Abstract
Iron is an essential trace element, but it is also toxic in excess, and thus mammals have developed elegant mechanisms for keeping both cellular and whole-body iron concentrations within the optimal physiologic range. In the diet, iron is either sequestered within heme or in various nonheme forms. Although the absorption of heme iron is poorly understood, nonheme iron is transported across the apical membrane of the intestinal enterocyte by divalent metal-ion transporter 1 (DMT1) and is exported into the circulation via ferroportin 1 (FPN1). Newly absorbed iron binds to plasma transferrin and is distributed around the body to sites of utilization with the erythroid marrow having particularly high iron requirements. Iron-loaded transferrin binds to transferrin receptor 1 on the surface of most body cells, and after endocytosis of the complex, iron enters the cytoplasm via DMT1 in the endosomal membrane. This iron can be used for metabolic functions, stored within cytosolic ferritin, or exported from the cell via FPN1. Cellular iron concentrations are modulated by the iron regulatory proteins (IRPs) IRP1 and IRP2. At the whole-body level, dietary iron absorption and iron export from the tissues into the plasma are regulated by the liver-derived peptide hepcidin. When tissue iron demands are high, hepcidin concentrations are low and vice versa. Too little or too much iron can have important clinical consequences. Most iron deficiency reflects an inadequate supply of iron in the diet, whereas iron excess is usually associated with hereditary disorders. These disorders include various forms of hemochromatosis, which are characterized by inadequate hepcidin production and, thus, increased dietary iron intake, and iron-loading anemias whereby both increased iron absorption and transfusion therapy contribute to the iron overload. Despite major recent advances, much remains to be learned about iron physiology and pathophysiology.
Keywords: anemia, ferritin, hemochromatosis, hepcidin, iron deficiency, iron overload, iron physiology, transferrin
INTRODUCTION
The field of iron homeostasis is a very active one, and each year thousands of papers are published in this area. The goal of this paper is not to provide a comprehensive overview of iron metabolism but to offer some basic information on cellular and body iron physiology and pathophysiology, to summarize where the field stands at the present time, and to highlight some of the remaining unanswered questions that require further investigation.
Although some of the proteins that are involved in iron homeostasis, such as ferritin and transferrin, have been known for many decades, much of our understanding of the molecular basis of iron metabolism has come in the past 15–20 y. A particularly prolific 5-y period from 1996 to 2001 led to the identification of many of the key proteins in this field, including the iron-import protein divalent metal-ion transporter 1 (DMT1), the iron export protein ferroportin 1 (FPN1), and the “master” regulator of iron homeostasis, the liver-derived peptide hepcidin (1). Although a number of important discoveries have been made since that time, much of what we have learned over the past 15 y has added layers of complexity onto the basic transport and regulatory processes that were established earlier. The unraveling of the regulation of hepcidin has been particularly dominant in this field. These molecular advances have underpinned our understanding of whole-body physiology and pathophysiology, but in turn, the analysis of iron-related disorders has been a major driving force in defining the molecular mechanisms.
BASIC IRON PHYSIOLOGY
Dietary iron
After birth, and excluding exogenous therapeutic sources, all iron enters the body from the diet. Dietary iron is usually considered as either heme or nonheme iron (2). Nonheme iron is abundant in foods of both animal and plant origins and is the dominant form of iron in plants. Nonheme iron is found in a wide variety of forms and includes soluble iron, iron in low–molecular-weight complexes, storage iron in ferritin, and iron in the catalytic centers of a wide range of other proteins. Much of this iron is not tightly sequestered, and consequently its bioavailability can be affected by a range of dietary constituents and luminal factors. The low pH of the stomach and proximal small intestine helps to keep iron in a soluble form, thereby making it available for absorption. Small organic acids such as citric acid and ascorbic acid also help to keep nonheme iron in a reduced and soluble form and can greatly enhance its absorption. Other dietary components, notably plant-derived phytates, tannins, and polyphenols, can bind nonheme iron and impede its absorption. In contrast, heme iron is tightly sequestered within a protoporphyrin ring and is not accessible to the factors that influence nonheme iron. As a consequence, heme iron tends to be absorbed more efficiently and its absorption is less dependent on the composition of the diet. Most heme iron in the diet is from myoglobin and hemoglobin and is animal derived.
Intestinal iron absorption
Iron is absorbed by the mature enterocytes of the midupper villus and mainly in the small intestine (3) (Figure 1). Although small amounts of iron can be absorbed by the more distal parts of the gastrointestinal tract, the proximal parts of the small intestine (the duodenum and first part of the jejunum) are particularly adapted for this role.
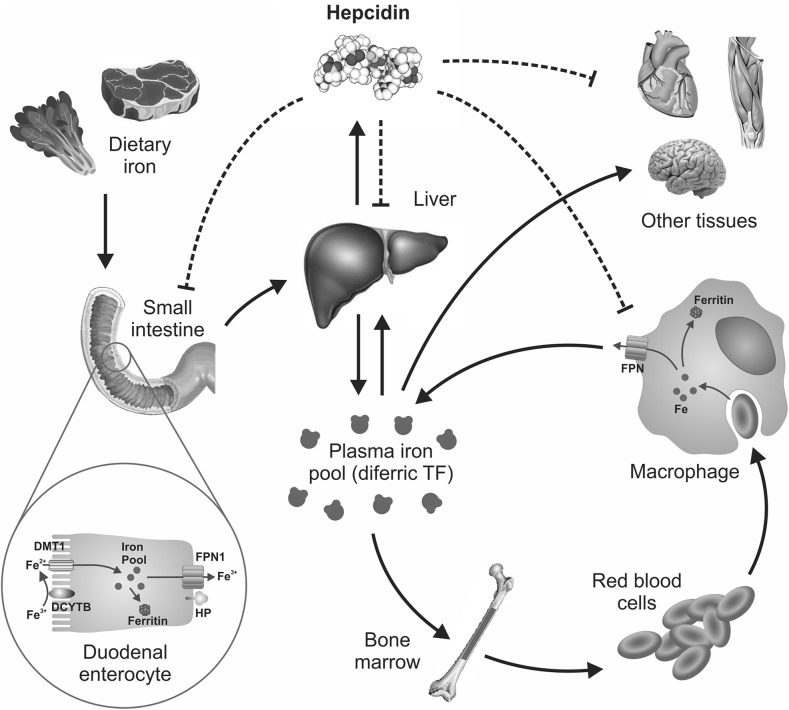
FIGURE 1.
Body iron homeostasis. Iron is present in the diet in both heme and nonheme forms. Although the mechanisms underlying heme absorption are poorly understood, nonheme iron enters the circulation after traversing the enterocyte apical membrane via DMT1 and the basolateral membrane via FPN1. Iron binds to plasma TF and is distributed to tissues throughout the body. Quantitatively, most iron is used by immature red blood cells in the bone marrow for hemoglobin production. Senescent erythrocytes are phagocytosed by macrophages, and the iron is released from catabolized hemoglobin and re-enters the circulation. The liver-derived peptide hepcidin plays a critical role in the regulation of body iron intake and distribution by binding to plasma membrane FPN1 on enterocytes, macrophages, and most body cells and facilitating its internalization and degradation. Hepcidin, in turn, is regulated by body iron demand. Thus, when the body is iron deficient, hepcidin concentrations are low, thereby favoring iron absorption and delivery to the plasma from storage sites; but when the body is iron replete, a higher hepcidin concentration reduces iron absorption and impairs iron release from stores. DCYTB, duodenal cytochrome b; DMT1, divalent metal-ion transporter 1; FPN, ferroportin; HP, hephaestin; TF, transferrin.
To move from the lumen of the intestine into the bloodstream, iron must cross both the apical brush-border membrane and the basolateral membrane of enterocytes (Figure 1). Nonheme iron traverses the brush-border membrane via DMT1 (4). This transporter requires ferrous iron (Fe2+) as a substrate, but most dietary iron is in the ferric (Fe3+) form. Thus, iron needs to be reduced before it can be absorbed. Duodenal cytochrome B is one potential brush-border reductase (5), but there are likely to be others. If iron in the enterocyte is not immediately required by the body, it becomes sequestered in the cell within the iron-storage protein ferritin only to be lost from the body once the enterocyte is sloughed at the end of its several day life span. If the iron is required, it can be exported rapidly across the enterocyte basolateral membrane via FPN1 (6). The efficiency of the basolateral export of iron is greatly enhanced by the copper-dependent iron oxidase hephaestin, which converts newly transported Fe2+ to the Fe3+ form (7).
Very little is known definitively about the absorption of heme iron (3). It is presumed to bind to the enterocyte brush border intact and, then, is likely endocytosed, but the molecular details of this process are unknown to our knowledge. Once within the enterocyte, it is considered that iron is released from heme through the action of heme oxygenases and that this iron is subsequently exported from the cells via FPN1 (i.e., the same pathway as nonheme iron). Iron may also traverse the small intestine in other forms [e.g., as ferritin (8)], but again, the mechanisms involved are unknown to our knowledge.
Systemic iron transport and its delivery to tissues
Recently absorbed iron (or iron that is released from storage sites) is bound to plasma transferrin which distributes it around the body to sites of utilization (9) (Figure 1). Each transferrin molecule can bind ≤2 atoms of iron. Under normal circumstances, only ∼30% of the iron-binding sites on the plasma transferrin pool are occupied at any one time [i.e., a transferrin saturation (TSAT) of 30%], thereby providing considerable buffering capacity against the appearance of potentially toxic non–transferrin-bound iron (NTBI). However, in iron-loading diseases, transferrin frequently becomes saturated, and the concentration of NTBI can be dangerously high (10). TSAT can provide a useful index of iron supply to the bone marrow, and TSAT less than ∼16% is correlated with a reduced production of new red blood cells (RBCs) (11). However, the mechanism involved is more complex than a simple restriction of iron supply for heme (and, hence, hemoglobin) production and also involves the influence of low iron (among other signals) on various signaling pathways in developing erythroid cells (12).
Diferric transferrin delivers iron to cells by binding to transferrin receptor (TfR) 1 on the plasma membrane (9) (Figure 2). The transferrin-TfR1 complex is internalized via clathrin-mediated endocytosis. The endosome is acidified, and a combination of low pH, a conformation change in transferrin that is associated with its binding to its receptor, and a reduction of transferrin-bound Fe3+ via an enzyme of the 6-transmembrane epithelial antigen of the prostate family of reductases (6-transmembrane epithelial antigen of the prostate 3 in the case of immature erythroid cells) releases iron from transferrin (13). This iron moves into the cytoplasm across the endosomal membrane via DMT1 (14). The fate of this iron depends on the iron requirements of the cell. If iron is needed for metabolic functions, it may traffic directly to sites of utilization such as the mitochondria. If iron is not immediately required, it may be sequestered within the iron-storage protein ferritin for later use. The cell may also divest itself of iron via export through FPN1. This export pathway can be invoked as a safety valve if very large amounts of iron accumulate within the cell. In enterocytes, the efficiency of cellular iron export is enhanced by the iron oxidase hephaestin, but in most body cells, this role is fulfilled by the circulating hephaestin homolog ceruloplasmin (15).
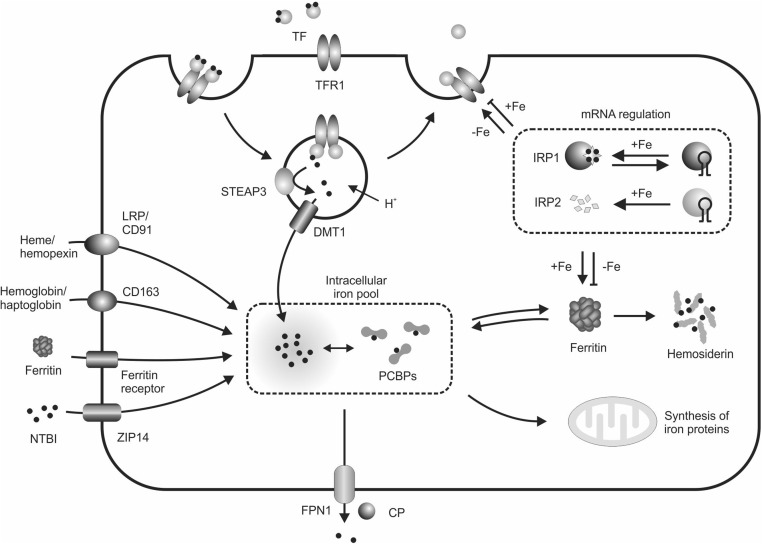
FIGURE 2.
Cellular iron homeostasis. Cells can take up iron in a variety of forms, but all nucleated cells have the capacity to use TF-bound iron. Diferric TF binds to TFR1 on the plasma membrane, and the complex is internalized in endosomes. Acidification of the endosome, which is accompanied by iron reduction by a member of the STEAP family of proteins, releases iron from TF, which subsequently moves across the endosomal membrane via DMT1 and into the cytoplasm. The precise form of this cytosolic iron pool is unclear, but at least some iron is bound by PCBP proteins that act as iron chaperones. These complexes can deliver iron to newly synthesized iron-containing proteins (although whether they can deliver iron to mitochondria is unclear) and to the iron-storage protein ferritin. Iron in excess of cellular needs may be exported through FPN1 with the iron oxidase CP increasing the efficiency of this process. Some cell types can take up iron in other forms, including NTBI, or iron that is contained within ferritin, heme, or hemoglobin. Cellular iron intake and storage are regulated by the iron-responsive element and IRP system such that low iron concentrations favor the synthesis of more TFR1 and suppress ferritin expression, whereas high cellular iron leads to an increase in ferritin concentrations and a decrease in TFR1. CP, ceruloplasmin; DMT1, divalent metal-ion transporter 1; FPN1, ferroportin 1; IRP, iron regulatory protein; LRP, LDL-receptor–related protein; mRNA, messenger RNA; NTBI, non–transferrin-bound iron; PCBP, poly(rC)-binding protein; STEAP, 6-transmembrane epithelial antigen of the prostate; TF, transferrin; TFR1, transferrin receptor 1; ZIP14, Zrt/Irt-like protein 14.
Although diferric transferrin is an iron source that can be used by most, and perhaps all, body cells, it is not the only source of cellular iron (Figure 2). NTBI can be taken up very efficiently by many cell types, and it has recently been shown that Zrt/Irt-like protein 14 is an important NTBI transporter (16). There is some evidence that ferritin can deliver its iron to cells as well (17). In pathologic situations that are characterized by hemolysis, released heme and hemoglobin can bind to circulating hemopexin and haptoglobin, respectively, and these complexes can also be taken up by certain cell types (18). These pathways are important for salvaging and reutilizing iron. The relative importance of different iron-uptake pathways varies between cell types. For example, immature erythroid cells are almost completely dependent on the TfR1-mediated endocytosis of diferric transferrin, but the liver utilizes a range of other pathways as well (9).
Intracellular iron trafficking and storage
Free iron in solution is quite toxic, and in biological situations, it is almost always tightly sequestered by proteins (9). It would be expected that the cytoplasm would contain iron-binding proteins, or chaperones, to move iron around the cell. However, it is only relatively recently that the poly(rC)-binding proteins have been identified as intracellular iron chaperones (19) (Figure 2). The poly(rC)-binding proteins have been shown to deliver iron to ferritin and several enzymes, but it is likely that other, as yet undiscovered, chaperones also exist within cells.
Iron storage is a critical component of cellular iron homeostasis, which enables iron to be sequestered in a nontoxic form but also provides a reservoir from which iron can be used for future metabolic needs. Ferritin is the major intracellular iron-storage protein (20) (Figure 2). Ferritin is a large protein that consists of 24 subunits that are arranged to form a spherical shell with a large central cavity. Pores in the protein shell enable the entry and exit of iron, and a single ferritin molecular can hold ≤4500 atoms of iron. This high capacity, combined with the fact that the expression of ferritin greatly increases when cellular iron concentrations rise, provides the cell with an enormous ability to sequester iron. When high concentrations of iron-laden ferritin accumulate within the cell, the ferritin molecules aggregate, and ultimately, these aggregates fuse with lysosomes. This process leads to the degradation of ferritin, and the resulting mixture of Fe3+ cores and peptides is known as hemosiderin (20). This form of storage iron is particularly prominent in the cells of patients with iron-loading diseases. Iron can be efficiently mobilized from both ferritin and hemosiderin when it is required elsewhere in the body. Another important feature of ferritin is that small amounts are secreted from the cell, and the amount that is secreted strongly correlates with the concentration of intracellular iron. This association makes serum ferritin concentrations a readily measured and accurate indicator of body iron stores (20).
Regulation of cellular iron homeostasis
Cellular iron homeostasis is tightly regulated to maximize the iron supply when the cell has an iron deficit and to restrict the iron supply and promote storage when the cell is iron replete. This regulation can occur at different levels, but the best-understood system is the iron-dependent binding of iron regulatory proteins (IRPs) IRP1 and IRP2 to stem-loop structures [iron-responsive elements (IREs)] in the untranslated regions (UTRs) of the messenger RNAs (mRNAs) encoding various iron-related proteins (21) (Figure 2). When cellular iron concentrations are low, the IRPs are in their mRNA-binding conformations. The binding of IRPs to the 5′ UTR of the ferritin mRNA blocks translation, thus ensuring that little ferritin is produced when iron storage is not required. Concurrently, the IRPs bind to IREs in the 3′ UTR of the TfR1 mRNA, thereby protecting the message from degradation and enabling more TfR1 to be expressed on the cell membrane. This effect, in turn, enables the cell to maximize its iron intake. When cellular iron concentrations are high, the IRPs do not bind to IREs, thereby allowing translation of the ferritin mRNA to proceed and exposing the TfR1 mRNA to degradation. These conditions limit cellular iron uptake and promote storage. A range of other mRNAs contain IREs, including DMT1, FPN1, 5-aminolevulinic acid synthase 2, and hypoxia inducible factor 2α (21). The net effect of these changes is to ensure that the cells mount an appropriate physiologic response to variations in intracellular iron concentrations.
The fact that concentrations of ferritin and TfR1 vary with iron status has important diagnostic implications. It was noted previously that small amounts of ferritin are secreted from storage sites, and thus, the concentration of ferritin in the serum reflects that amount of stored iron. Small amounts of TfR1 can also be present in the serum because the extracellular domain of the protein can be proteolytically cleaved at the plasma membrane. The resulting soluble TfR is proportional to the complement of cell surface TfR1 in the body and is a useful index of iron deficiency (ID) because TfR1 concentrations rise when iron concentrations are low (22, 23).
Other modes of regulation have been less well studied but are very worthy of further attention. At the transcriptional level, factors such as hypoxia (24), cytokines, and hormones (25, 26) are known to regulate various iron-related genes. Even iron itself will exert some degree of regulation on various iron-related genes (27).
Systemic iron homeostasis
The exquisite regulation of iron homeostasis at the cellular level is replicated at the whole-body level. If an individual requires more iron, more iron will be mobilized from body iron stores and there will be an increase in intestinal iron absorption. However, if the body is iron replete, these processes will be downregulated. Extensive work over the past 15 y has shown that this regulation is mediated by the small liver-derived peptide hepcidin (1) (Figure 1). Variations in body iron demand are communicated to the liver and this, in turn, modulates the expression of hepcidin, which is encoded by the hepcidin antimicrobial peptide gene. Hepcidin is distributed via the circulation to its target sites where it binds to the iron export protein FPN1. Thus, FPN1 acts as the hepcidin receptor. This complex is ubiquitinated, internalized, and degraded with the consequence that the capacity of iron to be released into the circulation is impeded. Thus, when the body is iron replete, hepcidin concentrations are high, and the iron supply to the plasma is reduced; however, when iron demands are high, hepcidin concentrations are reduced, and more iron enters the circulation. Major iron-exporting cells include macrophages, which recycle heme-derived iron from senescent RBCs; hepatocytes, which are the major site of iron storage; and intestinal enterocytes, which facilitate dietary intake of iron. However, most cells are able to export iron through FPN1 and, thus, are potential hepcidin targets.
Hepcidin regulation has been studied extensively but nevertheless remains incompletely understood. The bone morphogenetic protein-SMAD regulatory pathway is central to hepcidin regulation, and it is likely the pathway through which the iron-dependent regulation of hepcidin occurs (1), but inflammatory cytokines (28), hypoxia (24), and several other factors (25, 26) also can activate signaling pathways that lead to alterations in hepcidin transcription. Increased iron stores and inflammation enhance hepcidin expression, whereas reduced iron stores and hypoxia lower expression.
Organ-specific iron metabolism
Different organs have divergent iron requirements, and many of the details of these differences are only now being appreciated at the molecular level. For example, considerable recent progress has been made in understanding iron homeostasis in the heart (29). Even within individual tissues, different cell types differ in their capacity to take up various forms of iron and vary in their capacity to store iron. The erythroid marrow has the highest iron requirement of any body tissue because a very large amount of iron is required for hemoglobin synthesis in new RBCs (30). Tissues with a high proliferative capacity (e.g., the rapidly dividing cells of the intestinal crypts) also require relatively large amounts of iron and, like immature erythroid cells, express abundant TfR1 on their plasma membrane (31). Thus, when the iron supply becomes limiting, the iron distribution to tissues becomes prioritized with erythropoiesis being particularly well protected (32). This situation is also seen in pregnancy when the iron supply to the fetus is prioritized at the expense of the mother (33).
Iron and the microbiome
A contemporary overview of iron homeostasis would be incomplete without some consideration of the influence of iron on the microbiome. The intestinal microbiome influences a broad range of metabolic and physiologic processes (34). Most bacteria require iron for growth and survival, and some bacteria have an absolute requirement for iron for survival. Consequently, multiple studies have now shown that iron fortification can alter the microbial profile in the gut, thereby promoting the growth of potentially pathogenic enterobacteria species at the expense of protective lactobacilli and bifidobacteria species (35, 36). Any consideration of iron supplementation or fortification needs to take into account their effects on the microbiome and potential downstream consequences.
PATHOPHYSIOLOGY OF IRON
Under normal physiologic conditions, body iron amounts range from ∼3 to 5 g. Deviations from this range can lead to either ID or iron overload and can have pathologic consequences (10, 37). An insufficient iron supply may impair the synthesis of essential iron-containing proteins that are required for normal cellular physiology and result in a range of adverse consequences (38). In contrast, excess iron may catalyze reactions that produce reactive oxygen species (10) and consequently cause oxidative damage to cells and tissues that can lead to tissue fibrosis and organ dysfunction long term.
ID
ID is one of the world’s most common nutritional deficiencies and affects >2 billion people (37). Many of these individuals are so iron deficient that RBC production is impaired, thus resulting in anemia. Insufficient dietary intake is the major cause of ID (37), and low iron bioavailability in a plant-based diet enhances susceptibility. In rare cases, ID can result from genetic disturbances in iron homeostasis. In particular, mutations in the TMPRSS6 gene, which encodes an upstream regulator of hepcidin, lead to iron-refractory ID anemia (39).
The treatment of mild ID can be achieved simply by increasing the amount of bioavailable iron in the diet (e.g., through increased red-meat iron intake) or by taking supplements (37, 40). Commonly used iron supplements, including ferrous sulfate, ferrous gluconate, and ferrous fumarate (41), can effectively replenish a depleted iron pool but, at high doses, can lead to significant gastrointestinal side effects (42). Effective parenteral iron supplements are also available (41), and in cases of severe ID, an intravenous iron infusion may be warranted to deliver a large amount of iron quickly.
Iron overload
Most primary iron overload has a genetic basis (43). Primary iron-overload syndromes, often referred to as hemochromatosis, result from mutations in genes that are involved in the transport of iron or its regulation. In contrast, secondary iron overload usually comes from transfusions that are used to treat certain inherited disorders such as the iron-loading anemias (e.g., β-thalassemia). Five main types of hemochromatosis are recognized that result from mutations in the genes encoding the hemochromatosis protein (type 1), hemojuvelin (type 2A), hepcidin (type 2B), TfR2 (type 3), and FPN1 (type 4) (43). Most of these are uncommon-to-rare conditions, but hemochromatosis protein–related hemochromatosis is relatively common in populations of Northern European origin (frequencies from 1:80 to 1:200) (44). The common feature of these disorders is reduced or negligible hepcidin expression, which results in an inability to limit the absorption of dietary iron as the body iron load increases. Iron accumulates in many organs, most notably in the liver, heart, and pancreas (10). Clinical consequences of this accumulation include hepatic fibrosis and cirrhosis, increased risk of hepatocellular carcinoma, cardiomyopathy, arthritis, and diabetes.
Iron overload has also been associated with other genetic defects (e.g., atransferrinemia, aceruloplasminemia, and Friedreich ataxia), chronic disorders (e.g., chronic liver disease and porphyria cutanea tarda), surgery (e.g., portacaval shunting), or disease treatments (e.g., transfusion therapy for myelodysplastic syndromes) (45). Furthermore, excess iron can influence the severity of a range of other diseases such as fatty liver disease (46), cystic fibrosis (47), and a wide range of neurologic disorders (48).
The treatment of iron overload depends on the underlying disease. Hemochromatosis is typically treated via highly effective and relatively inexpensive phlebotomy (10) with the removal of ∼0.5 g Fe/L blood. Subsequently, iron is mobilized from storage for erythropoiesis to replace removed RBCs. In iron-loading anemias, such as the thalassemias, phlebotomy is not appropriate, and therefore iron is removed with the use of iron-binding drugs that are known as iron chelators (49). After iron chelators bind iron, the iron-chelator complex is excreted in the urine or feces.
Because the hepcidin-FPN axis plays such an important role in regulating body iron intake and distribution, therapeutic approaches targeting this system have shown considerable potential (50, 51). Hepcidin mimetics or agents that increase hepcidin antimicrobial peptide gene expression have shown promise in the treatment of hemochromatosis and β-thalassemia, whereas hepcidin antagonists or agents that block the gene synthesis have application in the treatment of the anemia of inflammation. A number of these agents are now in clinical trials.
Anemia of chronic disease
The anemia of chronic disease or the anemia of inflammation is the second most common form of anemia after that caused by ID (52). In response to inflammatory situations such as infection, cancer, and various chronic inflammatory states, the plasma iron concentration decreases, and macrophages and other cell types sequester iron. If this condition persists, the iron supply to the erythroid marrow can be compromised, and anemia may result. At the molecular level, much of the reduction in plasma iron that accompanies the anemia of chronic disease can be explained by the stimulation of hepcidin production by proinflammatory cytokines. Higher hepcidin concentrations reduce cell-surface FPN1 and result in iron sequestration within cells. However, FPN1 expression may also be reduced independently of hepcidin (53) during inflammation.
CONCLUSIONS AND FUTURE RESEARCH DIRECTIONS
Our understanding of the role played by iron in biological systems has advanced enormously over the past 2 decades and continues to occupy a large number of researchers worldwide. Despite these advances, much information remains to be learned. A summary of some of the areas in which more work is required is provided in Table 1. These areas range from a better understanding of the molecular mechanisms underlying the function of known proteins of iron metabolism and the identification of new players to how the roles of these proteins are integrated at the whole-body level. Even relatively well-studied normal physiologic processes such as intestinal iron absorption, erythropoiesis, and iron supply during pregnancy and infancy remain incompletely understood. Defining the integration of iron metabolism under normal physiologic conditions is an essential forerunner to understanding how these processes are disturbed under pathological conditions. Although the main iron-related diseases have been known and broadly understood for some time, the potential adverse role iron can play in a range of other disorders (e.g., neurologic, cardiac, renal, and hepatic diseases) is increasingly recognized, and with our more sophisticated understanding of iron homeostasis, we are now much better placed to work toward rational therapies for these conditions.
TABLE 1.
Some important areas for future research on iron homeostasis1
| Area | Comment |
| Molecules and cellular processes | |
| Identification of novel iron-related proteins | There are undoubtedly undiscovered molecules including novel transporters and regulators (e.g., the proteins involved in dietary heme-iron absorption). |
| Definition of structure and function relations of proteins of mammalian iron metabolism | Structural information on many iron-related proteins and particularly membrane transporters is grossly inadequate. |
| Intracellular iron trafficking and homeostasis | Identifying and studying the role of intracellular iron chaperones and iron delivery to key organelles (e.g., mitochondria) are important areas. |
| Relations between iron and other metals | Some links are widely appreciated (e.g., the copper-dependent iron oxidases,) but others require further investigation (e.g., the key iron-transport proteins also transport other metals). |
| Regulation of genes encoding iron-related proteins | Important regulatory mechanisms are well known (e.g., the IRE-IRP system), but our understanding of transcriptional regulators and their mechanisms of action remains limited. |
| Factors regulating hepcidin | Some pathways are well defined, but others, such as the roles played by HFE and TfR2, require delineation. |
| Physiology | |
| Effects of subtle changes in iron status | Much of our current understanding of iron homeostasis is based on the study of extremes (e.g., ID vs. iron loading). |
| Consequences of supraphysiologic iron intake | Defining the effects of supplementation of iron-replete individuals has important practical implications. |
| Organ-specific iron homeostasis | Different organs and different cell types within organs handle iron differently, and our understanding in this area remains limited. |
| Prioritization of iron supply to different organs | Some tissues are relatively protected from ID or relatively susceptible to iron loading. Understanding the mechanisms underlying this phenomenon is important. |
| Links between iron and the microbiome | How iron alters the gut microbiome (and potentially those of other sites) is an important consideration in any studies of supplementation, and the microbial community composition could affect body iron physiology. |
| Assessing iron status | |
| Measurement of subtle changes in iron status and their effects | The best methods for measuring small changes in iron status are poorly understood as is the physiologic relevance of these changes. |
| New indicators and combinations of indicators | Recent molecular advances might be useful to identify new iron-status indicators or new combinations of indicators. |
| Inflammation | Inflammation remains a major influence on key measures of iron status, and how it is taken into account is an important issue. This is increasingly a problem as the level of obesity in the community rises. |
| Genetic profile | Genetic influences on iron status and new genetic modifiers continue to be identified. Consideration of genetic profiling may be useful to assess the risk of iron-related disorders and the response to treatment. |
| Treatments | |
| Oral iron supplements | Despite recent advances, more efficient oral iron supplements with fewer adverse side effects are still required. |
| Iron removal | The suite of clinically approved iron chelators remains very limited, and better compounds and more efficient delivery methods are needed. |
| Personalized medicine and nutrition | Contemporary genetic and physiologic knowledge might be used to tailor iron-related treatments (whether they be iron supplementation, iron redistribution, or iron removal) to individuals, specific life stages, or specific clinical conditions more effectively. |
HFE, hemochromatosis protein; ID, iron deficiency; IRE, iron-responsive element; IRP, iron regulatory protein; TfR2, transferrin receptor 2.
Acknowledgments
The authors’ responsibilities were as follows—GJA: conceived, wrote, and edited the manuscript and had primary responsibility for its final content; DMF: wrote and edited the manuscript and prepared the figures; and both authors: read and approved the final manuscript. Neither author reported a conflict of interest related to the study.
Footnotes
Abbreviations used: DMT1, divalent metal-ion transporter 1; FPN1, ferroportin 1; ID, iron deficiency; IRE, iron-responsive element; IRP, iron regulatory protein; mRNA, messenger RNA; NTBI, non–transferrin-bound iron; RBC, red blood cell; TfR, transferrin receptor; TSAT, transferrin saturation; UTR, untranslated region.
REFERENCES
- 1.Ganz T. Systemic iron homeostasis. Physiol Rev 2013;93:1721–41. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter CE, Mahoney AW. Contributions of heme and nonheme iron to human nutrition. Crit Rev Food Sci Nutr 1992;31:333–67. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua BK, Vulpe CD, Anderson GJ. Intestinal iron absorption. J Trace Elem Med Biol 2012;26:115–9. [DOI] [PubMed] [Google Scholar]
- 4.Gunshin H, Fujiwara Y, Custodio AO, Direnzo C, Robine S, Andrews NC. Slc11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J Clin Invest 2005;115:1258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, et al. . An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 2001;291:1755–9. [DOI] [PubMed] [Google Scholar]
- 6.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab 2005;1:191–200. [DOI] [PubMed] [Google Scholar]
- 7.Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet 1999;21:195–9. [DOI] [PubMed] [Google Scholar]
- 8.Theil EC, Chen H, Miranda C, Janser H, Elsenhans B, Nunez MT, Pizarro F, Schumann K. Absorption of iron from ferritin is independent of heme iron and ferrous salts in women and rat intestinal segments. J Nutr 2012;142:478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frazer DM, Anderson GJ. The regulation of iron transport. Biofactors 2014;40:206–14. [DOI] [PubMed] [Google Scholar]
- 10.Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med 2012;366:348–59. [DOI] [PubMed] [Google Scholar]
- 11.Bainton DF, Finch CA. The diagnosis of iron deficiency anemia. Am J Med 1964;37:62–70. [DOI] [PubMed] [Google Scholar]
- 12.Bullock GC, Delehanty LL, Talbot AL, Gonias SL, Tong WH, Rouault TA, Dewar B, Macdonald JM, Chruma JJ, Goldfarb AN. Iron control of erythroid development by a novel aconitase-associated regulatory pathway. Blood 2010;116:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet 2005;37:1264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA 1998;95:1148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp P. The molecular basis of copper and iron interactions. Proc Nutr Soc 2004;63:563–9. [DOI] [PubMed] [Google Scholar]
- 16.Jenkitkasemwong S, Wang CY, Coffey R, Zhang W, Chan A, Biel T, Kim JS, Hojyo S, Fukada T, Knutson MD. SLC39A14 is required for the development of hepatocellular iron overload in murine models of hereditary hemochromatosis. Cell Metab 2015;22:138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelvan D, Fibach E, Meyron-Holtz EG, Konijn AM. Ferritin uptake by human erythroid precursors is a regulated iron uptake pathway. Blood 1996;88:3200–7. [PubMed] [Google Scholar]
- 18.Anderson GJ, Frazer DM. Hepatic iron metabolism. Semin Liver Dis 2005;25:420–32. [DOI] [PubMed] [Google Scholar]
- 19.Leidgens S, Bullough KZ, Shi H, Li F, Shakoury-Elizeh M, Yabe T, Subramanian P, Hsu E, Natarajan N, Nandal A, et al. . Each member of the poly-r(C)-binding protein 1 (PCBP) family exhibits iron chaperone activity toward ferritin. J Biol Chem 2013;288:17791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theil EC. Ferritin: the protein nanocage and iron biomineral in health and in disease. Inorg Chem 2013;52:12223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson N, Pantopoulos K. The IRP/IRE system in vivo: insights from mouse models. Front Pharmacol 2014;5:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms K, Kaiser T. Beyond soluble transferrin receptor: old challenges and new horizons. Best Pract Res Clin Endocrinol Metab 2015;29:799–810. [DOI] [PubMed] [Google Scholar]
- 23.Speeckaert MM, Speeckaert R, Delanghe JR. Biological and clinical aspects of soluble transferrin receptor. Crit Rev Clin Lab Sci 2010;47:213–28. [DOI] [PubMed] [Google Scholar]
- 24.Peyssonnaux C, Nizet V, Johnson RS. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle 2008;7:28–32. [DOI] [PubMed] [Google Scholar]
- 25.Guo W, Bachman E, Li M, Roy CN, Blusztajn J, Wong S, Chan SY, Serra C, Jasuja R, Travison TG, et al. . Testosterone administration inhibits hepcidin transcription and is associated with increased iron incorporation into red blood cells. Aging Cell 2013;12:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y, Zhang S, Wang L, Li J, Qu G, He J, Rong H, Ji H, Liu S. Estrogen regulates iron homeostasis through governing hepatic hepcidin expression via an estrogen response element. Gene 2012;511:398–403. [DOI] [PubMed] [Google Scholar]
- 27.Anderson GJ, Vulpe CD. Mammalian iron transport. Cell Mol Life Sci 2009;66:3241–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol 2015;15:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakhal-Littleton S, Wolna M, Carr CA, Miller JJ, Christian HC, Ball V, Santos A, Diaz R, Biggs D, Stillion R, et al. . Cardiac ferroportin regulates cellular iron homeostasis and is important for cardiac function. Proc Natl Acad Sci USA 2015;112:3164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finch C. Regulators of iron balance in humans. Blood 1994;84:1697–702. [PubMed] [Google Scholar]
- 31.Anderson GJ, Powell LW, Halliday JW. Transferrin receptor distribution and regulation in the rat small intestine. Effect of iron stores and erythropoiesis. Gastroenterology 1990;98:576–85. [DOI] [PubMed] [Google Scholar]
- 32.Guiang SF III, Georgieff MK, Lambert DJ, Schmidt RL, Widness JA. Intravenous iron supplementation effect on tissue iron and hemoproteins in chronically phlebotomized lambs. Am J Physiol 1997;273:R2124–31. [DOI] [PubMed] [Google Scholar]
- 33.Milman N. Iron prophylaxis in pregnancy–general or individual and in which dose? Ann Hematol 2006;85:821–8. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 2016;22:1079–89. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann MB, Chassard C, Rohner F, N’Goran EK, Nindjin C, Dostal A, Utzinger J, Ghattas H, Lacroix C, Hurrell RF. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d’Ivoire. Am J Clin Nutr 2010;92:1406–15. [DOI] [PubMed] [Google Scholar]
- 36.Jaeggi T, Kortman GA, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Timmerman HM, Swinkels DW, Tjalsma H, et al. . Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 2015;64:731–42. [DOI] [PubMed] [Google Scholar]
- 37.Zimmermann MB, Hurrell RF. Nutritional iron deficiency. Lancet 2007;370:511–20. [DOI] [PubMed] [Google Scholar]
- 38.Beard JL, Durward C. The liabilities of iron deficiency Anderson GJ, McLaren GD, editors. Iron physiology and pathophysiology in humans New York: Springer; 2012. p. 283–302. [Google Scholar]
- 39.Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, et al. . Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet 2008;40:569–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasricha SR, Flecknoe-Brown SC, Allen KJ, Gibson PR, McMahon LP, Olynyk JK, Roger SD, Savoia HF, Tampi R, Thomson AR, et al. . Diagnosis and management of iron deficiency anaemia: a clinical update. Med J Aust 2010;193:525–32. [DOI] [PubMed] [Google Scholar]
- 41.Lopez A, Cacoub P, Macdougall IC, Peyrin-Biroulet L. Iron deficiency anaemia. Lancet 2016;387:907–16. [DOI] [PubMed] [Google Scholar]
- 42.Cancelo-Hidalgo MJ, Castelo-Branco C, Palacios S, Haya-Palazuelos J, Ciria-Recasens M, Manasanch J, Perez-Edo L. Tolerability of different oral iron supplements: a systematic review. Curr Med Res Opin 2013;29:291–303. [DOI] [PubMed] [Google Scholar]
- 43.Anderson GJ. Mechanisms of iron loading and toxicity. Am J Hematol 2007;82:1128–31. [DOI] [PubMed] [Google Scholar]
- 44.Barton JC, Edwards CQ, Acton RT. HFE gene: structure, function, mutations, and associated iron abnormalities. Gene 2015;574:179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deugnier Y, Turlin B. The pathology of hepatic iron overload In: Anderson GJ, McLaren GD, editors. Iron physiology and pathophysiology in humans. New York: Springer; 2012. p. 345–83. [Google Scholar]
- 46.Aigner E, Theurl I, Theurl M, Lederer D, Haufe H, Dietze O, Strasser M, Datz C, Weiss G. Pathways underlying iron accumulation in human nonalcoholic fatty liver disease. Am J Clin Nutr 2008;87:1374–83. [DOI] [PubMed] [Google Scholar]
- 47.Reid DW, Anderson GJ, Lamont IL. Cystic fibrosis: ironing out the problem of infection? Am J Physiol Lung Cell Mol Physiol 2008;295:L23–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farina M, Avila DS, da Rocha JB, Aschner M. Metals, oxidative stress and neurodegeneration: a focus on iron, manganese and mercury. Neurochem Int 2013;62:575–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivieri NF, Brittenham GM. Management of the thalassemias. Cold Spring Harb Perspect Med 2013;3:a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chua K, Fung E, Micewicz ED, Ganz T, Nemeth E, Ruchala P. Small cyclic agonists of iron regulatory hormone hepcidin. Bioorg Med Chem Lett 2015;25:4961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arezes J, Nemeth E. Hepcidin and iron disorders: new biology and clinical approaches. Int J Lab Hematol 2015;37 Suppl 1:92–8. [DOI] [PubMed] [Google Scholar]
- 52.Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med 2005;352:1011–23. [DOI] [PubMed] [Google Scholar]
- 53.Guida C, Altamura S, Klein FA, Galy B, Boutros M, Ulmer AJ, Hentze MW, Muckenthaler MU. A novel inflammatory pathway mediating rapid hepcidin-independent hypoferremia. Blood 2015;125:2265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]