Abstract
Importance
A major challenge for drug development in neurodegenerative diseases is that adequately powered efficacy studies with meaningful end points typically require several hundred participants and long durations. Prion diseases represent the archetype of brain diseases caused by protein misfolding, the most common subtype being sporadic Creutzfeldt-Jakob disease (sCJD), a rapidly progressive dementia. There is no well-established trial method in prion disease.
Objective
To establish a more powerful and meaningful clinical trial method in sCJD.
Design, Setting, and Participants
A stratified medicine and simulation approach based on a prospective interval-cohort study conducted from October 2008 to June 2014. This study involved 598 participants with probable or definite sCJD followed up over 470 patient-years at a specialist national referral service in the United Kingdom with domiciliary, care home, and hospital patient visits. We fitted linear mixed models to the outcome measurements, and simulated clinical trials involving 10 to 120 patients (no dropouts) with early to moderately advanced prion disease using model parameters to compare the power of various designs.
Main Outcomes and Measures
A total of 2681 assessments were done using a functionally orientated composite end point (Medical Research Council Scale) and associated with clinical investigations (brain magnetic resonance imaging, electroencephalography, and cerebrospinal fluid analysis) and molecular data (prion protein [PrP] gene sequencing, PrPSc type).
Results
Of the 598 participants, 273 were men. The PrP gene sequence was significantly associated with decline relative to any other demographic or investigation factors. Patients with sCJD and polymorphic codon 129 genotypes MM, VV, and MV lost 10% of their function in 5.3 (95% CI, 4.2-6.9), 13.2 (95% CI, 10.9-16.6), and 27.8 (95% CI, 21.9-37.8) days, respectively (P < .001). Simulations indicate that an adequately powered (80%; 2-sided α = .05) open-label randomized trial using 50% reduction in Medical Research Council Scale decline as the primary outcome could be conducted with only 120 participants assessed every 10 days and only 90 participants assessed daily, providing considerably more power than using survival as the primary outcome. Restricting to VV or MV codon 129 genotypes increased power even further. Alternatively, single-arm intervention studies (half the total sample size) could provide similar power in comparison to the natural history cohort.
Conclusions and Relevance
Functional end points in neurodegeneration need not require long and very large clinical studies to be adequately powered for efficacy. Patients with sCJD may be an efficient and cost-effective group for testing disease-modifying therapeutics. Stratified medicine and natural history cohort approaches may transform the feasibility of clinical trials in orphan diseases.
Human prion diseases are fatal neurodegenerative conditions, which may occur sporadically, be inherited by coding mutation in the prion protein gene (PRNP), or be acquired from iatrogenic or dietary exposure to prions.1 Approximately 90 to 120 people are diagnosed as having prion disease each year in the United Kingdom.2 Sporadic Creutzfeldt-Jakob disease (sCJD) is the most common form (approximately 85%) and the most obvious target for clinical trials. Typically, sCJD manifests as a rapidly progressive dementia with myoclonus and other neurological signs. The median clinical duration is approximately 4 months, although forms with short (few weeks) and long (>2 years) durations are well recognized.3 The fundamental pathogenic process in prion disease is the conversion of host cellular prion protein (PrPC) to abnormally folded multimeric forms (PrPSc and other disease-related forms) by seeded polymerization, a process that is thought to be widely shared in neurodegeneration.4 Prion diseases are particularly tractable for drug development, with clear target validation and availability of suitable cellular and animal models. Indeed, laboratory animals are naturally susceptible to prion infections, providing increased confidence that therapeutics shown to be active in preclinical models may translate to humans. Passive immunotherapy with monoclonal antibodies targeting PrPC has been shown to be effective in mouse models,5 and humanized antibodies developed for clinical trial6 and small-molecule therapeutics, which suppress prion replication and delay or prevent disease progression in mice, have been reported from multiple laboratories.7–9 However, to our knowledge, there is no effective therapeutic that has been shown to modify the course of the human disease.
In response to the challenges faced during Medical Research Council (MRC) PRION-110 and other human trials,11,12 including the lack of a validated outcome measure and the paucity of natural history data, we have studied UK patients with prion disease to optimize clinical trial methods and develop a resource for open-label studies. This work was done as part of an ongoing prospective interval-cohort study, the National Prion Monitoring Cohort (cohort study), which began recruiting in 2008. In preparation for clinical trials in sCJD, we used item-response modeling to develop a functionally oriented rating scale (MRC Prion Disease Rating Scale, or MRC Scale) with favorable statistical properties and weighted the items to represent the most impactful domains reported by patients and caregivers.13
Stratified medicine involves identifying strata within a disease and deepening the understanding of the mechanisms underpinning these strata. The purpose of this is to allow better targeting of treatments to specific disease pathways and identification of treatments effective for particular groups of patients to reduce heterogeneity and improve the power of clinical trials. Here we took a stratified experimental medicine approach in prion disease with the aim of determining the key factors that are associated with functional decline in sCJD and might be included in future clinical trial models from a wide range of demographic, clinical, genetic, investigation, and molecular parameters. A common amino acid polymorphism at position 129 of the prion protein gene (PRNP) is the most important genetic susceptibility factor in prion disease and also affects propagation of distinct human prion strains by conformational selection.4,14–16 A combination of disease etiology, severity, and genetic stratification with PRNP codon 129 proved to be an extremely powerful approach in our trial model, resulting in estimated sample sizes for adequately powered and meaningful clinical trials, which are substantially smaller than those required in other more common neurodegenerative diseases.
Methods
Patients
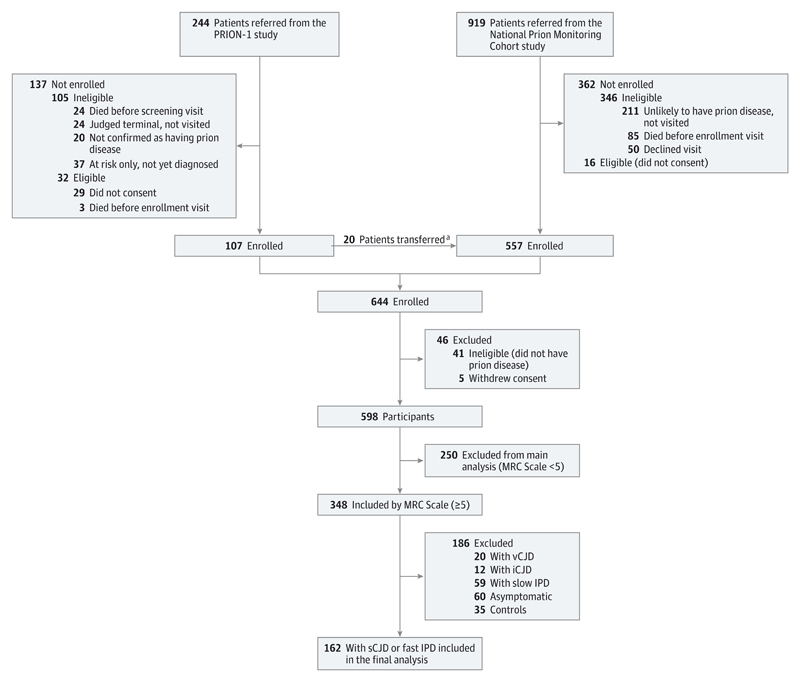
From 2004, UK neurologists were asked by the chief medical officer of the Department of Health, England, to refer all patients with suspected prion disease jointly to the National CJD Research and Surveillance Unit and to the National Health Service National Prion Clinic. Communication between both units several times per week ensures exchange of patient details referred to one of the units. Eighty-five percent of patients were visited by the National Prion Clinic within 5 days of referral. Details of enrollment into the PRION-1 Trial and cohort study have been published.10,13 In brief, the cohort aimed to enroll all symptomatic patients with prion disease in the United Kingdom including all patients with probable or definite sCJD, variant CJD, iatrogenic CJD, and inherited prion disease, according to updated diagnostic criteria.17 In addition, patients thought to have prion disease but not meeting formal criteria could be enrolled following review by an expert panel. Patients were enrolled at home, hospitals, and other health care settings around the United Kingdom from 2008 onwards (Figure 1). This current study was conducted from October 2008 to June 2014.
Figure 1. Flowchart of Enrollment and Selection of Patient Subset for Main Study.
iCJD indicates iatrogenic Creutzfeldt-Jakob disease; IPD, inherited prion disease; MRC, Medical Research Council; sCJD, sporadic Creutzfeldt-Jakob disease; vCJD, variant Creutzfeldt-Jakob disease.
a Twenty patients were enrolled in the cohort study after PRION-1 was completed.
Consent and Ethics
Informed consent was obtained directly from study participants or from relatives, caregivers, or Independent Mental Capacity Advocates as appropriate. Ethical approval was obtained from the Scotland A Research Ethics Committee (cohort) or the Eastern Research Ethics Committee (PRION-1).
The eAppendix in the Supplement provides the stratification and assessment schedule, molecular analysis, investigations, statistical analysis, and details of trial simulations.
Results
Baseline Characteristics and Completion of Investigations
We chose broad enrollment criteria to ensure that the cohort study was representative of prion disease in the United Kingdom; enrollments through June 2014 are included in these analyses (Table 1). Ninety-seven percent of eligible visited patients joined the study and less than 1% withdrew consent prior to death. A total of 2681 assessments were done over 470 patient-years of study. Diagnostic accuracy was very good, and 94% of those recruited met criteria for probable human prion disease. Autopsy was done in 60%, which confirmed prion disease in all cases, and a molecular diagnosis by gene test or biopsy was achieved in 22%, resulting in 70% with a definite diagnosis overall.
Table 1. Baseline Characteristics.
| Characteristic | Patients With MRC Scale <5 at Enrollment | Patients With MRC Scale ≥5 at Enrollment | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stratum 1 | Stratum 2 | Stratum 3 | ||||||||||
| Included in Model | Excluded From Model | |||||||||||
| sCJD | vCJD | iCJD | Fast IPD | Slow IPD | sCJD | Fast IPD | vCJD | iCJD | Slow IPD | Asymptomatic | Control | |
| Enrolled | 217 | 6 | 4 | 15 | 8 | 147 | 15 | 20 | 12 | 59 | 60 | 35 |
| Age at enrollment, median (range), y | 68 (46-88) | 54 (26-56) | 48 (27-51) | 65 (54 -78) | 47 (40-65) | 65 (39-83) | 55 (38-86) | 25 (14-50) | 46 (37-51) | 46 (26-69) | 39 (20-88) | 47 (23-75) |
| Sex | ||||||||||||
| Male | 78 | 2 | 3 | 7 | 3 | 81 | 8 | 15 | 11 | 26 | 22 | 17 |
| Female | 139 | 4 | 1 | 8 | 5 | 66 | 7 | 5 | 1 | 33 | 38 | 18 |
| Time between enrollment and first symptoms, median (IQR), mo | 3 (2-5) | 9 (7-22) | 5 (3-12) | 3 (2-4) | 36 (16-51) | 6 (3-9) | 7 (4-12) | 6 (5-11) | 8 (5-12) | 30 (14-62) | NA | NA |
| MRC Scale at enrollment [0-20] | ||||||||||||
| No. assessed | 215 | 5 | 4 | 15 | 8 | 147 | 15 | 20 | 12 | 59 | 60 | 35 |
| Median (IQR) | 1 (0-2) | 3 (2-4) | 3 (2-4) | 1 (0-2) | 2 (1-2) | 10 (7-14) | 14 (10-19) | 15 (11-18) | 16 (12-18) | 18 (12-19) | 20 (20-20) | 20 (20-20) |
| MMSE score at enrollment [0-30] | ||||||||||||
| No. assessed | 164 | 1 | 3 | 13 | 5 | 124 | 14 | 19 | 12 | 54 | 56 | 35 |
| Median (IQR) | 0 | 0 | 0 | 0 | 0 | 14 (6-20) | 18 (14-27) | 19 (14-23) | 21 (17-28) | 20 (15-28) | 30 (29-30) | 30 (30-30) |
| Rankin score at enrollment [0-5] | ||||||||||||
| No. assessed | 216 | 5 | 4 | 15 | 8 | 146 | 15 | 19 | 12 | 58 | 58 | 35 |
| Asymptomatic (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 56 | 35 |
| No or slight symptoms (1/2) | 0 | 0 | 0 | 0 | 0 | 10 | 3 | 2 | 4 | 18 | 1 | 0 |
| Moderate disability (3) | 0 | 0 | 0 | 0 | 0 | 31 | 6 | 6 | 1 | 23 | 0 | 0 |
| Moderate to severe disability (4) | 7 | 2 | 0 | 2 | 1 | 73 | 4 | 10 | 6 | 11 | 0 | 0 |
| Severe disability (5) | 209 | 3 | 4 | 13 | 7 | 32 | 2 | 1 | 1 | 3 | 1 | 0 |
| Median (IQR) | 5 | 5 (4-5) | 5 | 5 | 5 | 4 (3-4) | 3 (3-4) | 4 (3-4) | 4 (2-4) | 3 (2-3) | 0 | 0 |
| Codon 129 | ||||||||||||
| No. assessed | 170 | 6 | 4 | 14 | 8 | 139 | 15 | 20 | 12 | 57 | 33 | 0 |
| MM | 113 | 6 | 1 | 13 | 4 | 46 | 9 | 19 | 2 | 32 | 21 | … |
| MV | 25 | 0 | 2 | 1 | 3 | 56 | 6 | 1 | 10 | 22 | 11 | … |
| VV | 32 | 0 | 1 | 0 | 1 | 37 | 0 | 0 | 0 | 3 | 1 | … |
Abbreviations: ellipses, no data available for this group; iCJD, iatrogenic Creutzfeldt-Jakob disease; IPD, inherited prion disease; IQR, interquartile range; MMSE, Mini-Mental State Examination; MRC, Medical Research Council; NA, not applicable; sCJD, sporadic Creutzfeldt-Jakob disease; vCJD, variant Creutzfeldt-Jakob disease.
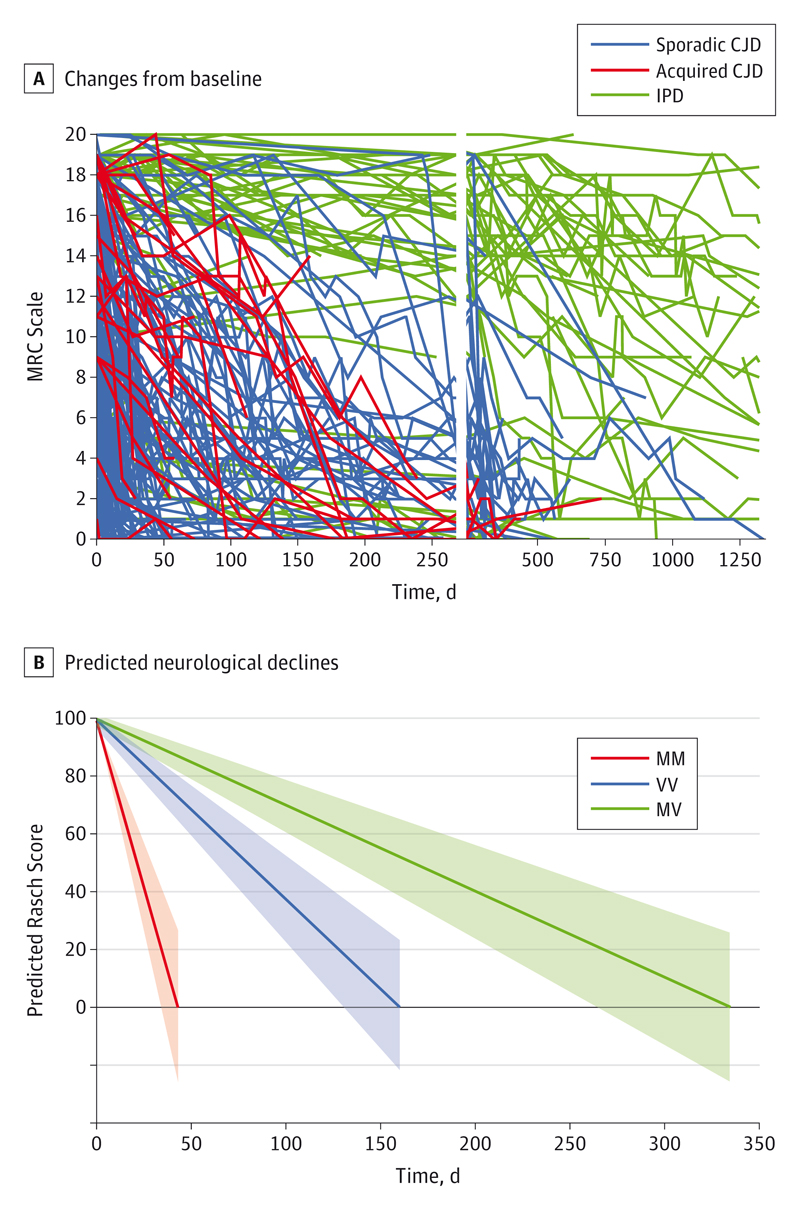
Visualization of MRC Scale measurements showed distinct patterns of decline (Figure 2A) namely: (1) rapid decline over weeks or a few months (mostly patients with sCJD, but including some acquired prion disease and specific inherited prion disease mutations [stratum 1, eAppendix in the Supplement]); (2) slow decline over years (almost exclusively patients with inherited prion disease typically associated with the GSS phenotype [stratum 2, eAppendix in the Supplement]); and (3) a small proportion of patients with rapid decline followed by prolonged survival at high levels of neurodisability, essentially a comatose state (MRC Scale <3), plausibly due to exceptional supportive care.
Figure 2. Spaghetti Plots and Graph of Rasch Score.
A, Spaghetti plots showing changes from baseline over time in all cohort patients. Individual patient trajectories are shown for sporadic Creutzfeldt-Jakob disease (CJD), acquired CJD, and inherited prion disease (IPD). This chart prompted initial decisions about patient selection for modeling: slowly progressive IPD and acquired CJD were excluded as were serial measurements of the Medical Research Council (MRC) Scale less than 3 (see the Baseline Characteristics and Completion of Investigations section). The vertical gap represents a change in the time scale (x-axis). B, Predicted neurological declines from best possible score by codon 129 subtype. This chart shows model-predicted mean declines (lines) and 95% CIs (bands) in stratum 1 patients with 3 different codon 129 genotypes at PRNP. The Rasch score approximates to 5 times the MRC Scale in Figure 2A.
The principal aim of this analysis was to establish a trial model based on repeated measurements of the MRC Scale. Therefore, we excluded 3 groups of patients from further analyses. First, because sCJD is by far the most common category of prion disease and given the vastly different time scales over which functional decline occurred in stratums 1 and 2 (Figure 2A), we decided to focus on rapidly progressive forms of prion disease, therefore excluding stratum 2. In stratum 1 patients, rates of decline were significantly slower for acquired prion disease vs sCJD (P < .001). Second, because incidence of acquired disease is currently very low, we excluded this etiological category from further analyses. As rapidly progressive forms of inherited prion disease may not be diagnosed for several weeks after prion disease itself is first identified, these patients were included, although certain mechanisms of action of experimental therapeutics may require them to be excluded from a prospective clinical trial. Third, because many patients are diagnosed as having CJD at advanced stages of neurodisability, when irreversible neuronal death has already occurred, we excluded patients with severe disease at presentation (MRC Scale <5).
Rates of MRC Scale Decline in Different Groups Defined by Codon 129
In the best-fitting linear mixed model in 154 patients with rapidly progressing prion disease (sCJD and fast-progressing inherited prion disease) (see eFigure 1 in the Supplement for individual fits), baseline MRC Scale depended only on age, being 4.1 Rasch units lower for every 10 years older (P < .008; Table 2).18 Subsequent decline was strongly associated with codon 129 genotype (interaction P < .001); predicted declines from the best possible score of 100 (20/20 MRC Scale) are shown in Figure 2B. Patients with sCJD/fast inherited prion disease and polymorphic codon 129 genotypes MM, VV, and MV lost 10% of their function measured by the MRC Scale in 5.3 (95% CI, 4.2-6.9), 13.2 (95% CI, 10.9-16.6), and 27.8 (95% CI, 21.9-37.8) days, respectively. There was no evidence that codon 129 genotype significantly affected the MRC Scale measurement at enrollment (in the final modeled sample, P = .11). However, in addition to these population-level differences in the mean declines, codon 129 genotype also significantly affected how variable individual patients were around these means (differential random effects and residual errors, P < .001). Although enrollment scores varied similarly between MM and non-MM genotype patients, subsequent declines varied significantly more in MM than non-MM, and residual errors (variability not explained by the underlying trajectories) were also significantly greater (Table 2).
Table 2. Model for MRC Scale Decline in Trial-Relevant Populationa.
| Factor | Estimate (95% CI) | P Value | P Value for Heterogeneity/Interaction vs Reference Category |
|---|---|---|---|
| Estimates of Baseline and Decline | |||
| Baseline, aged 65 y | |||
| MM | 53.19 (47.82 to 58.55) | NA | NA |
| MV | 57.88 (53.85 to 61.90) | NA | .17 |
| VV | 50.89 (45.60 to 56.18) | NA | .55 |
| Per 10 y older at enrollment | −4.12 (−7.14 to −1.10) | .008 | NA |
| Decline, per d | |||
| MM | −1.90 (−2.36 to −1.45) | <.001 | NA |
| MV | −0.36 (−0.83 to 0.11) | <.001 | <.001 |
| VV | −0.76 (−1.24 to −0.28) | <.001 | <.001 |
| Estimates of Individual Variability | |||
| MM | |||
| Baseline | 17.17 (13.31 to 22.17) | NA | NA |
| Decline | 1.36 (0.95 to 1.95) | NA | NA |
| Correlation (baseline, decline) | 0.38 (0.00 to 0.66) | NA | NA |
| Residual error | 12.80 (10.91 to 15.02) | NA | NA |
| Non-MM | |||
| Baseline | 14.75 (12.52 to 17.37) | NA | NA |
| Decline | 0.33 (0.24 to 0.46) | NA | NA |
| Correlation (baseline, decline) | 0.24 (−0.02 to 0.47) | NA | NA |
| Residual error | 9.20 (8.57 to 9.87) | NA | NA |
Abbreviations: MRC, Medical Research Council; NA, not applicable.
Model includes 824 measurements from 154 patients with sporadic Creutzfeldt-Jakob disease and patients with fast IPD in stratum 1 with MRC Scale of 5 or greater at enrollment. Reference category aged 65 years, MM.
Once the main model had been constructed and the potential effects of enrollment function, age, sex, disease category, and codon 129 had been considered, we went on to explore whether the following additional investigation factors had important effects on baseline or decline in MRC Scale: clinical phenotype as assessed by the visiting physician (7 types, sCJD only; eAppendix in the Supplement); the presence or absence of signal change in the basal ganglia, cortex, or thalamus on brain magnetic resonance imaging (MRI); molecular strain type (PrPSc types 1-3 using the London classification19); the presence of periodic sharp wave complexes or background abnormalities on routine electroencephalographic recordings; the presence or absence of 14-3-3 protein in routine cerebrospinal fluid (CSF) analysis; or the concentration of CSF S100b (eTable in the Supplement). Four of these factors affected decline in MRC Scale when considered alone (clinical category; n = 159; P < .001), molecular strain type20 (n = 63; P = .03), periodic sharp wave complexes (n = 147; P = .009), and CSF S100b (n = 124; P = .046), but either these effects disappeared after adjusting for the effect of codon 129 subtype on decline (periodic sharp wave complexes; adjusted P = .33) or effects weakened with the effect of codon 129 subtype remaining similar to Table 2 and much stronger in magnitude. In the multivariate analysis, higher levels of CSF S100b, a quantitative measurement of a glial protein routinely used to assist diagnosis of CJD, were associated with more rapid decline (P = .005), although not at a statistical level beyond that which might be expected as a result of performing tests of multiple hypotheses. Therefore, the PrP gene sequence appears to be a profoundly important predictor of decline relative to other demographic or investigation factors.
Power Calculations Using the MRC Scale
We used the observed enrollment frequency of codon 129 subtypes (33% MM, 43% MV, and 24% VV), distribution of ages (mean [SD], 65.1 [9.4] years), and population and individual rates of decline as estimated in Table 2 to simulate cohorts of patients who might be enrolled into clinical trials of an investigational agent that reduced the rate of decline by (proportionately) 0%, 25%, or 50%. We assumed a fixed percentage reduction in decline given the large absolute differences in decline observed (Table 2); therefore, we estimated the effect of the intervention using a test for the percentage reduction (see the eAppendix in the Supplement for details). We compared the power of different sized trials to detect effects, based on estimating the effect of an intervention on the MRC Scale and survival.
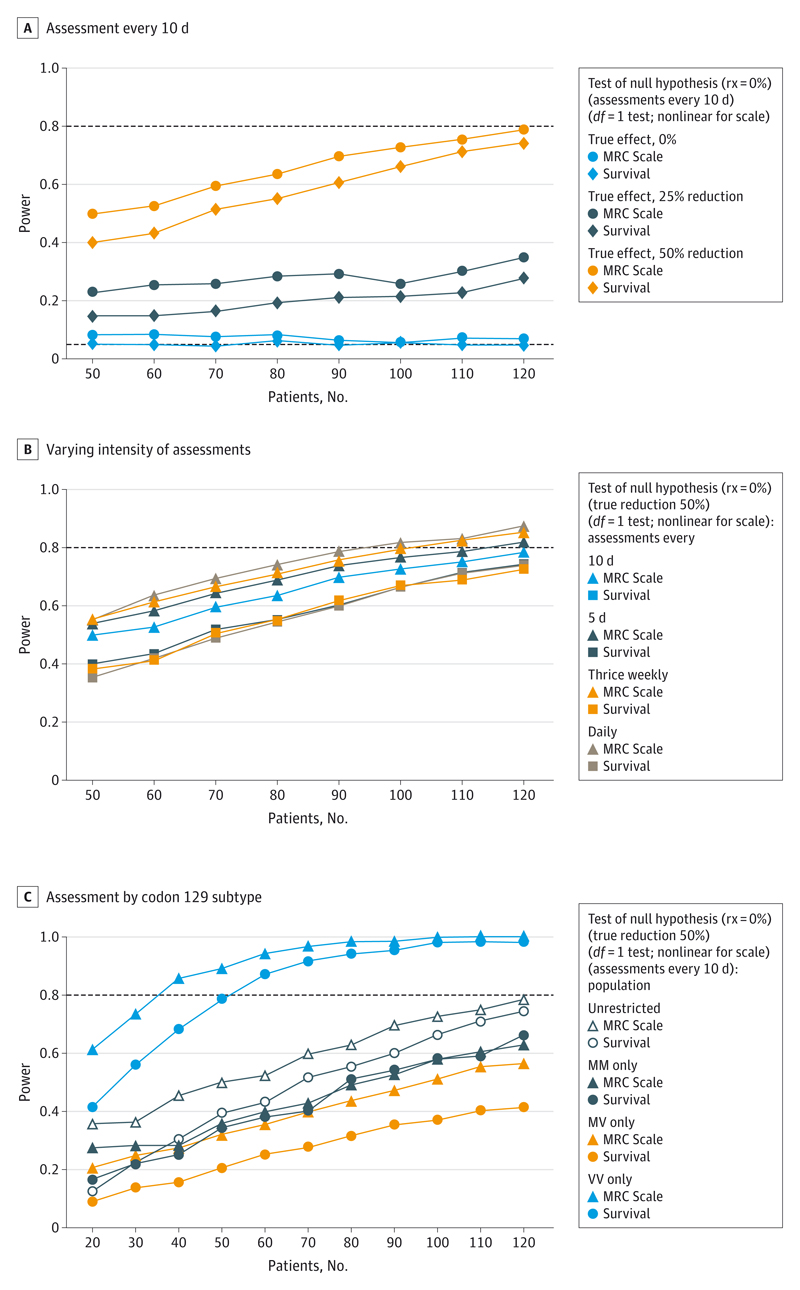
First, we considered a randomized trial (which could be either placebo controlled or open label, although the possibility of reporting bias in an open-label trial would need to be carefully considered). Figure 3A shows the power of such a trial, with a total of 50 to 120 fast-progressing patients (MM, MV, and VV) randomized 1:1 control to intervention, and assessed every 10 days, to detect a 25% or 50% reduction in decline in the MRC Scale. A total of 120 patients would need to be randomized to provide 80% power to detect a 50% reduction in decline; the same number of patients provides 5% to 10% greater power from using the repeated MRC Scale measurements than a survival end point on the same population.
Figure 3. Power for Different Sizes of Randomized Trial.
A, Graph shows assessments every 10 days, recruiting from the fast-progressing population (MM, MV and VV) and with varying intervention efficacy. When the true reduction is 0% (blue line), 5% of tests would be expected to fall below a P < .05 by chance (black dotted line). B, Graph shows intervention has a 50% reduction in decline, recruiting from the fast-progressing population (MM, MV, and VV), with varying number of assessments. Survival is estimated in the same manner regardless of the number of times that assessments are made; therefore, variation in the estimates of power for a survival end point reflect only sampling variation. C, Graph shows intervention has a 50% reduction in decline, with assessments every 10 days: varying recruitment from the entire fast-progressing population or according to codon 129 subtype.
The MRC Scale requires only 2 minutes to acquire over the telephone and is highly acceptable to patients and their caregivers: therefore, we considered the effect of more frequent measurements on power (Figure 3B). The same power as achieved with 120 randomized patients measured every 10 days could be achieved by 110 patients every 5 days, 100 patients thrice weekly, or 90 patients daily, as a consequence of improved precision of estimation of rates of decline. Therefore, power gains for daily measurement over a survival end point were 13% to 20% for the same number of patients.
Last, given the substantially greater variability observed in rates of decline in the MM subgroup, we considered the effect on power of restricting a trial to different codon 129 subtypes (Figure 3C). Because the VV subgroup have less variability than the MM subgroup, but faster declines than the MV subgroup, only 40 randomized VV patients provided more than 80% power to detect 50% reductions in decline. However, at the observed population frequencies, 40 VV patients would be expected from a cohort of 167 patients in total, which would have provided greater power overall.
An alternative approach would be a single-arm openlabel study with comparisons made to the cohort natural history data. Such nonrandomized comparisons have well-recognized limitations, the most important being changes in external management. Simulations of an existing fixed cohort of 200 individuals tested once every 10 days with a new cohort receiving treatment demonstrate that similar power could be achieved with around half the randomized patients (eFigure 2 in the Supplement).
Discussion
In this study, we identified a key predictor of functional decline in rapidly progressive forms of prion disease, codon 129 genotype, enabling us to assess the potential gains from a stratified medicine approach to future clinical trials. Through use of natural history data collected in a prospective observational cohort study, we have optimized a clinical trial model to increase power to detect clinically relevant effects on a premortality end point measuring functional decline that is judged highly relevant by patients and caregivers. Our 2 key findings are, first, that genetic variation in the prion protein gene has a profound influence on the rate of functional decline in rapidly progressive human prion disease and is, therefore, likely the essential factor on which to stratify a trial model. Second, that a combination of factors, including the use of a bespoke functionally orientated rating scale, the use of frequent assessments by telemedicine, the rapidly progressive nature of prion disease, and the stratification by codon 129, contribute to the potential for remarkably powerful clinical trials.
Despite the fact that codon 129 was known to be an important modifier of susceptibility and phenotype of prion diseases, we were surprised by the magnitude of the effects we observed on functional decline after diagnosis. The homotypic protein-protein interactions in patients with PRNP homozygous prion disease may occur faster than heterotypic interactions in heterozygous individuals,14 resulting in more rapid propagation of prions and/or generation of neurotoxic forms of abnormal PrP.21 Further, codon 129 is known to be important in determining prion strain selection.4 Prion strains, analogous to strains of bacteria or viruses, are associated with distinct types of misfolded PrP and distinct and transmissible clinicopathological features. Therefore, it is likely that some of the distinction between the rates of decline of codon 129 methionine and valine homozygotes are related to strain selection by codon 129. The PrPSc type, which is a measure of abnormal PrP conformation and strain, did provide some limited additional value beyond codon 129 alone but was only able to be measured in approximately 40% of the early and rapidly progressive CJD subset and always retrospectively after autopsy, seriously limiting its use as a factor in clinical trials. The lack of a detectable role of PrPSc type may relate to the fact that this is highly correlated with codon 129 genotype and that, while the autopsy rate in the study was high, many patients were alive at the data freeze and missing data remain substantial. We note that variation in the slopes of decline of codon 129 methionine homozygous patients was greater than for other genotypes, suggesting that strain diversity/permissibility may be greatest in this genotype.
The last decade has seen the advent of genome-wide technologies that have driven the discovery of many risk factors in neurodegeneration.22 Although examples are emerging,23 phenotypic heterogeneity is less well understood, in part because the imperative to study large sample sizes has come at the expense of well-characterized clinical cohorts. Clinical trials in common neurodegenerative diseases do not typically stratify by genetic factors. One exception is APOE in some studies of Alzheimer disease immunotherapy, although this decision was primarily driven by an increased susceptibility to amyloid-related imaging abnormalities in APOE4 carriers.24 The potent genetic effects we observed in this study suggest the potential of using genetic modifiers to improve power in clinical trials in neurodegeneration more widely.
There were some limitations of this study. A concern about avoiding making a diagnostic error, and a sense that it was only important to diagnose treatable disorders, means that referral to specialist centers often occurs late in the clinical course of human prion disease, when confirmatory tests are complete. Consequently, most patients enrolled in our cohort study had advanced neurodisability at enrollment. As a rough rule of thumb to simplify decision making, patients with a brain MRI scan suggestive of prion disease, who are either able to walk (even with help) or speak some words, should be referred promptly, particularly when clinical trials are active, because these patients are highly likely to have prion disease and MRC Scale of 5 or greater. Although this is by far the largest study of its type, to our knowledge, we were still underpowered to detect modifying effects of certain factors, particularly for rare combinations of PrPSc type and codon 129 genotype, which might be associated with atypical clinical courses.
Biomarker analysis is a highly active area of research in neurodegeneration and is a key component of the cohort study. However, in prion disease, where diagnostic accuracy (once referred) is high and progression rapid, the need for a surrogate marker of progression is less compelling than in slowly progressive disease syndromes. In the multivariate analysis, standard diagnostic MRI, electroencephalography, and CSF biomarkers did not add significantly to codon 129 subtypes for predicting the slopes of functional decline. The analysis of quantitative MRI parameters, which seem promising as predictors of the severity of microscopic tissue pathology, was beyond the scope of this report.25 The propensity of abnormal PrP seeds to trigger the polymerization of PrP in vitro, for example, by the protein misfolding cyclic amplification or real-time quaking-induced conversion reactions,26,27 have potential as highly responsive therapeutic biomarkers in CSF and will be important adjuncts to future trials.
Conclusions
Functional end points in other neurodegenerative diseases typically require several hundred or thousands of patients for adequately powered studies.28 Here we established that repeated measurements of a functional scale in only 120 patients with prion disease has the capacity to provide an adequately powered 2-arm study and that further reduction in patient number (by 10-30) may be possible by increasing the frequency of assessment. The MRC Scale also has advantages in that it is not affected by personal decisions regarding end-of-life care (including percutaneous endoscopic gastrostomy feeding and antibiotics), which can add substantial variation in small trials using a survival end point, obscuring treatment differences. It is now dogmatic that common neurodegenerative disorders share fundamental molecular mechanisms with prion disease, often referred to as prion-like mechanisms.29 Immunotherapeutics and small molecules have been developed that can clear prion infection from cultured neuronal cells and cure infection or greatly impede it in vivo.5,8,9 The prognosis for rapidly progressive prion disease is particularly severe and the unmet need is great, analogous to advanced cancer, which has proven to be a very effective test bed for novel therapeutics. We hope that our study will modify the view of human prion disease as an orphan disorder futile for clinical study into a tractable disorder for testing of therapeutics.
Supplementary Material
Key Points.
Question: Can clinical trial methods in Creutzfeldt-Jakob disease (CJD) be improved?
Findings: Use of everyday patient functions as a clinical trial outcome would make the finding of a therapeutic effect more meaningful. We modeled repeated measurements of the Medical Research Council Scale in CJD after excluding severely affected patients. The important determinant of the rate of deterioration in function was the genotype at polymorphic codon 129 of the prion protein gene. Using this genetic cofactor and frequent telemeasurements, statistical power in simulated trials was improved.
Meaning: Stratified medicine and natural history cohort approaches may transform the feasibility of clinical trials in CJD.
Acknowledgments
Funding/Support: The cohort study was funded by the Department of Health (England), Medical Research Council, and University College London Hospitals/University College London National Institutes for Health Research Biomedical Research Centre. This study was funded by the Medical Research Council, Department of Health (England), and the University College London /University College London Hospitals Biomedical Research Centre.
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Dr Mead had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drs Mead, Burnell, Walker, and Collinge contributed equally.
Study concept and design: Mead, Burnell, Rudge, Walker, Collinge.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Mead, Burnell, Rudge, Walker, Collinge.
Critical revision of the manuscript for important intellectual content: Mead, Burnell, Lowe, Thompson, Lukic, Porter, Carswell, Kaski, Kenny, Mok, Bjurstrom, Franko, Gorham, Druyeh, Wadsworth, Jaunmuktane, Brandner, Hyare, Rudge.
Statistical analysis: Mead, Burnell, Walker.
Obtained funding: Mead, Collinge.
Administrative, technical, or material support: Mead.
Study supervision: Mead, Rudge, Collinge.
Conflict of Interest Disclosures: Dr Collinge is a director and Drs Wadsworth and Collinge are shareholders of D-Gen Ltd, an academic spinout company working in the field of prion disease diagnosis, decontamination, and therapeutics. No other disclosures were reported.
Additional Contributions: We thank all the individuals, their caregivers, and families, who took part in the PRION-1 and cohort studies as well as UK neurologists and the National CJD Research and Surveillance Unit for referring patients. We thank officials at the Department of Health, Medical Research Council, co-chairs of the PRION-1 and cohort steering committee, and our colleagues at the National CJD Research and Surveillance Unit for establishing the National CJD referral arrangements, without which these studies would not have been possible. We thank all past and present colleagues at the National Prion Clinic (National Hospital for Neurology and Neurosurgery, Queen Square, London, England). Ray Young and Richard Newton, paid employees of the Medical Research Council Prion Unit, assisted with figure design.
References
- 1.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.National Creutzfeldt-Jakob Disease Surveillance Unit. Seventeenth Annual Report 2008: Creutzfeldt-Jakob Disease Surveillance in the UK. Edinburgh, Scotland: National Creutzfeldt-Jakob Disease Surveillance Unit; 2009. pp. 1–42. [Google Scholar]
- 3.Pocchiari M, Puopolo M, Croes EA, et al. Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004;127(pt 10):2348–2359. doi: 10.1093/brain/awh249. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318(5852):930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 5.White AR, Enever P, Tayebi M, et al. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature. 2003;422(6927):80–83. doi: 10.1038/nature01457. [DOI] [PubMed] [Google Scholar]
- 6.Klyubin I, Nicoll AJ, Khalili-Shirazi A, et al. Peripheral administration of a humanized anti-PrP antibody blocks Alzheimer’s disease Aβ synaptotoxicity. J Neurosci. 2014;34(18):6140–6145. doi: 10.1523/JNEUROSCI.3526-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trevitt CR, Collinge J. A systematic review of prion therapeutics in experimental models. Brain. 2006;129(pt 9):2241–2265. doi: 10.1093/brain/awl150. [DOI] [PubMed] [Google Scholar]
- 8.Wagner J, Ryazanov S, Leonov A, et al. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013;125(6):795–813. doi: 10.1007/s00401-013-1114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silber BM, Rao S, Fife KL, et al. Pharmacokinetics and metabolism of 2-aminothiazoles with antiprion activity in mice. Pharm Res. 2013;30(4):932–950. doi: 10.1007/s11095-012-0912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinge J, Gorham M, Hudson F, et al. Safety and efficacy of quinacrine in human prion disease (PRION-1 Study): a patient-preference trial. Lancet Neurology. 2009;8(4):334–344. doi: 10.1016/S1474-4422(09)70049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haïk S, Marcon G, Mallet A, et al. Doxycycline in Creutzfeldt-Jakob disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2014;13(2):150–158. doi: 10.1016/S1474-4422(13)70307-7. [DOI] [PubMed] [Google Scholar]
- 12.Geschwind MD, Kuo AL, Wong KS, et al. Quinacrine treatment trial for sporadic Creutzfeldt-Jakob disease. Neurology. 2013;81(23):2015–2023. doi: 10.1212/WNL.0b013e3182a9f3b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson AG, Lowe J, Fox Z, et al. The Medical Research Council Prion Disease Rating Scale: a new outcome measure for prion disease therapeutic trials developed and validated using systematic observational studies. Brain. 2013;136(pt 4):1116–1127. doi: 10.1093/brain/awt048. [DOI] [PubMed] [Google Scholar]
- 14.Palmer MS, Dryden AJ, Hughes JT, Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature. 1991;352(6333):340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 15.Mead S, Poulter M, Uphill J, et al. Genetic risk factors for variant Creutzfeldt-Jakob disease: a genome-wide association study. Lancet Neurol. 2009;8(1):57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collinge J. Variant Creutzfeldt-Jakob disease. Lancet. 1999;354(9175):317–323. doi: 10.1016/S0140-6736(99)05128-4. [DOI] [PubMed] [Google Scholar]
- 17.Zerr I, Kallenberg K, Summers DM, et al. Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain. 2009;132(pt 10):2659–2668. doi: 10.1093/brain/awp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.RUMM Laboratory. RUMM2030: Rasch Unidimensional Measurement Model [computer software] Perth, Western Australia, Australia: RUMM Laboratory; 2008. [Google Scholar]
- 19.Hill AF, Joiner S, Wadsworth JD, et al. Molecular classification of sporadic Creutzfeldt-Jakob disease. Brain. 2003;126(pt 6):1333–1346. doi: 10.1093/brain/awg125. [DOI] [PubMed] [Google Scholar]
- 20.Wadsworth JD, Powell C, Beck JA, et al. Molecular diagnosis of human prion disease. Methods Mol Biol. 2008;459:197–227. doi: 10.1007/978-1-59745-234-2_14. [DOI] [PubMed] [Google Scholar]
- 21.Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470(7335):540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 22.Singleton AB, Hardy J, Traynor BJ, Houlden H. Towards a complete resolution of the genetic architecture of disease. Trends Genet. 2010;26(10):438–442. doi: 10.1016/j.tig.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruchaga C, Kauwe JSK, Mayo K, et al. Alzheimer’s Disease Neuroimaging Initiative. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001101. e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sperling R, Salloway S, Brooks DJ, et al. Amyloid-related imaging abnormalities in patients with Alzheimer’s disease treated with bapineuzumab: a retrospective analysis. Lancet Neurol. 2012;11(3):241–249. doi: 10.1016/S1474-4422(12)70015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddique D, Hyare H, Wroe S, et al. Magnetization transfer ratio may be a surrogate of spongiform change in human prion diseases. Brain. 2010;133(10):3058–3068. doi: 10.1093/brain/awq243. [DOI] [PubMed] [Google Scholar]
- 26.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411(6839):810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 27.Atarashi R, Satoh K, Sano K, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011;17(2):175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 28.Green RC, Schneider LS, Amato DA, et al. Tarenflurbil Phase 3 Study Group Effect of tarenflurbil on cognitive decline and activities of daily living in patients with mild Alzheimer disease: a randomized controlled trial. JAMA. 2009;302(23):2557–2564. doi: 10.1001/jama.2009.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.