Abstract
Introduction
The aim of this study was to investigate the expression levels of microRNA-182 and microRNA-183 and their association with clinicopathological features in patients with osteosarcoma.
Material and methods
Total RNA was purified from samples and noncancerous bone tissues and then quantitative real-time polymerase chain reaction was applied to evaluate the expression levels of microRNAs, and their relationship with clinicopathological features and survival in osteosarcoma patients.
Results
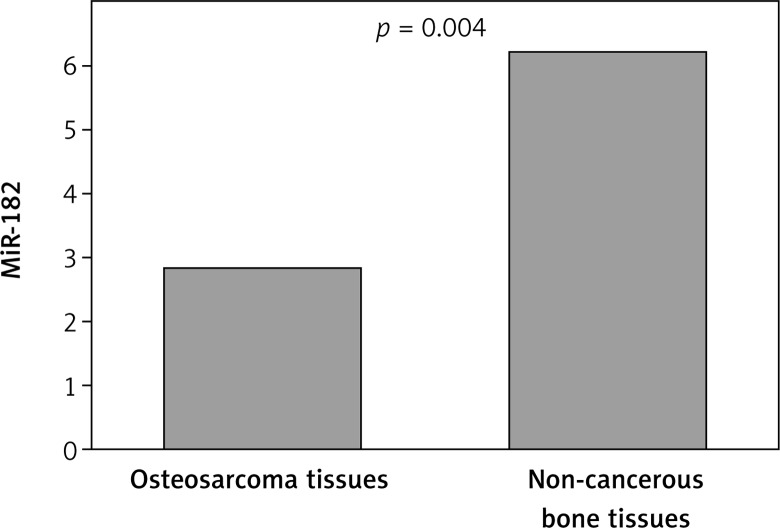
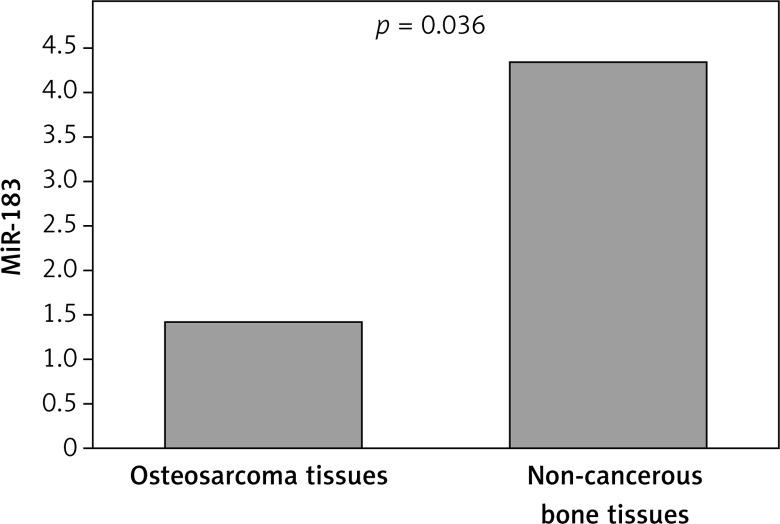
Our findings showed that expression of MiR-182 was clearly lower in osteosarcoma bone tissue (mean ± SD: 2.84 ±.07) compared with noncancerous bone tissues (6.23 ±1.72, p = 0.004). On the other hand, lower expression of MiR-183 was seen in osteosarcoma bone tissue (1.43 ±0.59) when compared with normal tissues (4.36 ±2.47, p = 0.036). Decreased expression of MiR-182 was clearly correlated with advanced clinical stage (p = 0.001), metastasis or recurrence (p = 0.024), and large tumor size (p = 0.032). Decreased expression of MiR-183 was associated with advanced TNM stage (p = 0.004), and metastasis or recurrence (p = 0.002). A multivariate Cox proportional hazards model revealed that low expression of MiR-182 and MiR-183 (p = 0.02; p = 0.016), TNM stage (p = 0.04), and metastasis or recurrence (p = 0.03) were significantly associated with poor survival as independent prognostic factors.
Conclusions
These findings suggest that MiR-182 and MiR-183 may be associated with progression and metastasis of osteosarcoma.
Keywords: tumor, oncology, pathology, marker, expression
Introduction
Osteosarcoma commonly occurs in children and young adults as a primary bone malignancy [1–3]. It has been reported that 60% of malignant bone tumors occur in the first 2 decades of life. Nevertheless, a large number of osteosarcoma cases have responded poorly to chemotherapy. Furthermore, it has been suggested that the patients are at risk of distant metastasis or local relapse after curative surgery and chemotherapy [4, 5]. Despite the progress in therapeutic targets, they remain unsatisfactory for most osteosarcoma patients with metastasis or recurrent osteosarcoma. Therefore, discovery of new specific therapeutic targets may provide effective management of the disease. MicroRNAs belong to the class of small non-coding RNAs [6], and may act as a significant marker for prognosis and detection of cancer [7–9]. It has been suggested that microRNAs are correlated with cell fate specification, cellular proliferation, differentiation and apoptosis through alteration of the targets’ expression [7, 10, 11].
Dysregulation of different microRNAs has been suggested in the context of osteosarcoma [12–15]. However, the role of MiRNAs in development of osteosarcoma remains ambiguous, and further studies are needed.
Therefore, our aim was to evaluate the expression pattern of MiR-182 and MiR-183 in human osteosarcoma and their association with clinicopathological factors.
Material and methods
Samples
Forty paired tissue samples of osteosarcoma and noncancerous bone tissue were collected from different hospitals in Tehran, Iran between March 2010 and February 2014 and were confirmed by the Research Ethics Committee. The diagnosis and the histological grading were approved by an independent pathologist. All the specimens were stored in liquid nitrogen after surgical operation until use. The clinicopathological features are presented in Table I.
Table I.
Relationship of microRNA expression with clinicopathological factors of patients with osteosarcoma
| Characteristic | N | MiR-182 expression | MiR-183 expression | P-value (MiR-182) |
P-value (MiR-183) |
||
|---|---|---|---|---|---|---|---|
| Low 21 | High 19 | Low 26 | High 14 | ||||
| Gender: | |||||||
| Male | 26 | 12 | 14 | 17 | 9 | 0.601 | 0.312 |
| Female | 14 | 9 | 5 | 9 | 5 | ||
| Age: | |||||||
| ≤ 40 | 29 | 15 | 14 | 19 | 10 | 0.523 | 0.414 |
| > 40 | 11 | 6 | 5 | 7 | 4 | ||
| Tumor diameter [cm]: | |||||||
| ≤ 5 | 24 | 14 | 10 | 15 | 9 | 0.032 | 0.423 |
| > 5 | 16 | 7 | 9 | 11 | 5 | ||
| Metastasis or recurrence: | |||||||
| No | 23 | 9 | 14 | 13 | 10 | 0.024 | 0.002 |
| Yes | 17 | 12 | 5 | 13 | 4 | ||
| Differentiation: | |||||||
| High and moderate | 21 | 10 | 11 | 12 | 9 | 0.123 | 0.113 |
| Poor | 19 | 11 | 8 | 14 | 5 | ||
| TNM stage: | |||||||
| I + II | 25 | 9 | 16 | 14 | 11 | 0.001 | 0.004 |
| III + IV | 15 | 12 | 3 | 12 | 3 | ||
Quantitative real-time PCR
The total RNA was isolated from frozen samples using TRIzol reagent based on the manufacturer’s instructions. Gene-specific primers were used to synthesize cDNA from the TaqMan MicroRNA assays and reagents from the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Real-time polymerase chain reaction was carried out to determine the expression level of microRNAs using an Invitrogen kit with the Rotor-gene 6000 system (Qiagen). The primers were used from the TaqMan MiRNA assays. The relative amount of microRNAs was normalized with the U6 gene as an internal reference. ΔΔCt (ΔΔCt = ΔCt tumor samples – ΔCt control sample) was calculated to qualify the expression rate of MiR-182 and MiR-183.
Statistical analysis
We used SPSS 18.0 software for statistical analysis (SPSS Inc., USA). Differences between groups were evaluated using the χ2 test. Survival analysis was performed using the log-rank test and Kaplan-Meier method. A Cox proportional hazards model was performed to evaluate prognostic values of clinicopathological factors. Differences were considered statistically significant when p < 0.05.
Results
Our findings showed that expression of MiR-182 was clearly lower in osteosarcoma bone tissue (mean ± SD: 2.84 ±0.07) compared with noncancerous bone tissues (6.23 ± 1.72, p = 0.004; Figure 1), On the other hand, lower expression of MiR-183 was observed in osteosarcoma bone tissue (1.43 ±0.59) when compared with normal tissues (4.36 ±2.47, p = 0.036; Figure 2). According to the median expression level of the two microRNAs, we categorized the patients into low and high expression groups. The correlations between clinicopathological factors and expression of the two microRNAs in high and low expression groups are summarized in Table I.
Figure 1.
Relative expression level of MiR-182 between osteosarcoma bone tissue and noncancerous bone tissues
Figure 2.
Relative expression level of MiR-183 between osteosarcoma bone tissue and noncancerous bone tissues
The results showed that decreased expression of MiR-182 was clearly correlated with advanced clinical stage (p = 0.001), metastasis or recurrence (p = 0.024), and large tumor size (p = 0.032). No significant difference was found between MiR-182 and age (p = 0.523), gender (p = 0.601), or differentiation (p = 0.123) (Table I). Decreased expression of MiR-183 was associated with advanced TNM stage (p = 0.004), and metastasis or recurrence (p = 0.002). There was no significant correlation of MiR-183 with other clinical factors (Table I).
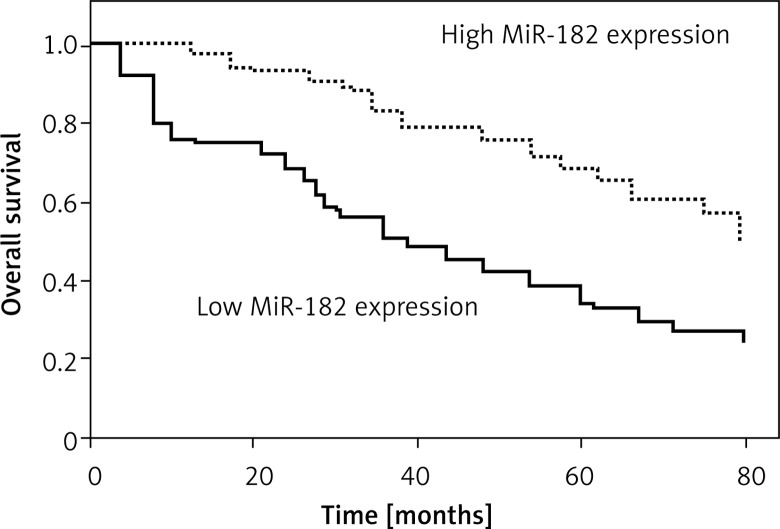
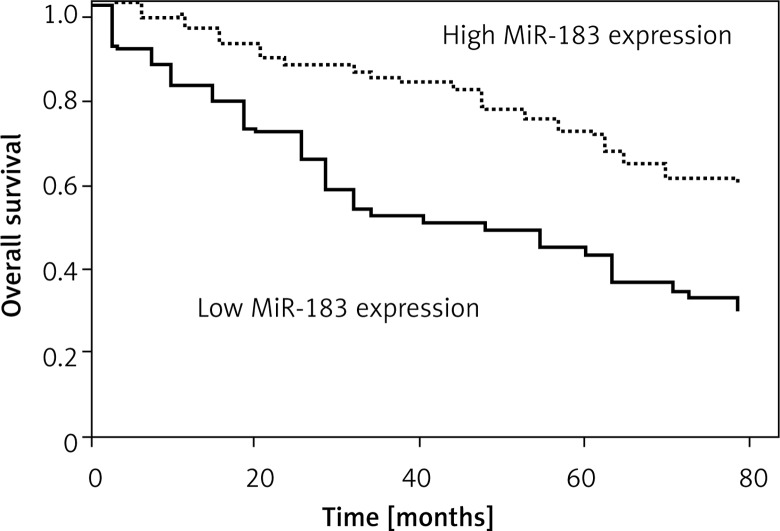
Kaplan-Meier survival and log-rank analysis were performed to evaluate the association of microRNA expression with survival of patients. As shown in Figures 3 and 4, the decreased expression of these microRNAs was strongly correlated with shorter overall survival (log-rank test p = 0.035; p = 0.029).
Figure 3.
Correlation of MiR-182 expression with survival time in patients with osteosarcoma
Figure 4.
Correlation of MiR-183 expression with survival time in patients with osteosarcoma
The multivariate Cox proportional hazards model revealed that low expression of MiR-182 and MiR-183 (p = 0.02; p = 0.016), TNM stage (p = 0.04), and metastasis or recurrence (p = 0.03) were significantly associated with poor survival as independent prognostic factors (Tables II and III).
Table II.
Multivariate analysis of the correlation of prognosis miR-182 with clinicopathological factors
| Clinicopathological characteristics | HR | 95% CI | P-value |
|---|---|---|---|
| Gender | 0.535 | 0.752–1.827 | 0.63 |
| Age | 1.231 | 0.663–2.531 | 0.52 |
| Anatomic location | 1.027 | 0.231–1.863 | 0.68 |
| TNM stage | 2.754 | 2.024–7.612 | 0.04 |
| Tumor size [cm] | 3.129 | 1.934–9.723 | 0.03 |
| Metastasis or recurrence | 4.374 | 3.432–12.026 | 0.03 |
| Differentiation | 1.748 | 1.631–4.931 | 0.09 |
| MiR-182 level | 3.852 | 1.473–10.432 | 0.02 |
Table III.
Multivariate analysis of the correlation of prognosis miR-183 with clinicopathological factors
| Clinicopathological characteristics | HR | 95% CI | P-value |
|---|---|---|---|
| Gender | 0.53 | 0.732–2.135 | 0.64 |
| Age | 1.102 | 0.543–2.983 | 0.54 |
| Anatomic location | 1.231 | 0.257–3.172 | 0.51 |
| TNM stage | 2.755 | 2.247–7.482 | 0.04 |
| Tumor size [cm] | 3.012 | 1.543–10.452 | 0.001 |
| Metastasis or recurrence | 4.618 | 3.155–13.026 | 0.01 |
| Differentiation | 1.892 | 1.821–4.367 | 0.08 |
| MiR-183 level | 4.183 | 1.643–11.521 | 0.02 |
Discussion
The role of MiRNAs in development of osteosarcoma remains ambiguous, and discovery of new specific therapeutic targets may provide effective management of the disease. Dysregulation of different MiRNAs has been previously suggested in many kinds of tumor [13, 14, 16, 17]. In the current study, we evaluated the expression pattern of MiR-182 and MiR-183 in relation to osteosarcoma.
Our findings showed that expression of MiR-182 and MiR-183 was clearly lower in osteosarcoma bone tissue compared with noncancerous bone tissues. Our findings suggest that their down-regulation is correlated with the progression of osteosarcoma and low expression of mentioned MiRNAs can contribute to tumor occurrence and development. Aberrant regulation of MiR-182 has been reported in many kinds of malignancies, including gastric cancer, lung, bladder, endometrial, prostate, colon and breast cancers, ovarian carcinoma, pediatric acute leukemia and melanoma [18, 19]. The tumor suppressor role of MiR-182 or oncogene has been suggested in different human malignancies. For instance, MiR-182 inhibits proliferation and invasion by targeting CTTN in lung adenocarcinoma cells [20], and could act as a tumor suppressor in lung tumor by targeting RGS17 [21]. In addition, the over-expression of MiR-182 has been previously reported, and also it was indicated that MiR-182 can play a role as an oncogene in different human cancers including melanoma [18].
Previous studies have suggested that MiR-183 can inhibit cell migration by targeting ITGB1 in neurosensory organs [22]. Moreover, MiR-183 can play a key role in initiation of tumor and progression in hepatocellular carcinoma cells [23]. Nevertheless, further investigations are needed to identify the role of MiR-183 in pathogenesis of osteosarcoma. Our results indicated that low expression of MiR-182 and MiR-183 was correlated with tumor progression in osteosarcoma. Decreased expression of MiR-182 was clearly correlated with advanced clinical stage, metastasis or recurrence, and high tumor size. Decreased expression of MiR-183 was associated with advanced TNM stage, and metastasis or recurrence.
It has been reported that MiR-182 was decreased in osteosarcoma tissues and cell lines. Up-regulation of MiR-182 can inhibit tumor growth, invasion and migration. Subsequent findings showed that TIAM1 was a direct target of MiR-182 in osteosarcoma cells [24]. On the other hand, it has been found that ectopic overexpression of MiR-183 repressed the expression levels of ezrin and markedly inhibited the motility and invasion of osteosarcoma cells. Recently, further studies suggested that MiR-183 may act as a tumor suppressor in the metastasis of osteosarcoma via downregulation of ezrin expression [25].
Kaplan-Meier survival and log-rank analysis indicated that decreased expression of these microRNAs was strongly correlated with shorter overall survival, which might be related to prognosis of osteosarcoma. The multivariate Cox proportional hazards model revealed that low expression of MiR-182 and MiR-183, TNM stage, and metastasis or recurrence were significantly associated with poor survival as independent prognostic factors.
In conclusion, our result showed that MiR-182 and MiR-183 may be associated with progression and metastasis of osteosarcoma. Further studies are needed to clarify the role of these markers.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Tang J, Shen L, Yang Q, Zhang C. Overexpression of metadherin mediates metastasis of osteosarcoma by regulating epithelial-mesenchymal transition. Cell Proliferation. 2014;47:427–34. doi: 10.1111/cpr.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, Cai M, Fu D, Chen K, Sun M, Cai Z. Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell Physiol Biochem. 2012;30:1481–90. doi: 10.1159/000343336. [DOI] [PubMed] [Google Scholar]
- 4.Wang XH, Cai P, Wang MH, Wang Z. microRNA25 promotes osteosarcoma cell proliferation by targeting the cell cycle inhibitor p27. Mol Med Rep. 2014;10:855–9. doi: 10.3892/mmr.2014.2260. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohdaira H, Sekiguchi M, Miyata K, Yoshida K. Micro-RNA-494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 2012;45:32–8. doi: 10.1111/j.1365-2184.2011.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Droshafor endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 9.Finnerty JR, Wang WX, Hébert SS. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatri R, Subramanian S. MicroRNA-135b and its circuitry networks as potential therapeutic targets in colon cancer. Frontiers Oncol. 2013;3:268. doi: 10.3389/fonc.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng B, Liang L, Wang C, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–83. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 12.Jin J, Cai L, Liu ZM, Zhou XS. MiRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev. 2013;14:3681–4. doi: 10.7314/apjcp.2013.14.6.3681. [DOI] [PubMed] [Google Scholar]
- 13.Wu XJ, Li Y, Liu D, Zhao LD, Bai B, Xue MH. MiR-27a as an oncogenic microRNA of hepatitis B virus-related hepatocellular carcinoma. Asian Pac J Cancer Prev. 2013;14:885–9. doi: 10.7314/apjcp.2013.14.2.885. [DOI] [PubMed] [Google Scholar]
- 14.Zhao G, Cai C, Yang T. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8:e53906. doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Gao K, Lin J. MicroRNA-100 inhibits osteosarcoma cell proliferation by targeting Cyr61. Tumor Biol. 2014;35:1095–100. doi: 10.1007/s13277-013-1146-8. [DOI] [PubMed] [Google Scholar]
- 16.Tang M, Lin L, Cai H, Tang J, Zhou Z. MicroRNA-145 downregulation associates with advanced tumor progression and poor prognosis in patients suffering osteosarcoma. Onco Targets Ther. 2013;6:833–8. doi: 10.2147/OTT.S40080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey N, Das F, Ghosh-Choudhury N, et al. MicroRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS One. 2012;7:e37366. doi: 10.1371/journal.pone.0037366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X. MicroRNA-182 targets cAMP-responsive element binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279:1252–60. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Liu T, Huang Y, Liu J. microRNA-182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein. Int J Mol Med. 2011;28:381–8. doi: 10.3892/ijmm.2011.679. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Fang R, Li C, Li L, Li F, Ye X. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Comm. 2010;396:501–7. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem. 2010;285:5461–547. doi: 10.1074/jbc.M109.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Fu H, Xu C, et al. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Lv G, Zhou S, et al. The downregulation of MiR-182 is associated with the growth and invasion of osteosarcoma cells through the regulation of TIAM1 expression. PLoS One. 2015;10:e0121175. doi: 10.1371/journal.pone.0121175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Zhao H, Guo M, Zhao G, et al. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med. 2012;30:1013–20. doi: 10.3892/ijmm.2012.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]