Abstract
Loss of IKAROS in committed B cell precursors causes a block in differentiation while at the same time augments aberrant cellular properties, such as bone marrow stromal adhesion, self-renewal and resistance to glucocorticoid-mediated cell death. B cell acute lymphoblastic leukemias originating from these early stages of B cell differentiation and associated with IKAROS mutations share a high-risk cellular phenotype suggesting that deregulation of IKAROS-based mechanisms causes a highly malignant disease process.
Recent studies show that IKAROS is critical for the activity of super-enhancers at genes required for pre-BCR signaling and differentiation, working either downstream of or in parallel with B cell master regulators such as EBF1 and PAX5. IKAROS also directly represses a cryptic regulatory network of transcription factors prevalent in mesenchymal and epithelial precursors that includes YAP1, TEAD1/2, LHX2 and LMO2, and their targets, which are not normally expressed in lymphocytes. IKAROS prevents not only expression of these “extra-lineage” transcription factors but also their co-operation with endogenous B cell master regulators, such as EBF1 and PAX5, leading to the formation of a de novo for lymphocytes super-enhancer network. IKAROS coordinates with the Polycomb repression complex (PRC2) to provide stable repression of associated genes during B cell development. However, induction of regulatory factors normally repressed by IKAROS starts a feed-forward loop that activates de novo enhancers and elevates them to super-enhancer status, thereby diminishing PRC2 repression and awakening aberrant epithelial-like cell properties in B cell precursors.
Insight into IKAROS-based transcriptional circuits sets new paradigms for cell differentiation but also provides new approaches for classifying and treating high-risk human B-ALL that originates from these early stages of B cell differentiation
Keywords: IKAROS, pre-B cell differentiation, super-enhancers, Polycomb repression, leukemia
Introduction
B cell differentiation is characterized by a progressive loss in capacity to differentiate into alternative cell fates with a stepwise gain in immunoglobulin gene rearrangements that is coupled to the induction of discrete downstream signaling pathways (Figure 1). The desired outcome is activation of pre-BCR signaling that supports not only precursor cell expansion but also differentiation (reviewed by [1]). The survival and proliferation of committed B cell precursors are normally constrained by an additional requirement for IL-7R signaling at the expansion phase (late pro-B/ large pre-B) followed by a second phase of pre-BCR signaling that mediates differentiation to a quiescent B cell precursor (small pre-B cell). This is concomitant with rearrangement of the immunoglobulin light chain locus (Igκ), followed by B cell repertoire selection and maturation (Figure 1).
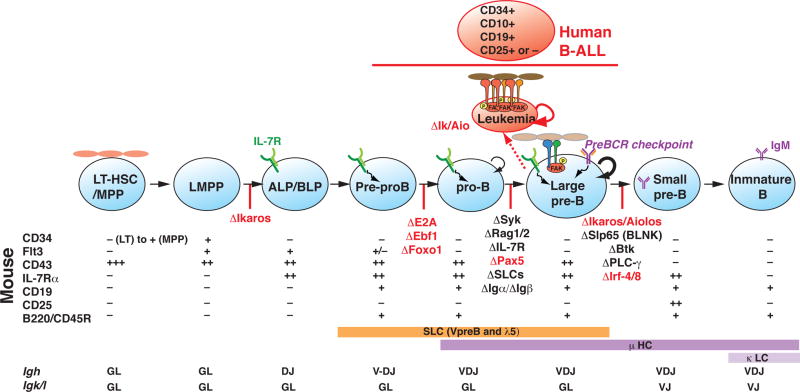
Figure 1. Developmental progression from HSC to immature B cell.
Expression of cell surface markers that define discrete stages of B cell differentiation in mouse and human are shown together with Igh and Igk gene rearrangements. The stage-specific arrest in differentiation caused by mouse genetic mutations in transcription and signaling factors is shown as red perpendicular lines on the developmental progression. Germ-line deletion of Ikzf1 arrest lymphocyte differentiation at the LMPP stage, whereas, conditional inactivation of Ikzf1 downstream of the LMPP arrests at the large pre-B stage. IKAROS deficient large pre-B cells exhibit leukemic potential upon transplantation into a NOD/SCID host. Surface phenotype of human B-ALL and its origins from the early stages of B cell differentiation are shown. Loss of IKAROS augments integrin signaling, weak stromal adhesion properties and self-renewal normally displayed by wild type large pre-B cells.
Acquisition of lymphoid lineage potential in multi-potent progenitors and subsequent restriction into the B cell lineage is the collective work of a network of transcription factors (e.g. IKAROS, E2A, EBF1, FOXO1, PAX5 and IRF4) acting at discrete steps of this cellular process to induce and reinforce the molecular mechanisms that support this differentiation program (reviewed by [2–4]). Loss of function in individual regulators arrests differentiation at discrete steps both prior to and after B cell lineage commitment, arguing for their non-redundant participation in this process (Figure 1).
B-cell acute lymphoblastic leukemia (B-ALL) is a malignancy of B cell precursor lymphocytes that affects both children and adults. Human leukemias that derive from these early stages of differentiation (Figure 1) frequently harbor activating mutations in signaling factors and inactivating mutations in transcription factors highlighting the importance of balancing these activities for progression through the leukemia-prone stages of B cell differentiation [5–8]. Loss-of function or hypomorphic mutations in genes including PAX5, TCF3/E2A, EBF1, IKZF1 (Ikaros) and IKZF3 (the AIOLOS member of the IKAROS family) are common in B cell precursor ALL cases, suggesting that disruption of B-cell differentiation contributes to the pathogenesis of this disease [5]. However, mutations in IKZF1 (IKAROS) appear to be uniquely associated with B-ALL that is refractory to standard treatment normally effective in 85% of the cases [9]. This suggests that IKAROS may serve a unique role in differentiation that when disrupted elevates B-ALL to a high-risk status. Here, we discuss recent insights into the role of IKAROS in B cell precursor differentiation.
Balancing self-renewal, proliferation and B cell differentiation
Assembly of a functional pre-BCR signaling complex in the late pro-B cell/ large pre-B cell is followed by a massive proliferative expansion supported by pre-BCR and IL-7R signaling [10–14]. This serves as a key checkpoint for productive Igh recombination events that generate a large pool of pre-B cell precursors in which Igκ recombination and B cell receptor selection (BCR) takes place (Figure 1). Although the stromal dependence of early B cell progenitors for growth (e.g. pre-pro-B and pro-B) is well established, pre-B cell expansion is thought to be stromal independent [15–17]. However, although large pre-B cells can undergo limited proliferative expansion in the presence of IL-7 they can only be propagated by a stromal adherent fraction [18]. Nonetheless, the stromal adherent phase of wild type (WT) large pre-B cells is transient and their capacity for stromal re-attachment and self-renewal is limited [18].
The ability of WT large pre-B cells to tightly associate with bone marrow stroma has shed light on the puzzle of how pre-BCR signaling switches B cell precursors from proliferation to a differentiation mode. Signaling pathways such as MAPK and PI3K that support proliferation and survival are active in WT stromal-adherent large pre-B cells but become attenuated upon detachment [18]. On the other hand, induction of pre-BCR signaling adaptors (i.e. BLNK) and Ca2+ signaling, that support differentiation is observed upon stromal detachment. The transcription profiles of cultured stromal-adherent and non-adherent large pre-B cells are also in line with the respective profiles of bone marrow ex vivo isolated large and small pre-B cells [18]. Thus it appears that during normal differentiation a transient pre-B cell-stromal interaction is in place to support a limited self-renewing expansion and to prevent signaling that promotes differentiation.
Loss of IKAROS (in combination with its family member AIOLOS) arrests differentiation at the stromal-adherent self-renewing large pre-B cell (Figure 1) [18,19]. Loss of IKAROS does not cause a mere block in differentiation but also a dramatic gain in cell adhesion. Although WT adherent pre-B cells are limited in their capacity to re-adhere to stroma (~20%), the majority of IKAROS deficient pre-B cells can do so (~85%) [18]. The increase in stromal re-adhesion correlates with a dramatic increase in self-renewal and proliferative capacity and are likely supported by a number of induced genes involved in cell adhesion, extracellular matrix, actin cytoskeleton, formation of neuronal projections and pluripotency, not normally expressed in lymphocytes. Stromal detachment causes an anoikis-type of cell death in IKAROS deficient primary pre-B cells, whereas it induces the differentiation and survival of WT pre-B cells, indicating that the two cell types rely on distinct survival mechanisms [18].
Surprisingly, the strong proliferative capacity displayed by IKAROS deficient large pre-B cells is independent of pre-BCR signaling. Although surface expression of the pre-BCR is increased, expression of downstream tyrosine kinase effectors (i.e. SYK, LYN, FYN and BLK) is attenuated. In spite of an apparent lack in pre-BCR signaling, MAPK activity is elevated [18]. An increase in the expression of integrins and downstream signaling effectors is central to the adhesion and survival of IKAROS deficient pre-leukemic pre-B cells [18] and to the adhesion properties of IKAROS deficient leukemic pre-B cells [18,20]. Treatment with a small molecule inhibitor that blocks activity of the focal adhesion tyrosine kinase (FAK) reduces stromal adhesion and induces apoptosis of IKAROS deficient pre-B cell in vivo and in vitro [18]. Moreover, integrin signaling in combination with cytokine receptor signaling (i.e. IL7R) can substitute for the stromal-dependent survival and proliferation of Ikaros deficient pre-B cells [18]. Importantly, a combination therapy with inhibitors for the ABL and FAK kinases shows promise in reducing adhesion properties and leukemia initiating frequencies in human B-ALL [21].
The role of IKAROS in promoting pre-B cell differentiation is reminiscent of its earlier role in multi-potent progenitors where it is induces the potential for lymphoid differentiation by priming transcription of lymphoid lineage-promoting genes [22,23]. There is also a significant overlap in genes and pathways that are negatively regulated by IKAROS both prior to (in the HSC and LMPP) and after B cell lineage commitment (in the large pre-B)[18,23] (Figure 2). Thus IKAROS is engaged in promoting differentiating and moderating self-renewal from the onset of hematopoiesis in multi-potent progenitors through B cell lineage-committed precursors by participating into similar regulatory mechanisms. Deregulation of these mechanisms produces arrested differentiation intermediates with exceptional properties such as cell adhesion and self-renewal.
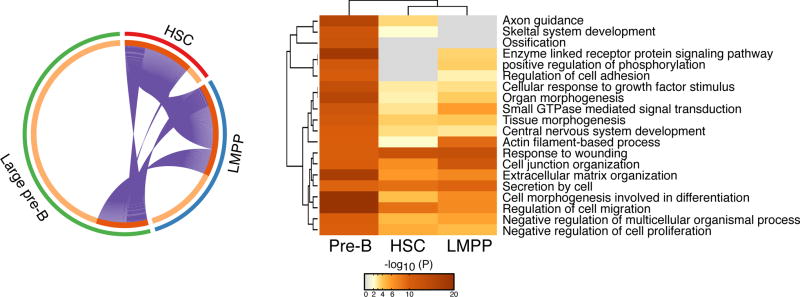
Figure 2. Repression of niche-interactions from the HSC to the large pre-B cell.
Comparative analysis of genes that were previously shown to be up-regulated upon loss of IKAROS in the HSC, LMPP and large pre-B cell [23,24]. A circos diagram depicting overlap between up-regulated genes (purple connecting lines) in IKAROS deficient HSC, LMPP and large pre-B cells is shown on the left side. A heatmap of functional pathways supported by the IKAROS-repressed genes in these cell types is shown on the right side. The color intensity of the heatmap is a measure of the p-value that indicates the enrichment for a given pathway in the corresponding list of up-regulated genes provided by a given experimental condition (e.g. in HSC, LMPP or large pre-B cell)
A silent landscape of enhancers marked by IKAROS
IKAROS is located primarily at enhancer locations in stromal adherent large pre-B cells, suggesting that it controls differentiation by regulating the activity of these sites [24].
In their majority IKAROS enrichment sites are present in a relatively restricted chromatin environment that is marked by low levels of enhancer-prevalent histone modifications (e.g. H3K4me1 and H3K4me2) and lack of histone modifications that correlate with transcriptional activity (e.g. H3K27Ac). The chromatin profile at these IKAROS-bound enhancers is therefore indicative of an inactive enhancer status. These inactive enhancers also show little occupancy by B cell-specific transcription factors (e.g. PAX5, EBF1), or general transcription factors (e.g. RNApII, MED1), and are in the vicinity of transcriptionally inactive genes [24]. Remarkably, the majority of these enhancers acquire a highly permissive chromatin in IKAROS deficient B cell precursors, are highly enriched with RNApII and MED1, and are associated with induction of genes that include core regulatory components of cell adhesion, actin cytoskeleton, small GTPases, Ras signal transduction and transcription (Figure 3) [24].
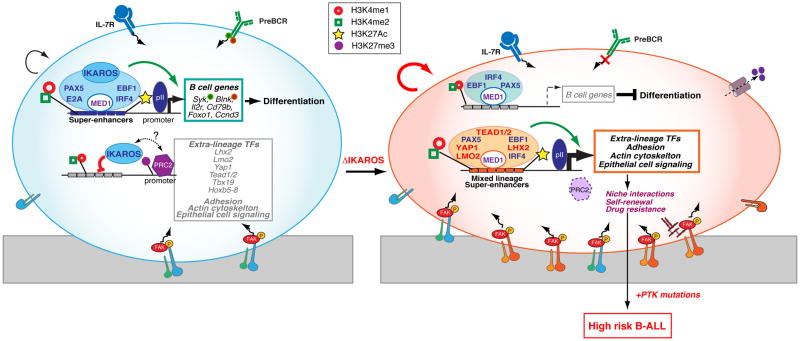
Figure 3. Reciprocal regulation of enhancers and B cell lineage fidelity.
IKAROS sets a highly permissive chromatin environment that defines super-enhancers at genes involved in pre-BCR signaling and differentiation. IKAROS also associates with the inactive enhancers of genes that support pathways such as cell adhesion, self-renewal and drug resistance normally affiliated with a non-lymphoid cell identity. Among the repressed IKAROS gene targets are master transcription regulators prevalent in non-lymphoid cells that upon IKAROS loss become induced and co-operate with B cell transcription factors to activate a de novo super-enhancer repertoire that supports the aberrant cellular properties of the mutant B cell precursors. Notably, these aberrant properties manifested in primary IKAROS pre-B cells are likely responsible for the high-risk status of leukemias caused by activating mutations in tyrosine kinases (e.g. BCR-ABL, JAK2-PDGFR).
Loss of IKAROS is not the only contributing factor to the induction of these normally repressed regulatory domains and their associated genes. A group of extra-lineage transcription master regulators that are directly repressed by IKAROS in B cell precursors are strongly enriched at de novo enhancers that frequently cluster in a super-enhancer configuration in mutant cells (Figure 3). Super-enhancers are defined as clusters of binding sites for master transcription regulators, are highly enriched for active histone modifications and the Mediator complex, and are responsible for expression of genes that specify a new cell identity [25–27]. Among the group of extra-lineage transcription factors were; LMO2, associated with hematopoietic stem cell self-renewal, early hematopoietic lineage decisions as well as T and B cell precursor leukemias [28,29]; YAP1 and TEAD (1/2), the nuclear effectors of the Hippo pathway implicated in growth and self-renewal in non-lymphoid cell types and responsible for super-enhancer activation through recruitment of the Mediator and the CDK9 elongation complex [30,31]; and LHX2 a pioneer factor in neuronal and skin epithelial stem cells [32–34]. Notably, B cell master regulators (e.g. EBF1, PAX5, IRF4) that are normally excluded from the repressed enhancer landscape in WT large pre-B cells were also highly enriched at the de novo enhancer and super-enhancers in IKAROS deficient pre-B cells. As this new network of transcription factors supports its own expression, loss of IKAROS sets in motion a powerful feed-forward regulatory loop that results in the rapid induction of both “extra-lineage” transcription factors and their downstream targets (Figure 3)[24].
Knockdown studies of either “extra-lineage” or B cell transcription factors in IKAROS deficient pre-B cells confirmed that they work together to induce expression of genes associated with these de novo regulatory elements (Figure 4). Thus IKAROS not only represses expression of “extra-lineage” transcription factors but also restricts B cell transcriptional regulators from accessing the regulatory elements of lineage-inappropriate genes. Intermingling of these normally distinct control regimens in the same cell can lead to induction of an aberrant network of super-enhancers with dire consequences for B cell differentiation.
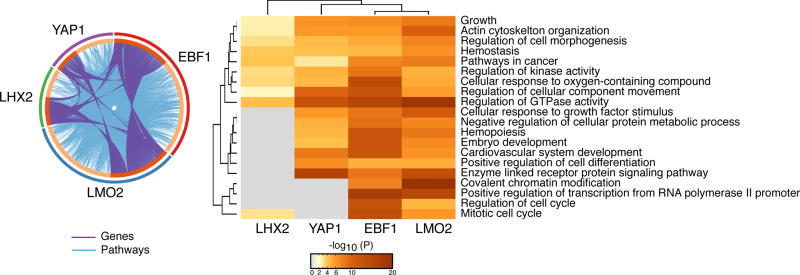
Figure 4. A hybrid “extra-lineage-B cell” transcription factor network is responsible for the IKAROS loss-of-function genetic effects.
The knockdown effect of individual “extra-lineage” and B cell transcription factors on genes up-regulated in IKAROS deficient large pre-B cells is shown. A circos diagram depicting the overlap of YAP1, EBF1, LMO2 and LHX2 on inducing the genes up-regulated (connected purple lines) in IKAROS deficient pre-B cells is shown. The effect on up-regulated functional pathways associated with these genes is also shown (connected light blue lines) in the same diagram. A heatmap of functional pathways supported by the normally IKAROS-repressed genes and their dependency on YAP1, EBF1, LMO2 and LHX2 for expression is shown on the right side (as described in Figure 2).
Collaboration between IKAROS and Polycomb in B cell precursors
The majority of genes repressed by IKAROS in pre-B cells are also occupied by the Polycomb complex (PRC2), and in most cases loss of IKAROS results in Polycomb eviction (Figure 3) [24]. However, IKAROS is mostly located at enhancers whereas PRC2 complex activity is found at the promoters of these genes suggesting an indirect functional interplay between the two types of factors. PRC2 eviction correlates with de novo induction of local super-enhancers upon IKAROS loss, suggesting a functional antagonism between strong enhancers (transcription) and PRC activity and support for an indirect IKAROS-PRC2 interaction [24].
In the model presented in Figure 3, IKAROS and the PRC2 complex may contribute independently to the repression of a stem-epithelial transcriptional program in large pre-B cells. Such a regulatory scenario occurs during development of the Drosophila embryo, with HUNCHBACK, the Drosophila homologue of IKAROS [35, 36 McCarty, 2003 #8426], initiating repression of the HOX gene locus and with Polycomb maintaining repression throughout development [37–39].
Less but not least; defining pre-B cell differentiation super-enhancers
A smaller fraction of IKAROS sites in pre-B cells is associated with clusters of highly permissive, transcriptionally active enhancers (Figure 3). These sites are also highly enriched for B cell transcription factors, such as EBF1, PAX5, E2A and IRF4, as well as for the RNAPII and the Mediator complex, and are associated with genes that serve as key regulators of pre-B cell differentiation thereby satisfying the criteria for being super-enhancers [24]. Key components of pre-BCR signaling, such as Sykb, CD79b, Blnk, Foxo1 and Ccnd3, whose loss-of-function mutations have been reported to arrest B cell differentiation [40–45], were associated with IKAROS super-enhancers and were down-regulated in IKAROS deficient pre-B cells (Figure 3)[24].
Importantly, these pre-B cell super-enhancers were functionally defined by IKAROS, as both their highly permissive chromatin and strong effect on transcription were attenuated upon loss of IKAROS despite the continued presence of other B cell master transcription regulators, such as EBF1 and PAX5, as well as the Mediator complex at these sites (Figure 3). Other B cell transcription factors may be important for the initial induction of this gene expression program during development but IKAROS is essential at this stage. IKAROS’ role in the pre-B cell is to establish a highly permissive chromatin environment and promote super-enhancer activities and gene expression levels required for the pre-B cell transition.
Conclusions
The transcriptional regulatory mechanisms supported by IKAROS in B cell precursors set a new paradigm for regulation of lineage progression and fidelity but also provide new insights into the making of high-risk human B-ALL.
It appears that all of the early B cell master transcriptional regulators are engaged in promoting differentiation and preventing alternative cell fates. Nonetheless, the mode of action of such factors can dramatically change in a leukemia setting in a manner that is dictated by the chromatin environment. EBF1 normally supports B cell differentiation acting through a B cell-specific chromatin landscape, however it can be redirected to the new chromatin landscape that arises upon IKAROS loss and by doing so it promotes an altered high-risk B cell precursor cell identity. Thus carefully evaluating the sequence of events surrounding the inactivation of a key transcriptional regulator such as IKAROS in the context of the transcriptional network through which it operates is critical for deciphering the networks driving normal differentiation and how they are utilized for malignant transformation. Do IKAROS mutations, normally considered a leukemia poor prognostic factor lose this value when combined with EBF1 mutations?
It is important to establish whether ectopic expression of extra-lineage transcription factors normally up-regulated upon loss of IKAROS is sufficient to override wild-type levels of IKAROS activity and induce the cryptic regulatory network in B cell precursors. Such studies may add new prognostic factors in the classification of high-risk B-ALL with intact IKAROS activity. Do these factors also negatively impact B cell differentiation and does their inactivation restore the original differentiation process?
Although loss of IKAROS reprograms the transcription and epigenetic landscape in support of an “altered” pre-B cell fate, IKAROS deficient pre-B cells are not leukemic and are sensitive to an anoikis-type of cell death. Activating mutations in tyrosine kinases affiliated with growth factor receptor signaling provide stromal independent survival of IKAROS deficient pre-B cells and are responsible for transition to a highly aggressive leukemic state. Such leukemia-associated signaling pathways may utilize the new epigenetic and transcription environment established by loss of IKAROS to achieve transformation. Transcription factors such as STAT5 whose activity is induced by leukemogenic signaling may engage the de novo enhancer landscape in the absence of Ikaros, hyper-activating and perhaps expanding the de novo super-enhancer repertoire in support of a genetic program that underscores high-risk leukemia.
Continued effort on these basic research endeavors with special applications in human disease holds promise in not only setting new paradigms in cell differentiation but also in providing new therapies for leukemia intervention, tailored to the epigenetic and transcriptional circuits of individual leukemic clones.
Key bullet points.
Loss of IKAROS blocks differentiation at the pre-B cell proliferative stage and confers epithelial-like cell properties including self-renewal.
IKAROS defines and activates super-enhancers that support pre-BCR signaling and pre-B cell differentiation genes.
IKAROS represses extra-lineage transcription factors and their gene targets by inactivating their enhancers, acting in concert with Polycomb located at the promoters of these genes.
Extra-lineage transcription factors co-operate with endogenous B cell regulators to establish a de-novo super-enhancer repertoire that evicts Polycomb from the promoters of these genes.
IKAROS loss induces an extra-lineage-B cell transcriptional network that confers high-risk properties to B cell precursors prior to leukemic transformation
Acknowledgments
We would like to thank Dr. Bruce Morgan for critical comments on the manuscript.
Financial support and sponsorship
Efforts of the Georgopoulos lab in understanding lymphocyte development and leukemogenesis have been supported by 5R01CA162092, 5R01CA190964 and R21AI124326. K.G. is an MGH scholar supported by J. de Gunzburg.
Footnotes
Conflicts of interest
We declare no conflict of interest.
References
- 1.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 2.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Bryder D, Sigvardsson M. Shaping up a lineage--lessons from B lymphopoesis. Curr Opin Immunol. 2010;22:148–153. doi: 10.1016/j.coi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22:161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122:3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 10.Marshall AJ, Fleming HE, Wu GE, Paige CJ. Modulation of the IL-7 dose-response threshold during pro-B cell differentiation is dependent on pre-B cell receptor expression. J Immunol. 1998;161:6038–6045. [PubMed] [Google Scholar]
- 11.Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 12.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corfe SA, Paige CJ. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin Immunol. 2012;24:198–208. doi: 10.1016/j.smim.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda T, Sanjo H, Pages G, Kawano Y, Karasuyama H, Pouyssegur J, Ogata M, Kurosaki T. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28:499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Kierney PC, Dorshkind K. B lymphocyte precursors and myeloid progenitors survive in diffusion chamber cultures but B cell differentiation requires close association with stromal cells. Blood. 1987;70:1418–1424. [PubMed] [Google Scholar]
- 16.Hayashi S, Kunisada T, Ogawa M, Sudo T, Kodama H, Suda T, Nishikawa S, Nishikawa S. Stepwise progression of B lineage differentiation supported by interleukin 7 and other stromal cell molecules. J Exp Med. 1990;171:1683–1695. doi: 10.1084/jem.171.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolink A, Haasner D, Nishikawa S, Melchers F. Changes in frequencies of clonable pre B cells during life in different lymphoid organs of mice. Blood. 1993;81:2290–2300. [PubMed] [Google Scholar]
- 18**.Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15:294–304. doi: 10.1038/ni.2821. Conditional inactivation of the IKAROS DNA-binding domain after the lymphoid primed multi-potent progenitor (LMPP) arrests after B cell lineage commitment, at the large pre-B cell stage. Loss in IKAROS DNA binding attenuates pre-BCR signaling and augments cell adhesion-dependent growth, survival and self-renewal. In particular, integrin signaling and its downstream effector, the focal adhesion kinase (FAK), are induced at the transcriptional and protein activity level. Importantly, FAK inhibition effectively interferes with the adhesion and survival of IKAROS deficient pre-B cells both in vivo and in vitro . FAK inhibition also interferes with adhesion of IKAROS leukemic cells that spontaneously develop upon adoptive transfer into NOD/SCID hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, Payne-Turner D, Althoff MJ, Song G, Chen SC, et al. Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell. 2015;28:343–356. doi: 10.1016/j.ccell.2015.07.016. This study shows that Ikzf1 alterations in mouse models of BCR-ABL1 leukemia induce stem cell-like features and poor responsiveness to ABL kinase inhibitor therapy. However, retinoids ameliorate these phenotypes in part by inducing expression of wild-type IKZF1, suggesting potential for therapeutic appliaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Churchman ML, Evans K, Richmond J, Robbins A, Jones L, Shapiro IM, Pachter JA, Weaver DT, Houghton PJ, Smith MA, et al. Synergism of FAK and tyrosine kinase inhibition in Ph+ B-ALL. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86082. This study shows that FAK inhibition attenuates the aberrant cell adhesion and stem cell–like properties of mouse leukemias generated by combining Ph+ and IKAROS DNA binding domain mutations. Importantly, it shows that a combined therapy with FAK and ABL inhibitors improves outcome in Ph+ leukemia mouse models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Hu Y, Zhang Z, Kashiwagi M, Yoshida T, Joshi I, Jena N, Somasundaram R, Emmanuel AO, Sigvardsson M, Fitamant J, et al. Superenhancer reprogramming drives a B-cell-epithelial transition and high-risk leukemia. Genes Dev. 2016;30:1971–1990. doi: 10.1101/gad.283762.116. IKAROS is described as a positive and negative regulator of super-enhancer networks that define B cell identity. It is shown that IKAROS is critical for establishing the activity of super-enhancers associated with key pre-BCR signaling and differentiation genes. At the same time IKAROS co-operates with the Polycomb complex to repress a transcriptional network comprised of both "extra-lineage and B cell master regulators. Induction of this "B cell-extra-lineage" transcription network activates a de novo repertoire of super-enhancers affiliated with a stem cell epithelial-like genetic program that is closely associated with the high-risk properties of B-ALL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Cell. Vol. 153. Elsevier Inc.; 2013. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. In; pp. 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. This study shows that the constituent elements of super-enhancers are bound by transcription factors that respond to multiple signaling pathways during development. The clustered structure of super-enhancer elements augments responsiveness to these signals during differentiation and is responsible for determining cell identity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers J, Rabbitts TH. LMO2 at 25 years: a paradigm of chromosomal translocation proteins. Open Biol. 2015;5:150062. doi: 10.1098/rsob.150062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60:328–337. doi: 10.1016/j.molcel.2015.09.001. This study shows that in human liver carcinoma cells, YAP binds to a relatively small subset of TEAD-bound enhancers and super-enhancers and promotes transcriptional elongation by recruiting the Mediator complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvatierra J, Lee DA, Zibetti C, Duran-Moreno M, Yoo S, Newman EA, Wang H, Bedont JL, de Melo J, Miranda-Angulo AL, et al. The LIM homeodomain factor Lhx2 is required for hypothalamic tanycyte specification and differentiation. J Neurosci. 2014;34:16809–16820. doi: 10.1523/JNEUROSCI.1711-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. Demonstrates that the dynamic regulation of super-enhancers by pioneer master regulators is responsible for key events in differentiation such as lineage priming, commitment and plasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. Embo J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 37.Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimell MJ, Simon J, Bender W, O'Connor MB. Enhancer point mutation results in a homeotic transformation in Drosophila. Science. 1994;264:968–971. doi: 10.1126/science.7909957. [DOI] [PubMed] [Google Scholar]
- 39.Muller J, Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 1992;11:3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng AM, Rowley B, Pao W, Hayday A, Bolen JB, Pawson T. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 41.Gong S, Nussenzweig MC. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 42.Minegishi Y, Rohrer J, Coustan-Smith E, Lederman HM, Pappu R, Campana D, Chan AC, Conley ME. An essential role for BLNK in human B cell development. Science. 1999;286:1954–1957. doi: 10.1126/science.286.5446.1954. [DOI] [PubMed] [Google Scholar]
- 43.Pappu R, Cheng AM, Li B, Gong Q, Chiu C, Griffin N, White M, Sleckman BP, Chan AC. Requirement for B cell linker protein (BLNK) in B cell development. Science. 1999;286:1949–1954. doi: 10.1126/science.286.5446.1949. [DOI] [PubMed] [Google Scholar]
- 44.Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, Aifantis I. A unique function for cyclin D3 in early B cell development. Nat Immunol. 2006;7:489–497. doi: 10.1038/ni1324. [DOI] [PubMed] [Google Scholar]
- 45.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]