Abstract
There has been no indication to test for BRCA1/2 in children (with the rare exception of Fanconi anemia) as screening begins in adult years and there is a potential to induce anxiety related to adult-onset cancers. However, in the setting of pediatric cancer, with increasing utility and frequency of companion tumor-normal sequencing without regard for phenotype and with BRCA1/2 included in tumor profiling panels, germline mutations in BRCA1/2 and other DNA damage repair genes have been found. When mutations in these genes are revealed, there are implications for immediate family members. Here we present two children in whom BRCA2 mutations identified through tumor sequencing prompted parental genetic testing and medical action. These cases illustrate the potential importance of including a matched normal DNA sample when performing tumor profiling of pediatric cancer patients to ensure optimal care.
Keywords: neuroblastoma, osteosarcoma
CASE PRESENTATION
Patient 1
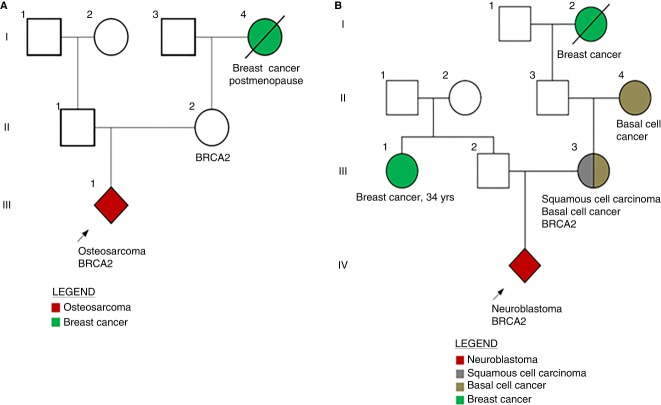
An 11-yr-old boy with localized osteosarcoma in the left proximal humerus presented with intermittent pain in the affected arm and underwent a definitive resection of a biopsy-proven osteosarcoma. Mutation panel sequencing of the tumor was performed by Foundation Medicine that revealed a BRCA2 Q2354* (Frampton et al. 2013). Given the potential implications of a germline BRCA2 mutation for family members, parental blood samples were drawn, revealing the patient's 40-yr-old mother was found to carry the BRCA2 mutation. This finding led to her decision for a risk-reductive mastectomy and bilateral salpingo oophorectomy. Although the patient's grandmother had a history of breast cancer, her age of onset did not raise suspicion for a predisposition to cancer or indicate germline testing at the time of her diagnosis (Fig. 1A).
Figure 1.
Children identified harboring BRCA2 mutations prompting maternal testing and care. (A) Patient 1. (B) Patient 2.
Patient 2
A 2 yr-old boy with global developmental delay presented with a 3- to 4-wk history of respiratory symptoms. Chest computed tomography (CT) revealed a 9.3 cm × 8.7 cm mediastinal mass. An iodine-123-meta-iodobenzylguanidine (MIBG) scan showed uptake compatible with neuroblastoma that was histologically confirmed.
Foundation Medicine tumor sequencing revealed a BRCA2 p.Y2215fs* mutation. In-house paired tumor-normal sequencing, MSK-IMPACT was also performed utilizing a control germline blood sample for tumor variant calling. Of note, the practice at Memorial Sloan Kettering Cancer Center (MSKCC) is to not report germline mutations detected by MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) unless specific consent to return germline results has been provided. In this instance, MSK-IMPACT testing did not report the BRCA2 p. Y2215fs mutation; prompting the suspicion that the BRCA2 mutation reported by Foundation Medicine was constitutional. The boy's family history was notable for his mother having been diagnosed with squamous cell carcinoma at age 34 yr, his maternal grandmother diagnosed with basal cell carcinoma at age 40 yr, his maternal great grandmother diagnosed with breast cancer in her 50s, and a paternal aunt diagnosed with breast cancer at age 34 yr (Fig. 1B). Parental testing revealed the child's mother harbored the medically actionable BRCA2 p.Y2215fs* mutation, which subsequently led her to undergo magnetic resonance imaging (MRI) and mammography breast screening and to seek consultation with breast and gynecologic specialists.
TECHNICAL ANALYSIS
In Patient 1, the BRCA2 p.Q2354* germline mutation identified through tumor testing (performed by Foundation Medicine) and, in Patient 2, the BRCA2 p. Y2215fs* germline mutation identified by tumor testing (performed by Foundation Medicine and MSKCC IMPACT testing) were confirmed by targeted site-specific testing at the commercial laboratory GeneDx (Frampton et al. 2013; Cheng et al. 2015; Susswein et al. 2016).
VARIANT INTERPRETATION
The Q2354* variant in BRCA2 is entered five times in ClinVar and annotated as pathogenic (Landrum et al. 2016). The variant is linked with Database for Short Genetic Variations (dbSNP): 80358936 and the National Center for Biotechnology Information (NCBI) 1000 Genomes browser rs80358936. This specific truncation mutation is not annotated by the Evidenced-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA), the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA), or the Breast Cancer Information Core (BIC) (Genomes Project et al. 2010; Spurdle et al. 2012). ENIGMA describes the variant as predicted to encode a truncated nonfunctional protein. Moreover, the mutation is reported in the literature as occurring in a Spanish patient with breast cancer, and the commercial laboratory InVitae has asserted this variant as pathogenic (Duran et al. 2003; Hirsch et al. 2004).
The p.Y2215fs* variant in BRCA2 is predicted to encode a truncated nonfunctional protein and termination mutations at this codon (Y2215*) are annotated three times as pathogenic in ClinVar (Landrum et al. 2016). Termination variants at this codon are linked with dbSNP: 80358892, and the NCBI 1000 Genomes browser rs80358892 and the BIC have asserted termination variants at this location as pathogenic (Genomes Project et al. 2010). However, this specific truncation mutation is not annotated in the CIMBA (Spurdle et al. 2012; Rebbeck et al. 2016).
SUMMARY
Here we present two childhood cancer patients, for whom the results of tumor sequencing prompted parental testing for germline BRCA2 mutations, leading to genetic counseling and medical action in both mothers harboring the germline BRCA2 variants.
BRCA1 and BRCA2 are proven autosomal dominant breast, ovarian, pancreatic, and prostate cancer predisposition genes (Neuhausen et al. 1996; Kauff et al. 2001; Pritchard et al. 2016). Pediatric phenotypes are known to result when a child inherits biallelic mutations in BRCA2, and rarely in BRCA1 (i.e., the recessive disorder Fanconi anemia) (D'Andrea and Grompe 1997, 2003; Howlett et al. 2002; Sawyer et al. 2015).
In pediatrics, the only indication to test for BRCA1/2 has been in the context of Fanconi anemia given the potential to induce anxiety related to adult-onset cancers (Green et al. 2013; Sawyer et al. 2015). Recently, with increasing companion tumor-normal sequencing performed, without regard for phenotype and with BRCA1/2 included in tumor profiling panels, germline mutations in these and other DNA damage genes have been detected (Table 1; Dite et al. 2010; Walsh et al. 2014; Mody et al. 2015; Zhang et al. 2015; Parsons et al. 2016; Schrader et al. 2016). Pathogenic cancer predisposition gene mutations were found in 95/1120 (8.5%) pediatric cancer patients participating in the Pediatric Cancer Genome Project (PCGP), as compared to 1.1% of persons in the 1000 Genomes Project and 0.6% of the participants in the autism study (Daly et al.; Downing et al. 2012; Genomes Project et al. 2015). However, given that a true incidence study comparing pediatric cancer patients with healthy controls has yet to occur and a uniform sequencing approach has not been deployed in all pediatric studies to date, it remains to be determined if heterozygous mutations in BRCA2 detected in pediatric cancer patients are driver or passenger events (Offit et al. 2003; Zhang et al. 2015). Notably, the second most commonly mutated gene in the PCGP was BRCA2 (n = 6), and BRCA2 mutations may be enriched in children cancer survivors compared with controls (Zhang et al. 2015; Wang et al. 2017).
Table 1.
Review of germline BRCA1/2 mutations detected in pediatric sequencing studies
| Tumor type | Gene | Chr | Location | HGVS DNA ref | HGVS protein | Variant type | Germline platform | Allele frequency (normal) | Target coverage | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Osteosarcoma | BRCA2 | Chr 13 | 32354913 | G | p.Q2354* | Nonsense | Sanger | 0.46 | 500× | This study |
| Neuroblastoma | BRCA2 | Chr 13 | 32340999 | C | p.Y2215fs | Frameshift | Sanger | 0.48 | 500× | This study |
| Neuroblastoma | BRCA1 | Chr 17 | 43124028 | G | p.E23fs | Frameshift | WES | 0.43 | 272× | Parsons et al. 2016 |
| Anaplastic medulloblastoma | BRCA1 | Chr 17 | 43094833 | GT | p.V233fs | Frameshift | WES | 0.5 | 272× | Parsons et al. 2016 |
| Ewing sarcoma | BRCA2 | Chr 17 | 32332757 | G | p.D427fs | Frameshift | WES | 0.6 | 272× | Parsons et al. 2016 |
| Acute lymphoblastic leukemia | BRCA1 | Chr 17 | 41245200 | AT | p.I783fs | Frameshift | WES | 0.43 | >30× | Zhang et al. 2015 |
| Acute lymphoblastic leukemia | BRCA2 | Chr 13 | 32945092 | G | p.W2830_E20splice | Splice | WES | 0.51 | >30× | Zhang et al. 2015 |
| Acute lymphoblastic leukemia | BRCA2 | Chr 13 | 32968863 | C | p.Y3098* | Nonsense | WES | 0.51 | >30× | Zhang et al. 2015 |
| Medulloblastoma | BRCA2 | Chr 13 | 32929213 | C | p.P2408fs | Frameshift | WGS | 0.21 | >30× | Zhang et al. 2015 |
| Medulloblastoma | BRCA2 | Chr 13 | 32953932 | T | p.L3000* | Nonsense | WGS | 0.5 | >30× | Zhang et al. 2015 |
| Neuroblastoma | BRCA2 | Chr 13 | 32945092 | G | p.W2830_E20splice | Splice | WES | 0.43 | >30× | Zhang et al. 2015 |
| Rhabdomyosarcoma | BRCA2 | Chr 13 | 32911681 | G | p.Q1063_S1064fs | Frameshift | WGS | 0.37 | >30× | Zhang et al. 2015 |
HGVS, Human Genome Variation Society.
Regardless, when mutations in these genes are revealed, there are implications for immediate family members. Standard of care dictates that testing for BRCA1/2 mutations commences at age 25 yr in at-risk individuals with known familial mutations or family histories described as per the National Comprehensive Cancer Network (NCCN) guidelines and in some instances younger if this information will impact family planning. This standard is based on the premise that BRCA1/2 mutations underpin cancers that arise in adulthood and the clinical benefits of this knowledge result from screening and risk-reductive surgeries occurring in adulthood (Brooks et al. 2006). However, results from recent tumor-normal sequencing studies in pediatrics give pause and raise questions regarding the benefit of testing siblings of children diagnosed with cancer harboring BRCA mutations (Table 1).
In view of recent pediatric sequencing studies detecting children with germline BRCA1/2 mutations across solid and liquid tumors, and the cases described here, pediatric oncologists should be linked with avenues to adult care for the potential screening, risk-reductive interventions (such as surgery), and family planning options currently available for parents of children carrying BRCA1/2 mutations. For our patients, detection of previously unidentified heritable mutations prompted medical action and provided potentially lifesaving information.
It is important to note that numerous mutational platforms exist wherein tumors are profiled without corresponding normal DNA samples, rendering it difficult to assign certain mutations as somatic or germline. Moreover, in some institutions, germline mutations are subtracted from reporting if consent has not been obtained and may not be realized. These cases illustrate the importance that a corresponding normal DNA be reported to ensure accurate profiling and optimal care.
ADDITIONAL INFORMATION
Data Deposition and Access
Both variants described in this manuscript were deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) and were assigned the submission numbers SCV000564127 and SCV000564128 (Landrum et al. 2016).
Ethics Statement
Patients reported here provided written consent permitting use of molecular and phenotypic description under MSKCC's institutional review board (IRB)-approved studies #06-107 and #12-245.
Acknowledgment
We thank Joseph Olechnowicz for editorial assistance.
Author Contributions
M.F.W. and P.M. conceived the study. M.F.W., J.K., M.H., and K.O. performed clinical interpretation of the germline variants. All authors reviewed and drafted the manuscript.
Funding
This research was funded in part through the National Institutes of Health/ National Cancer Institute (NIH/NCI) Cancer Center Support Grant P30 CA008748. M.F.W. receives research funding through the V Foundation Scholar Grant, Niehaus Center for Inherited Genomics, and the Crawford Fund for Pediatric Genomics.
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Brooks GA, Stopfer JE, Erlichman J, Davidson R, Nathanson KL, Domchek SM. 2006. Childhood cancer in families with and without BRCA1 or BRCA2 mutations ascertained at a high-risk breast cancer clinic. Cancer Biol Ther 5: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. 2015. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M, Sutcliffe J, Gibbs R, Buxbaum J, Schellenberg G. NIMH Data Archive. National Institute of Mental Health, Bethesda, MD; http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000298.v2.p2 10.15154/1163515 Accessed March 28, 2017. [DOI] [Google Scholar]
- D'Andrea AD, Grompe M. 1997. Molecular biology of Fanconi anemia: implications for diagnosis and therapy. Blood 90: 1725–1736. [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. 2003. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3: 23–34. [DOI] [PubMed] [Google Scholar]
- Dite GS, Whittemore AS, Knight JA, John EM, Milne RL, Andrulis IL, Southey MC, McCredie MR, Giles GG, Miron A, et al. 2010. Increased cancer risks for relatives of very early-onset breast cancer cases with and without BRCA1 and BRCA2 mutations. Br J Cancer 103: 1103–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. 2012. The Pediatric Cancer Genome Project. Nat Genet 44: 619–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran M, Esteban-Cardeñosa E, Velasco E, Infante M, Miner C. 2003. Mutational analysis of BRCA2 in Spanish breast cancer patients from Castilla-Leon: identification of four novel truncating mutations. Hum Mutat 21: 448. [DOI] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. 2013. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31: 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond KE, et al. 2013. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch B, Shimamura A, Moreau L, Baldinger S, Hag-alshiekh M, Bostrom B, Sencer S, D'Andrea AD. 2004. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood 103: 2554–2559. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. [DOI] [PubMed] [Google Scholar]
- Kauff ND, Scheuer L, Robson ME, Glogowski E, Kelly B, Barakat R, Heerdt A, Borgen PI, Davis JG, Offit K. 2001. Insurance reimbursement for risk-reducing mastectomy and oophorectomy in women with BRCA1 or BRCA2 mutations. Genet Med 3: 422–425. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44: D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody RJ, Wu YM, Lonigro RJ, Cao X, Roychowdhury S, Vats P, Frank KM, Prensner JR, Asangani I, Palanisamy N, et al. 2015. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 314: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen S, Gilewski T, Norton L, Tran T, McGuire P, Swensen J, Hampel H, Borgen P, Brown K, Skolnick M, et al. 1996. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet 13: 126–128. [DOI] [PubMed] [Google Scholar]
- Offit K, Levran O, Mullaney B, Mah K, Nafa K, Batish SD, Diotti R, Schneider H, Deffenbaugh A, Scholl T, et al. 2003. Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anemia. J Natl Cancer Inst 95: 1548–1551. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, Kerstein RA, Gutierrez S, Petersen AK, Bavle A, et al. 2016. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol 10.1001/jamaoncol.2015.5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, et al. 2016. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, Friebel TM, Mitra N, Wan F, Chen S, Andrulis IL, Apostolou P, Arnold N, Arun BK, Barrowdale D, et al. 2016. Inheritance of deleterious mutations at both BRCA1 and BRCA2 in an international sample of 32,295 women. Breast Cancer Res 18: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Tian L, Kahkönen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian Genomics; FORGE Canada Consortium, Majewski J, Dyment DA, Innes AM, et al. 2015. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 5: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, Ni A, Thomas T, Benayed R, Ashraf A, et al. 2016. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol 2: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Healey S, Devereau A, Hogervorst FBL, Monteiro ANA, Nathanson KL, Radice P, Stoppa-Lyonnet D, Tavtigian S, Wappenschmidt B, et al. 2012. ENIGMA-evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 33: 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J, Booker JK, Cremona ML, et al. 2016. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 18: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MF, Wu G, Edmonson M, Gruber TA, Easton J, Yergeau D, Vadodaria B, Ma XT, Chen X, Mcgee R, et al. 2014. Incidence of germline mutations in cancer-predisposition genes in children with hematologic malignancies: a report from the pediatric cancer genome project. Blood 124: 127. [Google Scholar]
- Wang Z, Wilson CL, Easton J, Hedges D, Liu Q, Wu G, Rusch M, Edmonson M, Levy S, Lanctot JQ, et al. 2017. Germline mutations in cancer predisposition genes and risk for subsequent neoplasms among long-term survivors of childhood cancer in the St. Jude Lifetime Cohort. In Proceedings of the 107th Annual Meeting of the American Association for Cancer Research; 2017 Apr 1–5; AACR; 2017, Washington, DC. Philadelphia, PA. Abstract 3001 [Google Scholar]
- Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, Hedges D, Ma X, Zhou X, Yergeau DA, et al. 2015. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 373: 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]