Abstract
GLE1 encodes a protein important for mRNA export and appears to play roles in translation initiation and termination as well. Pathogenic variants in GLE1 mutations have been associated with lethal contracture syndrome and lethal arthrogryposis with anterior horn cell disease; phenotypes reported in individuals include fetal akinesia and a severe form of motor neuron disease, typically presenting with prenatal symptoms and perinatal lethality. In this article, we identified biallelic missense mutations in GLE1 by trio whole-exome sequencing in an individual affected with congenital motor weakness and contractures as well as feeding and respiratory difficulties. Muscle biopsy was consistent with anterior horn cell disease and supported the pathogenicity of the sequence variants. Importantly, this individual survived past the perinatal period with respiratory support and currently demonstrates age-appropriate cognition and slow but steady motor developmental progress. We propose that pathogenic variants in GLE1 can be associated with a nonperinatal lethal motor phenotype, and affected individuals can demonstrate motor skill progression, unlike prototypical anterior horn cell diseases such as spinal muscular atrophy.
Keywords: abnormality of the lower motor neuron, arthrogryposis multiplex congenita, fetal akinesia sequence, generalized muscle weakness, generalized neonatal hypotonia
INTRODUCTION
Whole-exome sequencing (WES) is increasingly being used to obtain a molecular diagnosis for rare, congenital diseases. In many cases, WES has resulted in the broadening of the clinical spectrum associated with disorders caused by variants in particular genes. With WES data, we are seeing more examples of allelic heterogeneity caused by variants in the same gene (Dyment et al. 2015; Thevenon et al. 2016), as well as uncovering new genes associated with particular disorders (Valencia et al. 2015). This has led to the consideration of the use of WES earlier in the diagnostic process (Shashi et al. 2014; Retterer et al. 2016; Stark et al. 2016).
A diverse group of disorders is associated with congenital arthrogryposis related to fetal akinesia deformation sequence (FADS) (Hall 2009). Two of such disorders, first described in Finnish families, are lethal congenital contracture syndrome 1 (LCCS1; OMIM 253310) and lethal arthrogryposis and anterior horn disease (LAAHD; OMIM 611890). These disorders are both associated with pathogenic variants in the gene GLE1 (Nousiainen et al. 2008), an important regulator of gene expression (Folkmann et al. 2013). Autosomal recessive LCCS1 is one of the most severe forms of arthrogryposis associated with FADS, with about 50 cases described in the literature (Herva et al. 1985, 1988; Vuopala and Herva 1994). Pathological analysis reveals a lack of anterior horn motor neurons, atrophy of the ventral spinal cord, and the near absence of skeletal muscles, as well as pulmonary hypoplasia (Herva et al. 1985, 1988; Vuopala and Herva 1994). Affected cases present with intrauterine growth restriction (IUGR), decreased fetal movement, fetal hydrops, polyhydramnios, joint contractures, and facial anomalies, typically resulting in prenatal demise (Herva et al. 1985). LAAHD, which has been described in 15 cases, presents with a similar but milder phenotype, with most infants surviving for up to 20 d after birth (Vuopala et al. 1995). IUGR and hydrops were less frequently observed or mild. Infants with LAAHD also demonstrated changes in their skeletal muscles with varying severity ranging from minor variation in fiber diameter to groups of atrophic fibers with hypertrophic type I fibers, and anterior horn cell motor neurons were degenerated and decreased (Vuopala et al. 1995). Recently, Smith et al. (2017) reported on two brothers with compound heterozygous splicing variants in GLE1; one brother passed away at the age of 2 wk but the other brother is currently alive at age 12 yr, suggesting that milder disease can be part of the phenotypic spectrum associated with GLE1.
Here we report a case of an individual who demonstrated features of fetal akinesia sequence at birth with contractures including talipes equinovarus, as well as motor weakness and hypotonia, which led to recurrent apneas and respiratory arrest. Trio WES revealed compound heterozygosity in two likely pathogenic missense variants in GLE1. Skeletal muscle histologic features were consistent with anterior horn cell disease and support the pathogenicity of missense variants. We discuss the clinical characteristics and developmental progression in this individual and highlight the favorable outcomes that can be associated with individuals bearing GLE1 mutations, as well as the expanded phenotypic spectrum with GLE1-associated disease.
RESULTS
Clinical Presentation and Family History
The patient is the third child of healthy nonconsanguineous parents of Caucasian descent with some Native American ethnicity on the paternal side. The pregnancy was complicated by decreased fetal movement and breech presentation. She was born prematurely at 36 wk gestation by vaginal delivery after successful eversion, with some fetal decelerations noted during labor. The baby was apneic at birth and required positive pressure ventilation and continuous positive airway pressure. Apgar scores were 4 at 1 min and 8 at 5 min.
Her birth parameters were head circumference 33.5 cm (70th percentile), weight 2675 g (50th percentile), and length 46 cm (50th percentile). She was noted to have mild dysmorphic facial features (Fig. 1A,B), including somewhat prominent forehead, depressed nasal bridge, low-set ears, and excess nuchal folds. She had clenched fists and her thumbs were biphalangeal but appeared finger-like. She had ulnar deviation of her fingers. She had a single palmar crease on her right hand and short palmar creases on her left. Bilateral talipes equinovarus were present. Her nipples were widely spaced. Neurologic examination was notable for alert mental status, low muscle tone, and weak facial, truncal, and extremity muscles with minimal spontaneous movement. She had flexion contractures of her upper and lower extremities with underlying low passive tone and preserved deep tendon reflexes.
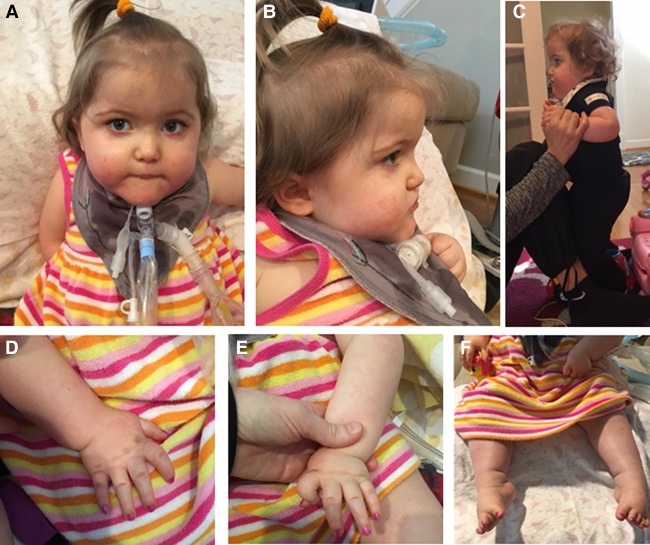
Figure 1.
Pictures of individual at age 24–28 mo. (A,B) Facial profile showing lack of facial expression and dysmorphic features including prominent forehead and low-set ears, (C) standing with assistance, (D) right hand showing mild right index finger contracture, (E) left hand showing more severe left index finger contracture, and (F) bilateral talipes equinovarus.
Over the first several weeks of life, she had repeated apneic episodes associated with increased secretions, eventually requiring intubation. Evaluation of airway patency was unremarkable aside from mild left bronchomalacia. Persistent hypotonia, weakness, and associated poor secretion handling necessitated tracheostomy at age 4 mo. She had a gastrostomy tube placed at age 1 mo to provide a means for feeding and Nissen fundoplication at age 3 mo after diagnosis of severe gastroesophageal reflux. Within a few weeks of birth, the flexion contractures improved, but her hypotonia and weakness persisted. She continued to have normal mental status with good social and cognitive development. Her spontaneous facial and extremity movements remained very weak but did improve slightly over time. Reflexes remained present and symmetric.
The patient had a comprehensive developmental evaluation at age 18 mo. Cognitive and receptive language skills were commensurate with the 11- to 12-mo level, indicating mild-to-moderate delays relative to age-mates. Assessment of cognitive skills was difficult because hand contractures made toy manipulation difficult. Expressive language was not assessed because of the presence of tracheostomy. Motor skills continued to be her greatest area of delay, especially gross motor skills; she was able to sit independently if placed in sitting position but could not get into sitting position independently and was primarily wheelchair-bound. She underwent a subsequent developmental assessment at age 26 mo. There was no evidence of developmental regression, and, in fact, she exhibited remarkable developmental progression across all domains, but particularly in cognitive and language development. Her cognitive and receptive language development had progressed to the average-to-low average range for her age with skills commensurate with the 22- to 23-mo level. She also made considerable progress in expressive language development with skills estimated to be low average for her age when use of sign language for communication was considered. She had mastered more than 15 signs and had begun to combine two or three signs into short sentences. In terms of gross motor development, she continued to have equinovarus deformities, but was able to pull to standing with some assistance and use a stander with ankle foot orthotics in place (Fig. 1C,F). Fine motor development was improved given improvement in finger contractures although she used her right hand more than her left because of left arm weakness and left index finger contracture, which remained more severe compared with the right index finger (Fig. 1D,E). She was also toilet-trained for bowel movements.
She continued to rely solely on gastrostomy tube feedings with good weight gain. At age 26 mo, her weight was 13.9 kg (85%), height was 80.1 cm (3%), and head circumference was 47.3 cm (39%). She had been tolerating weans of at least a few hours off her ventilator in the absence of respiratory illnesses.
Other procedures performed as part of her workup included a brain magnetic resonance imaging at age 1 wk, which showed a small germinal matrix hemorrhage and punctate areas of restriction diffusion in the deep white matter consistent with mild hypoxic ischemic encephalopathic changes that were attributed to her code event secondary to apnea; these changes were improved on subsequent brain MRIs. Echocardiogram in infancy showed normal heart anatomy with a small patent foramen ovale. Electromyography (EMG) in the first month of life was normal with no myotonia or fibrillations, and creatine kinase was borderline low (67 with reference range 70–320 U/l). Myotonic dystrophy type 1 gene triplet repeat analysis (Greenwood Genetics Center) returned normal. Biochemical testing all returned normal, including mitochondrial sequencing (Baylor Miraca Genetics Laboratories) and testing for congenital disorders or glycosylation (Mayo Medical Laboratories and Emory Genetics Laboratories). SNP chromosomal microarray (Duke Cytogenetics Laboratory) also returned normal. The neurology service was initially concerned about a congenital myasthenic syndrome, but genetic testing (Congenital Myasthenic syndrome Next-generation sequencing panel, Medical Neurogenetics) was normal and a trial of an anti-acetylcholinesterase agent gave inconclusive results. Ophthalmologic examination at age 21 mo was normal.
Her family history is negative for neuromuscular diseases, perinatal respiratory issues, stillbirths, or recurrent miscarriages. There is no known Finnish ancestry. We have provided a summary of clinical features present in our patient and comparison with previously reported cases of GLE1-associated disease (Table 1).
Table 1.
Summary of phenotypes associated with biallelic GLE-1 variants
| Source | Pathogenic alteration | Mortality | Hydrops | Decreased fetal movements or fetal akinesis | Joint Contractures | Pulmonary hypoplasia | Weakness and hypotonia |
|---|---|---|---|---|---|---|---|
| This study (one case) | c.1808G>T p.Arg603Leu and c.1997G>T p.Gly666Val | Alive at age 29 mo | − | + | + | - | + |
| Smith et al. 2017 (one family with two cases) | c.100-7_1003delTCTCT p.Asp34_Lys107del and c.1882-2A>G p.Val238_Asnfs*2 | One died at age 2 wk; one alive at age 12 yr | − | NR | + | NR | + |
| Ellard et al. 2015 (one family with two cases) | c.1706G>A p.Arg569His and c.1849G>A p.Val617Met | Pregnancies terminated | NR | NR | + | NR | NA |
| Nousiainen et al. 2008 | c.433-10A>G p.Thr144_Glu145insProPheGln (homozygous) c.433-10A>G p.Thr144_Glu145insProPheGln and c.1706G>A p.Arg569His (LCCS1 phenotype) | Prenatal death, pregnancy terminations or died during/right after delivery | + (Severe 12/16) | + (Some pregnancies) | + (16/16) | + (11/16) | NA |
| c.433-10A>G p.Thr144_Glu145insProPheGln and c.1849G>A p.Val617Met c.433-10A>G p.Thr144_Glu145insProPheGln and c.2051T>C p.Ile684Thr (LAAHD phenotype) | Perinatal lethal (3 stillborn, 5 died within 1 h, 6 died within a few days, 1 survived for 20 d) | + (Mild 2/15) | + (4/15) | + (15/15) | + (6/15) | NA/NR |
Phenotypes for LCCS1 and LAAHD were obtained from references cited in the Nousiainen et al. (2008) paper or references therein: Herva et al. (1985) for LCCS1 and Vuopala et al. (1995) for LAAHD.
NR, not reported; NA, not applicable.
Trio Whole-Exome Sequencing
Trio WES revealed that the patient was compound heterozygous for two likely pathogenic variants in the GLE1 gene (Genbank: AF058922). The maternally inherited c.1808G>T (p.Arg603Leu) variant is a nonconservative amino acid substitution that occurs at a position that is conserved in mammals. The paternally inherited c.1997G>T (p.Gly666Val) variant is a conservative amino acid substitution in a position that is conserved across species. Both p.Arg603Leu and p.Gly666Val are predicted to be damaging by PolyPhen, MutationTaster, PhyloP, and CADD, whereas SIFT predicted the changes would be tolerated. The p.Arg603Leu variant was detected in Exome Aggregation Consortium (ExAc; http://exac.broadinstitute.org), with an allele frequency of <1/10,000. The p.Gly666Val variant is novel and not present in ExAC, the Database of Single-Nucleotide Polymorphisms (http://www.ncbi.nlm.nih.gov/ SNP/), 1000 Genomes (1000G; http://www.1000genomes.org/), or Exome Variant Server (http://evs.gs.washington.edu/EVS/). The proband's brothers were tested for segregation analysis of the variants. One brother did not carry either variant, whereas the other brother only carried the p.Gly666Val variant. Both brothers are healthy and did not have any congenital problems.
Skeletal Muscle Biopsy
Thigh muscle biopsy at age 1 mo showed abnormal variation in fiber diameter, with frequent hypertrophied or atrophied fibers (Fig. 2). The normal skeletal muscle fiber diameter for babies between birth and 3 mo is ∼15–17 μm, and the fibers in our patient's muscle varied from 5 to 40 μm. Cytochrome oxidase (COX) showed uneven staining overall, with numerous weak fibers, but no ragged red or ragged blue fibers were seen on trichrome or SDH staining, respectively. The COX-weak fibers were generally atrophic, and hypertrophied fibers showed dense, granular staining. No target structures were seen on NADH or COX. Nonspecific enolase showed positive staining in numerous, very atrophic fibers. Motor end plates were seen, peripheral nerve twigs were myelinated, and muscle spindles were present. On myosin heavy chain (MHC) fast and slow fiber typing, there was no convincing neurogenic grouping, grouped atrophy, or type-specific atrophy. Developmental and neonatal MHC staining revealed positive staining that indicates fiber regeneration. On electron microscopy, there were mild, nonspecific mitochondrial changes (balling, vacuolation, and thickening of cristae with increased matrix material) and a mild increase in loose glycogen. The overall impression was of a chronic, active neuromuscular insult, showing features of ongoing denervation without evidence of reinnervation. These findings are consistent with anterior horn cell disease.
Figure 2.

Light microscopic findings on frozen sections of thigh muscle biopsy done at 1 mo of age. (A) H&E staining shows moderate variation in fiber diameter, with clusters of atrophic fibers <10 μm, and frequent fiber hypertrophy up to 40 μm (normal 15–17 μm). 20× objective. (B) MHC “fast” immunostaining shows brown staining of type 2 fibers. This stain highlights clusters of atrophic fibers, but there is no definitive grouped atrophy. Type 2 fibers are in the majority, but atrophy and hypertrophy are seen in both fiber types. 20× objective. (C) Nonspecific enolase is up-regulated in numerous of the atrophic fibers (dark brown staining), indicating acute denervation of those fibers. 20× objective. (D) COX staining shows uneven staining, with dense, granular staining in hypertrophied fibers, and very weak activity in atrophic fibers. 40× objective. Scale bars: A–C = 100 μm; D = 50 μm.
DISCUSSION
We present a case of a patient, currently close to age 2.5 yr, with two likely pathogenic, missense variants in GLE1 identified from WES, who was born with contractures, hypotonia, weakness, and recurrent apneas and who has survived with stable respiratory status after undergoing tracheostomy. Histologic studies on muscle are consistent with anterior horn cell disease, similar to previously reported anterior horn cell pathology seen in LAAHD (Vuopala et al. 1995), further corroborating that the GLE1 variants are causative for the patient's clinical features. Importantly, in contrast with all the cases reported in the literature, she is cognitively appropriate for age and is able to communicate with signs, and she is continuing to make steady progress with motor milestones. The progression in motor development seen in our patient is similar to the case reported by Smith et al. (2017). Both our patient and the surviving brother reported in Smith et al. (2017) underwent early tracheostomy and mechanical ventilation, as well as gastrostomy feeding, suggesting that early medical interventions may help to prolong the life of these patients. However, unlike the case reported by Smith et al. (2017), our patient does not have cerebral atrophy, microcephaly or seizures, which may indicate phenotypic variability, genotype–phenotype differences, or a separate clinical etiology for these features.
Nousiainen et al. (2008) discovered that the majority of cases affected by LCCS1 (28/29 families) were homozygous for the FinMajor mutation (c.433-10A>G) located in intron 3 of GLE1 gene, which creates a new splice acceptor site, and this new cDNA is predicted to cause the insertion of three amino acids in the coiled-coil domain of GLE1 (Thr144_Glu145insProPheGln). One case with LCCS1 was found to be compound heterozygous for the FinMajor mutation and c.1706G>A (p.Arg569His) variant in GLE1. Patients with LAAHD, on the other hand, are all compound heterozygous for the FinMajor mutation in GLE1 and another missense mutation (either c.1849G>A (p.Val617Met) or c.2051T>C(p.Ile684Thr)), implicating that homozygotes for the FinMajor mutation appear to have more severe phenotypes than compound heterozygotes. More recently, Ellard et al. (2015) reported on a family with two pregnancies affected with fetal akinesia, which they attributed to compound heterozygosity for the p.Val617Met and p.Arg569His variants. We have provided a summary of the cases of GLE1-associated disease reported in the literature (Table 1).
GLE1 acts as an mRNA export factor (Watkins et al. 1998), but appears to have roles in both initiation and termination of protein translation in eukaryotes (Bolger et al. 2008). Studies of the FinMajor mutation in a zebrafish model suggest that apoptosis of neuronal precursors and defects in motor axon arborization likely explain the LCCS1 motor neuron defects (Jao et al. 2012).
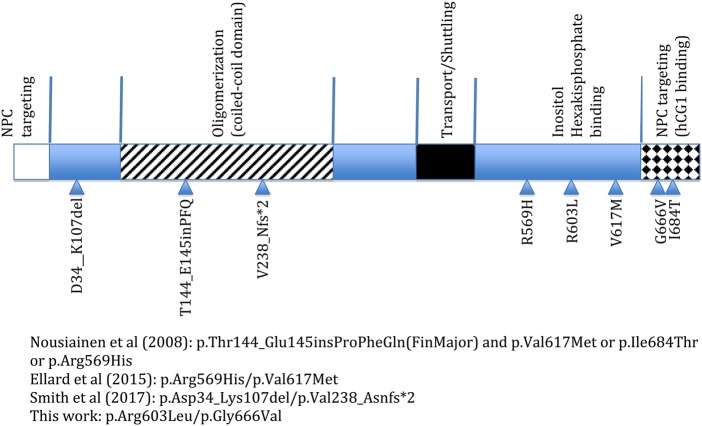
The GLE1 protein consists of different domains (Fig. 3; Folkmann et al. 2014). The coiled-coil domain, which is disrupted by the FinMajor mutation, is important for GLE1 oligomerization (Folkmann et al. 2013). Other domains necessary for function include the inositol hexakisphosphate (IP6) binding domain, important for mRNA export (Fig. 3; Alcázar-Román et al. 2006, Weirich et al. 2006). The GLE1 gene codes for two different protein isoforms, GLE1A and GLE1B. The GLE1B isoform contains a 43 amino acid segment, not found in GLE1A, which is important in the localization of GLE1 to the nuclear pore complex (NPC) by mediating binding to the nucleoporin hCG1 (Fig. 3; Kendirgi et al. 2005).
Figure 3.
Schematic of the GLE1 protein domains and the variants associated with autosomal recessive disease, as reported by the referenced articles. Checkered region at the carboxyl terminus is found in the GLE1B isoform but not in the GLE1A isoform. NPC, nuclear pore complex.
The GLE1 variants in this case are located near the carboxy-terminus end of the protein. The p.Arg603 residue is located in the region of the GLE1 protein implicated in IP6 binding, and this residue is present in both transcripts of GLE1. The p.Gly666 residue, however, is present only in the GLE1B isoform, within the 43-amino-acid segment important for hCG1 interaction. It is plausible that our patient's phenotype was milder compared with the perinatal forms of the disease because GLE1 oligomerization is not disrupted, and there is residual GLE1 activity within the oligomeric complexes for function. Further studies are needed to elucidate the roles that these variants have on protein localization and function.
In summary, we have described variants of GLE1 associated with congenital arthrogryposis in a patient, who, despite weakness and hypotonia, is progressing in motor skills and exhibits cognitive, language, and social development that is commensurate with age expectations. Her severe oropharyngeal hypotonia necessitated a tracheostomy but she is currently able to tolerate a few hours without mechanical ventilation. We propose that GLE1-associated disease should be on the differential in a child with fetal akinesia, joint contractures, and congenital but nonprogressive muscle disease. This report, similar to Smith et al.’s (2017) report, expands the clinical heterogeneity associated with GLE1 mutations and demonstrates the utility of WES in the diagnosis of disorders, especially ones with atypical presentations. Similar to cases reported in the literature, the increasing use of WES has uncovered more genetic disorders with extensive allelic heterogeneity (e.g., variants in ASAH1 associated with epilepsy with varying onset of spinal muscular atrophy; Dyment et al. 2015). In view of this, it is critically important that clinical decisions based on WES results should take into consideration the evolving phenotypic heterogeneity seen in disorders that have been characterized in the past. This is especially pertinent in the case of ultrarare disorders, in which limited information about the associated disease is known and the genotype–phenotype correlation is not well understood.
METHODS
Whole-Exome Sequencing
Trio WES was performed on exons captured with the Clinical Research Exome Kit (Agilent Technologies) and sequenced using an Illumina HiSeq4000 sequencing system with 2 × 150-bp reads, with other methodology and variant interpretation as previously described (Tanaka et al. 2015). Please see Table 2 for sequencing coverage. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957). Bioinformatic analysis revealed that the proband was compound heterozygous for the maternally inherited c.1808G>T (p.Arg603Leu) and paternally inherited c.1997G>T (p.Gly666Val) missense variants in GLE1. Bioinformatic filtering did not identify any other homozygous, compound heterozygous, X-linked or de novo variants that were consistent with the proband's phenotype. Candidate variants were validated and segregation confirmed by Sanger sequencing of DNA. Primers used in Sanger sequencing are listed in the Supplemental Table.
Table 2.
Sequencing coverage for whole-exome sequencing
| Average coverage | Proband | 95.79 |
| Mother | 95.55 | |
| Father | 95.35 | |
| Sequence read lengths | 2 × 150-bp, paired-end | |
| Coverage for variants | Mean coding sequence coverage: 109.94× |
Skeletal Muscle Biopsy
Skeletal muscle tissue was obtained by open biopsy of the right thigh during laparoscopic gastrostomy tube placement. Frozen sections of the muscle biopsy tissue stained with H&E were used to assess fiber diameter using an ocular micrometer. Frozen sections were stained with antibodies to MHCs fast, slow, developmental, and neonatal (Leica Biosystems). Enzyme histochemical staining was done on frozen sections using standard protocols.
ADDITIONAL INFORMATION
Data Deposition and Access
Variant data has been deposited to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) by GeneDx under accession numbers c.1997G>T|SCV000566649.1 and c.1808G>T|SCV000566648.1. WES data is not available as patient consent was not obtained for deposition.
Ethics Statement
Written consent for the case report and pictures was obtained from the individual's parents. This activity was reviewed by the Duke University Health System IRB and declared exempt with protocol ID Pro00084952.
Acknowledgments
We thank the family for their participation.
Author Contributions
Q.K.-G.T. contributed to drafting the manuscript and analysis and interpretation of data. A.M.-R. contributed to the acquisition, analysis, and interpretation of data and the critical revision of the manuscript. J.J. contributed to acquisition, analysis, and interpretation of data (sequencing) and the critical revision of the manuscript. K.E.G. contributed to analysis and interpretation of data (developmental assessment) and the critical revision of the manuscript. C.E.P. contributed to analysis and interpretation of data (neurological assessment) and the critical revision of the manuscript. A.F.B. contributed to analysis and interpretation of data (muscle biopsy) and the critical revision of the manuscript. Y.-h.J. contributed to analysis and interpretation of data and the critical revision of the manuscript.
Funding
Q.K.-G.T. was funded by a Genzyme-supported fellowship. J.J. is an employee of GeneDx. Y.-H.J. is supported by National Institutes of Health (NIH) grants MH098114, HD077197, MH104316. HD088007, and HD087795.
Competing Interest Statement
J.J. is an employee of GeneDx.
Referees
Judith G. Hall
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Alcázar-Román AR, Tran EJ, Guo S, Wente SR. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716. [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR. 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment DA, Tétreault M, Beaulieu CL, Hartley T, Ferreira P, Chardon JW, Marcadier J, Sawyer SL, Mosca SJ, Innes AM, et al. 2015. Whole-exome sequencing broadens the phenotypic spectrum of rare pediatric epilepsy: a retrospective study. Clin Genet 88: 34–40. [DOI] [PubMed] [Google Scholar]
- Ellard S, Kivuva E, Turnpenny P, Stals K, Johnson M, Xie W, Caswell R, Lango Allen H. 2015. An exome sequencing strategy to diagnose lethal autosomal recessive disorders. Eur J Hum Genet 23: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Collier SE, Zhan X, Aditi, Ohi MD, Wente SR. 2013. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell 155: 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Dawson TR, Wente SR. 2014. Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure–function analysis. Adv Biol Regul 54: 74–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JG. 2009. Pena–Shokeir phenotype (fetal akinesia deformation sequence) revisited. Birth Defects Res A Clin Mol Teratol 85: 677–694. [DOI] [PubMed] [Google Scholar]
- Herva R, Leisti J, Kirkinen P, Seppänen U. 1985. A lethal autosomal recessive syndrome of multiple congenital contractures. Am J Med Genet 20: 431–439. [DOI] [PubMed] [Google Scholar]
- Herva R, Conradi NG, Kalimo H, Leisti J, Sourander P. 1988. A syndrome of multiple congenital contractures: neuropathological analysis on five fetal cases. Am J Med Genet 29: 67–76. [DOI] [PubMed] [Google Scholar]
- Jao LE, Appel B, Wente SR. 2012. A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development 139: 1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Rexer DJ, Alcázar-Román AR, Onishko HM, Wente SR. 2005. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol Biol Cell 16: 4304–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen HO, Kestilä M, Pakkasjärvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. 2008. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet 40: 155–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, et al. 2016. Clinical application of whole-exome sequencing across clinical indications. Genet Med 18: 696–704. [DOI] [PubMed] [Google Scholar]
- Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB. 2014. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med 16: 176–182. [DOI] [PubMed] [Google Scholar]
- Smith C, Parboosingh JS, Boycott KM, Bonnemann CG, Mah JK; Care4Rare Canada Consortium, Lamont RE, Micheil Innes A, Bernier FP. 2017. Expansion of the GLE1-associated arthrogryposis multiplex congenita clinical spectrum. Clin Genet 91: 426–430. [DOI] [PubMed] [Google Scholar]
- Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, Yeung A, Peters H, Mordaunt D, Cowie S, et al. 2016. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med 18: 1090–1096. [DOI] [PubMed] [Google Scholar]
- Tanaka AJ, Cho MT, Millan F, Juusola J, Retterer K, Joshi C, Niyazov D, Garnica A, Gratz E, Deardorff M, et al. 2015. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am J Hum Genet 97: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, St-Onge J, Steinmetz A, Huet F, Chouchane M, et al. 2016. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet 89: 700–707. [DOI] [PubMed] [Google Scholar]
- Valencia CA, Husami A, Holle J, Johnson JA, Qian Y, Mathur A, Wei C, Indugula SR, Zou F, Meng H, et al. 2015. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center's experience. Front Pediatr 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuopala K, Herva R. 1994. Lethal congenital contracture syndrome: further delineation and genetic aspects. J Med Genet 31: 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuopala K, Ignatius J, Herva R. 1995. Lethal arthrogryposis with anterior horn cell disease. Hum Pathol 26: 12–19. [DOI] [PubMed] [Google Scholar]
- Watkins JL, Murphy R, Emtage JL, Wente SR. 1998. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc Natl Acad Sci 95: 6779–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.