Abstract
Elfamycins are a relatively understudied group of antibiotics that target the essential process of translation through impairment of EF-Tu function. For the most part, the utility of these compounds has been as laboratory tools for the study of EF-Tu and the ribosome, as their poor pharmacokinetic profile and solubility has prevented implementation as therapeutic agents. However, due to the slowing of the antibiotic pipeline and the rapid emergence of resistance to approved antibiotics, this group is being reconsidered. Some researchers are using screens for novel naturally produced variants, while others are making directed, systematic chemical improvements on publically disclosed compounds. As an example of the latter approach, a GE2270 A derivative, LFF571, has completed phase 2 clinical trials, thus demonstrating the potential for elfamycins to become more prominent antibiotics in the future.
Keywords: Elfamycin, antibiotics, EF-Tu, GTPase, kirromycin, enacylocin IIa, pulvomycin, GE2270 A
Graphical abstract
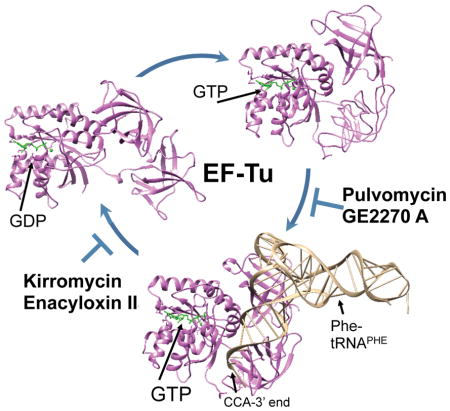
Elfamycins, a relatively understudied group of antibiotics, target the essential process of translation through impairment of EF-Tu function. The utility of these compounds has mainly been as laboratory tools for the study of EF-Tu and the ribosome. However, due to the slowing of the antibiotic pipeline and the rapid emergence of resistance to approved antibiotics, this collection of drugs is being reconsidered for their potential to become clinically utilized antibiotics. (PBD IDs: 1EXM; 1TTT; 1TUI)
Introduction
Translation of mRNA transcripts into proteins is the vital cellular function provided by the ribosome. After initiation, the ribosome relies on the elongation factor-thermo unstable (EF-Tu) to deliver subsequent amino acyl-tRNAs (aa-tRNA) to the A-site of the programmed ribosome in order to elongate the newly synthesized protein. EF-Tu is a G protein, possessing an intrinsic ability to hydrolyze GTP to GDP (GTPase activity) and contributes to the overall fidelity of translation. This hydrolysis occurs during a process termed ‘decoding,’ and provides a level of proofreading between the codon-anticodon pairing of the tRNA and the mRNA transcript.
EF-Tu forms a complex with GTP and an aa-tRNA (Figure 1A). Upon binding the A-site of the ribosome and achieving the appropriate conformation, EF-Tu hydrolyzes GTP to GDP, loses affinity for the aa-tRNA/ribosome, and dissociates. Antibiotics can disrupt EF-Tu function by either preventing its association with the aa-tRNA or impairing dissociation away from the ribosome after hydrolysis of GTP to GDP – the signal which normally allows EF-Tu to leave and the next round of decoding to proceed.
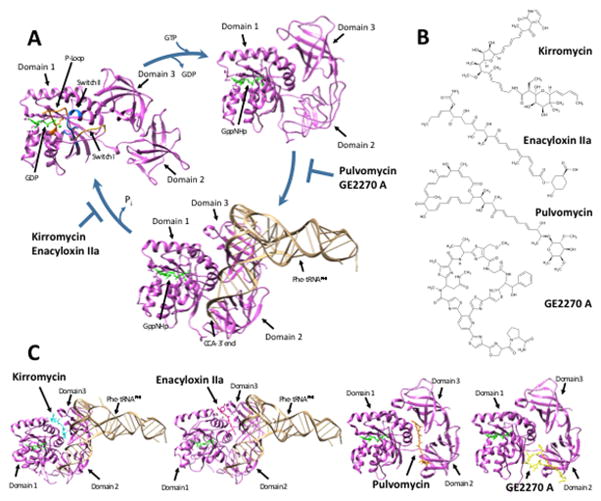
Figure 1. Crystal Structures and Chemical Structures of EF-Tu and its inhibitors.
(A) Crystal structures of the EF-Tu cycle. EF-Tu from Thermus aquaticus is indicated in purple, while bound GDP, GTP analogue, and/or magnesium ion are indicated in green. Top right: Crystal structure of active EF-Tu bound to GppNHp, a non-hydrolyzable GTP analogue (PDB ID: 1EXM). Bottom: Crystal structure of active EF-Tu bound to GppNHp and Phe-tRNAPhe. Bound Phe-tRNAPhe is indicated in tan (PDB ID: 1TTT). Top left: After accommodation of the tRNA into the ribosomal A site, GTP is hydrolyzed to GDP. Structure of inactive EF-Tu bound to GDP (PDB ID: 1TUI). P-loop indicated in orange, Switch I in yellow, and Switch II in blue. Images drawn using Chimera (UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF). (B) Chemical structures of EF-Tu inhibitors, drawn using ChemSketch (ACD/Chemsketch). (C) Crystal structures of inhibitors bound to EF-Tu. First: Kirromycin binds between domain 1 and 3 in the crystal structure of the EF-Tu:GppNHp:Phe-tRNAPhe complex. Escherichia coli EF-Tu activated with a non-hydrolyzable GTP analogue, GppHNp, bound to Phe-tRNAPhe and kirromycin. In the model, EF-Tu is indicated in purple, GppNHp in green, Phe-tRNAPhe in tan, and kirromycin in cyan (PDB ID: 1OB2). Second: Enacyloxin IIa binds between domain 1 and 3 in the crystal structure of the EF-Tu:GppNHp:Phe-tRNAPhe complex. T. aquaticus EF-Tu activated with a non-hydrolyzable GTP analogue, GppHNp, bound to Phe-tRNAPhe and enacyloxin IIa. In the model, EF-Tu is indicated in purple, GppNHp in green, Phe-tRNAPhe in tan, and enacyloxin IIa in magenta (PDB ID: 1OB5). Third: Pulvomycin binds at the interface of EF-Tu’s three domains in the crystal structure of EF-Tu:GppNHp complex. T. thermophilus EF-Tu activated with a non-hydrolyzable GTP analogue, GppHNp, bound to pulvomycin. In the model, EF-Tu is indicated in purple, GppNHp in green, and pulvomycin in orange (PDB ID: 2C78). Fourth: GE2270 A binds between domains 1 and 2 in the crystal structure of EF-Tu:GppNHp complex. T. thermophilus EF-Tu activated with a non-hydrolyzable GTP analogue, GppHNp, bound to GE2270 A. In the model, EF-Tu is indicated in purple, GppNHp in green, and GE2270 A in yellow (PDB ID: 2C77). Images drawn using Chimera (UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF).
There are four main families of EF-Tu inhibitors (Figure 1B); the prototypes of these families are kirromycin, enacylocin IIa, pulvomycin, and GE2270 A. These four share little structural similarity, but can be grouped into two main mechanisms of action. Kirromycin and enacyloxin IIa prevent EF-Tu:GDP from dissociating from the ribosome after it’s enzymatic activity has been realized, thus trapping EF-Tu on the ribosome and preventing the next round of elongation. Conversely, pulvomycin and GE2270 A inhibit the formation of the EF-Tu:GTP and aa-tRNA ternary complex, thus preventing EF-Tu from associating with the ribosome and performing it’s enzymatic activity. These compounds collectively have been given the designation ‘elfamycins,’ for their ability to target prokaryotic elongation factor Tu (EF-Tu), and are defined by their target, rather than a conserved structure. With development of resistance to other classic antibiotics, interest has renewed in inhibitors of EF-Tu.
EF-Tu GTPase activity
EF-Tu belongs to the G protein family, a collection of GTPase enzymes that bind guanosine nucleotides (GTP and GDP) and possess the intrinsic ability to hydrolyze GTP to GDP. The overall structure of EF-Tu consists of three domains (Figure 1A). Domain 1, or the G domain, is largely responsible for the GTPase activity of EF-Tu (Parmeggiani et al., 1987). This domain is often called the Ras-like domain due to its similarity to the eukaryotic G protein, Ras (Jurnak, 1985). Domains 2 and 3 form anti-parallel beta-barrels that allosterically regulate the activity of domain 1, including an enhanced affinity for GDP over GTP (Parmeggiani et al., 1987). Moreover, domain 2 has been shown to enhance the GTPase activity of EF-Tu (Nock et al., 1995). Domain 3 can be phosphorylated at Thr-382 by the toxin Doc of bacteriophage P1 toxin-antitoxin system phd-doc (Castro-Roa et al., 2013). This phosphorylation blocks translation elongation, which ultimately leads to cell death. Interestingly, kirromycin has been found to block phosphorylation by Doc, potentially through steric hindrance; this suggests that the P1 toxin/antitoxin system for maintenance of phage in the bacterial chromosome may mimic the effects of bacterial antibiotic kirromycin (Cruz et al., 2014).
GTPases are often described as molecular switches. Binding of GTP renders EF-Tu in an ‘on’ conformation, while GTP hydrolysis turns EF-Tu ‘off.’ EF-Tu, like other G proteins, contains a GTPase fold in domain 1 which allows for binding of the guanosine nucleotide (Sprang, 1997). This fold consists of 5 loops, designated G-1 through G-5, that are similar to folds observed in other nucleotide binding proteins (Sprang, 1997). The G-1 loop, also called the P-loop (Figure 1A; orange), is responsible for making contact with the β-phosphate of the guanosine nucleotide (Kjeldgaard et al., 1996, Dahl et al., 2006). Upon GTP hydrolysis, mobile elements called switch regions undergo a conformational change. The G-2 loop corresponds to switch I (Figure 1A; yellow) which participates in binding a magnesium ion important for stabilizing the guanosine nucleotide in the G protein fold (Abel et al., 1996). The G-3 loop and following α-helix corresponds to switch II (Figure 1A; blue) which binds the magnesium ion and γ-phosphate of GTP (Knudsen et al., 2001).
GTP binding alters the conformation of EF-Tu to increase its affinity for aa-tRNAs (Louie & Jurnak, 1985). GTP binding induces EF-Tu to become more compact as switch II engages the γ-phosphate of GTP and domain 2 shifts closer to domain 1, as seen in Figure 1A with the non-hydrolyzable GTP analog GppNHp (Kavaliauskas et al., 2012). As seen in Figure 1A, the movement of switch II causes domain 3 to pack against switch II and opens a cleft between domains 1 and 2 for both the 5′- and 3′-ends of the aa-tRNA to bind (Nissen et al., 1995). The tRNA acceptor stem contacts the switch regions in domain 1 while the tRNA T stem binds domain 3 (Nissen et al., 1995). The resulting EF-Tu:GTP:aa-tRNA ternary complex is then able to bind the mRNA programmed ribosome.
Upon forming a complex with the ribosome, the EF-Tu:GTP:aa-tRNA undergoes a conformational change that increases the rate of GTP hydrolysis (Mesters et al., 1994, Pape et al., 1998). This conformational change and subsequent increase in GTPase activity is dependent on and preceded by codon-anticodon pairing (Rodnina et al., 1995, Blanchard et al., 2004). Upon establishing a codon-anticodon interaction, a domain closure occurs in the 30S ribosomal subunit which alters the tRNA conformation at the acceptor end, disorders switch I, and breaks a hydrophobic gate between Val20 (P-loop) and Ile60 (switch I) to allow GTPase hydrolysis catalyzed by His84 (switch II) (Escherichia coli numbering) (Schmeing et al., 2009). His84 is believed to serve as a base to activate the water molecule responsible for nucleophilic attack on the γ-phosphate in the GTP hydrolysis reaction (Daviter et al., 2003). While free EF-Tu:GTP:aa-tRNA has a slow GTPase rate in the absence of the ribosome (Gromadski & Rodnina, 2004), upon binding the ribosome and achieving the appropriate conformation, the GTPase rate of EF-Tu is greatly accelerated and contributes to the fidelity of decoding.
Antibiotics targeting EF-Tu
Given the important role of EF-Tu in decoding and translational fidelity, microbial viability can be disrupted by inhibiting EF-Tu activity. In contrast to ribosome-targeting antibiotics such as gentamicin, which induce aberrant protein translation by disrupting the fidelity of codon-anticodon pairing while still allowing synthesis of the growing peptide chain to progress, elfamycins halt protein synthesis elongation altogether by stalling the progression of the ribosome in the elongation cycle (Wolf et al., 1972, Cetin et al., 1996). Over 30 antibiotics have been found to bind EF-Tu, the majority of which were discovered in actinomycetes. While these compounds effectively inhibit the activity of bacterial EF-Tu, they show relatively little toxicity against the mitochondrial elongation factor as assessed in a mitochondrial translation system (Zhang et al., 2005). Kirromycin was shown to be less inhibitory to mitochondrial translation than several representative macrolides (Zhang et al., 2005) and have a 200-fold difference in inhibition when challenged against mitochondrial vs E. coli ribosomes (Zhang et al., 2005). Below the four main classes of EF-Tu antibiotics are discussed; MICs against representative strains are presented in Table 1.
Table 1.
Minimum Inhibitory Concentrations (MICs) for Elfamycin Antibiotics
| Kirromycin | MIC (μg/mL) | Reference |
|---|---|---|
| Gram-Positive | ||
| Enterococcus faecium NB05001 | 2 | (Leeds et al., 2011) |
| Enterococcus faecium NB05019 | 2 | (Leeds et al., 2011) |
| Staphylococcus aureus NB01001 | >32 | (Leeds et al., 2011) |
| Enterococcus faecalis NB04004 | >32 | (Leeds et al., 2011) |
| Enterococcus faecalis NB04006 | >32 | (Leeds et al., 2011) |
| Gram-Negative | ||
| Moraxella catarrhalis ATCC 8176 | 0.06 | (Tavecchia et al., 1996) |
| Haemophilus influenzae type B ATCC 19418 | 4 | (Tavecchia et al., 1996) |
| Neisseria gonorhoeae ISM68/126 | 0.06 | (Tavecchia et al., 1996) |
| Escherichia coli SKF 12140 | >128 | (Tavecchia et al., 1996) |
|
| ||
| Enacyloxin IIa | ||
|
| ||
| Gram-Positive | ||
| Enterococcus hirae ATCC 8043 | 1 | (Watanabe et al., 1982) |
| Staphylococcus aureus 209 P | 50 | (Watanabe et al., 1982) |
| Gram-Negative | ||
| Acinetobacter baumannii OXA23 clone 2 | 3 | (Mahenthiralingam et al., 2011) |
| Burkholderia dolosa LMG 18943 | 7.5 | (Mahenthiralingam et al., 2011) |
| Pseudomonas aeruginosa NCTC 12903 | >100 | (Mahenthiralingam et al., 2011) |
|
| ||
| Pulvomycin | ||
|
| ||
| Gram-Positive | ||
| Staphylococcus aureus ATCC 29213 | 2 | (McKenzie et al., 2010) |
| Enterococcus faecalis ATCC 29212 | 4 | (McKenzie et al., 2010) |
| Staphylococcus aureus CMRSA - 1 | 32 | (McKenzie et al., 2010) |
| Enterococcus faecalis ATCC 51299 | 32 | (McKenzie et al., 2010) |
| Gram-Negative | ||
| Burkholderia cepacia C3865 | 8 | (McKenzie et al., 2010) |
| Pseudomonas aeruginosa PAO1 | 32 | (McKenzie et al., 2010) |
| Acinetobacter baumannii ATCC 17978 | 32 | (McKenzie et al., 2010) |
| Escherichia coli NU14 | 32 | (McKenzie et al., 2010) |
| Klebsiella pneumoniae HQ142423 | >128 | (McKenzie et al., 2010) |
|
| ||
| GE2270 A | ||
|
| ||
| Gram-Positive | ||
| Clostridium difficile L1363 ATCC9689 | 0.03 | (Selva et al., 1991) |
| Enterococcus faecium NB05001 | 0.03 | (Leeds et al., 2011) |
| Enterococcus faecium NB05019 | 0.06 | (Leeds et al., 2011) |
| Enterococcus faecalis L149 ATCC7080 | 0.13 | (Selva et al., 1991) |
| Staphylococcus aureus L165 Tour | 0.25 | (Selva et al., 1991) |
| Staphylococcus aureus NB01001 | 0.25 | (Leeds et al., 2011) |
| Enterococcus faecalis NB04004 | 0.25 | (Leeds et al., 2011) |
| Gram-Negative | ||
| Pseudomonas aeruginosa L4 ATCC 10145 | >128 | (Selva et al., 1991) |
| Klebsiella pneumoniae L142 ISM | >128 | (Selva et al., 1991) |
| Neisseria gonorhoeae L997 ISM68/126 | 32 | (Selva et al., 1991) |
| Chlamydia trachomatis | >128 | (Selva et al., 1991) |
| Escherichia coli L47 SKF12140 | >128 | (Selva et al., 1991) |
Kirromycin
Kirromycin was discovered to target EF-Tu in 1972 (Wolf & Zahner, 1972). The soil-dwelling Streptomyces family of bacteria are natural producers kirromycin, which was discovered in Streptomyces collinus Tü 365 (Wolf & Zahner, 1972) but is also produced in Streptomyces ramocissimus (Tieleman et al., 1997) (antibiotic originally described as mocimycin; later found to be identical to kirromycin (Maehr et al., 1973)). Additional antibiotics with similar structures to the linear polyketide antibiotic kirromycin (Figure 1B) include aurodox (Dolle & Nicolaou, 1985) (referred to as X-5108 in early publications), discovered in Streptomyces goldiniensis (Berger et al., 1973); efrotomycin (Dolle & Nicolaou, 1985); phenelfamycin A–C, E, F, discovered in 1988 (Jackson et al., 1988); and phenelfamycin G and H, discovered in 2011 (Brotz et al., 2011).
Kirromycin has a narrow spectrum against a few Gram-positive bacteria as well as a few Gram-negatives (Table 1) (Tavecchia et al., 1996). Kirromycin can interact with EF-Tu in both its GDP and GTP bound form (Pingoud et al., 1978), though kirromycin shows higher affinity for EF-Tu:GTP. As shown in Figure 1C, kirromycin binds the EF-Tu:GDP complex between domain 1 and domain 3, thereby inducing EF-Tu to maintain the conformation it would adopt when GTP is bound (Vogeley et al., 2001). This leaves EF-Tu in a constitutive ‘on’-like state, even when bound to GDP, resulting in the ternary complex remaining bound to the ribosome after GTP hydrolysis. Since EF-Tu dissociation does not occur after hydrolysis of GTP, the kirromycin-bound EF-Tu prevents the incoming of subsequent EF-Tu:GTP:aa-tRNA complexes (Vogeley et al., 2001), which results in a ribosome ‘traffic jam’ and cessation of protein synthesis.
Enacyloxin IIa
Enacyloxin IIa is the only EF-Tu antibiotic to be discovered in non-actinomycetes. It is a linear polyenic antibiotic (Figure 1B) produced by Frateuria sp. W-315 (previously belonging to the Gluconobacter genus). Both this bacterial strain and its antibiotic product were identified and characterized through an antifungal screen in 1982 (Watanabe et al., 1982). While the newly isolated compound was found to be only slightly active against fungi and not active at all against yeast, fortuitously the screen was extended to gram-positive and gram-negative bacteria. There it was found to be active against a wide range of gram-positive and gram-negative bacteria (Watanabe et al., 1982) (Table 1). Like kirromycin, enacyloxin IIa inhibits EF-Tu function by preventing dissociation from the ribosome. Structural studies have indicated enacyloxin IIa binds EF-Tu between the domain 1 and domain 3 (Figure 1C) and its binding site overlaps with kirromycin (Parmeggiani et al., 2006b). As seen with kirromycin, enacyloxin IIa binding to the EF-Tu:GDP complex induces a conformation similar to the GTP bound EF-Tu (Parmeggiani et al., 2006b) and therefore this conformation prevents EF-Tu:GDP from dissociating from the ribosome (Parmeggiani et al., 2006b) resulting in disruption of protein synthesis, yielding a bacteriostatic mechanism of inhibition (Watanabe et al., 1991).
Pulvomycin
Pulvomycin was first discovered in 1957 and is produced by Streptoverticillium netropsis and Streptomyces albosporeus var. labilomyceticus (Zief et al., 1957). Pulvomycin is active against some gram-positive and gram-negative bacteria (Table 1), and of particular note, the intrinsically highly antibiotic-resistant opportunistic pathogen Burkholderia cepacia (McKenzie et al., 2010). Upon binding EF-Tu, pulvomycin makes contact with all three domains (Figure 1C), binding at the interface of domain 1 and domain 3 and extending to contact domain 2 (Parmeggiani et al., 2006a). Pulvomycin binding locks EF-Tu into its GTP-bound state and prevents association with aa-tRNAs by masking the sites necessary for contact with both the 3′ and 5′ ends of the aa-tRNA (Parmeggiani et al., 2006a), thereby inhibiting protein synthesis.
GE2270 A
GE2270 A, a thiopeptide, was discovered in 1991 as a product from the rare actinomycete genus Planobispora rosea ATCC53773 (Selva et al., 1991). GE2270 is naturally produced by P. rosea in 10 different forms with various methylation states and activities, but GE2270 A is the form with the highest antibacterial activity (Selva et al., 1995). GE2270 A is active against a wide range of gram-positive bacteria (Selva et al., 1991) (Table 1) and demonstrates a similar effect on EF-Tu as pulvomycin, though its binding site and structure differs. As seen in Figure 1C, GE2270 A binds to domain 2 and makes some contact with domain 1 of EF-Tu, disrupting the binding of the aa-tRNA 3′ end (Parmeggiani et al., 2006a). Unlike pulvomycin, GE2270 A does not interfere with the interaction of the aa-tRNA 5′ end (Parmeggiani et al., 2006a). But like pulvomycin, GE2270 A widens the interface between domain 1 and 2, preventing GTP hydrolysis (Parmeggiani et al., 2006a). Despite having similar mechanistic outcomes, pulvomycin-resistant forms of EF-Tu retain sensitivity to GE2270 (Zeef et al., 1994).
Production of elfamycins in bacteria
Several peptide antibiotics, such as vancomycin (Recktenwald et al., 2002) and daptomycin (Robbel & Marahiel, 2010), are synthesized nonribosomally, meaning that their synthesis is not dependent on mRNA but rather a series of template-free nonribosomal peptide synthetases (NRPSs) that assemble peptides not directly inscribed in the bacterial genome (Sieber & Marahiel, 2005). Kirromycin, a large linear polyketide, is also non-ribosomally synthesized from a precursor molecule of acetyl-CoA (Weber et al., 2008). The biosynthesis gene clusters involved in creating kirromycin from this precursor were first identified in Streptomyces as modular polyketide synthases (PKS) and nonribosomal peptide synthetases (Weber et al., 2003). Modular polyketide synthases involved in kirromycin production are large, multifunctional enzymes that can catalyze all the steps necessary for production of polyketides, characterizing them as Type I PKSs, while nonribosomal peptide synthetases catalyze regiospecific and stereospecific reactions to assemble peptides. A more detailed study on kirromycin biosynthesis confirmed the genes involved in synthesis by gene disruption and monitoring for loss of antibiotic production by HPLC/MS, as well as radiolabeling precursors for confirmation of biosynthesis enzyme function through monitoring position of the radiolabel in the completed compound (Weber et al., 2008). Enacyloxin IIa synthesis is interesting in that Frateuria W-315 actually secretes a different form of the compound outside the bacterial cell; enacyloxin IVa is released into the culture fluid, which is then dehydrogenated at C-15 by the enzyme enacyloxin oxidase (ENX oxidase), therefore becoming enacyloxin IIa (Oyama et al., 1994). GE2270, a thiopeptide, was hypothesized to also be generated in a similar nonribosomal manner but is actually ribosomally originated (Morris et al., 2009). A genetic mining strategy was devised to identify genes involved in the production of the thiopeptide backbone: this was accomplished by designing primers against the predicted nucleotide coding sequence when using the amino acid sequence of the antibiotic as a template. This identified cluster of chromosomally-encoded genes was designated tpdA-tpdG, for thiopeptide. Ancilliary tpd genes encode enzymes required for maturing the precursor peptide, as well as introducing modifications specific to the particular thiopeptide (Morris et al., 2009). Most elfamycins, however, are not ribosomally originated as discussed above.
Production of EF-Tu, the target, in bacteria
EF-Tu is the most abundant protein produced in the bacterial cell (Kavaliauskas et al., 2012). Most proteobacteria, such as E. coli, encode two copies of the gene for EF-Tu (Lathe & Bork, 2001); tufA and tufB (van der Meide et al., 1983, Bosch et al., 1983). In Salmonella and related species, these are widely separated on the chromosome (700 kb on opposite sides of the origin of replication). Both are at the end of operons, are 99% identical at the coding level, and produce near-identical proteins (Hughes, 1990) differing only in their carboxy-terminal amino acid. In E. coli and Salmonella, approximately two-thirds of cellular EF-Tu was shown to be expressed from the tufA copy of the gene, while only one-third of total cellular EF-Tu came from tufB (van der Meide et al., 1982). However, when tufA was inactivated, tufB expression increased to produce expression equaling two-thirds of the typical amount seen in the cell; this implies that tufB can sense and compensate for cellular levels by doubling its normal production (Hughes, 1990). The autoregulatory mechanism of tufB was shown to be due to the tufB 5′UTR mediating Rho-dependent transcriptional termination in response to rate of translation elongation (as dictated by EF-Tu levels in the cell) (Brandis et al., 2016).
A natural question to ask when bacteria produce antibiotics is how they avoid self-intoxication. Interestingly, it was discovered in the early 1990s that S. ramocissimus (a producer strain for kirromycin) encodes three copies of the tuf gene (tuf1-3; producing EF-Tu1-EF-Tu3), but only one (tuf1) yields the standard EF-Tu protein sequence in appreciable quantities (Vijgenboom et al., 1994). This standard version is constitutively expressed and is sensitive to kirromycin. In 2007, it was shown that S. ramocissimus produces a minor quantity of EF-Tu from tuf3 in exponential phase, and this version of EF-Tu (65% amino acid homology to tuf1) is resistant to kirromycin (as well as pulvomycin and GE2270 A) (Olsthoorn-Tieleman et al., 2007). While this seemed like a plausible resistance mechanism for circumventing the toxicity associated with producing kirromycin, antibiotic synthesis actually occurs in stationary phase. Therefore, the induction of tuf3 does not correlate with kirromycin production and further did not respond to kirromycin induction (Olsthoorn-Tieleman et al., 2007), eliminating the possibility of this third EF-Tu copy allowed for compound production without self-intoxication. In addition, antibiotic-sensitive EF-Tu displays dominance in a mixed population of sensitive and resistant EF-Tu, rendering an additional resistant copy ineffective. Thus, while the presence of tuf3 is an attractive explanation for resistance in the producer strain, the real mechanism has yet to be determined.
In a survey of P. rosea, the producer strain for GE2270 A also possessing three copies of EF-Tu, EF-Tu1 was discovered to have accumulated a number of mutations, any combination of which could have lead to the observed resistance of the resulting EF-Tu against GE2270 A (Mohrle et al., 1997). Individual assessment of each mutation in an E. coli wild-type EF-Tu background lead to the conclusion that G257S and G275A (E. coli numbering) were the only natural mutations in P. rosea EF-Tu1 that conferred resistance to GE2270 A by allowing productive interactions of the EF-Tu:GTP:GE2270 A complex with the ribosome (Zuurmond et al., 2000). However, the other copies of EF-Tu in the producer strain remain sensitive to the produced antibiotic.
Recent work with enacyloxin in Frateuria showed that like kirromycin, the producer strain of this antibiotic does not encode a resistant copy of EF-Tu to avoid self-intoxication. This was demonstrated when EF-Tu purified from Frateuria sp. W-315 was inhibited by enacyloxin IIa in a poly(U)-dependent poly(Phe) synthesis assay at similar levels as susceptible EF-Tu, showing that intrinsic resistance in the coding sequence for EF-Tu is not the mechanism utilized by Frateuria sp. W-315 to avoid self-intoxication (Crechet et al., 2016). The mechanism by which Frateuria is therefore resistant to the enacyloxin it produces remains an intriguing question. As mentioned above, enacyloxin IVa is secreted from the cell and is processed extracellularly to its final form of enacyloxin IIa. This precursor secretion may therefore be the mechanism by which Frateuria sp. W-315 avoids inhibiting its own EF-Tu. Similarly, the actinomyces WAC5292 was reported to possess an ABC transporter, FacT, which protects the producer from self-intoxication from the kirromycin-like antibiotic factumycin rather than a mechanism of EF-Tu alteration (Thaker et al., 2012).
Elfamycin resistance
Of major concern to any clinically implemented antibiotic is bacterial resistance. Studies examining the generation of elfamycin-resistant forms of EF-Tu in bacterial populations are complicated by the fact that E. coli, and most other proteobacteria (Lathe & Bork, 2001), contain two virtually identical copies encoding for EF-Tu as stated above. It was recognized that in some species, such as Salmonella, either copy (but not both) is dispensable for cell viability (Hughes, 1990). However, it took another ten years of research before it was accepted that either copy is dispensable in E. coli as well (Zuurmond et al., 1999).
Inactivating one copy of the gene encoding EF-Tu enabled researchers to more easily identify EF-Tu mutants that conferred elfamycin resistance, as inactivating one copy of the tuf genes allows for a homogenous pool of mutant EF-Tu proteins in the cell, all coded by the one remaining copy of tuf. If the population were to be mixed, the wild-type copy of the protein (that which is sensitive to the effect of the antibiotic) would be dominant when challenged with kirromycin or enacyloxin IIa (i.e. trapped on the ribosome, and physically blocking the resistant copy from performing it’s enzymatic role). Earlier techniques, such as that developed by Zeef and Bosch in 1993 (Zeef & Bosch, 1993), circumvented this problem by inactivating tufB in E. coli using Mu phage insertion, then mutagenizing tufA before using a recombinant phage M13mp to deliver the mutagenized tufA to the chromosome. Resistant mutants were subsequently isolated and identified. Together these efforts identified several mutational ‘hot spots’ in the single copy of EF-Tu that are responsible for kirromycin resistance. The strategy of inactivating one copy of tuf allowed a rush of studies identifying EF-Tu amino acid substitutions that conferred resistance to this understudied group of compounds. These studies, done in the late 1990s, are cited in modern reviews as tables of resistance mutations in EF-Tu; an example being Olsthoorn-Tieleman’s (Olsthoorn-Tieleman et al., 2007) reporting of Abdulkarim’s (Abdulkarim et al., 1994) findings of mutations causing resistance to kirromycin. However, this citation and others fail to mention the caveat that the original studies identifying these mutations were done in strains that contained one inactivated copy of EF-Tu (Abdulkarim et al., 1994, Mesters et al., 1994). Overall, resistance is easy to generate in a laboratory with a sensitized background strain (single tuf gene) and reveals valuable information as to binding mechanisms of elfamycins, but the emergence of resistance in a natural environment against a bacterial strain with two (or three) copies of the tuf genes has not sufficiently been addressed.
In contrast to the sensitive-dominant nature of mixed EF-Tu populations against kirromycin, when a mixed population of EF-Tu susceptible and resistant bacterial culture is challenged with GE2270 A, the resistant is dominant. This antibiotic prevents ternary complex formation, instead of trapping an existing complex, meaning that the resistant form of EF-Tu is still able to access free ribosomes (Mohrle et al., 1997). Although pulvomycin also prevents ternary complex formation, resistance to pulvomycin and GE2270 A do not appear to be interchangeable (Zeef et al., 1994, Mohrle et al., 1997).
In addition to the complication of multiple gene copies of EF-Tu, compound permeability issues further obfuscate the question of bacterial resistance to the elfamycins. Several groups posit that the narrow spectrum of action of elfamycins is based on permeability barriers to some cells (Zeef et al., 1994), whereas others counter that EF-Tu itself is resistant (Kraal et al., 1995, Miele et al., 1994) and that there are no permeability barriers. Either way, the poor pharmacokinetics of compounds such as GE2270 have been stated to ultimately render these drugs unsuitable for clinical use (Flinspach et al., 2014, Just-Baringo et al., 2014), but have not prevented them from being useful as laboratory tools in crystallography and EF-Tu function studies (see below).
Interestingly, the mechanism of streptomycin resistance gave a hint as to a second mechanism by which a cell could naturally acquire kirromycin resistance. Mutations in rpsL (ribosome small subunit, protein S12) confer high-level resistance to the aminoglycoside streptomycin. Similarly, the same rpsL500 allele (R53L; E. coli numbering) conferring resistance to streptomycin was shown to bypass the dominant effect of sensitive EF-Tu in a bacterium where one tuf copy is resistant and the other is sensitive (Tubulekas et al., 1991). This is accomplished by the mutant S12 protein preferentially interacting with the resistant copy of EF-Tu, thus saturating ribosomal complexes with a resistant form of EF-Tu through exclusion of the sensitive version of the protein. While this mechanism of resistance only requires one copy of tuf to be mutated, not both, there is still a second mutation necessary in a separate gene (rpsL), and thus it is still necessary for the cell to acquire mutations in two separate gene loci. As shown for other antibiotics, such as the quinolones (Huseby et al., 2017), it is achievable for a cell to acquire several mutations to enable resistance. While it is therefore feasible for natural elfamycin resistance to develop, multiple protein mutations need to occur in combination to acquire resistance, potentially making elfamycins clinically preferential to a class of antibiotics that only requires a single mutation to achieve resistance.
Utility of elfamycins in the study of ribosomal function
Elfamycins have proved to be useful laboratory reagents, allowing for significant advances in several areas of ribosomal biology. Cryo-electron microscopy is a powerful tool for solving ribosome structure, and kirromycin has been used to trap EF-Tu (Stark et al., 2002) and aa-tRNA (Valle et al., 2003) in action, allowing for uniform particles that can be analyzed by single-particle reconstruction for determination of structure. These have provided insights into EF-Tu GTPase activity and subsequent EF-Tu dissociation from the ternary complex, two events critical for tRNA accommodation into the peptidyl transferase center of the ribosome (Schuette et al., 2009, Villa et al., 2009, Valle et al., 2002). Recently kirromycin was used to help achieve an improved high-resolution cryo-electron microscopy structure of the 70S ribosome complex (Fischer et al., 2015). The antibiotic stalled the ternary complex EF-Tu:GDP:Phe-tRNAPhe on the 70S ribosome which, when analyzed by aberration-corrected and computational sorting analysis, then yielded a model of high enough resolution for the visualization of all 35 rRNA modifications (i.e. base methylation, ribose methylation, pseudouridylation) for the first time (Fischer et al., 2015). This accurate visualization of modifications as small as a single methyl group provides valuable information. For example, single modifications can impact antibiotic sensitivity: loss of methylation of ribosomal rRNA base A2503 (E. coli numbering) contributes to resistance against the broad-spectrum gram-positive pathogen antibiotic linezolid. A high-resolution structure allows for mechanistic determination of the effect of this loss, and revealed that an absence of methylation at this rRNA base destabilized the stacking interaction with A2059, subsequently disrupting the binding site for the antibiotic.
Kirromycin’s ability to trap aa-tRNAs on the ribosome also contributed to a surprising discovery about transfer-messenger RNAs (tmRNAs; dual-function RNA that helps stalled ribosomes) (Miller & Buskirk, 2014). When kirromycin was added to Phe-tRNAPhe on the translating ribosome, peptidyl transfer rate was inhibited 1000-fold, as to be expected. However, when tmRNA and its accessory protein SmpB replaced the charged tRNA, rate of peptidyl transfer was only inhibited 40-fold, meaning peptide transfer to tmRNA is relatively resistant to kirromycin when compared to canonical tRNAs. This suggests that the tmRNA-SmpB complex is released from EF-Tu more easily than canonical tRNAs, presumably when in its GTP-bound confirmation, not the GDP confirmation typically seen when tRNA is released from EF-Tu. Since kirromycin blocks the conformational change accompanying GTP hydrolysis, using kirromycin to show that peptidyl transfer is relatively unaffected in the presence of the compound was a useful laboratory tool that helped show that the conformational change in EF-Tu accompanying GTP hydrolysis may not be important in tmRNA’s mechanism of action (Miller & Buskirk, 2014).
As stated previously, pulvomycin can stabilize the EF-Tu:EF-Ts complex; it naturally follows that this elfamycin has played a role in studying the mechanism of the guanine exchange catalyzed during their interaction. A series of five x-ray crystallographic structures were determined for intermediate complexes in this exchange, which allowed for a schematic representation to be built, detailing the guanine nucleotide exchange reaction (Thirup et al., 2015). Crystals of EF-Tu:EF-Ts complexed with a GppNHp, a nonhydrolyzable analog of GTP (EF-Tu:GppNHp:EF-Ts), were of significantly higher resolution when formed in the presence of pulvomycin than those formed without. In addition, pulvomycin allowed for the intermediate complex just prior to the release of EF-Ts to be solved. Pulvomycin prevents the movement of EF-Tu domain 1, thus trapping EF-Tu:Mg2+:GTP with EF-Ts for formation of crystals (Thirup et al., 2015). These examples illustrate some of the multiple uses of pulvomycin during crystallographic studies of the EF-Tu cycle. Elfamycins are useful tools for the study of cell physiology beyond just translation, as well. Kirromycin, for example, was used in a study of the interaction between EF-Tu and MreB, a protein that impacts cell shape (Defeu Soufo et al., 2010); this study determined that EF-Tu also impacts cell shape, with kirromycin being used to determine that EF-Tu activity in translation is independent from colocalization activity with MreB. Thus, elfamycins are not only useful in the context of new antibiotics for clinical treatment, but also as laboratory tools allowing for advancement in studies of the ribosome, translation, and cell physiology.
Modern resurgence and future outlook
The efficacy of many antibiotics in clinical use are challenged by the rapid spread of antibiotic resistance alleles between strains and prevalence of multi-drug resistant strains in the community; elfamycins do not currently have resistance determinants in widespread circulation between bacterial populations, making them an attractive option for a revival in research and development towards clinical implementation. Interest seems to be largely abandoned since the peak in studies in the late 1990s; perhaps the spectrum of activity was deemed too narrow to be of widespread use in combating infection, or perhaps due to problems with poor compound solubility (Tocchetti et al., 2013) and pharmacokinetics (Flinspach et al., 2014). However, the antibiotic pipeline has steadily slowed down in recent years (Cooper & Shlaes, 2011). Second-generation derivatives offer an opportunity to improve on the natural compounds, such as was done with second and third generation β-lactams, second through fifth generation cephalosporins, and second through fourth generation quinolones. In addition, no current clinically used drugs target EF-Tu, which reduces the risk of pre-existing cross-resistance to any newly deployed elfamycin.
Several groups have fortunately taken the initiative on improving elfamycins. One method for improvement over current options is to screen nature for effective compounds already in existence in the microbial world. In 2011, two novel, naturally produced derivatives of previously-known elfamycins (Brotz et al., 2011) were identified from microbes in Malaysia. These new compounds were designated phenelfamycin G and H, and differ from those previously described by the presence of a hydroxyl group at C-30. However, these compounds were demonstrated to have a very narrow range of activity; namely, only against Propionibacterium acnes. While this limits the potential for treatment of clinically significant pathogens, it does represent an opportunity to treat cosmetic or chronic acne without disruption of the gut microflora. Another group, in 2012, rediscovered factumycin from serpentine soil in Santa Clara hills, Cuba (Thaker et al., 2012). This previously known elfamycin, similar to kirromycin, was newly found to be produced by the actinomycete strain WAC5292 and was demonstrated to have activity against the human pathogen Acinetobacter baumannii. Most interesting about this compound is the finding that multidrug resistant strains of A. baumannii were actually more susceptible to factumycin than non-multidrug resistant strains, and that factumycin could be used in combination with certain other antibiotics (ex. penicillin G, daptomycin) to further increase activity against multi-drug resistant strains when tested in vitro (Thaker et al., 2012).
Work is also being undertaken on improving the quality of these antibiotics through rational design. One group has exacted synthetic modifications on GE2270 A in 2015 which resulted in an improved derivative named NAI003 (Fabbretti et al., 2015). This compound has also been demonstrated to have strong activity against P. acnes, again without targeting other commensal bacteria; targeted antibiotics which leave the microbiome unaltered appear to be a bigger health benefit than previously thought (Hajela et al., 2015), making these modifications attractive from a clinical standpoint. Another group has combined the head moiety from enacyloxin IIa with the tail of kirromycin, and thus increased the binding affinity for EF-Tu of the resulting enacyloxin IIa derivative (Parmeggiani et al., 2006b). The pharmaceutical company Novartis has designed and synthesized 4-aminothiazolyl analogs of GE2270 A (LaMarche et al., 2012). They have added functional groups that increase compound solubility while simultaneously facilitating passage through the bacterial membrane; the resulting antibiotic was named LFF571. These efforts increased solubility from barely measurable, to >10 mg/mL. These modifications were further demonstrated to increase clinical relevance by being tested in a hamster model of Clostridium difficile infection. The successful protection of hamsters from C. difficile for 21 days represents a promising route of treatment of infection in humans. Currently, LFF571 has completed Phase 2 clinical trials (Mullane et al., 2015). This multi-center trial (ClinicalTrials.gov identifier: NCT01232595) examined LFF571 against primary episodes or first reoccurrences of moderate C. difficile infections by randomly assigning patients to 125 mg vancomycin or 200 mg LFF571 four times daily for a total of 10 days. 90.6% of the LFF571-treated patients achieved a clinical cure to infection, whereas 78.3% of the vancomycin-treated patients reached clinical cure. Tolerances to the antibiotics were generally similar, as was rate of relapse in infection (Mullane et al., 2015); this represents a large step forward in bringing an elfamycin to the clinic.
An interesting follow-up study with LFF571 examined C. difficile toxin production at sub-inhibitory concentrations of antibiotic. Sub-inhibitory concentrations are known to change transcription activity, with some responses being: increased virulence factor production, decreased carbon catabolism, and an increase in prophage gene expression (Davies et al., 2006). Of concern is reports that subinhibitory concentrations of vancomycin and metronidazole can actually increase C. difficile toxin production when grown in culture (Gerber et al., 2008). In contrast, LFF571 was shown to decrease toxin expression; it was hypothesized that this was due to LFF571’s effects on inhibiting protein translation, which would be consistent with the effects of several other translation-inhibiting compounds (Sachdeva & Leeds, 2015). Overall, LFF571 and NAI003, and the approach taken to arrive at these compounds, are promising for the implementation of the elfamycin family as a new group of antibiotics in an era where options are rapidly being depleted.
Concluding statement
Elfamycins are an interesting and underappreciated group of antibiotics that target a clinically unprecedented protein, the essential translation factor EF-Tu. Most bacteria have multiple copies of the gene encoding EF-Tu, and sensitive forms of the protein are typically dominant over resistant forms, making elfamycins attractive antibiotics in an era when the numbers of clinical options available are rapidly declining. With a renewed effort in increasing compound solubility and permeability, several clinical trials are already underway to begin to utilize these antibiotics in a manner beyond just the laboratory and into the clinic for the treatment of patients.
Acknowledgments
SMP was supported in part by a training grant from the National Institute of Allergy and Infectious Diseases of the NIH to Emory University (T32AI106699, Antimicrobial Resistance and Therapeutic Discovery Training Program). Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
We would like to thank Dr. William Shafer and Dr. Eryn Bernardy for review and critical comments of the manuscript.
References
- Abdulkarim F, Liljas L, Hughes D. Mutations to kirromycin resistance occur in the interface of domains I and III of EF-Tu.GTP. FEBS letters. 1994;352:118–122. doi: 10.1016/0014-5793(94)00937-6. [DOI] [PubMed] [Google Scholar]
- Abel K, Yoder MD, Hilgenfeld R, Jurnak F. An alpha to beta conformational switch in EF-Tu. Structure. 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- ACD/Chemsketch. Advanced Chemistry Development, Inc; Toronto, ON, Canada: 2010. http://www.acdlabs.com. [Google Scholar]
- Berger J, Lehr H, Teitel S, Maehr H, Grunberg E. A new antibiotic X-5108 of Streptomyces origin. I. Production, isolation and properties. The Journal of antibiotics. 1973;26:15–22. doi: 10.7164/antibiotics.26.15. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nature structural & molecular biology. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Bosch L, Kraal B, Van der Meide PH, Duisterwinkel FJ, Van Noort JM. The elongation factor EF-Tu and its two encoding genes. Prog Nucleic Acid Res Mol Biol. 1983;30:91–126. doi: 10.1016/s0079-6603(08)60684-4. [DOI] [PubMed] [Google Scholar]
- Brandis G, Bergman JM, Hughes D. Autoregulation of the tufB operon in Salmonella. Mol Microbiol. 2016;100:1004–1016. doi: 10.1111/mmi.13364. [DOI] [PubMed] [Google Scholar]
- Brotz E, Kulik A, Vikineswary S, Lim CT, Tan GY, Zinecker H, Imhoff JF, Paululat T, Fiedler HP. Phenelfamycins G and H, new elfamycin-type antibiotics produced by Streptomyces albospinus Acta 3619. The Journal of antibiotics. 2011;64:257–266. doi: 10.1038/ja.2010.170. [DOI] [PubMed] [Google Scholar]
- Castro-Roa D, Garcia-Pino A, De Gieter S, van Nuland NA, Loris R, Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9:811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin R, I, Krab M, Anborgh PH, Cool RH, Watanabe T, Sugiyama T, Izaki K, Parmeggiani A. Enacyloxin IIa, an inhibitor of protein biosynthesis that acts on elongation factor Tu and the ribosome. The EMBO journal. 1996;15:2604–2611. [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- Crechet JB, Malosse C, Hountondji C. EF-Tu from the enacyloxin producing Frateuria W-315 strain: Structure/activity relationship and antibiotic resistance. Biochimie. 2016;127:59–69. doi: 10.1016/j.biochi.2016.04.019. [DOI] [PubMed] [Google Scholar]
- Cruz JW, Rothenbacher FP, Maehigashi T, Lane WS, Dunham CM, Woychik NA. Doc toxin is a kinase that inactivates elongation factor Tu. The Journal of biological chemistry. 2014;289:7788–7798. doi: 10.1074/jbc.M113.544429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl LD, Wieden HJ, Rodnina MV, Knudsen CR. The importance of P-loop and domain movements in EF-Tu for guanine nucleotide exchange. The Journal of biological chemistry. 2006;281:21139–21146. doi: 10.1074/jbc.M602068200. [DOI] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. Journal of molecular biology. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3163–3168. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle RE, Nicolaou KC. Total Synthesis of Elfamycins - Aurodox and Efrotomycin .1. Strategy and Construction of Key Intermediates. Journal of the American Chemical Society. 1985;107:1691–1694. [Google Scholar]
- Fabbretti A, He CG, Gaspari E, Maffioli S, Brandi L, Spurio R, Sosio M, Jabes D, Donadio S. A derivative of the thiopeptide GE2270A highly selective against Propionibacterium acnes. Antimicrobial agents and chemotherapy. 2015 doi: 10.1128/AAC.05155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- Flinspach K, Kapitzke C, Tocchetti A, Sosio M, Apel AK. Heterologous expression of the thiopeptide antibiotic GE2270 from Planobispora rosea ATCC 53733 in Streptomyces coelicolor requires deletion of ribosomal genes from the expression construct. PLoS One. 2014;9:e90499. doi: 10.1371/journal.pone.0090499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Walch C, Loffler B, Tischendorf K, Reischl U, Ackermann G. Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J Med Microbiol. 2008;57:776–783. doi: 10.1099/jmm.0.47739-0. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Molecular cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hajela N, Ramakrishna BS, Nair GB, Abraham P, Gopalan S, Ganguly NK. Gut microbiome, gut function, and probiotics: Implications for health. Indian J Gastroenterol. 2015;34:93–107. doi: 10.1007/s12664-015-0547-6. [DOI] [PubMed] [Google Scholar]
- Hughes D. Both genes for EF-Tu in Salmonella typhimurium are individually dispensable for growth. Journal of molecular biology. 1990;215:41–51. doi: 10.1016/S0022-2836(05)80093-2. [DOI] [PubMed] [Google Scholar]
- Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, Hughes D. Mutation Supply and Relative Fitness Shape the Genotypes of Ciprofloxacin-Resistant Escherichia coli. Mol Biol Evol. 2017;34:1029–1039. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Karwowski JP, Theriault RJ, Maus ML, Kohl WL. Phenelfamycins, a novel complex of elfamycin-type antibiotics. I. Discovery, taxonomy and fermentation. The Journal of antibiotics. 1988;41:1293–1299. doi: 10.7164/antibiotics.41.1293. [DOI] [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985;230:32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Just-Baringo X, Albericio F, Alvarez M. Thiopeptide antibiotics: retrospective and recent advances. Mar Drugs. 2014;12:317–351. doi: 10.3390/md12010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaliauskas D, Nissen P, Knudsen CR. The busiest of all ribosomal assistants: elongation factor Tu. Biochemistry. 2012;51:2642–2651. doi: 10.1021/bi300077s. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M, Nyborg J, Clark BF. The GTP binding motif: variations on a theme. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1996;10:1347–1368. [PubMed] [Google Scholar]
- Knudsen C, Wieden HJ, Rodnina MV. The importance of structural transitions of the switch II region for the functions of elongation factor Tu on the ribosome. The Journal of biological chemistry. 2001;276:22183–22190. doi: 10.1074/jbc.M102186200. [DOI] [PubMed] [Google Scholar]
- Kraal B, Zeef LA, Mesters JR, Boon K, Vorstenbosch EL, Bosch L, Anborgh PH, Parmeggiani A, Hilgenfeld R. Antibiotic resistance mechanisms of mutant EF-Tu species in Escherichia coli. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1995;73:1167–1177. doi: 10.1139/o95-126. [DOI] [PubMed] [Google Scholar]
- LaMarche MJ, Leeds JA, Amaral A, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Ding J, Dzink-Fox J, Gamber G, Jain A, Lee K, Lee L, Lister T, McKenney D, Mullin S, Osborne C, Palestrant D, Patane MA, Rann EM, Sachdeva M, Shao J, Tiamfook S, Trzasko A, Whitehead L, Yifru A, Yu D, Yan W, Zhu Q. Discovery of LFF571: an investigational agent for Clostridium difficile infection. Journal of medicinal chemistry. 2012;55:2376–2387. doi: 10.1021/jm201685h. [DOI] [PubMed] [Google Scholar]
- Lathe WC, 3rd, Bork &P. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS letters. 2001;502:113–116. doi: 10.1016/s0014-5793(01)02639-4. [DOI] [PubMed] [Google Scholar]
- Leeds JA, LaMarche MJ, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Dzink-Fox J, Gangl E, Jain A, Lee L, Lilly M, Manni K, Mullin S, Neckermann G, Osborne C, Palestrant D, Patane MA, Raimondi A, Ranjitkar S, Rann EM, Sachdeva M, Shao J, Tiamfook S, Whitehead L, Yu D. In vitro and in vivo activities of novel, semisynthetic thiopeptide inhibitors of bacterial elongation factor Tu. Antimicrobial agents and chemotherapy. 2011;55:5277–5283. doi: 10.1128/AAC.00582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A, Jurnak F. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5′-triphosphate-aminoacyl-tRNA complexes. Biochemistry. 1985;24:6433–6439. doi: 10.1021/bi00344a019. [DOI] [PubMed] [Google Scholar]
- Maehr H, Leach M, Yarmchuk L, Stempel A. Antibiotic X-5108. V. Structures of antibiotic X-5108 and mocimycin. J Am Chem Soc. 1973;95:8449–8450. doi: 10.1021/ja00806a043. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, Boaisha O, Paine J, Knight D, Challis GL. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria Genomic Island. Chemistry & biology. 2011;18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- McKenzie NL, Thaker M, Koteva K, Hughes DW, Wright GD, Nodwell JR. Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. The Journal of antibiotics. 2010;63:177–182. doi: 10.1038/ja.2010.13. [DOI] [PubMed] [Google Scholar]
- Mesters JR, Potapov AP, de Graaf JM, Kraal B. Synergism between the GTPase activities of EF-Tu.GTP and EF-G.GTP on empty ribosomes. Elongation factors as stimulators of the ribosomal oscillation between two conformations. Journal of molecular biology. 1994;242:644–654. doi: 10.1006/jmbi.1994.1614. [DOI] [PubMed] [Google Scholar]
- Miele A, Goldstein BP, Bandera M, Jarvis C, Resconi A, Williams RJ. Differential susceptibilities of enterococcal species to elfamycin antibiotics. Journal of clinical microbiology. 1994;32:2016–2018. doi: 10.1128/jcm.32.8.2016-2018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Buskirk AR. An unusual mechanism for EF-Tu activation during tmRNA-mediated ribosome rescue. RNA. 2014;20:228–235. doi: 10.1261/rna.042226.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrle VG, Tieleman LN, Kraal B. Elongation factor Tu1 of the antibiotic GE2270A producer Planobispora rosea has an unexpected resistance profile against EF-Tu targeted antibiotics. Biochemical and biophysical research communications. 1997;230:320–326. doi: 10.1006/bbrc.1996.5947. [DOI] [PubMed] [Google Scholar]
- Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- Mullane K, Lee C, Bressler A, Buitrago M, Weiss K, Dabovic K, Praestgaard J, Leeds JA, Blais J, Pertel P. Multicenter, randomized clinical trial to compare the safety and efficacy of LFF571 and vancomycin for Clostridium difficile infections. Antimicrobial agents and chemotherapy. 2015;59:1435–1440. doi: 10.1128/AAC.04251-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Nock S, Grillenbeck N, Ahmadian MR, Ribeiro S, Kreutzer R, Sprinzl M. Properties of isolated domains of the elongation factor Tu from Thermus thermophilus HB8. European journal of biochemistry / FEBS. 1995;234:132–139. doi: 10.1111/j.1432-1033.1995.00132.x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn-Tieleman LN, Palstra RJ, van Wezel GP, Bibb MJ, Pleij CW. Elongation factor Tu3 (EF-Tu3) from the kirromycin producer Streptomyces ramocissimus Is resistant to three classes of EF-Tu-specific inhibitors. Journal of bacteriology. 2007;189:3581–3590. doi: 10.1128/JB.01810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama R, Watanabe T, Hanzawa H, Sano T, Sugiyama T, Izaki K. An extracellular quinoprotein oxidase that catalyzes conversion of enacyloxin IVa to enacyloxin IIa. Biosci Biotechnol Biochem. 1994;58:1914–1917. doi: 10.1271/bbb.58.1914. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. The EMBO journal. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani A, I, Krab M, Okamura S, Nielsen RC, Nyborg J, Nissen P. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry. 2006a;45:6846–6857. doi: 10.1021/bi0525122. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, I, Krab M, Watanabe T, Nielsen RC, Dahlberg C, Nyborg J, Nissen P. Enacyloxin IIa pinpoints a binding pocket of elongation factor Tu for development of novel antibiotics. The Journal of biological chemistry. 2006b;281:2893–2900. doi: 10.1074/jbc.M505951200. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, Swart GW, Mortensen KK, Jensen M, Clark BF, Dente L, Cortese R. Properties of a genetically engineered G domain of elongation factor Tu. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:3141–3145. doi: 10.1073/pnas.84.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A, Urbanke C, Wolf H, Maass G. The binding of kirromycin to elongation factor Tu. Structural alterations are responsible for the inhibitory action. European journal of biochemistry / FEBS. 1978;86:153–157. doi: 10.1111/j.1432-1033.1978.tb12294.x. [DOI] [PubMed] [Google Scholar]
- Recktenwald J, Shawky R, Puk O, Pfennig F, Keller U, Wohlleben W, Pelzer S. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiol-Sgm. 2002;148:1105–1118. doi: 10.1099/00221287-148-4-1105. [DOI] [PubMed] [Google Scholar]
- Robbel L, Marahiel MA. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. The Journal of biological chemistry. 2010;285:27501–27508. doi: 10.1074/jbc.R110.128181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. The EMBO journal. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Leeds JA. Subinhibitory concentrations of LFF571 reduce toxin production by Clostridium difficile. Antimicrobial agents and chemotherapy. 2015;59:1252–1257. doi: 10.1128/AAC.04436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. The EMBO journal. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva E, Beretta G, Montanini N, Saddler GS, Gastaldo L, Ferrari P, Lorenzetti R, Landini P, Ripamonti F, Goldstein BP, et al. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. The Journal of antibiotics. 1991;44:693–701. doi: 10.7164/antibiotics.44.693. [DOI] [PubMed] [Google Scholar]
- Selva E, Ferrari P, Kurz M, Tavecchia P, Colombo L, Stella S, Restelli E, Goldstein BP, Ripamonti F, Denaro M. Components of the GE2270 complex produced by Planobispora rosea ATCC 53773. The Journal of antibiotics. 1995;48:1039–1042. doi: 10.7164/antibiotics.48.1039. [DOI] [PubMed] [Google Scholar]
- Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annual review of biochemistry. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Tavecchia P, Marazzi A, Dallanoce C, Trani A, Ciciliato I, Ferrari P, Selva E, Ciabatti R. Synthesis and biological evaluation of new fragments from kirromycin antibiotic. The Journal of antibiotics. 1996;49:1249–1257. doi: 10.7164/antibiotics.49.1249. [DOI] [PubMed] [Google Scholar]
- Thaker MN, Garcia M, Koteva K, Waglechner N, Sorensen D, Medina R, Wright GD. Biosynthetic gene cluster and antimicrobial activity of the elfamycin antibiotic factumycin. Medchemcomm. 2012;3:1020–1026. [Google Scholar]
- Thirup SS, Van LB, Nielsen TK, Knudsen CR. Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu. J Struct Biol. 2015;191:10–21. doi: 10.1016/j.jsb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Tieleman LN, van Wezel GP, Bibb MJ, Kraal B. Growth phase-dependent transcription of the Streptomyces ramocissimus tuf1 gene occurs from two promoters. Journal of bacteriology. 1997;179:3619–3624. doi: 10.1128/jb.179.11.3619-3624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchetti A, Maffioli S, Iorio M, Alt S, Mazzei E, Brunati C, Sosio M, Donadio S. Capturing linear intermediates and C-terminal variants during maturation of the thiopeptide GE2270. Chemistry & biology. 2013;20:1067–1077. doi: 10.1016/j.chembiol.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Tubulekas I, Buckingham RH, Hughes D. Mutant ribosomes can generate dominant kirromycin resistance. Journal of bacteriology. 1991;173:3635–3643. doi: 10.1128/jb.173.12.3635-3643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, GT, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. The EMBO journal. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- van der Meide PH, Vijgenboom E, Dicke M, Bosch L. Regulation of the expression of tufA and tufB, the two genes coding for the elongation factor EF-Tu in Escherichia coli. FEBS letters. 1982;139:325–330. doi: 10.1016/0014-5793(82)80881-8. [DOI] [PubMed] [Google Scholar]
- van der Meide PH, Vijgenboom E, Talens A, Bosch L. The role of EF-Tu in the expression of tufA and tufB genes. European journal of biochemistry / FEBS. 1983;130:397–407. doi: 10.1111/j.1432-1033.1983.tb07166.x. [DOI] [PubMed] [Google Scholar]
- Vijgenboom E, Woudt LP, Heinstra PW, Rietveld K, van Haarlem J, van Wezel GP, Shochat S, Bosch L. Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology. 1994;140(Pt 4):983–998. doi: 10.1099/00221287-140-4-983. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu.GDP and aurodox. The Journal of biological chemistry. 2001;276:17149–17155. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Izaki K, Takahashi H. New polyenic antibiotics active against gram-positive and -negative bacteria. I. Isolation and purification of antibiotics produced by Gluconobacter sp. W-315. The Journal of antibiotics. 1982;35:1141–1147. doi: 10.7164/antibiotics.35.1141. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Suzuki T, Izaki K. New polyenic antibiotics active against gram-positive and gram-negative bacteria. V. Mode of action of enacyloxin IIa. The Journal of antibiotics. 1991;44:1457–1459. doi: 10.7164/antibiotics.44.1457. [DOI] [PubMed] [Google Scholar]
- Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chemistry & biology. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Weber T, Welzel K, Pelzer S, Vente A, Wohlleben W. Exploiting the genetic potential of polyketide producing Streptomycetes. J Biotechnol. 2003;106:221–232. doi: 10.1016/j.jbiotec.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Wolf H, Zahner H. Metabolic products of microorganisms. 99. Kirromycin. Archiv fur Mikrobiologie. 1972;83:147–154. [PubMed] [Google Scholar]
- Wolf H, Zahner H, Nierhaus K. Kirromycin, an inhibitor of the 30 S ribosomal subunits function. FEBS letters. 1972;21:347–350. doi: 10.1016/0014-5793(72)80199-6. [DOI] [PubMed] [Google Scholar]
- Zeef LA, Bosch L. A technique for targeted mutagenesis of the EF-Tu chromosomal gene by M13-mediated gene replacement. Molecular & general genetics : MGG. 1993;238:252–260. doi: 10.1007/BF00279554. [DOI] [PubMed] [Google Scholar]
- Zeef LA, Bosch L, Anborgh PH, Cetin R, Parmeggiani A, Hilgenfeld R. Pulvomycin-resistant mutants of E. coli elongation factor Tu. The EMBO journal. 1994;13:5113–5120. doi: 10.1002/j.1460-2075.1994.tb06840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ging NC, Komoda T, Hanada T, Suzuki T, Watanabe K. Antibiotic susceptibility of mammalian mitochondrial translation. FEBS letters. 2005;579:6423–6427. doi: 10.1016/j.febslet.2005.09.103. [DOI] [PubMed] [Google Scholar]
- Zief M, Woodside R, Schmitz H. Pulvomycin. Antibiotics & chemotherapy. 1957;7:384–386. [PubMed] [Google Scholar]
- Zuurmond AM, Martien de Graaf J, Olsthoorn-Tieleman LN, van Duyl BY, Morhle VG, Jurnak F, Mesters JR, Hilgenfeld R, Kraal B. GE2270A-resistant mutations in elongation factor Tu allow productive aminoacyl-tRNA binding to EF-Tu.GTP.GE2270A complexes. Journal of molecular biology. 2000;304:995–1005. doi: 10.1006/jmbi.2000.4260. [DOI] [PubMed] [Google Scholar]
- Zuurmond AM, Rundlof AK, Kraal B. Either of the chromosomal tuf genes of E. coli K-12 can be deleted without loss of cell viability. Molecular & general genetics : MGG. 1999;260:603–607. doi: 10.1007/s004380050934. [DOI] [PubMed] [Google Scholar]