Abstract
Pork meat is estimated to be responsible for 10–20% of human salmonellosis cases in Europe. Control strategies at the farm could reduce contamination at the slaughterhouse. One of the targeted sectors of production is maternity, where sows could be Salmonella reservoirs. The aim of this study was to assess the dynamics of shedding of Salmonella in terms of variation in both shedding prevalence and strains excreted during gestation in Quebec’s maternity sector. The evolution of the fecal microbiota of these sows during gestation was also assessed to detect bacterial populations associated with these variations. A total of 73 sows both at the beginning and the end of the gestation were randomly selected and their fecal matter was analyzed. Salmonella detection was conducted using a method that includes two selective enrichment media (MSRV and TBG). Nine isolates per positive samples were collected. Among the 73 sows tested, 27 were shedding Salmonella. Sows in the first third of their gestation shed Salmonella significantly more frequently (21/27) than those in the last third (6/46) (χ2 P < 0.05). The shedding status of 19 of the sows that were previously sampled in the first third of their gestation was followed, this time in the last third of their gestation, which confirmed reduction of shedding. Using 16S rRNA gene sequencing and qPCR, significant differences between the fecal flora of sows at the beginning and the end of the gestation, shedding Salmonella or not and with different parity number were detected. Using MaAsLin, multiple OTUs were found to be associated with the time of gestation, the status of Salmonella excretion and parity number. Some of the identified taxa could be linked to the reduction of the shedding of Salmonella at the end of gestation. In this study, we showed that the level of Salmonella shedding was variable during gestation with significantly higher shedding at the beginning rather than at the end of gestation. We also observed for the first time a significant change in the microbiota during sow gestation and identified interesting taxa which could be linked to a reduced Salmonella shedding.
Keywords: Salmonella, sows, fecal microbiota, gestation, excretion
Introduction
In Canada, Salmonella enterica is estimated to cause 269.26 infections per 100,000 inhabitants each year, confirming this pathogen as a public health priority (Thomas et al., 2013). Contamination in humans causes gastroenteritis and occurs from the consumption or mishandling of contaminated meats. The economic impact of productivity losses and medical care expenses caused by salmonellosis is estimated to reach 3.3 billion dollars in the US alone each year (Hoffmann et al., 2012). In Canada, the part of salmonellosis that are caused by consumption of pork products is not known. However, in Europe, it has been estimated that between 10 and 20% of all salmonellosis cases are due to the consumption of contaminated pork meat (EFSA Panel on Biological Hazards [BIOHAZ], 2010).
Salmonella can contaminate swine on the farm and most of the serotypes can be carried asymptomatically in their intestinal tract, gut-associated lymphoid tissue and tonsils, and enter slaughterhouses with the animals where it can contaminate the meat (Boyen et al., 2008). Salmonella can enter the farm through multiple pathways, such as contaminated feeds or employees (Funk and Gebreyes, 2004). One of the most important sources of new strains on a farm is the introduction of already contaminated animals (Funk and Gebreyes, 2004; Gotter et al., 2012). Once on the farm it can be very difficult to eliminate using regular washing and disinfection methods and residual strains can contaminate newly arrived Salmonella free animals (Argüello et al., 2011; Dewaele et al., 2012). In certain parts of Canada up to 60% of the swine farms are contaminated (Rajic et al., 2005; Farzan et al., 2008) with some studies showing 25% of contaminated animals (Wilkins et al., 2010). Since it has been shown that the entrance of contaminated pigs into slaughterhouses is linked with an increased risk for the contamination of carcasses, a reduction at the first stages of the production could be an important step in the reduction of the contamination of the meat (Letellier et al., 2009; EFSA Panel on Biological Hazards [BIOHAZ], 2010).
Canadian sows in the provinces of Saskatchewan and Alberta have been shown to be highly contaminated by Salmonella, with levels of shedding higher than swine in fattening at 38% compared to 25% respectively (Wilkins et al., 2010). These contaminated sows are believed to be important reservoirs of contamination during the maternity phase where Salmonella could be transmitted to the piglets, spreading contamination to later stages of production (Belœil et al., 2004; EFSA Panel on Biological Hazards [BIOHAZ], 2010; Hill et al., 2015). Variation in the percentage of sows excreting Salmonella during the maternity production cycle has already been observed (Nollet et al., 2005b; Magistrali et al., 2011). However, information is still scarce on the dynamics of shedding by sows during gestation, on the real impact of contamination on the rest of production and the factors that are responsible for this variation.
Sequencing technologies have helped to show the importance of microbiota in fighting the colonization of possibly harmful bacteria by competitive exclusion, stimulation of immunity and the production of antimicrobial substances. In the case of Salmonella, it has been shown that short chain fatty acids (SCFA, e.g., butyric acid), that are by-products of the microbiota’s digestion of complex sugar, can reduce virulence of Salmonella by lowering the activation of the Salmonella Pathogenicity Island-1 (SPI-1) (Gantois et al., 2006). Studies have also shown that swine excreting Salmonella had a different microbiota than swine that were not excreting (Fravalo et al., 2013; Lebel et al., 2016). Similarly, it has been shown that swine with different levels of Salmonella shedding after an experimental infection had different microbiota compositions (Bearson et al., 2013). Interestingly, in other species, fecal microbiota has been shown to vary during gestation (Collado et al., 2008; Koren et al., 2012). However, this variation, and its possible impact on the shedding of Salmonella, has not been studied in sows.
The aim of this study was to assess the dynamics of Salmonella shedding by sows in terms of variation in both shedding prevalence and strains excreted during gestation in an industrial setting in Quebec. The evolution of the fecal microbiota of these sows during gestation was also assessed in order to detect bacterial populations that could be associated with the observed variations.
Materials and Methods
Sampling
All animal experimentations were approved by the ethics committee of the Faculty of Veterinary Medicine of the University of Montreal, certificate number 14-Rech-1714. The protocol was approved by the ethics committee of the Faculty of Veterinary Medicine of the University of Montreal. A total of 73 sows at various gestation stages and parity (between 1 and 8; Table 1) were randomly selected from a breeding farm known for its frequent Salmonella contamination. All sows were fed with the same feed during all the gestation. For each selected sow, 100 g of fresh fecal matter was collected and analyzed. After the first sampling, 19 out of the 73 sows that were sampled at the beginning of their gestation (first 50 days) were sampled a second time at the end of this period (last 50 days). For each sampled sow, 1 g of feces was subsampled in the farm and immediately frozen in liquid nitrogen for 16S rRNA gene amplicon MiSeq sequencing.
Table 1.
Number of sows sampled in each parity and their Salmonella shedding status.
| Parity | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Number of sows | 21 | 8 | 11 | 8 | 10 | 7 | 5 | 3 |
| % of excreting sows | 29 | 50 | 36 | 25 | 40 | 57 | 20 | 67 |
Detection
Salmonella detection was conducted using a method adapted from the method described by De Busser et al. (2013). The method was optimized by collecting isolates from multiple selective enrichment media as well as from two migration distances on MSRV to obtain the best possible description of the eventual variety of strains contained in the samples. First the samples (100 g) were pre-enriched in buffered peptone water (BPW) (1:10 w/v, 18 h, 37°C) (Biokar diagnostic, Beauvais, France). Modified Semi-Solid Rappaport-Vassiliadis Agar (MSRV) (24–48 h, 42°C) (Lab M, Heywood, United Kingdom) and Tetrathionate Brilliant Green Bile Broth (TBG) (24 h, 42°C) (BD Difco, Franklin Lakes, NJ, United States) selective enrichment media were then used in parallel and further inoculated on Brilliant Green Sulfa Agar (BGS) (BD Difco, Franklin Lakes, NJ, United States). Two BGS plates were inoculated from the MSRV from two different regions of the migration zone (far from and near the inoculation spot) and one from the TBG. A maximum of nine (3 per BGS plate) isolates per positive samples were collected. Suspect colonies on BGS were confirmed as Salmonella using triple sugar iron agar slants (Lab M, Heywood, United Kingdom) and urea agar slants (Lab M, Heywood, United Kingdom) followed by seroagglutination using Salmonella O antiserum Poly A-I + Vi (Statens Serum Institute, Denmark).
After isolation, the motility of the isolates originating from samples that were only positive for TBG and from samples where isolates from multiple genotypes were detected with MRSV was confirmed. Each isolate was grown individually for 24 h at 37°C in 10 ml of BPW. Then 100 μl of the enrichment broth was inoculated on the middle of an MSRV plate and incubated for 24 h at 42°C before the migration zone was observed. Each of the isolate was tested in triplicate.
Percentages of Salmonella positive animals at the beginning and the end of gestation were compared using Fisher’s exact test for the first random sampling and McNemar’s test for the second sampling where each sow was sampled twice (GraphPad v6, Prism, LaJolla, CA, United States). The link between shedding and the parity of sows was also assessed using a logistic regression (R 3.4.1).
Genotypic Characterization
Each isolate was genotyped using a high resolution melt (HRM) technique adapted from the method described by Bratchikov and Mauricas (2011). The precise melting curves of three genomic regions, two CRISPRs (cr1 and cr2) and one VNTR (yohm), were analyzed after PCR amplification using a LightCycler 96 real time PCR (Roche diagnostics, Mannheim, Germany). The combined analysis of these three curves was associated with an HRM type. A variation in the melting curve profile from one of these three regions was considered to reveal a new HRM type. Representative isolates of each of the different types were serotyped by the veterinary epidemiosurveillance laboratory of the Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec.
16S rRNA Gene Sequencing
Total bacterial DNA was extracted using mechanical and chemical lysis followed by a phenol/chloroform purification. Five hundred μg of the frozen fecal content was added to 500 μl of lysis buffer [Tris-HCl 500 mM pH 8, EDTA 100 mM pH 8, NaCl 100mM, SDS 1% (w/v)] with 500 mg of 0.1 mm glass beads. Cells in the samples were mechanically lysed two times using MP Fastprep (Santa Ana, MP Biomedicals) for 40 s at 6 m/s. Samples were kept 5 min on ice between runs. Lysates were centrifuged 15 min at 18,000 g to remove beads and cell debris. DNA was extracted from the supernatant using a standard phenol/chloroform purification protocol (Thibodeau et al., 2015). Final DNA concentration was measured using the Qubit 3.0 broad range assay (Fisher Scientific, Ottawa, ON, Canada). Purified DNA samples were stored at -20°C for further analysis. The extraction was also conducted on negative controls without fecal matter.
16S rRNA gene amplicon sequencing libraries were prepared following the Illumina MiSeq protocol (Illumina, 2013). A 292 bp segment of the v4 region of the 16S rRNA gene was amplified using primers targeting the total bacterial and archaeal population (515F_Ill and 806R_Ill) (Caporaso et al., 2012). 12.5 ng of DNA was pre-amplified in a final 25 μl reaction using KAPA HiFi HotSart ReadyMix (KAPA Biosystems, Wilmington, DE, United States). The amplification was carried out for 25 cycles with a denaturation step at 95°C for 30 s, an annealing step at 55°C for 30 s, and an elongation step at 72°C for 45 s ending with a final elongation of 10 min at 72°C. Each sample was then indexed using the Nextera XT index kit (Illumina). Five microliter of the PCR product was amplified in a 20 μl reaction containing 12.5 μl of KAPA HiFi HotSart ReadyMix (KAPA Biosystems, Wilmington, DE, United States), 2.5 μl of each index primer (10 nM) and 2.5 μl of purified water. This second amplification was carried out for 8 cycles with a denaturation step at 95°C for 30 s, an annealing step at 55°C for 30 s, and an elongation step at 72°C for 30 s ending with a final elongation of 5 min at 72°C. After each PCR step, PCR product purification was conducted using Agencourt AMpure XP beads (Beckman Coulter, Brea, CA, United States). The purified PCR products were diluted to 5 nM. Fifty-one samples were pooled (5 μl of each product) and sequenced by the Illumina MiSeq sequencing system using the MiSeq Reagent Kit v2 (500 cycles).
All sequences were cleaned and analyzed using Mothur v.1.39.5 following an adapted version of the MiSeq standard operation procedure available online1 (accessed August 2015) (Kozich et al., 2013). First, the two complementary sets of reads were combined for each sample after removal of the primers. Sequences that were too long or that contained ambiguity were discarded. Identical sequences were grouped to reduce the necessary processing power. These unique sequences were then aligned using a Mothur adapted version of the SILVA database (silva.seed.v119).2 Chimeras where removed using UCHIME3. The sequences were clustered into operational taxonomic units (OTU) at genetic distance dissimilarity of 3% and then classified using the Mothur formatted Ribosome database project (RDP) trainset version 14.4 Raw reads for each sow fecal microbiota analyzed in this study are available through the NCBI SRA database under accession SRP100939.
For alpha diversity analysis, indexes were calculated in Mothur using a subsample size consisting of the lowest number of sequences in samples with 1000 iterations. The average number of OTUs, the estimation of the portion of the diversity covered by our subsampling (coverage), the diversity of the OTUs found in the samples (inverted Simpson’s index) and their evenness (Shannon evenness) was measured. Results were compared between groups using ANOVA and Student’s t-test and relation with parity rank using linear regression with a significance level of 0.05. For beta diversity analysis, a distance matrix comparing all the samples was created with a subsample using the same number of sequences as previously used and using Jaccard and Bray-Curtis dissimilarity index. These results were visualized using Non-metric multidimensional scaling (NMDS) graphs and the beta diversity of the different groups were statistically compared using the AMOVA test. Furthermore, OTUs that were associated with the different sow groups were identified using the Multivariate Association with Linear Models (MaAsLin) method (Morgan et al., 2012).
Real Time Quantitative PCR of Specific Bacterial Populations
Real time PCR was conducted for specific populations on all samples: Enterobacteria (Castillo et al., 2006), Bifidobacteria (Matsuki et al., 2004) and Lactobacilli (Castillo et al., 2006) – classical indicators in microbiome analysis – as well as Lachnospiraceae (Wilson et al., 2014), which was identified as an interesting population because of its known role as a butyrate producer. Standard curves were built by PCR using known quantities of each targeted gene. All reactions were conducted using 10 ng of DNA using EvaGreen mastermix (Montréal Biotech, Montréal, QC, Canada) for a total reaction of 20 μl in a LightCycler 96 real time PCR (Roche diagnostics, Mannheim, Germany). Results were expressed in log of copies of the gene per 10 ng of total DNA. Levels of these populations were compared using the Student’s t-test and ANOVA (GraphPad v6, Prism, La Jolla, CA, United States).
Results
Salmonella Detection
Salmonella was detected in the feces of 37% (27 out of 73) of the sows that were sampled. Proportion of Salmonella excreting sows at the beginning of gestation (first 50 days of gestation) (21 out of 27, 78%) was significantly (Fisher’s exact, p < 0.05) higher than at the end of this period (last 50 days of gestation) (6 of 46, 13%). Similar results were obtained when sampling for a second time at the end of gestation the 19 sows that had already been sampled at the beginning of their gestation, with a statistically (McNemar’s, p < 0.05) lower proportion of positive samples at the end of the gestation (2 of 19, 11%) than at the beginning (13 of 19, 68%). Logistic regression showed no significant link between shedding and the parity of the sow (p > 0.05) (Table 1).
Of the 27 positive samples, 7 had discrepancies in the results when comparing enrichment by TBG and MSRV, with 3 samples only positive on MSRV and 4 samples only positive on TBG. All the isolates that were collected on TBG positive only samples were further tested for motility on MSRV. All isolates were motile and produced migration zones covering the whole MSRV after 24 h of incubation.
Salmonella Typing
Among the isolates 3 different HRM types were detected. The most prevalent profile (HRM type 1) was present in 25 out of the 27 positive samples that were collected and was most prevalent both at the beginning and the end of gestation (19 out of 21 and 6 out of 6 respectively) (Table 2). For this HRM type, 10 out of the 11 isolates that were randomly selected for serotype confirmation belonged to the S. Infantis serotype and one was an autoagglutinating strain of the partial antigenic formula O:r:1,5. The second most frequent profile (HRM type 2) was present in 3 out of the 27 positive samples and belonged to the Derby serotype (3 isolates confirmed by serotyping) and was only detected in animals at the beginning of gestation. Finally, the third profile (HRM type 3) was present in 2 of the 27 positive samples, was only detected at the beginning of the gestation and was found to belong to the Typhimurium serotype (2 isolate confirmed by serotyping). Three samples contained isolates of two different profiles. Two samples contained Salmonella from profiles 1 and 2 and one sample contained Salmonella from profiles 1 and 3. For all the samples that contained strains of multiple profiles, all the isolates from a single BGS (TBG, MSRV near the site of inoculation and MSRV at the edge of the migration front) were of the same type. However, for these three samples, HRM type of the isolates collected from the edge of the migration zone differed from those collected near the MSRV inoculation site. The motility of these isolates was assessed. All the tested isolates were motile and covered the MSRV plate after 24 h of incubation.
Table 2.
High resolution melt (HRM) types of Salmonella isolated from sows at the beginning and the end of gestation.
| HRM genes profiles | HRM types | Serotype | Beginning of gestation | End of gestation | ||
|---|---|---|---|---|---|---|
| CRISPR 1 | CRISPR 2 | Yohm | (Samples) | (Samples) | ||
| 1 | 1 | 1 | 1 | Infantis/O:r:1,5 | 19/21 | 6/6 |
| 2 | 2 | 2 | 2 | Derby | 3/21 | 0/6 |
| 3 | 3 | 3 | 3 | Typhimurium | 2/21 | 0/6 |
16S rRNA Gene Amplicon Sequencing
From this section onward, samples collected from sows that were sampled once and those from sows sampled twice were analyzed together to get the highest statistical power possible. Forty-seven samples were sequenced and 45 were retained after data analysis, containing a total of 4,770,924 sequences. Most of the sequences were bacterial sequences with 4,711,793 sequences (98.8%) followed by archaeal sequences with 59,129 sequences (1.3%) and 2 unclassifiable sequences (0.00004%).
A total of 3,377 OTUs were detected, with an average of 106,020 sequences per sample. The lowest values observed for a sample were 18,037 sequences and 490 OTUs and the highest were 560,410 sequences and 1,333 OTUs.
Rarefaction was conducted and showed that after subsampling each sample at 18,037 a good coverage was obtained for all the samples (Supplementary Figure S1).
For the Alpha diversity, a significant although small difference in coverage was measured when comparing samples based on their Salmonella shedding status while no differences where found when comparing the animals based on their time of gestation. When the sows were classified both on their Salmonella shedding status and time of gestation, no significant variation in alpha diversity was measured. (Table 3) Finally, no relationship was measured between the parity number and the different alpha diversity indices (data not shown).
Table 3.
Comparison of alpha-diversity indices across sow groups and according to Salmonella shedding status and time of gestation.
| Sow group | Salmonella status | Time of gestation | ||||||
|---|---|---|---|---|---|---|---|---|
| Salmonella Negative/Beginning | Salmonella Negative/End | Salmonella Positive/Beginning | Salmonella Positive/End | Positive | Negative | Beginning | End | |
| Coverage | 0.990 | 0.991 | 0.990 | 0.990 | 0.991a | 0.990b | 0.990 | 0.990 |
| OTUs | 688 | 637 | 682 | 676 | 680 | 642 | 679 | 647 |
| Inverted Simpson’s | 49 | 46 | 58 | 47 | 54 | 47 | 56 | 46 |
| Shannon evenness | 0.73 | 0.73 | 0.75 | 0.74 | 0.75 | 0.73 | 0.74 | 0.73 |
The means were based on 1000 subsampling of 18,037 sequences. On the same row a is significantly different from b. Significant p values of 0.05.
Bacterial beta-diversity was compared between groups. The similarity of the composition of communities at the OTU level was compared using the Jaccard and Bray-Curtis indexes and these distance matrices were plotted using NMDS on 2 axes when relevant (Figures 1A–C). Statistical variations in the composition of the microflora between the different groups were assessed using analysis of molecular variance (AMOVA) (Table 4). When splitting the sows into two groups, the time of gestation was revealed to have a significant effect on the beta-diversity in fecal content when using the Jaccard index. When the beta-diversity was compared based on the status of Salmonella shedding a significant difference was measured when using Bray-Curtis’ index. Analysis were also conducted by splitting the sows into four groups combining both their Salmonella shedding status and time of gestation. Significant differences were found for at least one test when comparing the group of sows shedding Salmonella at the beginning of gestation with the group composed of non-shedding sows at the end of gestation. Finally, the beta-diversity significantly varied in the samples collected from sows with a different number of parity when comparing sows with low (1–3) and high levels of parity (6–8) when using the Jaccard index.
FIGURE 1.
Non-metric multidimensional scaling (NMDS) plot illustrating sow fecal microbiome beta-diversity according to the time of gestation, Salmonella shedding status and number of gestation. (A) Blue = beginning of gestation; Red = end of gestation. (B) Red = shedding; Blue = no shedding. (C) Blue = low number of gestation (1–3); Green = medium number of gestation (4–5) Red = high number of gestation (6–8).
Table 4.
Beta-diversity analysis across sow groups and according to Salmonella shedding or time of gestation.
| Compared groups | AMOVA (p-value) | ||
|---|---|---|---|
| Jaccard | Bray-Curtis | ||
| Beginning/Salmo+ | Beginning/Salmo- | 0.554 | 0.044 |
| End/Salmo+ | 0.014 | ||
| End/Salmo- | <0.001 | ||
| Beginning/Salmo- | End/Salmo+ | 0.162 | 0.018 |
| End/Salmo- | 0.047 | ||
| End/Salmo+ | End/Salmo- | 0.289 | |
| Beginning | End | <0.001 | 0.077 |
| Salmo+ | Salmo- | 0.05 | 0.012 |
| Low parity (1–3) | High parity (6–8) | 0.012 | 0.141 |
| Low parity (1–3) | Medium parity (4–5) | 0.071 | |
| High parity (6–8) | Medium parity (4–5) | 0.191 | |
Based on a subsample of 18,037 sequences, there is a significant p-value of 0.0083 with Bonferroni correction when comparing 4 groups, 0.0166 when comparing three groups and 0.05 when comparing 2 groups. P-values under significant levels are in bold.
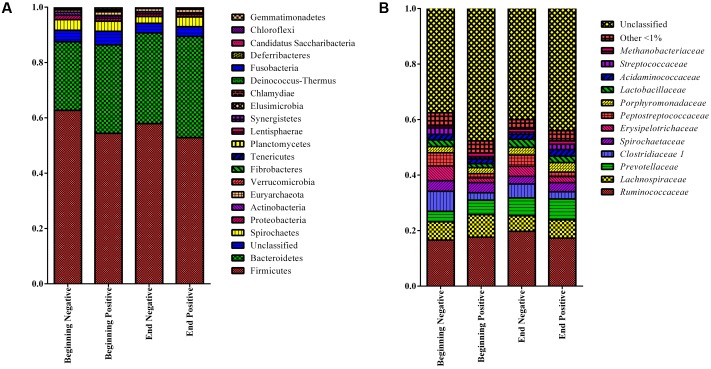
Stacked bar graphs of the taxa found at the phylum level showed that for all the groups, the Firmicutes represented 56.9% of the sequences in the samples followed by the Bacteroidetes at 31.5% while 3.5% of the sequences could not be classified at the phylum level (Figure 2A). For the RDP family level taxa (taxon level 5), 12 taxa were classified and present at a level of more than 1% of the sequences and represented 53.6% of the total sequences in all the groups while 42.3% of the sequences could not be classified at the family level (Figure 2B).
FIGURE 2.
Stacked bar graphs showing the relative abundance at the phylum (A) and family level (B) levels using RDP classification (only taxa representing more than 1% of the sequences) for the four groups of sows. Beginning: Sows in the 50 first days of gestation, End: Sows in the 50 last days of gestation, Negative: Sows not shedding Salmonella, Positive: Sows shedding Salmonella.
Variation in individual clades was assessed using MaAsLin with status of shedding (positive or negative), time of gestation (beginning or end) and parity number (1–8) as metadata. For the parity, 13 OTUs classified to 10 taxa were found to be negatively associated with this value, while 13 OTUs classified to 5 taxa were positively associated. For the Salmonella shedding status, 11 OTUs classified to 6 taxa were positively associated with shedding animals, while 3 OTUs classified to 2 taxa were associated with non-shedding animal. Finally, for the gestation time, 58 OTUs classified to 22 taxa were positively associated with animals at the beginning of the gestation, while 20 OTUs classified to 10 taxa were positively associated with animals at the end of the gestation (Supplementary Data S1).
Real Time Quantitative PCR for Specific Populations
A difference between the sows at the beginning of gestation and the end of gestation was detected for the Enterobacteria group with a mean of 0.61 log of copies more at the beginning of gestation (Student’s t-test, p < 0.05) (Figure 3A). Furthermore, a significant difference of 0.44 log more copies at the beginning of gestation than at the end was detected for the Lachnospiraceae family (Figure 3B). No differences were detected for the Lactobacilli or Bifidobacteria quantifications. When comparing the sows by Salmonella status or in groups that combine both shedding status and time of gestation, no significant differences were measured for the four tested populations.
FIGURE 3.
Enterobacteria (A) and Lachnospiraceae (B) 16S rRNA gene copies in sow fecal content at beginning and end of gestation. Horizontal bars represent the mean of each group, each point represent the fecal content of a single sow. ∗Indicates p < 0.05.
Discussion
In this study, we investigated the dynamics of shedding of Salmonella by sows in terms of variation of both shedding prevalence and type of strains excreted during gestation in an industrial setting in Quebec. We also evaluated the modification of the fecal microbiota during gestation and the possible link with the shedding of Salmonella.
High percentages of Salmonella shedding animals were detected on the farm with a total of 37% of sampled sows positive for the bacteria. These results are similar to what had already been measured in other provinces of Canada, with 38% of the individual samples found to be positive in Saskatchewan and Alberta (Wilkins et al., 2010). However, Wilkins’ study used samples of 10 g of feces and since it is known that fecal sample weight affects sensitivity, the real prevalence could have been underestimated (Funk et al., 2000; Wilkins et al., 2010). Our results confirm that sows are reservoirs of Salmonella.
When separating sows at the beginning and the end of gestation, we measured a significant variation of shedding, with higher number of animals shedding (78%) at the beginning when compared to the end (13 %). Description of variations of Salmonella shedding during the production cycle of sows is scarce. Some research has been done in an Italian study conducted in a farrow-to-finish farm that showed a low shedding of 0.6% in late gestation (2 weeks before parturition) and a high shedding of 26.5% in post-weaning (1 week after parturition) (Magistrali et al., 2011). Similar results were also found in Belgian herds were the prevalence of Salmonella shedding by sows was less than 10% during late gestation, around farrowing and during lactation and raised significantly 7 days after weaning (Nollet et al., 2005b). Hence it seems that the number of sows shedding Salmonella is at its highest when the sows are not in contact with the piglets.
The majority of positive samples (25 of the 27) from this farm contained isolates of HRM type that belong to the Infantis serovar. The isolates from this serovar could represent a major resident strain present on the farm. A possible association between this serotype and the maternity phase of the production has been described in Canadian farrow-to-finish farms (Wilkins et al., 2010). However, this serotype was not one of the 5 most frequently found in the samples submitted to the Public Health Agency of Canada for antimicrobial susceptibility testing, showing that this serotype is somewhat rare in human cases in Canada (Public Health Agency of Canada [PHAC], 2016). Also, detected at lower levels, were isolates of two other HRM types that belong to strains of the Derby and Typhimurium serovars. The low diversity of serotypes in the farm is similar to what was found in other studies in maternity and fattening units. In studies conducted in Europe, similar strains were found at different stages of production in farrow to finish farms (Nollet et al., 2005a), however, in studies carried out on multi-site production systems, like in the present study, the link between strains found in maternity and fattening units has not been clearly established. In these production systems, changes in the major strains present at different stages of production and appearance of new serotypes at the nursery and fattening stages were detected (Funk et al., 2001). This lack of a detected link in this type of production could be the result of the low sensitivity of the method to detect minor strains in samples containing many strains.
In this study, to get the highest sensitivity and the best snapshot of the different serotypes that were excreted by the sows, a modified Salmonella detection method was used. Since it is known that different serovars of Salmonella have different growth rates in selective enrichment media, using multiple media could help get better representativeness of the excreted strains by the sows (Gorski, 2012; De Busser et al., 2013). For seven samples, only one of the two selective enrichment media was positive. For four of these samples, only the TBG was positive. Since Salmonella detection on MSRV is based on the motility of the bacteria, these results could show that these isolates are non-motile. The motility of the isolates that were not detected on MSRV was tested, and all the isolates formed migration zones on the media covering the whole surface. Therefore, the lack of detection could not be explained by strains that are non-motile. It is possible that the lack of detection on either TBG or MRSV could be a result of the competitive flora present in the media after the first enrichment in BPW. The method used in this study also proved to be useful in three samples where strains of different serotypes were recovered at different distances of migration on the MSRV media. Interestingly, in two of these samples, because the isolate recovered from the MSRV at the inoculation site was different from those recovered on TBG and at the edge of the migration zone on MRSV, using the standard method would have resulted in a loss of information. The motility of these strains was also tested as previously described and with identical results. These results confirm the importance of using multiple enrichment methods and possibly multiple migration distances on MSRV when trying to get an accurate account of the different strains of Salmonella present on a farm.
In this study, to explain the mechanisms that are at play in the variation of shedding between the beginning and end of gestation, the microbiota of sows was studied. We described for the first time the beta diversity of the fecal microbiota of Salmonella excreting and non-excreting sows at the beginning and end of gestation.
The AMOVA statistical test showed that the gestation time, the Salmonella shedding status and the parity number significantly affected the populations found in the fecal flora. However, the gestation time and parity number variations were only significant using the Jaccard index which only considers the presence or absence of the OTUs while the status of Salmonella shedding variations were only significant when using the Bray-Curtis index which also considers the number of time the OTUs are found in the samples (Legendre and Legendre, 2012). This shows that while these factors all have significant impact on the flora they have different impacts on the microflora composition. Variation in the microbiota during gestation has already been described in other studies, mostly in human, with non-convergent conclusions. Two studies showed significant differences between the beginning and end of gestation in humans (first to third trimester) (Collado et al., 2008; Koren et al., 2012). However, another study showed no significant variation in the microbiota during gestation (DiGiulio et al., 2015). Similarly, Salmonella has been shown to be associated with variations in the flora of both challenged and naturally infected animals (Bearson et al., 2013; Borewicz et al., 2015). However, it is the first time that these modifications are measured in sows.
MaAsLin was used to identify biomarkers linked with the time of gestation, status of excretion and number of parity while considering confounding factors. Three OTUs negatively associated with the number of parity were classified to the genus level to Succinivibrio (1 OTU), Coprococcus (1 OTU), Blautia (1 OTU). While only one OTU positively associated with the number of parity was classified to the genus level at 63% to the Clostridium cluster IV. Most of the statistically significant taxa were associated with the time of gestation. Five OTUs that were positively associated with the end of the gestation, when Salmonella shedding is at it’s lowest, were classified at the genus level to Alloprevotella (2 OTUs), Prevotella (2 OTUs) and Clostridium cluster IV (1 OTU). All those genera are known short chain fatty acid producers and could affect the virulence of Salmonella. Indeed, it has been shown that butyrate, regulates the expression of SPI-1, reducing the invasiveness of Salmonella (Gantois et al., 2006). However, supplementing finishing pigs with butyric acid was tested with variable level of success. A study showing significant reduction of Salmonella levels in feces after sodium butyrate supplementation (Lynch et al., 2017) while another showed no effect in reducing Salmonella carriage under farm conditions (Walia et al., 2016). Ten OTUs that were negatively associated with the time of gestation, hence higher at the beginning of the gestation when Salmonella shedding is at its highest, were classified at the genus level. Most interestingly Bacteroides (1 OTU), Blautia (1 OTU), Coprococcus (1 OTU) and Ruminococcus (OTU 1) which are all known short chain fatty acid producers (Pryde et al., 2002; den Besten et al., 2013; Park et al., 2013; Rivière et al., 2016). Another study already showed a reduction of health-associated species such as butyrate producers, like Coprococcus and Ruminoccocus in the current study, in the last trimester of human gestation, coupled with an augmentation of the Proteobacteria which is frequently found in inflammation-associated dysbiosis (Koren et al., 2012). These results may seem unexpected in the context of the higher level of shedding of Salmonella at the beginning of the gestation, since as previously mentioned short chain fatty acids, such as butyric acid, are known to reduce Salmonella’s virulence and invasiveness. However, the Bacteroides and Blautia genus are acetate producers. This short chain fatty acid has been shown in vitro to activate virulence genes in Salmonella (Lawhon et al., 2002). Hence, the higher presence of the genus at the beginning of gestation could be linked with the higher excretion. The only significantly varying archaeal taxon in the study, the genus Methanomassiliicoccus, which is a methane producing microorganism, was also associated with the beginning of the gestation. These results show that the reduction of Salmonella shedding at the end of the gestation could be linked with specific taxa of short-chain fatty acid producers or that other factors such as hormonal and immunological changes are also in play. No OTUs positively associated with the excretion of Salmonella could be classified at the genus level. However, one interesting OTU was classified to the Enterobacteriaceae family. This result is coherent with the higher number of excreting animals and show that the conditions that are favorable for Salmonella shedding could also be favorable for other Enterobacteriaceae. Only one OTU associated with animals not shedding Salmonella was classified to the genus level to the Clostridium cluster XI. More research will be necessary to confirm causal relationships between the abovementioned taxa and Salmonella shedding.
We observed a higher abundance of Enterobacteria and of Lachnospiraceae in sows at the beginning of the gestation using qPCR. The Enterobacteria results are consistent with our results showing that this time point was also associated with a higher level of Salmonella shedding but also with the association between shedding animals and the Enterobacteriaceae family found with MaAsLin. However, since Salmonella is only found in low levels compared to other Enterobacteria this significant reduction must be the results of the reduction of multiple genus contained in this group. The Lachnospiraceae results also seem to be consistent with the sequencing results which showed that 3 OTUs classified to the Lachnospiraceae family and 1 Coprococcus OTU which is part of this family were enriched at the beginning of the gestation.
The cause and effect of these changes in the microbiota during gestation and Salmonella shedding are not well known. However, similarly to the important change in the microbiota between the beginning and the end of gestation observed here, it has been shown that the fecal microbiota varies greatly between the first and the third semester in pregnant women (Collado et al., 2008; Koren et al., 2012). These changes were shown to contribute to modification in the metabolism and immunity of the host, playing a role, for example, in weight gain associated with pregnancy (Nuriel-Ohayon et al., 2016). Similar modifications could be at play during sow gestation, resulting in lower Salmonella carriage later in gestation. As for the mechanisms that are responsible for these changes, it has been hypothesized that changes in the mucosal immune system or at the hormonal level could be responsible for the variation of certain bacterial populations (Koren et al., 2012). For the changes associated with Salmonella shedding, another studies have found that a natural infection by this bacteria can alter the microflora in swine (Borewicz et al., 2015). It is known that Salmonella possess multiple strategies to compete with the intestinal microflora (Thiennimitr et al., 2012). However, in swine, the mechanisms that are in cause are still not well known. Since Salmonella infection in swine is mostly asymptomatic, authors have hypothesized that in swine mechanisms other than inflammation could be responsible for this variation (Kim and Isaacson, 2017).
Conclusion
In the present study, we showed that gestating sows are a possible reservoir of Salmonella with high levels of contamination and that standard detection methods are not sensitive enough to describe the entire diversity of the Salmonella strains present on the farm. We also showed that the level of shedding was variable during gestation with significantly higher shedding at the beginning rather than at the end of gestation. We also observed for the first time a significant change in the microbiota during sow gestation and identified interesting taxa such as Alloprevotella, Prevotella and the cluster IV Clostridium, which could be linked to the reduction of Salmonella shedding.
Author Contributions
GL-G designed the experiments, did all experimentations, analyzed all results, discussed the results and wrote the manuscript. AL designed the experiments, discussed the results and revised the manuscript. ÉY designed the experiments, discussed the results and revised the manuscript. AT did some experimentations, discussed the results and revised the manuscript. PF designed the experiments, discussed the results and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC, No: 412247-2010), financial partners in the Industrial Research Chair in Meat Safety and the bourse de la Cité de la biotechnologie agroalimentaire, vétérinaire et agroenvironnementale. The authors would also like to thank F. Ménard Inc. and more particularly Benoît Laplante for their collaboration and their participation. Computations were made on the supercomputer Briarée from Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), the Ministère de l’Économie, de la Science et de l’Innovation du Québec (MESI) and the Fonds de recherche du Québec - Nature et Technologies (FRQ-NT).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02219/full#supplementary-material
DATA S1 | Complete list of features associated with gestation time, parity number and Salmonella shedding status.
FIGURE S1 | Rarefaction curve comparing the number reads with the average number of OTUs found in all sequenced samples of sows’ feces for 1000 iterations.
References
- Argüello H., Rubio P., Jaramillo A., Barrios V., García M., Carvajal A. (2011). “Evaluation of cleaning and disinfection procedures against Salmonella enterica at swine farms, transport and lairage facilities,” in Proceedings of International Safepork Congress, Maastrich, 254–256. [Google Scholar]
- Bearson S. M., Allen H. K., Bearson B. L., Looft T., Brunelle B. W., Kich J. D., et al. (2013). Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 16 330–340. 10.1016/j.meegid.2013.03.022 [DOI] [PubMed] [Google Scholar]
- Belœil P. A., Fravalo P., Fablet C., Jolly J. P., Eveno E., Hascoet Y., et al. (2004). Risk factors for Salmonella enterica subsp. enterica shedding by market-age pigs in French farrow-to-finish herds. Prev. Vet. Med. 63 103–120. 10.1016/j.prevetmed.2004.01.010 [DOI] [PubMed] [Google Scholar]
- Borewicz K. A., Kim H. B., Singer R. S., Gebhart C. J., Sreevatsan S., Johnson T., et al. (2015). Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLOS ONE 10:e0139106. 10.1371/journal.pone.0139106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F., Haesebrouck F., Maes D., Van Immerseel F., Ducatelle R., Pasmans F. (2008). Non-typhoidal Salmonella infections in pigs: a closer look at epidemiology, pathogenesis and control. Vet. Microbiol. 130 1–19. 10.1016/j.vetmic.2007.12.017 [DOI] [PubMed] [Google Scholar]
- Bratchikov M., Mauricas M. (2011). Development of a multiple-run high-resolution melting assay for Salmonella spp. genotyping HRM application for Salmonella spp. subtyping. Diagn. Microbiol. Infect. Dis. 71 192–200. 10.1016/j.diagmicrobio.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M., Martin-Orue S. M., Manzanilla E. G., Badiola I., Martin M., Gasa J. (2006). Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 114 165–170. 10.1016/j.vetmic.2005.11.055 [DOI] [PubMed] [Google Scholar]
- Collado M. C., Isolauri E., Laitinen K., Salminen S. (2008). Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 88 894–899. [DOI] [PubMed] [Google Scholar]
- De Busser E. V., Maes D., Houf K., Dewulf J., De Zutter L. (2013). Effect of the enrichment medium on the detection and diversity of Salmonella from porcine duodenal content. Foodborne Pathog. Dis. 10 182–188. 10.1089/fpd.2012.1268 [DOI] [PubMed] [Google Scholar]
- den Besten G., Van Eunen K., Groen A. K., Venema K., Reijngoud D.-J., Bakker B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54 2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele I., Van Meirhaeghe H., Rasschaert G., Vanrobaeys M., De Graef E., Herman L., et al. (2012). Persistent Salmonella Enteritidis environmental contamination on layer farms in the context of an implemented national control program with obligatory vaccination. Poult. Sci. 91 282–291. 10.3382/ps.2011-01673 [DOI] [PubMed] [Google Scholar]
- DiGiulio D. B., Callahan B. J., Mcmurdie P. J., Costello E. K., Lyell D. J., Robaczewska A., et al. (2015). Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. U.S.A. 112 11060–11065. 10.1073/pnas.1502875112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Panel on Biological Hazards [BIOHAZ] (2010). Scientific opinion on a quantitative microbiological risk assessment of Salmonella in slaughter and breeder pigs. EFSA J. 8 1547. [Google Scholar]
- Farzan A., Friendship R. M., Dewey C. E., Muckle A. C., Gray J. T., Funk J. (2008). Distribution of Salmonella serovars and phage types on 80 Ontario swine farms in 2004. Can. J. Vet. Res. 72 1–6. [PMC free article] [PubMed] [Google Scholar]
- Fravalo P., Lebel P., Longpré J., Laplante B., Yergeau É, Massé D., et al. (2013). “Salmonella associated taxonomic and functional changes in the pig digestive tract during application of feed associated mitigation option in production,” in Proceedings of the 2013 International Symposium on Salmonella and Salmonellosis, Saint-Malo. [Google Scholar]
- Funk J., Gebreyes W. (2004). Risk factors associated with Salmonella on swine farms. J. Swine Health Prod. 12 247–251. [Google Scholar]
- Funk J. A., Davies P. R., Nichols M. A. (2000). The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J. Vet. Diagn. Invest. 12 412–418. 10.1177/104063870001200504 [DOI] [PubMed] [Google Scholar]
- Funk J. A., Davies P. R., Nichols M. A. (2001). Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Vet. Microbiol. 83 45–60. 10.1016/S0378-1135(01)00404-7 [DOI] [PubMed] [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Hautefort I., Thompson A., et al. (2006). Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72 946–949. 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski L. (2012). Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLOS ONE 7:e34722. 10.1371/journal.pone.0034722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter V., Klein G., Koesters S., Kreienbrock L., Blaha T., Campe A. (2012). Main risk factors for Salmonella-infections in pigs in north-western Germany. Prev. Vet. Med. 106 301–307. 10.1016/j.prevetmed.2012.03.016 [DOI] [PubMed] [Google Scholar]
- Hill A. A., Simons R. R., Kelly L., Snary E. L. (2015). A farm transmission model for Salmonella in pigs, applicable to EU Member States. Risk Anal. 36 461–481. 10.1111/risa.12356 [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Batz M. B., Morris J. G., Jr. (2012). Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 75 1292–1302. 10.4315/0362-028X.JFP-11-417 [DOI] [PubMed] [Google Scholar]
- Illumina (2013). 16S Metagenomic Sequencing Library Preparation [Online]. Illumina. Available: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf [Google Scholar]
- Kim H. B., Isaacson R. E. (2017). Salmonella in swine: microbiota interactions. Annu. Rev. Anim. Biosci. 5 43–63. 10.1146/annurev-animal-022516-022834 [DOI] [PubMed] [Google Scholar]
- Koren O., Goodrich J. K., Cullender T. C., Spor A., Laitinen K., Backhed H. K., et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150 470–480. 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina Sequencing platform. Appl. Environ. Microbiol. 79 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D., Maurer R., Suyemoto M., Altier C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46 1451–1464. 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- Lebel P., Letellier A., Longpre J., Laplante B., Yergeau E., Fravalo P. (2016). Feed presentation options in swine early fattening mitigates Salmonella shedding and specifically modulates the faecal microbiota. J. Appl. Microbiol. 122 30–39. 10.1111/jam.13305 [DOI] [PubMed] [Google Scholar]
- Legendre P., Legendre L. F. (2012). Numerical Ecology. Amsterdam: Elsevier. [Google Scholar]
- Letellier A., Beauchamp G., Guévremont E., D’allaire S., Hurnik D., Quessy S. (2009). Risk factors at slaughter associated with presence of Salmonella on hog carcasses in Canada. J. Food Prot. 72 2326–2331. 10.4315/0362-028X-72.11.2326 [DOI] [PubMed] [Google Scholar]
- Lynch H., Leonard F. C., Walia K., Lawlor P. G., Duffy G., Fanning S., et al. (2017). Investigation of in-feed organic acids as a low cost strategy to combat Salmonella in grower pigs. Prev. Vet. Med. 139 50–57. 10.1016/j.prevetmed.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Magistrali C. F., D’avino N., Ciuti F., Cucco L., Maresca C., Paniccià M., et al. (2011). Longitudinal study of fecal Salmonella shedding by sows Estudio longitudinal de excreción de Salmonella fecal en hembras Étude longitudinale sur l’excrétion fécale de Salmonella par des truies. J. Swine Health Prod. 19 326–330. [Google Scholar]
- Matsuki T., Watanabe K., Fujimoto J., Kado Y., Takada T., Matsumoto K., et al. (2004). Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70 167–173. 10.1128/AEM.70.1.167-173.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan X. C., Tickle T. L., Sokol H., Gevers D., Devaney K. L., Ward D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13 R79 10.1186/gb-2012-13-9-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet N., Houf K., Dewulf J., Duchateau L., De Zutter L., De Kruif A., et al. (2005a). Distribution of Salmonella strains in farrow-to-finish pig herds: a longitudinal study. J. Food Prot. 68 2012–2021. [DOI] [PubMed] [Google Scholar]
- Nollet N., Houf K., Dewulf J., Kruif A. D., Zutter L. D., Maes D. (2005b). Salmonella in sows: a longitudinal study in farrow-to-finish pig herds. Vet. Res. 36 645–656. 10.1051/vetres:2005022 [DOI] [PubMed] [Google Scholar]
- Nuriel-Ohayon M., Neuman H., Koren O. (2016). Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 7:1031 10.3389/fmicb.2016.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-K., Kim M.-S., Bae J.-W. (2013). Blautia faecis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 63 599–603. 10.1099/ijs.0.036541-0 [DOI] [PubMed] [Google Scholar]
- Pryde S. E., Duncan S. H., Hold G. L., Stewart C. S., Flint H. J. (2002). The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217 133–139. 10.1111/j.1574-6968.2002.tb11467.x [DOI] [PubMed] [Google Scholar]
- Public Health Agency of Canada [PHAC] (2016). Canadian Antimicrobial Resistance Surveillance System Report 2016. Available at: https://www.canada.ca/en/public-health/services/publications/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-report-2016.html [Google Scholar]
- Rajic A., Keenliside J., Mcfall M. E., Deckert A. E., Muckle A. C., O’connor B. P., et al. (2005). Longitudinal study of Salmonella species in 90 Alberta swine finishing farms. Vet. Microbiol. 105 47–56. 10.1016/j.vetmic.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau A., Fravalo P., Yergeau É, Arsenault J., Lahaye L., Letellier A. (2015). Chicken caecal microbiome modifications induced by campylobacter jejuni colonization and by a non-antibiotic feed additive. PLOS ONE 10:e0131978. 10.1371/journal.pone.0131978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P., Winter S. E., Bäumler A. J. (2012). Salmonella, the host and its microbiota. Curr. Opin. Microbiol. 15 108–114. 10.1016/j.mib.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. K., Murray R., Flockhart L., Pintar K., Pollari F., Fazil A., et al. (2013). Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathog. Dis. 10 639–648. 10.1089/fpd.2012.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia K., Argüello H., Lynch H., Leonard F. C., Grant J., Yearsley D., et al. (2016). Effect of feeding sodium butyrate in the late finishing period on Salmonella carriage, seroprevalence, and growth of finishing pigs. Prev. Vet. Med. 131 79–86. 10.1016/j.prevetmed.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Wilkins W., Rajić A., Waldner C., Mcfall M., Chow E., Muckle A., et al. (2010). Distribution of Salmonella serovars in breeding, nursery, and grow-to-finish pigs, and risk factors for shedding in ten farrow-to-finish swine farms in Alberta and Saskatchewan. Can. J. Vet. Res. 74 81–90. [PMC free article] [PubMed] [Google Scholar]
- Wilson C. M., Loach D., Lawley B., Bell T., Sims I. M., O’Toole P. W., et al. (2014). Lactobacillus reuteri 100-23 modulates urea hydrolysis in the murine stomach. Appl. Environ. Microbiol. 80 6104–6113. 10.1128/AEM.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DATA S1 | Complete list of features associated with gestation time, parity number and Salmonella shedding status.
FIGURE S1 | Rarefaction curve comparing the number reads with the average number of OTUs found in all sequenced samples of sows’ feces for 1000 iterations.