Paget disease), and metastatic adenocarcinoma (from the gastrointestinal tract or other gynecologic organs) and melanoma.
Post–Genome Pathology Analyses
Subsequent to the genomic analysis favoring a diagnosis of mammary-type carcinoma (described below), follow-up validation stains were performed at the BCCA. This validation work was carried out on the initial vulvar biopsy and on a repeat aspirate from the left supraclavicular lymph node.
IHC Validation—Vulvar Biopsy
The initial vulvar biopsy was reviewed at the BCCA, and additional IHC was done. This revealed that the tumor was negative for ER, PAX8, GCDFP-15 and mammaglobin, and focally positive for vimentin. The negative CEA, GCDFP-15 and PAS-diastase were against a diagnosis of adenocarcinoma arising from EMPD. The negative CK5 was against squamous cell carcinoma. The negative ER, GCDFP, and mammaglobin were against luminal A/B-types of primary MLAV. The negative PAX8 and CK20 were against metastatic gynecologic and lower gastrointestinal tract carcinomas, respectively. The negative MART-1 and S100 were against melanoma. The BCCA established that based on the clinical, histologic, and immune-phenotype findings, the patient had a poorly differentiated carcinoma.
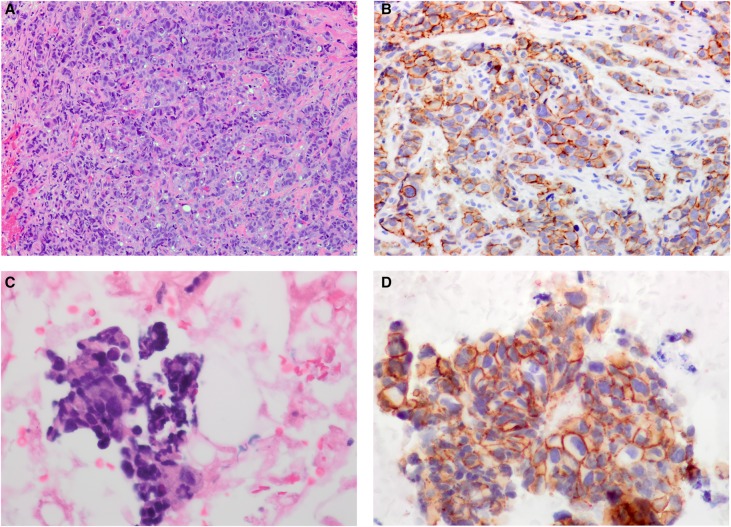
IHC on the initial vulvar biopsy for HER2, the protein product of the ERBB2 gene, was equivocal (score 2+; Fig. 1). This prompted reflex testing for HER2 amplification with fluorescent in situ hybridization (FISH) testing. FISH showed HER2/CEP17 ratio of 2.0 with 20 cells counted and an average HER2 copy number per cell of 6.35, with an average CEP 17 copy number per cell of 3.2. This is considered to be HER2 positive as per the 2013 ASCO/CAP guidelines, which require “complete, intense staining” of the circumferential membranes of >10% of tumor cells (Wolff et al. 2013).
Figure 1.
Histopathology of the MLAV. (A) The biopsy of the vulvar mass shows a poorly differentiated tumor composed of nests and cords of pleomorphic tumor cells (20× magnification). (B) The HER2 immunostain on the biopsy of the vulvar mass is equivocal, compatible with score 2+ based on predominantly incomplete, weak and moderate membrane staining within >10% of tumor cells (20× magnification). (C) The fine needle aspirate of the supraclavicular lymph node shows clusters of pleomorphic tumor cells consistent with metastatic mammary-like adenocarcinoma (40× magnification). (D) The HER2 immunostain of the supraclavicular lymph node shows tumor cells with complete, intense membrane staining in >10% of tumor cells compatible with score 3+ (40× magnification).
Validation—Repeat FNA of the Recurrence Site
An initial FNA of the supraclavicular lymph node showed poorly cohesive irregular glandular clusters of pleomorphic malignant cells consistent with metastatic adenocarcinoma. IHC of a repeat FNA of this lymph node demonstrated that the sample was strongly positive for GATA3, a relatively recent marker for mammary carcinoma (Miettinen et al. 2014). HER2 IHC was positive (score 3+) based on 30% of tumor cells showing strong circumferential membranous staining. ER was negative, as was the case with the initial vulvar biopsy. Therefore, the clinical, pathologic, immunophenotypic and genetic findings for the initial vulvar biopsy and supraclavicular lymph node metastasis were consistent with high-grade, ER–, HER2+ MLAV.
Exclusion of EMPD as a Differential Diagnosis
HER2 overexpression can also be observed in adenocarcinomas arising from EMPD. A 2005 study of patients with mammary and extramammary Paget disease observed coexpression of ERBB2 and AR in 88% (51/58) of the cases with mammary Paget (Liegl et al. 2005); the genomic analysis for this case also found high expression of ERBB2 and AR. With this observation in mind, invasive carcinoma arising from primary EMPD, rare anogenital tumors with proposed precursors that include Toker cells, pluripotent germinative cells, eccrine or apocrine glands, and mammary-like glands (Willman et al. 2005; Kazakov et al. 2011), were considered for differential diagnosis during the validation pathology workup for the recurrence. The positive staining for CK7, GATA3, and HER2 and the negative staining for ER overlap with previous reports of EMPD (De Leon et al. 2000; Perrotto et al. 2010; Richter et al. 2010; Morbeck et al. 2017). However, CEA, a nonspecific immunostain that is positive in most cases of primary EMPD (Brown and Wilkinson 2002), was negative in this patient's tumor. Furthermore, the overlying epidermis was not seen in the vulvar biopsy. Thus, the presence of pagetoid cells, which are necessary for the diagnosis of Paget disease and carcinomas arising therein, could not be assessed. This further supports the diagnosis of MLAV.
Genomic Analyses
The recurrence sample was submitted for whole-genome and transcriptome-sequencing and analyses. In the absence of standard treatment options for the patient, the aim of this exercise was to (a) identify potentially actionable genomic targets and (b) clarify the diagnosis and evaluate the validity of the initial diagnosis as vulvar adenocarcinoma. A constitutional blood sample and tumor from the lymph node biopsy were sequenced to a redundant sequence coverage depth of 43-fold and 90-fold, respectively. A transcriptome of 291 million sequence reads was also generated from the same tumor sample. The genomic and transcriptomic findings indicated the cancer was a mammary-like adenocarcinoma (see Results—Genomic Analyses). The association of specific genomic events and transcriptomic changes with breast cancer was confirmed upon detailed literature review and integrative analysis.
RESULTS
Genomic Analyses
Single-Nucleotide Variants
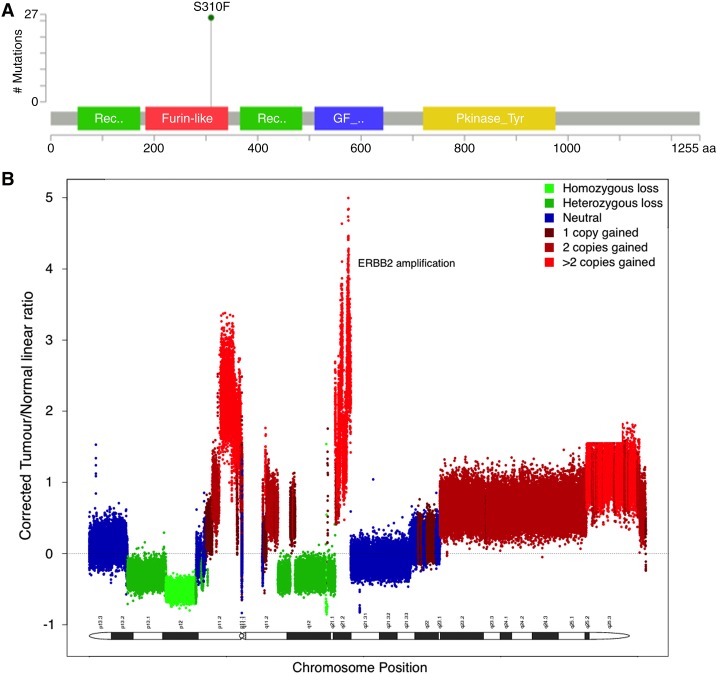
Comparison of sequencing results from the tumor and constitutional sample detected 375 nonsynonymous single-nucleotide variants (SNVs) and 15 insertion/deletion events (indels), 16 of which were considered to be of biological and/or clinical interest (summarized in Table 1). Notably, a previously described gain of function (GoF) mutation was observed in ERBB2 (p.S310F). The S310F mutation has been identified in several cancers, including breast, lung, and ovarian (Herter-Sprie et al. 2013). The mutation focus is highlighted in Figure 2A. Loss-of-function (LoF) mutations were also observed in tumor suppressor genes TP53 and RB1. Variants of unknown significance (VUS) were noted in PIK3CA, AKT3, and GNAS (Table 1).
Table 1.
Genomic findings: single nucleotide variants of interest
| Gene | Chr | DNA change | AA change | Protein change | COSM | SNV variant | Ref/Ref_RNA | Alt/Alt_RNA |
|---|---|---|---|---|---|---|---|---|
| AKT3 | 1 | 244006441 C>A | c.W11L | Trp11Leu | NA | VUS | 113/11 | 18/0 |
| ERBB2 | 17 | 37868208 C>T | S310F | Ser310Phe | 48358 | GoF | 26/625 | 165/4181 |
| GNAS | 20 | 57430298 C>G | R33G | Arg660Gly | NA | VUS | 39/4 | 7/0 |
| MAP3K12 | 12 | 53877268 C>T | R493Q | Arg493Gln | 1147056 | 62/44 | 7/4 | |
| OR14A16 | 1 | 247978827 G>T | L69I | Leu69Ile | 283503 | 133/– | 19/– | |
| PAF1 | 19 | 39876915 G>A | E438K | Glu438Lys | 87803 | 37/208 | 10/76 | |
| PCDHA6 | 5 | 140208403 G>A | E243K | Glu243Lys | 1062027 | 38/0 | 25/0 | |
| PIK3CA | 3 | 178938934 G>A | E726K | Glu276Lys | 87306 | VUS | 40/24 | 69/102 |
| PPM1B | 2 | 44428594 A>G | R86G | Arg86Gly | 188967 | 72/232 | 20/71 | |
| RB1 | 13 | 49033844 C>T | R661W | Arg661Trp | 861 | LoF | 14/59 | 35/294 |
| SLCO3A1 | 15 | 92669422 G>A | V436I | Val436Ile | 71259 | 37/21 | 18/9 | |
| SNTG2 | 2 | 1271197 G>C | D380H | Asp380His | 459649 | 57/– | 13/– | |
| THBS2 | 6 | 169629714 C>T | D738N | Asp738Asn | 595355 | 53/102 | 8/0 | |
| TP53 | 17 | 7577082 G>A | E286K | Glu286Lys | 99924 | LoF | 13/24 | 30/230 |
| UPF3A | 13 | 115047496 G>C | V70L | Val70Leu | 130084 | 15/25 | 9/0 | |
| ZNF830 | 17 | 33289390 C>T | Q269* | Gln269* | 705542 | 39/74 | 15/51 | |
| ZXDB | X | 57618845 G>A | E122K | Glu122Lys | 225105 | 22/16 | 11/2 | |
| ZXDB | X | 57618849 A>C | E123A | Glu123Ala | 225106 | 21/17 | 11/3 |
The key SNVs are listed, along with details on the counts of the reads spanning each gene in the tumor genome and transcriptome.
The rows in bold highlight the genes of key interest in the genomic analysis.
AA, amino acid; ALOH, amplification with loss of heterozygosity; Alt, coverage of alternative allele; Alt_RNA, RNA reads mapping alternative allele; Amplif, amplification; BCNA, balanced amplification; Chr, chromosome; COSM, COSMIC Mutation ID (numeric ID listed; query as COSMxxxxx); DLOH, deletion with loss of heterozygosity; GoF, gain of function; HET, heterozygous; LoF, loss of function; NLOH, neutral with loss of heterozygosity; Ref, coverage of reference allele; Ref_RNA, RNA reads mapping reference allele; SNV, single-nucleotide variant; VUS, variant of unknown significance.
–, No RNA-seq reads mapped to this gene.
Figure 2.
ERBB2 overexpression. (A) A lollipop plot showing the coordinate of the S310F gain-of-function mutation observed in this case. (B) A plot of the copy-number landscape of Chromosome 17. The ERBB2 copy-number gain is indicated.
Copy-Number Variants
A triploid model with an estimated 68% tumor content was inferred based on the prediction of allelic imbalance and loss of heterozygosity in the sample (see Methods). Copy-number variants are estimated with respect to this ploidy model. Focal copy-number amplifications were detected on Chromosomes 2, 8, 9, 17 and X. Of particular interest, copy-number gains were observed for ERBB2, AKT3, PIK3CA, CDK1, CCNB1, and AR. Loss of heterozygosity (LOH) events were detected for the tumor suppressors BRCA2, RB1, and TP53, resulting from a single copy loss. Furthermore, RB1 and TP53 had LoF mutations in the two remaining (homozygous) copies. These findings are summarized in Table 2, and the ERBB2 amplification is highlighted in Figure 2B. Copy-number variants associated with other SNVs are listed in Supplemental Table 2.
Table 2.
Genomic findings: copy number variants of interest
| Gene | Chr | Copy change versus ploidy corrected model (i.e., 2n = 2) | Copy type | TCGA expression percentilea | Fold expression changeb |
|---|---|---|---|---|---|
| AKT3 | 1 | +1 (HET) | Gain | 21 | −5.18 |
| ERBB2 | 17 | +8 (ALOH) | Amplif | 98 | 32.73 |
| GNAS | 20 | +1 (NLOH) | Gain | 2 | −1.55 |
| PIK3CA | 3 | +1 (HET) | Gain | 49 | −1.40 |
| RB1 | 13 | −1 (DLOH) | Loss | 88 | 1.76 |
| TP53 | 17 | −1 (DLOH) | Loss | 21 | −1.10 |
| AR | X | +8 (NLOH) | Amplif | 100 | 26.39 |
| BIRC5 | 17 | +5 (BCNA) | Amplif | 100 | 40.87 |
| BRCA2 | 13 | −1 (DLOH) | Loss | 94 | 2.21 |
| CDK12 | 17 | +8 (ALOH) | Amplif | 99 | 5.00 |
| CCNE2 | 8 | +17 (ALOH) | Amplif | 100 | 25.71 |
The key copy-number events in the tumor genome are listed, along with percentile values and fold changes calculated from the respective RPKMs.
The rows in bold highlight the genes of key interest in the genomic analysis.
AA, amino acid; ALOH, amplification with loss of heterozygosity; Alt, coverage of alternative allele; Alt_RNA, RNA reads mapping alternative allele; Amplif, amplification; BCNA, balanced amplification; Chr, chromosome; COSM, COSMIC Mutation ID (numeric ID listed; query as COSMxxxxx); DLOH, deletion with loss of heterozygosity; GoF, gain of function; HET, heterozygous; LoF, loss of function; NLOH, neutral with loss of heterozygosity; Ref, coverage of reference allele; Ref_RNA, RNA reads mapping reference allele; SNV, single-nucleotide variant; VUS, variant of unknown significance.
–, No RNA-seq reads mapped to this gene.
aCalculated against TCGA Breast Cancer Compendium.
bCalculated against Illumina BodyMap 2.0.
Structural Variants
De novo assembly of the genome and transcriptome was performed to identify structural rearrangements of potential biological and clinical significance. However, none were detected.
Transcriptome Analysis
A pairwise correlation analysis of the sample's transcriptome was undertaken to identify the closest correlated cancer type for the tumor from across 27 different tumor types available from The Cancer Genome Atlas (TCGA; see Methods) (The Cancer Genome Atlas Research Network 2013). The tumor sample correlated most strongly with the breast cancer data set (Fig. 3A). Based on this observation, we replicated the PAM50 test by selecting the PAM50 set of genes for correlating the sample's transcriptome against TCGA breast cancer samples with known BRCA molecular subtype status (PAM50). Consistent with the amplification and gain-of-function mutation in the ERBB2 gene, the tumor sample correlated the highest with the HER2 enriched and Luminal B subtypes (Fig. 3B).
Figure 3.
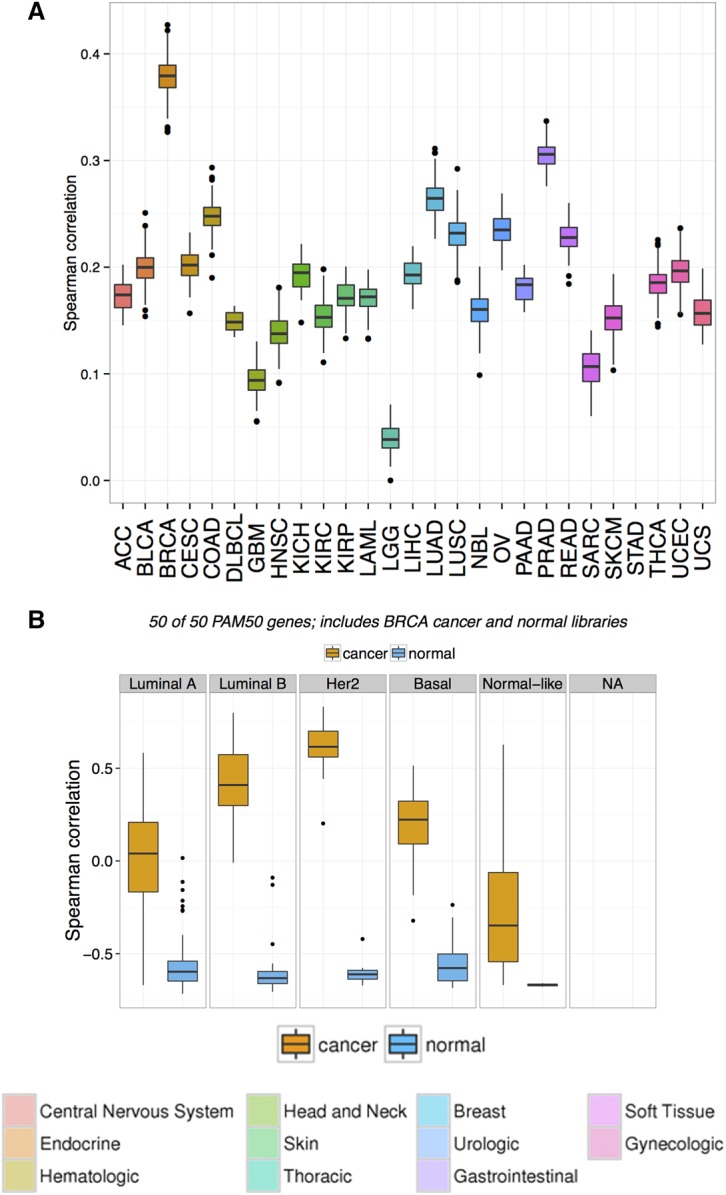
Correlation plot of the sample with TCGA data sets. (A) Boxplot distribution of the pairwise Spearman correlation of the recurrence biopsy with all TCGA samples, based on 3000 genes selected by ANOVA analysis (see Methods). The x-axis represents cancer types following TCGA conventions for naming. The highest correlation is observed with the TCGA Breast Cancer data set (BRCA). (B) Boxplot distribution of the pairwise Spearman correlation of the recurrence biopsy with the TCGA Breast Cancer data set, based on PAM50 subtypes (see Methods). The pairwise correlations with adjacent normal are shown to exclude any biases from normal contamination. The highest correlation is observed for HER2-enriched type, followed by Luminal B.
Based on the findings from the transcriptome-wide correlation analysis, the genomic events and RNA-level enrichments were considered against a background of breast cancer. A fold-change value for each gene was calculated against a normal breast tissue transcriptome (Illumina Human BodyMap 2.0) and a percentile rank of expression calculated in comparison to the breast cancer cohort from TCGA. Expression outliers were identified and evaluated in conjunction with mutational status, copy-number state, and known biological function. Nine genes of interest were identified having gains of more than three copies each, and also ranked in the 98th–100th percentile versus TCGA breast cancers (summarized in Table 2). Of particular interest among these genes were ERBB2 (98th percentile), CDK12 (99th percentile), AR (100th percentile), and CCNE2 (100th percentile). The extreme outlier expression of ERBB2 (33-fold overexpression, 98th percentile of BRCA) combined with the observed of the gain of function mutation (p.S310F) and estimated five copy gain (Fig. 2) further supported a diagnosis of a HER2+ mammary-like cancer and identified HER2 as a likely driver of the disease.
Mutational Signatures
WGS mutational data were compared against previously cataloged mutational signatures (Ju et al. 2017). A strong APOBEC signature was observed (Signatures 2 and 13). The APOBEC family of cytidine deaminases generates mutations of a specific pattern (the APOBEC signature mutation pattern), which has been reported in several cancers (Roberts et al. 2013). APOBEC activation has been associated with HPV; however, screening for microbial and viral sequences was negative for any microbial contaminants and no evidence for genomic integration of HPV was detected.
Clinical Interpretation
The reclassification of the cancer to a mammary-like adenocarcinoma and the overexpression of a clearly targetable protein (HER2) led to the patient being treated with standard therapies for metastatic HER2+ breast cancer. The patient received vinorelbine, trastuzumab, and pertuzumab, followed by capecitabine and lapatinib. The patient had a poor clinical response to all targeted therapies tried, at best achieving short-term disease stabilization but never achieving disease regression (summarized in Supplemental Table S1). The patient passed away 2 yr and 5 mo after her initial diagnosis.
DISCUSSION
Differential Diagnosis
The diagnosis and classification of vulvar adenocarcinomas is a complicated and understudied area, as this is a rare histologic subtype of vulvar cancers. The differential diagnoses include MLAV, adenocarcinoma arising from EMPD, mucinal carcinoma, Bartholin gland adenocarcinomas, and metastatic adenocarcinomas from various sites. In our case, the clinical, radiologic, and histologic features indicated a poorly differentiated (high-grade) primary vulvar adenocarcinoma. The patient was enrolled in the BCCA POG program upon the development of new metastases, with the two aims of characterizing the underlying genomics of this poorly differentiated cancer and identifying actionable therapeutic targets.
The bioinformatics analysis findings indicated the patient's tumor was most consistent with a HER2+ breast cancer profile. Additional genomic events, specifically the GoF S310F mutation in ERBB2, the LoF mutations combined with LOH in TP53 and RB1, and the coexpression of ERBB2 and AR at high levels, pointed to a mammary-like cancer. Post hoc histopathologic investigations on the initial biopsy and a repeat aspirate from the supraclavicular lymph node site corroborate the determinative finding of HER2 overexpression from the bioinformatics analysis, and supported the diagnosis of MLAV.
A recent IHC study showed that MLAV can be classified into four breast intrinsic subtypes, including a HER2+/ER− group (Tran et al. 2015; Tessier-Cloutier et al. 2017). Here we note that during the initial pathology workup for the vulvar biopsy for this cancer, the diagnosis of MLAV was not favored on the basis of ER, mammaglobin, and GCDFP-15 negativity. However, primary breast carcinomas with high nuclear grade are mostly ER− (Nadji et al. 2005), and a recent study has proposed GATA3 as a more sensitive marker for HER2+/ER− breast carcinomas than mammaglobin and GCDFP-15 (Tang et al. 2017). GATA3 was positive in the recurrence sample, which supports the conclusion that this tumor is mammary-type. The analyses presented herein are consistent with this body of evidence and confirm that ER negativity is consistent with the diagnosis of HER2+ MLAV.
Implications of Genomic Analyses
Here we highlight some interesting observations arising from the integrative genomic analysis of this case and their implications thereof on the selection of therapy. These findings also highlight the potential of this approach for identifying additional candidates for histopathology diagnostics.
AR overexpression is found in 60% of breast cancers and is generally observed more frequently in ER+ breast cancers than ER– ones—previous research has indicated a positive association with survival in ER+ breast cancers (Micello et al. 2010; Pietri et al. 2016). On the other hand, AR expression is significantly correlated with HER2 expression in ER– breast cancers, and a proliferative role for AR has been suggested recently in ER–, HER2+ patients (Micello et al. 2010; Pietri et al. 2016).
These recent findings support our observations, and additionally suggest the utility of considering AR expression in the diagnosis of mammary-like carcinomas.
A strong APOBEC signature is associated with HER2+ breast cancers and, because of its association with PDL1, has been positively correlated with response to immunotherapy in other cancers (Morganella et al. 2016). However, at the time of this analysis, immunotherapy was not available as an accessible line of treatment and was not pursued further (Mullane et al. 2016).
This case study demonstrates the utility of an integrative genome- and transcriptome-wide analysis to complement histopathology and to guide the selection of additional diagnostic tests, especially in scenarios where morphologic assessment of the tumor mass is insufficient in providing a confident diagnosis or where the immunophenotype of certain cancer types is not well known. An overexpression of CDK2 and a high-percentile expression of CCNE2 were observed in the recurrence sample's transcriptomic analysis. These have been suggested as potential resistance markers for trastuzumab (Scaltriti et al. 2011; Ichikawa et al. 2012; Tormo et al. 2017), and we can speculate on their potential role in rendering the treatment ineffective.
METHODS
Ultrasound-guided core-needle biopsies were obtained for the POG study. FISH assays and IHC were performed by the clinical laboratories at the BCCA according to established protocols. The rabbit monoclonal antibody to HER2 (clone 4B5; Ventana Medical Systems) was used for HER2 protein staining. Immunostaining was performed on the Ventana Benchmark Ultra automated system (Ventana Medical Systems) with 36 min of ULTRA CC1 before being incubated with the prediluted HER2 antibody for 8 min at 36°C. The ultraView DAB detection kit was used with an ultraWash step.
RNA and DNA were extracted and sequence libraries constructed using standard protocols (summarized in Table 3). Sequencing was performed on an Illumina HiSeq2500 platform at Canada's Michael Smith Genome Sciences Centre (GSC). One microgram each of DNA from normal blood and tumor biopsy were separately used as input to the GSC PCR (polymerase chain reaction)-free whole-genome sequencing protocol, and sequenced to 43× and 90× coverage, respectively. Of note, 1.725 μg of total RNA from the tumor was treated with the strand-specific messenger RNA sequencing protocol and sequenced to a total of 291 million reads. The reads were aligned to the GRCh37 reference human genome using BWA v0.5.7 (Li and Durbin 2010). Duplicate reads were marked using Picard (v1.38, https://github.com/broadinstitute/picard). Microbial and viral integration detection analysis was done using an in-house pipeline and BioBloom Tools (BBT) (Chu et al. 2014). WGS variants identified using SAMtools v0.1.7 mpileup (Li et al. 2009). The tumor and normal samples were compared with identify somatic events. SNVs were called using Strelka v0.4.62 (Saunders et al. 2012) and mutationSeq v1.0.2 (Ding et al. 2012). Strelka v0.4.62 was also used to called small insertions and deletions. The somatic variant annotation was done with the Ensemble database (v69), and the effect calculation was assisted by annotations from SnpEff 3.2 (Cingolani et al. 2012), COSMIC v64, and dbSNP v137. LOH events and tumor content were estimated with APOLLOH v0.1.1 (Ha et al. 2012). Copy-number variants were identified using CNAseq v0.0.6 (https://www.bcgsc.ca/platform/bioinfo/software/cnaseq).
Table 3.
Details of sequencing experiments
| Sample | Type | Input (μg) | Library protocol | Library | Coverage | Reads (total) |
|---|---|---|---|---|---|---|
| Biopsy tumor | DNA | 1 | PCR-Free WGS Library | P00401 | 90× | NA |
| Biopsy tumor | RNA | 1.725 | ssRNA-Seq Library | P00403 | NA | 291 million |
| Normal blood | DNA | 1 | PCR-Free WGS Library | P00381 | 43× | NA |
NA, not available; PCR, polymerase chain reaction; ssRNA, single-stranded RNA; WGS, whole-genome sequencing.
RNA-seq data were analyzed using JAGuaR v2.0.3 (Butterfield et al. 2014). The RNA-seq data were subsequently processed by an in-house pipeline for whole-transcriptome shotgun sequencing (WTSS) coverage analysis, to yield exon- and transcript-level read counts and normalized gene expression values (reads per kilobase of transcript per million mapped reads, RPKM). Gene-level RPKM values were then calculated based on a collapsed gene model. Fold change for each gene was calculated by dividing each gene's RPKM value against an average of the RPKM values for the gene in a compendium of adjacent normal tissue samples from the Illumina Human BodyMap 2.0 project. A percentile ranking of the RPKM of each gene against the compendium of breast cancer transcriptomes from TCGA was used to identify genes with aberrant expression and to prioritize genes of interest.
Expression correlation analysis for tumor typing was undertaken relative to the entire set of normal and tumor transcriptomes in TCGA. Two-way ANOVA was used to select a set of 3000 genes that were the most informative in explaining patterns of variance between tumor types within TCGA. A spearman correlation was calculated for this set of genes from the tumor sample against each TCGA sample. These pairwise correlations were clustered by the disease status of the TCGA samples (tumor or normal) and the type of cancer of the TCGA sample. The cancer set with the highest correlation was determined to be representative of the closest cancer type for the sample.
ADDITIONAL INFORMATION
Data Deposition and Access
The whole-genome sequencing and RNA-seq data for this case are available as .bam files from the European Genome-phenome Archive (EGA; www.ebi.ac.uk/ega/home) as part of the study EGAS00001001159, accession ID EGAD00001002590. Variants have been submitted to ClinVar (http:www.ncbi.nlm.nih.gov/clinvar/) and can be accessed under the accession numbers SCV000598654 and SCV000598653.
Ethics Statement
This work, including data deposition, was approved by the Research Ethics Board at the British Columbia Cancer Agency, protocol H14-00681. Written consent was obtained from the patient after discussion with their oncologist.
Acknowledgments
We gratefully acknowledge the participation of our patients and families, the POG team, and the generous support of the BC Cancer Foundation. The results published here are in whole or part based upon data generated by TCGA managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov.
Author Contributions
S.J.M.J., M.M., J.L., and A.K. contributed to the conception and design of the study. A.V.T. referred the patient to the study. Initial pathology at the Vancouver General Hospital was led by C.Z. Validation pathology was led by A.N.K. and assisted by K.C. and B.T.-C. A.M. contributed to the collection and assembly of data. M.J. and P.E. contributed to data analysis and interpretation. J.K.G., K.C., B.T.-C., M.J., A.V.T., and A.N.K. contributed to manuscript writing. All authors approved the final manuscript.
Funding
This work was supported by the British Columbia Cancer Foundation funding held by J.L. and M.M.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
References
- Abbott JJ, Ahmed I. 2006. Adenocarcinoma of mammary-like glands of the vulva: report of a case and review of the literature. Am J Dermatopathol 28: 127–133. [DOI] [PubMed] [Google Scholar]
- Alligood-Percoco NR, Kessler MS, Willis G. 2015. Breast cancer metastasis to the vulva 20 years remote from initial diagnosis: a case report and literature review. Gynecol Oncol Rep 13: 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HM, Wilkinson EJ. 2002. Uroplakin-III to distinguish primary vulvar Paget disease from Paget disease secondary to urothelial carcinoma. Hum Pathol 33: 545–548. [DOI] [PubMed] [Google Scholar]
- Butler B, Leath CA, Barnett JC. 2014. Primary invasive breast carcinoma arising in mammary-like glands of the vulva managed with excision and sentinel lymph node biopsy. Gynecol Oncol Case Rep 7: 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield YS, Kreitzman M, Thiessen N, Corbett RD, Li Y, Pang J, Ma YP, Jones SJM, Birol I. 2014. JAGuaR: junction alignments to genome for RNA-seq reads. PLoS One 9: e102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Sadeghi S, Raymond A, Jackman SD, Nip KM, Mar R, Mohamadi H, Butterfield YS, Robertson AG, Birol I. 2014. BioBloom tools: fast, accurate and memory-efficient host species sequence screening using bloom filters. Bioinformatics 30: 3402–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon ED, Carcangiu ML, Prieto VG, McCue PA, Burchette JL, To G, Norris BA, Kovatich AJ, Sanchez RL, Krigman HR, et al. 2000. Extramammary Paget disease is characterized by the consistent lack of estrogen and progesterone receptors but frequently expresses androgen receptor. Am J Clin Pathol 113: 572–575. [DOI] [PubMed] [Google Scholar]
- Del Pino M, Rodriguez-Carunchio L, Ordi J. 2013. Pathways of vulvar intraepithelial neoplasia and squamous cell carcinoma. Histopathology 62: 161–175. [DOI] [PubMed] [Google Scholar]
- Ding J, Bashashati A, Roth A, Oloumi A, Tse K, Zeng T, Haffari G, Hirst M, Marra MA, Condon A, et al. 2012. Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics 28: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha G, Roth A, Lai D, Bashashati A, Ding J, Goya R, Giuliany R, Rosner J, Oloumi A, Shumansky K, et al. 2012. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22: 1995–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung E. 1875. Ueber einen Fall von Mamma accessoria. Univ., Diss, Erlangen. [Google Scholar]
- Herter-Sprie GS, Greulich H, Wong K-K. 2013. Activating mutations in ERBB2 and their impact on diagnostics and treatment. Front Oncol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Sato F, Terasawa K, Tsuchiya S, Toi M, Tsujimoto G, Shimizu K. 2012. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS One 7: e31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Martincorena I, Gerstung M, Petljak M, Alexandrov LB, Rahbari R, Wedge DC, Davies HR, Ramakrishna M, Fullam A, et al. 2017. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 543: 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov DV, Spagnolo DV, Kacerovska D, Michal M. 2011. Lesions of anogenital mammary-like glands: an update. Adv Anat Pathol 18: 1–28. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegl B, Horn L-C, Moinfar F. 2005. Androgen receptors are frequently expressed in mammary and extramammary Paget's disease. Mod Pathol 18: 1283–1288. [DOI] [PubMed] [Google Scholar]
- McMaster J, Dua A, Dowdy SC. 2013. Primary breast adenocarcinoma in ectopic breast tissue in the vulva. Case Rep Obstet Gynecol 2013: 721696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micello D, Marando A, Sahnane N, Riva C, Capella C, Sessa F. 2010. Androgen receptor is frequently expressed in HER2-positive, ER/PR-negative breast cancers. Virchows Arch 457: 467–476. [DOI] [PubMed] [Google Scholar]
- Miettinen M, McCue PA, Sarlomo-Rikala M, Rys J, Czapiewski P, Wazny K, Langfort R, Waloszczyk P, Biernat W, Lasota J, et al. 2014. GATA3 a multispecific but potentially useful marker in surgical pathology—a systematic analysis of 2500 epithelial and nonepithelial tumors. Am J Surg Pathol 38: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbeck D, Tregnago AC, Netto GB, Sacomani C, Peresi PM, Osório CT, Schutz L, Bezerra SM, de Brot L, Cunha IW. 2017. GATA3 expression in primary vulvar Paget disease: a potential pitfall leading to misdiagnosis of pagetoid urothelial intraepithelial neoplasia. Histopathology 70: 435–441. [DOI] [PubMed] [Google Scholar]
- Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, Staaf J, Sieuwerts AM, Brinkman AB, Martin S, Ramakrishna M, et al. 2016. The topography of mutational processes in breast cancer genomes. Nat Commun 7: 11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullane SA, Werner L, Rosenberg J, Signoretti S, Callea M, Choueiri TK, Freeman GJ, Bellmunt J. 2016. Correlation of APOBEC mRNA expression with overall survival and PD-L1 expression in urothelial carcinoma. Sci Rep 6: 27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR. 2005. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol 123: 21–27. [DOI] [PubMed] [Google Scholar]
- Neto AG, Deavers MT, Silva EG, Malpica A. 2003. Metastatic tumors of the vulva: a clinicopathologic study of 66 cases. Am J Surg Pathol 27: 799–804. [DOI] [PubMed] [Google Scholar]
- PDQ Adult Treatment Editorial Board. 2017. Vulvar Cancer Treatment (PDQ®): Health Professional Version.
- Perrone G, Altomare V, Zagami M, Vulcano E, Muzii L, Battista C, Rabitti C, Muda AO. 2009. Breast-like vulvar lesion with concurrent breast cancer: a case report and critical literature review. In Vivo (Brooklyn) 23: 629–634. [PubMed] [Google Scholar]
- Perrotto J, Abbott JJ, Ceilley RI, Ahmed I. 2010. The role of immunohistochemistry in discriminating primary from secondary extramammary Paget disease. Am J Dermatopathol 32: 137–143. [DOI] [PubMed] [Google Scholar]
- Pietri E, Conteduca V, Andreis D, Massa I, Melegari E, Sarti S, Cecconetto L, Schirone A, Bravaccini S, Serra P, et al. 2016. Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr Relat Cancer 23: R485–R498. [DOI] [PubMed] [Google Scholar]
- Richter CE, Hui P, Buza N, Silasi D-A, Azodi M, Santin AD, Schwartz PE, Rutherford TJ. 2010. HER-2/NEU overexpression in vulvar Paget disease: the Yale experience. J Clin Pathol 63: 544–547. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Scaltriti M, Eichhorn PJ, Cortés J, Prudkin L, Aurac C, Jiménez J, Chandarlapaty S, Serra V, Prat A, Ibrahim YH, et al. 2011. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci 108: 3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SX, Yu BH, Xu XL, Cheng YF, Tu XY, Lu HF, Bi R, Sun XJ, Shui RH, Yang WT. 2017. Characterisation of GATA3 expression in invasive breast cancer: differences in histological subtypes and immunohistochemically defined molecular subtypes. J Clin Pathol 10.1136/jclinpath-2016-204137. [DOI] [PubMed] [Google Scholar]
- Tessier-Cloutier B, Asleh-Aburaya K, Shah V, McCluggage WG, Tinker A, Gilks CB. 2017. Molecular subtyping of mammary-like adenocarcinoma of the vulva shows molecular similarity to breast carcinomas. Histopathology 71: 446–452. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo E, Adam-Artigues A, Ballester S, Pineda B, Zazo S, González-Alonso P, Albanell J, Rovira A, Rojo F, Lluch A, et al. 2017. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep 7: 41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TA, Deavers MT, Carlson JA, Malpica A. 2015. Collision of ductal carcinoma in situ of anogenital mammary-like glands and vulvar sarcomatoid squamous cell carcinoma. Int J Gynecol Pathol 34: 487–494. [DOI] [PubMed] [Google Scholar]
- van der Putte SC. 1994. Mammary-like glands of the vulva and their disorders. Int J Gynecol Pathol 13: 150–160. [DOI] [PubMed] [Google Scholar]
- Willman JH, Golitz LE, Fitzpatrick JE. 2005. Vulvar clear cells of Toker: precursors of extramammary Paget's disease. Am J Dermatopathol 27: 185–188. [DOI] [PubMed] [Google Scholar]
- Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JMS, Bilous M, Fitzgibbons P, et al. 2013. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline update. J Clin Oncol 31: 3997–4013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.