Abstract
Introduction
Our aim was to determine the effect of the single nucleotide polymorphisms (SNP) –93G>A of the MLH1 gene (rs1800734) and Gly322Asp of the MSH2 gene (rs4987188) on the risk of colon cancer (CC) and identify any relationship with clinical factors.
Material and methods
The study included 144 unrelated patients with sporadic CC (71 males; mean age: 61.7 ±11 years) and 151 control patients (74 males; mean age: 63 ±11 years). DNA was extracted from peripheral blood lymphocytes, and genotypes were determined by polymerase chain reaction-restriction fragment length polymorphism.
Results
In our population, the homozygous G/G genotype of the –93G>AMLH1 gene increased the risk of sporadic CC (OR = 2.07; 95% CI: 1.11–3.83; p < 0.02). For A/G and A/A genotypes, the MLH1-93G>A polymorphism was significantly more common in women (p = 0.034). The SNP demonstrated differences in allele distribution according to the location of the tumor, i.e. right vs. left side (p = 0.014), and disease recurrence (p = 0.022). Significant differences were found in the occurrence of Gly322Asp of MSH2 with regard to primary and recurrent disease (p = 0.001).
Conclusions
The –93G>AMLH1 polymorphism plays an important role in evaluating the risk of sporadic CC. It can also be used as an indicator in some patients with left-sided and recurrent tumors. MSH2 Gly322Asp is a potential marker in patients with risk of recurrence.
Keywords: colorectal cancer, polymorphism, cancer risk, mismatch repair genes
Introduction
Colorectal cancer (CRC) is one of the major causes of cancer-associated morbidity and mortality in developing and well-developed countries. It is the second most prevalent cancer, and one that affects men and women almost equally. Approximately 600,000 deaths from CRC are estimated worldwide each year, representing 8% of all cancer deaths, which makes it the fourth leading cause of death among all malignancies [1].
Maintaining the integrity of genomic DNA is necessary for correct cell function. Genome disorders can contribute to the development of several malignancies, including CRC. The CRC can be initiated by DNA damage induced by chemical agents, smoking, alcohol consumption, and fat metabolism. A number of single nucleotide polymorphisms (SNP) have been identified in a group of more than 130 known DNA repair genes [2]. The presence of polymorphisms in repair genes may influence their efficiency to eliminate DNA damage and thereby increase the chance of cancer. The human mismatch repair (MMR) system repairs DNA misinformation or damage. Microsatellite regions are especially susceptible to mutations and polymorphisms due to slippage of DNA polymerase during DNA replication, and failure to excise these errors may lead to frameshift mutations in many genes.
MLH1, MSH2, MSH6, and PMS2 are major MMR genes implicated in genetic stability [3], with germline mutations in these MMR genes being responsible for hereditary non-polyposis colorectal cancer (HNPCC). The presence of single nucleotide polymorphisms (SNPs) in the MMR genes can also predispose the patient to non-familial CRC by contributing common genetic variants of CRC susceptibility in a given population [4]. However, although approximately 70% of sporadic cancer is caused by external factors, some studies suggest the possibility of a genetic predisposition associated with alterations in DNA repair genes, including the MMR genes. Furthermore, other studies suggest that the presence of MMR gene polymorphisms may have an influence on the late outcomes of treating CRC patients [5].
The aim of this study was to determine the effect of SNP –93G>A of the MLH1 gene and Gly322Asp of MSH2 on the risk of colon cancer in the studied population in comparison with healthy subjects, and to identify differences in allele distribution between different subgroups of colon cancer patients according to age, sex, tumor location, some pathological findings, staging and severity of the disease.
Material and methods
The recruited subjects comprised a case group of patients with histologically confirmed colon cancer and a control group of healthy, unrelated volunteers. DNA was isolated from peripheral blood lymphocytes obtained from blood samples taken from the subjects.
The study was conducted in accordance with the approval of the Local Ethics Committee of the Medical University of Lodz (no. RNN/693/14/KB). The subjects were fully informed of the nature of the study and each gave their written consent before taking part. The study design conforms with the Code of Ethics of the World Medical Association (Declaration of Helsinki).
The study group consisted of 144 unrelated patients (71 men and 73 women; mean age: 61.7 ±11) with pathologically confirmed colon cancer. None had any family history of any malignancies. The control group comprised 151 unrelated patients without any confirmed malignancies (74 men and 77 women; mean age: 63 ±11). All patients and controls were cross-matched according to age and gender. The patients in the study group were operated on between July and December 2014 in the university tertiary colorectal center for primary or recurrent disease. All surgical procedures were performed by a specialized surgical team according to colorectal cancer surgery guidelines.
We analyzed the influence of SNP in 93G>A MLH1 (rs1800734) and Gly322Asp MSH2 (rs4987188) and the risk of CC in a population in central Poland. Subgroups of colon cancer patients were formed to investigate the relationship between the distribution of MMR gene alleles according to selected groups of clinical factors.
Polymorphism analysis
hMLH1-93 G>A genotyping by PCR-RFLP
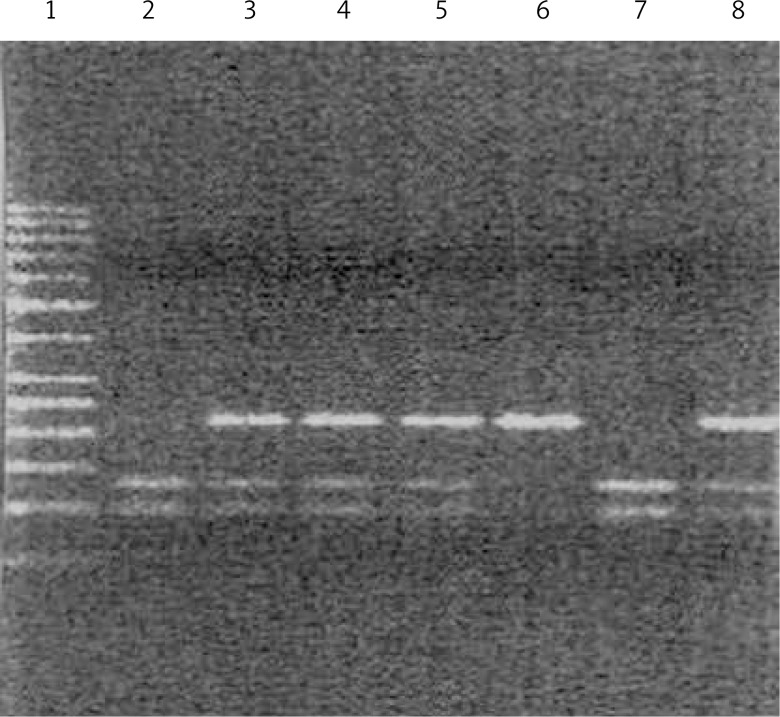
DNA was extracted from peripheral blood lymphocytes using DNA Blood Mini Kits (A&A Biotechnology, Gdynia, Poland). Genotypes were determined by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) approach. The PCR was carried out in a volume of 10 μl. The reaction mixture consisted of 100 ng of genomic DNA, 0.5 μmol of each primer and 3 U of Taq polymerase. The PCR cycling conditions consisted of an initial denaturation step of 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 56°C for 45 s and 72°C for 45 s, followed by a final extension stage of 72°C for 10 min. The 10 μl of specifically positive PCR products was digested overnight with 1 μl of PvuII (New England Bio Labs, Beverly, MA) at 37°C and digested DNA fragments were resolved on 3% agarose gel. The homozygous AA genotype gave undigested fragments of 387 bp while the homozygous GG genotype yielded two digested fragments of sizes 207 bp and 180 bp. The AG heterozygous condition revealed three bands sized 387 bp, 207 bp and 180 bp (Figure 1). The randomly selected PCR-amplified DNA samples for each genotype were cross-checked by DNA sequencing and the results were found to be 100% concordant.
Figure 1.
Analysis of genotype polymorphism –93G>A gene hMLH1 after hydrolysis with restriction enzyme PvuII. Track 1 marker tracks 6, homozygous AA (one band at 387 bp), lane 2, 7, heterozygous GG (two bands of size 180 bp, 207 bp) lane 3, 4, 5, 8 is heterozygous AG (three bands of the size 180 bp, 207 bp and 387 bp)
HMSH2 Gly322Asp
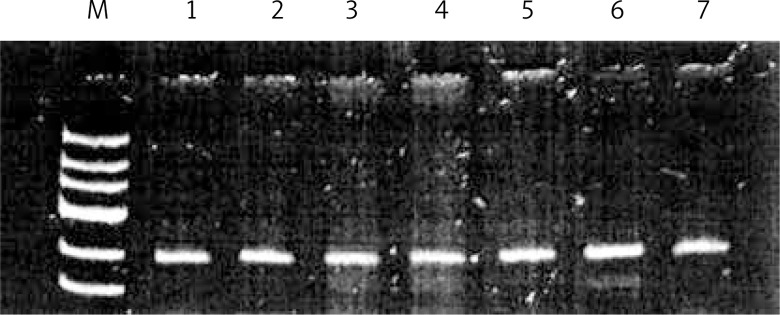
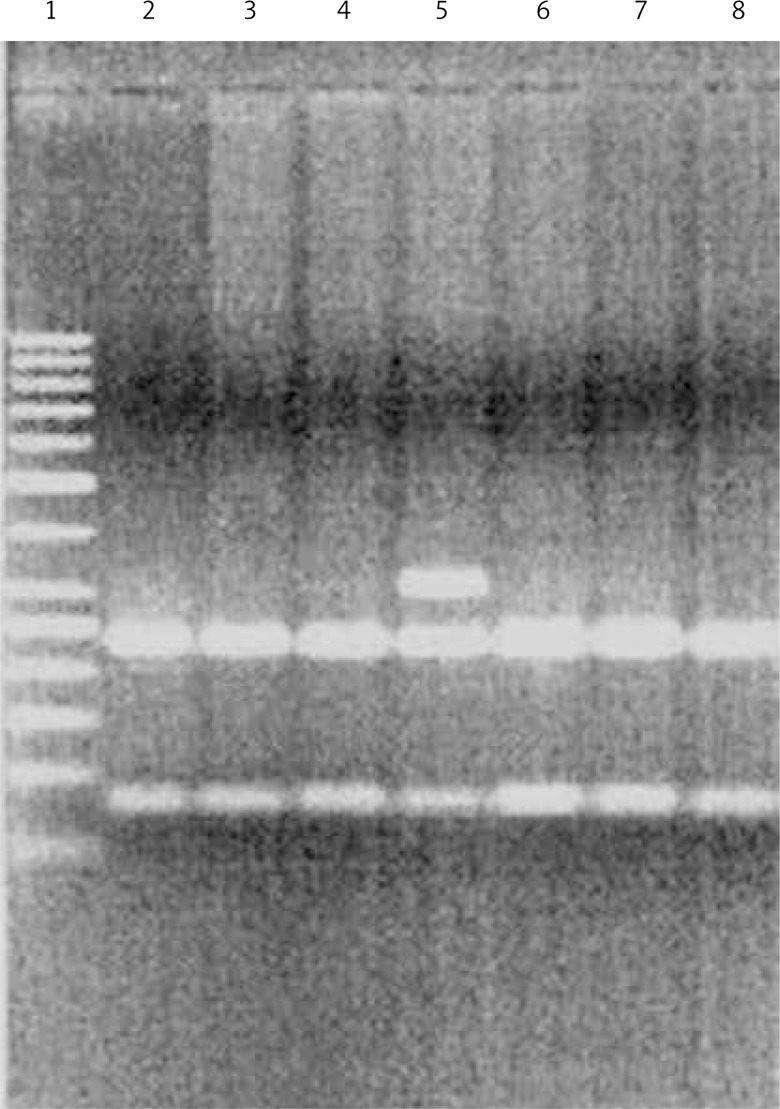
DNA was extracted from peripheral blood lymphocytes using DNA Blood Mini Kits (A&A Biotechnology, Gdynia, Poland). Genotypes were determined by PCR-RFLP. The procedure was carried out in a volume of 10 μl. The reaction mixture consisted of 100 ng of genomic DNA, 0.5 μmol of each primer and 3 U of Taq polymerase. The PCR cycling conditions consisted of an initial denaturation step of 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, at the annealing temperature of 60°C, and at 72°C for 30 s and then a final extension of 72°C for 5 min. The 10 μl of specifically positive 252 bp PCR products was digested overnight with 1 μl of HinfI (New England Bio Labs, Beverly, MA) at 37°C, and the digested DNA fragments were resolved on 3% agarose gel. The Asp allele was digested into 70 and 182 bp fragments, whereas the Gly variant remained intact (Figures 2, 3, Table I).
Figure 2.
Analysis of genotype polymorphism Gly322Asp gene hMSH2 after hydrolysis with restriction enzyme HinfI. Track 1 marker tracks 1, 2, 3, 4, 5, 6, 7 homozygous GG
Figure 3.
Analysis of genotype polymorphism Gly322Asp gene hMSH2 after hydrolysis with restriction enzyme HinfI. Track 1 marker, lane 5, heterozygous AG 2, 3, 4, 6, 7, 8, is heterozygous AA
Table I.
Primers, length of PCR products and restriction enzymes
| Polymorphism | Primers | Product length [bp] | Restriction enzyme | |
|---|---|---|---|---|
| MLH1 –93G>A (rs1800734) |
Sense Antisense |
5’-CCGAGCTCCTAAAAACGAAC-3’ 5’-CTGGCCGCTGGATAACTTC-3’ |
387 |
PvuII (37°C) |
| MSH2 Gly322Asp (rs4987188) |
Sense Antisense |
5’-GTTTTCACTAATGAGCTTGC-3’ 5’-AGTGGTATAATCATGTGGGT-3’ |
252 |
HinfI (37°C) |
Statistical analysis
The frequency of the analyzed polymorphisms was evaluated using the Hardy-Weinberg equilibrium test among CRC patients and case controls. The odds ratios (OR) and 95% confidence intervals (95% CI) were adjusted for age and gender. Means were compared by t-test or analysis of variance. Logistic regression analysis was performed to estimate the statistical association between SNP genetic variants and the risk of colon cancer. The χ2 test with Yates’ correction was used to analyze the distribution of the alleles according to age, gender, tumor location, grading, nodal involvement, distant metastases and recurrence. P-values were calculated as two-sided. Probabilities were considered significant at p-values less than 0.05.
Results
For MLH1 –93G>A, the G/G genotype was found in 74 patients with colon cancer (51.4%), A/G in 45 (31.2%) patients and A/A in 25 (17.4%) patients, while for the control subjects, G/G was found in 53 subjects (35.1%), A/G in 61 (40.4%) and A/A in 37 (24.5%). The G/G genotype was found to be associated with an increased risk of sporadic colon cancer in patients compared with control cases (OR = 2.07; 95% CI: 1.11–3.83; p < 0.02); however, no such influence was found for A/G heterozygotes (OR = 1.09; 95% CI: 0.58–2.05; p = 0.7).
For MSH2 Gly322Asp, neither homozygous A/A (OR = 1.44; 95% CI: 0.32–6.56; p = 0.7) nor A/G (OR = 2.70; 95% CI: 0.52–14.17; p = 0.2) was found to have any influence on the risk of sporadic cancer in the study population. A more detailed distribution of genotypes and alleles for the case group and control group is shown in Table II.
Table II.
MSH2 Gly322Asp and MLH1 -93G>A polymorphisms and the risk of colon cancer in studied population
| Genotype/allele | Case patients, n (%) | Control subjects, n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| Total | 144 (100) | 151 (100) | – | – |
| MSH2 Gly322Asp: | ||||
| G/G | 135 (93.8) | 146 (96.7) | 1.00 (referent) | |
| A/G | 5 (3.5) | 2 (1.3) | 2.70 (0.52–14.17) | 0.2 |
| A/A | 4 (2.7) | 3 (2.0) | 1.44 (0.32–6.56) | 0.7 |
| G | 275 (95) | 294 (97.4) | 1.00 (referent) | |
| A | 13 (5) | 8 (2.6) | 1.74 (0.71–4.26) | 0.2 |
| MLH1 – 93G>A: | ||||
| A/A | 25 (17.4) | 37 (24.5) | 1.00 (referent) | |
| A/G | 45 (31.2) | 61 (40.4) | 1.09 (0.58–2.05) | 0.7 |
| G/G | 74 (51.4) | 53 (35.1) | 2.07 (1.11–3.83) | 0.02 |
| A | 95 (33.0) | 135 (44.7) | 1.00 (referent) | |
| G | 193 (67.0) | 167 (55.3) | 1.66 (1.19–2.32) | 0.002 |
In the studies on colon cancer patients, the frequency of the MLH1-93G>A polymorphisms was higher in women than in men: 28 (38.4%) vs. 17 (23.9%) for A/G and 16 (21.9%) vs. 9 (12.7%) for A/A, and the distribution differed significantly (p = 0.034).
The relationship between tumor location and the distribution of the MLH1 –93G>A polymorphisms was also examined. In left-side colon tumors, the frequency of the A/G genotype was significantly higher than in right-sided tumors, 38 (39.6%) vs. 7 (14.6%), but the percentage of the A/A genotype was only slightly lower in left colon cancer, 16 (16.6%) vs. 9 (18.7%) (p = 0.014).
In recurrent disease, the frequency of the A/G genotype was significantly lower than in primary tumor, 1 (5.6%) vs. 44 (34.9%), but A/A homozygotes were more frequent in recurrent tumors; 7 (38.9%) vs. 18 (14.3%), p = 0.022.
For the MLH1 –93G>A polymorphism, no other significant differences were observed with regard to other pathological or clinical factors. All details are given in Table III.
Table III.
MLH1 –93G>A polymorphisms and analyzed clinical features of patients with non-familial colon cancer
| Clinical feature | N | MLH1 genotype, n (%) | Yates’ χ2 | P-value | |||
|---|---|---|---|---|---|---|---|
| GG | AG | AA | |||||
| Total | 144 | 74 (51.4) | 45 (31.2) | 25 (17.4) | N/A | N/A | |
| Gender: | |||||||
| Male | 71 | 45 (63.4) | 17 (23.9) | 9 (12.7) | 7.52 | 0.034 | |
| Female | 73 | 29 (39.7) | 28 (38.4) | 16 (21.9) | |||
| Age: | |||||||
| < 50 | 21 | 13 (61.9) | 5 (23.8) | 3 (14.3) | 0.523 | 0.769 | |
| > 50 | 123 | 61 (49.6) | 40 (32.5) | 22 (17.9) | |||
| Tumor location: | |||||||
| Right colon | 48 | 32 (66.7) | 7 (14.6) | 9 (18.7) | 8.47 | 0.014 | |
| Left colon | 96 | 42 (43.8) | 38 (39.6) | 16 (16.6) | |||
| Tumor grading: | |||||||
| Well | 15 | 9 (60.0) | 4 (26.7) | 2 (13.3) | |||
| Moderate | 114 | 58 (50.9) | 36 (31.6) | 20 (17.5) | 0.115 | 0.998 | |
| Poor | 15 | 7 (46.7) | 5 (33.3) | 3 (20.0) | |||
| Nodal involvement: | |||||||
| Yes | 98 | 49 (50.0) | 29 (29.6) | 20 (20.4) | 1.31 | 0.519 | |
| No | 46 | 25 (54.3) | 16 (34.8) | 5 (10.9) | |||
| Distant metastases: | |||||||
| Yes | 25 | 18 (72.0) | 5 (20.0) | 2 (8.0) | 3.81 | 0.149 | |
| No | 119 | 56 (47.1) | 40 (33.6) | 23 (19.3) | |||
| Recurrent disease: | |||||||
| Yes | 18 | 10 (55.5) | 1 (5.6) | 7 (38.9) | 7.63 | 0.022 | |
| No | 126 | 64 (50.8) | 44 (34.9) | 18 (14.3) | |||
Due to the small number of examples of A/G and A/A of polymorphism MSH2 Gly322Asp, the numbers were summarized. Higher frequencies of the A/G plus A/A genotypes of MSH2 Gly322Asp were found in recurrent than in primary disease: 4 (22.2%) vs. 5 (4.0%) (p = 0.001). No other clinical or pathological factors influenced the SNP genotype distribution (Table IV).
Table IV.
MSH2 Gly322Asp polymorphisms and analyzed clinical features of patients with non-familial colon cancer
| Clinical feature | N | MSH2 genotype, n (%) | Yates’ χ2 | P-value | |
|---|---|---|---|---|---|
| GG | AA + AG | ||||
| Total | 144 | 135 (93.8) | 9 (6.2) | N/A | N/A |
| Gender: | 0.42 | 0.518 | |||
| Male | 71 | 68 (95.8) | 3 (4.2) | ||
| Female | 73 | 67 (91.8) | 6 (8.2) | ||
| Age: | 0.76 | 0.856 | |||
| < 50 | 21 | 20 (95.2) | 1 (4.8) | ||
| > 50 | 123 | 115 (93.5) | 8 (6.5) | ||
| Tumor location: | 0.13 | 0.715 | |||
| Right colon | 48 | 44 (91.7) | 4 (8.3) | ||
| Left colon | 96 | 91 (94.8) | 5 (5.2) | ||
| Tumor grading: | 0.64 | 0.727 | |||
| Well | 15 | 14 (93.3) | 1 (6.7) | ||
| Moderate | 114 | 108 (94.7) | 6 (5.3) | ||
| Poor | 15 | 13 (86.7) | 2 (13.3) | ||
| Nodal involvement: | 0.08 | 0.781 | |||
| Yes | 98 | 92 (93.9) | 6 (6.1) | ||
| No | 46 | 43 (93.5) | 3 (6.5) | ||
| Distant metastases: | 0.73 | 0.394 | |||
| Yes | 25 | 22 (88.0) | 3 (12.0) | ||
| No | 119 | 113 (94.6) | 6 (5.4) | ||
| Recurrent disease: | 12.34 | 0.001 | |||
| Yes | 18 | 14 (77.8) | 4 (22.2) | ||
| No | 126 | 121 (96.0) | 5 (4.0) | ||
Discussion
This case-control study analyzes two SNPs of the MLH1 and MSH2 mismatch repair genes and their influence on malignant transformation in the human colon. It also investigates the associations between genetic characteristics, the clinical and pathological factors of the tumor and the genetic models of the studied SNPs.
Some rare constitutional mutations and methylations of MMR genes (including MLH1 and MSH2) are known to be primary causes of autosomal dominant disorders, such as hereditary non-polyposis colorectal cancer (HNPCC). However, the study focused on the properties of SNPs in patients with sporadic colon cancer, as the presence of SNPs in the MMR genes is associated with a higher risk of colon cancer, with low to moderate penetration in some populations with only sporadic tumors [6, 7].
As two transcription binding sites, NF-IL6 and GT-IIB, exist in this promoter region of the MLH1 gene, the –93G>A polymorphism is functional and may alter MLH1 transcription and expression, thereby changing DNA repair abilities. The studied SNP of the MLH1 gene is located within its promoter region, which accounts for the maximal activity of MLH1 transcription [8].
The –93G>A polymorphism was found to be associated with a higher risk of colon cancer in our studied population. Many previous studies have found SNPs to be associated with increased risk of CRC [9–11]; however, results across other studies have been inconclusive. A meta-analysis by Whiffin et al. found that the –93G>A polymorphism (heterozygous genotype) increased the risk of CRC in a population consisting of 14121 CRC cases and 10 890 controls [12]. Another meta-analysis by Pan et al. did not identify any such association, but the group included in the study seemed to have incorrect information: the control group included some individuals with no proven absence of CRC history, which could have influenced the results [13]. A meta-analysis of six case-controlled studies published in 2012 by Wang et al. found that the presence of the –93G>AMLH1 polymorphism in two genetic patterns, A/A vs. G/G and AA/AG vs. G/G, was significantly associated with a higher risk of CRC. The population included in the analysis was large and consisted of 17 791 CRC cases and 13 782 controls [8].
As in the present study, Martinez-Urueña et al. also found a statistically significant relationship between a homozygous –93G>A polymorphism in the MLH1 gene and the risk of sporadic CRC. In addition, the relationship was absent in patients with a family history of CRC [14].
The present study also tried to determine whether the presence of the Gly322Asp polymorphism of MSH2 was associated with a higher risk of CRC. Although no such association was reported in a previous study, the analysis was insufficient for such conclusions and the study group included HNPCC patients [15]. Smolarz et al. recently reported that one of the genetic variants of Gly233Asp decreased the risk of triple-negative breast cancer in a Polish population [16] – hence the choice of the same SNP for the present study, which is based on an analysis of colon cancer patients without a family history of cancer taken from a similar population. No significant association was found between any of the genetic models of Gly233Asp MSH2 and the risk of colon cancer in our group. Nevertheless, although no significant correlation was found, further studies are merited due to the fact that few such studies have been published, especially on sporadic colorectal cancer patients.
The present study attempts to analyze the distribution of polymorphism genotypes according to various clinical factors. In the 1990s, Bufill proposed division of the colon into a right and left-sided organ, believing that anatomical factors can influence the clinical differences observed in CRC tumors, as well as the known genetic, biological and pathological factors [17]. Many authors have since found that left- and right-sided colon cancer may be considered two different disease entities with differing disease spread, length of outcome and prognosis [18, 19]. A fuller understanding of the basis of these differences would be valuable in that it might influence the treatment or screening of patients with colon cancer. In accordance with other studies, our findings indicate that right and left-sided colon cancers differ with regard to the distribution of the genotype of the MLH1 gene SNP [5, 10, 20]; these relationships should also be taken into account in further studies. However, no such relationships were found between the distribution of the MSH2 Gly322Asp SNP and tumor location. Unfortunately, due to the paucity of analyses available in the literature, it is impossible to compare our findings with others.
We observed a relationship between the SNP genotype for MLH1 and sex, as well as with the presence of recurrent disease. The A allele was more common in the studied women. As, generally speaking, men are known to be slightly more likely to develop colorectal cancer than women, it is possible that the presence of the A allele, among other factors, can have a protective influence and reduce the risk of CRC in this group of individuals. What makes our findings particularly is the fact that the subjects are not divided according to gender in molecular research. The findings need to be evaluated on the basis of a larger study group.
We also found that patients with recurrent disease were significantly more likely to possess a copy of the A allele for the MLH1 gene SNP than patients with only a primary tumor. These results also should be re-evaluated using larger study groups, together with other factors as part of a multivariate analysis.
In the MSH2 Gly322Asp polymorphism, the frequency of the A allele was found to be higher in recurrent than in primary disease. However, due to the small number of cases in the group, further studies are necessary to gain a fuller picture of the situation.
One limitation of the study is its small sample size. However, the homogeneity of sporadic colon cancer cases and controls can be an advantage for genetic evaluation, especially in the analysis of polymorphisms. A greater insight is needed into the relationships between the SNPs of studied MMR genes and clinical factors. Although any further analyses should be based on larger sample sizes, the best study format for genetic assessments, precise clinical databases, are sometimes very difficult to obtain for population-based studies with large numbers of cases.
In conclusion, the MLH1 –93G>A polymorphism plays an important role in evaluating the risk of sporadic colon cancer not only in HNPCC patients. It may also be used as an indicator in some groups of patients with left-sided and recurrent tumors.
The MSH2 Gly322Asp polymorphism may also be used as a marker in patients with high risk of recurrent disease. However, this needs to be confirmed in further studies and by multivariate analysis.
Acknowledgments
Michal Mik and Lukasz Dziki equally contributed to this paper.
This study was supported by grant no. N N403 250340 and N N402 422138 from the Polish Ministry of Science and Higher Education.
The paper was presented during the 9th International European Federation for Colorectal Cancer, Vienna, Austria, 9–11 April, 2015
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics. CA Cancer J Clin. 2002;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Charames GS, Bapat B. Genomic instability and cancer. Curr Mol Med. 2003;3:589–96. doi: 10.2174/1566524033479456. [DOI] [PubMed] [Google Scholar]
- 3.Ford BN, Ruttan CC, Kyle VL, et al. Identification of single nucleotide polymorphisms in human DNA repair genes. Carcinogenesis. 2000;21:1977–81. doi: 10.1093/carcin/21.11.1977. [DOI] [PubMed] [Google Scholar]
- 4.Malhotra P, Anwar M, Kochhar R, et al. Promoter methylation and immunohistochemical expression of hMLH1 and hMSH2 in sporadic colorectal cancer: a study from India. Tumour Biol. 2014;35:3679–87. doi: 10.1007/s13277-013-1487-3. [DOI] [PubMed] [Google Scholar]
- 5.Nejda N, Iglesias D, Moreno Azcoita M, et al. A MLH1 polymorphism that increases cancer risk is associated with better outcome in sporadic colorectal cancer. Cancer Genet Cytogenet. 2009;193:71–7. doi: 10.1016/j.cancergencyto.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Fearnhead NS, Wilding JL, Winney B, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci USA. 2004;101:15992–7. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipkin SM, Rozek LS, Rennert G, et al. The MLH1 D132H variant is associated with susceptibility to sporadic colorectal cancer. Nat Genet. 2004;36:694–9. doi: 10.1038/ng1374. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Liu Y, Sima L, et al. Association between MLH1 -93G>a polymorphism and risk of colorectal cancer. PLoS One. 2012;7:e50449. doi: 10.1371/journal.pone.0050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell PT, Curtin K, Ulrich CM, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58:661–7. doi: 10.1136/gut.2007.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raptis S, Mrkonjic M, Green RC, et al. MLH1 -93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J Natl Cancer Inst. 2007;99:463–74. doi: 10.1093/jnci/djk095. [DOI] [PubMed] [Google Scholar]
- 11.Samowitz WS, Curtin K, Wolff RK, et al. The MLH1 -93 G>A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes Chromosomes Cancer. 2008;47:835–44. doi: 10.1002/gcc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiffin N, Broderick P, Lubbe SJ, et al. MLH1-93G>A is a risk factor for MSI colorectal cancer. Carcinogenesis. 2011;32:1157–61. doi: 10.1093/carcin/bgr089. [DOI] [PubMed] [Google Scholar]
- 13.Pan XM, Yang WZ, Xu GH, et al. The association between MLH1 -93 G>A polymorphism of DNA mismatch repair and cancer susceptibility: a meta-analysis. Mutagenesis. 2011;26:667–73. doi: 10.1093/mutage/ger032. [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Urueña N, Macías L, Pérez-Cabornero L, et al. Incidence of -93 MLH1 promoter polymorphism in familial and sporadic colorectal cancer. Colorectal Dis. 2013;15:e118–23. doi: 10.1111/codi.12112. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Stathopoulos P, Lindblom P, et al. MSH2 codon 322 Gly to Asp seems not to confer an increased risk for colorectal cancer susceptibility. Eur J Cancer. 1998;34:1981.. doi: 10.1016/s0959-8049(98)00217-2. [DOI] [PubMed] [Google Scholar]
- 16.Smolarz B, Makowska M, Samulak D, et al. Gly322Asp and Asn127Ser single nucleotide polymorphisms (SNPs) of hMSH2 mismatch repair gene and the risk of triple-negative breast cancer in Polish women. Fam Cancer. 2015;14:81–8. doi: 10.1007/s10689-014-9746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–88. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 18.Benedix F, Meyer F, Kube R, et al. Right- and left-sided colonic cancer – different tumour entities. Zentralbl Chir. 2010;135:312–7. doi: 10.1055/s-0030-1247471. [DOI] [PubMed] [Google Scholar]
- 19.Jess P, Hansen IO, Gamborg M, et al. BMJ Open. 2013. Danish Colorectal Cancer Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyakura Y, Tahara M, Lefor AT, et al. Haplotype defined by the MLH1-93G/A polymorphism is associated with MLH1 promoter hypermethylation in sporadic colorectal cancers. BMC Res Notes. 2014;24:835. doi: 10.1186/1756-0500-7-835. [DOI] [PMC free article] [PubMed] [Google Scholar]