Abstract
Objective
Insulin resistance has deleterious effects on cardiometabolic disease. We used Mendelian randomization analyses to clarify the causal relationships of insulin resistance on circulating blood-based metabolites to shed light on potential mediators of the insulin resistance to cardiometabolic disease relationship.
Research Design and Methods
We used 53 single nucleotide polymorphisms associated with insulin resistance from a recent genome-wide association study to explore their effects on circulating lipids and metabolites. We used published summary-level data from two genome-wide association studies (GWASs) of European individuals; data on the exposure (insulin resistance) were obtained from meta-GWASs of 188,577 individuals and data on the outcomes (58 metabolic measures assessed by NMR) were taken from a GWAS of 24,925 individuals.
Results
One standard deviation (SD) genetically elevated insulin resistance (equivalent to 55% higher geometric mean of fasting insulin, 0.89 mmol/L higher triglycerides and 0.46 mmol/L lower HDL-C) was associated with higher concentrations of all branched-chain amino acids, isoleucine (0.56 SD; 95%CI: 0.43, 0.70), leucine (0.42 SD; 95%CI: 0.28, 0.55) and valine (0.26 SD; 95%CI: 0.12, 0.39) as well as with higher glycoprotein acetyls (an inflammation marker; 0.47 SD; 95%CI: 0.32, 0.62) (P<0.0003 for each). Results were broadly consistent when using multiple sensitivity analyses to account for potential genetic pleiotropy.
Conclusions
We provide robust evidence that insulin resistance causally impacts on each individual branched-chain amino acid and inflammation. Taken together with existing studies, this implies that branched-chain amino acid metabolism lies on a causal pathway from adiposity and insulin resistance to type 2 diabetes.
Keywords: adiposity, insulin resistance, branched-chain amino acids, type 2 diabetes
Subject codes: Biomarkers, Mechanisms, Risk Factors, Type 2 Diabetes, Genetics
The obesity pandemic is a public health crisis leading to a dramatic surge in the incidence of type 2 diabetes mellitus (T2DM) and related diseases (e.g., cardiovascular diseases) (1). Adiposity, particularly visceral adiposity (2), is associated with insulin resistance (IR) and subsequent T2DM. Recent genetic studies employing the Mendelian randomization approach have shown adiposity traits (such as general adiposity, indexed by body mass index, and central adiposity, indexed by waist-to-hip ratio) to show causal relationships with blood pressure, lipids, coronary heart disease, stroke and diabetes (3–6). Furthermore, such studies have demonstrated that adiposity traits causally impact on insulin resistance (3,4,6). Insulin resistance is the clinical state of a reduced sensitivity to insulin, typically manifested as elevated levels of fasting insulin and often accompanied with higher levels of circulating triglycerides and lower levels of high-density lipoprotein cholesterol (HDL-C) (7).
Exploring the molecular mechanism by which IR leads to T2DM may help to identify biomarkers that could mediate the relationship, and provide novel opportunities for disease prevention. Recent studies have suggested that branched-chain amino acids (BCAAs) might play a role in the development of T2DM. Prospective observational studies show that higher levels of circulating BCAAs are positively associated with markers of insulin resistance (8) and risk of incident T2DM (9,10). Recent genetic studies have also implicated the metabolism of BCAAs in the development of diabetes (11).
Insulin resistance is a complex trait, which can be assessed by different metrics, including clamp/insulin suppression test (gold standard), insulin sensitivity test (based on OGTT), HOMA-IR and fasting insulin. The GENESIS consortium has published a GWAS of insulin sensitivity measured by clamp/insulin suppression test in a modest number of subjects (N = 5624) (12). However, the statistical power limits the findings of this study. Other metrics which can be more easily measured, such as fasting insulin or HOMA-IR, are often used in large-scale genetic and epidemiological studies. In the GWAS of fasting insulin conducted by Scott et al (up to 108,557 individuals) (13), they also tested the associations of insulin-associated SNPs with lipid traits. They found that majority of the insulin-associated SNPs were associated with HDL-C and/or TG and this pattern was not observed for those SNPs associated with fasting glucose or 2hour glucose. Subsequently, a genetic instrument was built for insulin resistance that used the 19 SNPs associated with fasting insulin, and restricted the instrument to those SNPs that were also associated with triglycerides and HDL-C (14). This instrument was recently adopted by Mahendran et al, and the results suggest that insulin resistance might be causal for circulating concentrations of BCAAs (15). More recently, Lotta et al considerably expanded the set of SNPs associated with three components of insulin resistance (higher fasting insulin, higher triglycerides and lower HDL-cholesterol), identifying 53 such SNPs and found that the SNPs in aggregate also associated with risks of CHD and T2DM (7).
Here we aim to: 1) assess the causal effects of insulin resistance employing these 53 SNPs recently identified from across the genome that associated with higher fasting insulin, higher triglycerides and lower HDL-C (7); 2) use multiple instruments and multiple sensitivity analysis as a means to detect and correct for potential genetic pleiotropy in order to ensure reliable findings; 3) expand the outcome measures from BCAAs to a comprehensive panel of amino acids (including alanine, glutamine, tyrosine and phenylalanine), lipoprotein subclasses, fatty acids, glycolysis-related measures and one inflammatory marker, which are established or emerging biomarkers for T2DM and cardiovascular diseases; 4) provide an overview of the potential causal pathways and mediator roles that insulin resistance places in the underlying association of adiposity with T2DM, by incorporating our findings into multiple strands of genetic evidence.
Research Design and Methods
We used published summary-level data from two GWA studies of European individuals (7,16). Data on the exposure (insulin resistance) were obtained from a meta-GWASs (meta-analysis of genome-wide association studies) of up to 188,577 individuals (7) and data on the outcome (58 circulating metabolic measures) were taken from a GWAS of up to 24,925 individuals (16). Characteristics of these GWASs are reported in Supplemental Tables 1 & 2.
Generation of Genetic Instruments
We used the 53 SNPs associated with an insulin resistance phenotype from Lotta et al (7). In brief, Lotta et al. conducted a meta-GWAS to identify SNPs that associated with an insulin resistance phenotype of: (i) higher fasting insulin adjusted for BMI; (ii) higher triglycerides; and, (iii) lower HDL-C at P<0.005 for each trait. The combined association with the triad of phenotypes have been proposed as a means to characterising the genetic architecture of insulin resistance (7). This meta-GWAS identified 53 SNPs, of which a subset of 25 loci had been previously associated with triglycerides or HDL-C at genome-wide significance, whereas the remaining 28 had not.
We used the 53 SNPs to generate a genetic instrument for IR. To conduct the Mendelian randomization (MR) analyses (17), we needed to obtain the association of SNPs with the exposure (insulin resistance) and also the associations with outcomes (metabolic measures). Lotta et al. (7) did not provide beta or SE for the associations of individual SNPs with the insulin resistance phenotype. To generate our own SNP to exposure estimate, we took the absolute value of the standardized beta coefficient for each of the 53 SNP associations with the individual components of the composite IR phenotype (i.e. fasting insulin adjusted for BMI, triglycerides and HDL-C) and meta-analysed the estimates together using a fixed-effect inverse-variance weighted method (data sources provided in Supplemental Table 3). We used this meta-analysed value as the SNP-exposure estimate for the summary-level MR analyses. Supplemental Fig. 1 shows the associations of the 53 individual SNPs for our insulin resistance trait with the three individual components. Most of the SNPs fell in a straight line (with a slope equal to 1), suggesting a similar contribution of the three traits to the ‘composite’ insulin resistance phenotype with the exception of rs1011685 (near LPL), which had a much weaker effect on insulin adjusted for BMI. We therefore conducted sensitivity analyses in which rs1011685 was excluded from the instrument.
Two-Sample Mendelian Randomization Analysis
We used data from Kettunen et al. (16) to obtain SNP-associations with metabolic measures. Summary data for 58 measures were used in this study, including 14 lipoprotein subclasses, 3 lipoprotein size measures, 9 total lipids, ApoA1, ApoB, 10 fatty acids related measures, 9 amino acids, one inflammation marker - glycoprotein acetyls and several other measures. These metabolic measures were quantified by a high-throughput NMR metabolomics platform using primarily fasting serum samples with an approximately 1:1 male-to-female ratio and age span of 20-60 years (Supplemental Table 2). We used a conventional inverse variance weighted (IVW) MR analysis, in which the SNP to outcome estimate is regressed on the SNP to exposure, with the y-axis intercept forced through the origin. The data used for the MR analyses are presented in Supplemental Tables 3 & 4.
Sensitivity Analyses
As the conventional IVW MR approach can be vulnerable to unbalanced horizontal pleiotropy (18), we conducted MR-Egger, weighted median and weighted mode-based MR analyses, which allow relaxation of some of the instrumental variable assumptions. The characteristics of these different MR methods are summarized in Supplemental Table 5. Overall, use of several MR methods that each makes different assumptions on the amount and type of genetic confounding is a useful strategy to assess the robustness of findings to potential violations of the instrumental variable assumptions (19).
In addition to the 53 SNP instrument, we: (i) removed the rs1011685 (near LPL), which, as described above, did not show consistent associations across individual phenotypes of insulin resistance; (ii) used the 28 SNPs reported in Lotta et al. (7) that were not in loci previously associated with triglycerides or HDL-C at genome-wide significance, and (iii) used 12 SNPs associated with fasting insulin (BMI adjusted) reported by MAGIC consortium (13). As fasting insulin is another marker of insulin resistance, consistent results of the primary analysis and sensitivity analysis (iii) would provide further confidence in concluding the causal role of insulin resistance on the circulating metabolites. Further, sensitivity analysis (ii & iii) are helpful in assessing the contribution of primarily lipid-associated SNPs on the casual effect estimates. In addition, to quantify whether the genetic instruments for IR associated with BMI, we regressed the associations of SNPs with insulin resistance against the associations of SNPs with BMI using summary-level data from GIANT consortium (20). A final step was to remove SNPs from the 53 SNP instrument that individually associated with BMI at P<0.001 using GIANT summary statistics (20) in order to clarify whether this materially altered the MR effect estimates.
Genetic effect estimates are presented as standard deviation (SD) differences in metabolite concentrations per 1-SD genetically higher insulin resistance. To gain insight into the association of the genetic instrument with its individual components, we quantified the association of a 1-SD higher genetically higher insulin resistance on fasting insulin from the MAGIC consortium (13), and the blood lipids HDL-C and triglycerides from the Global Lipids Genetics Consortium (21). We used a two-sided P<0.001 (= 0.05/58; multiple testing correction) to denote evidence of an association.
All analyses were conducted in R.
Results
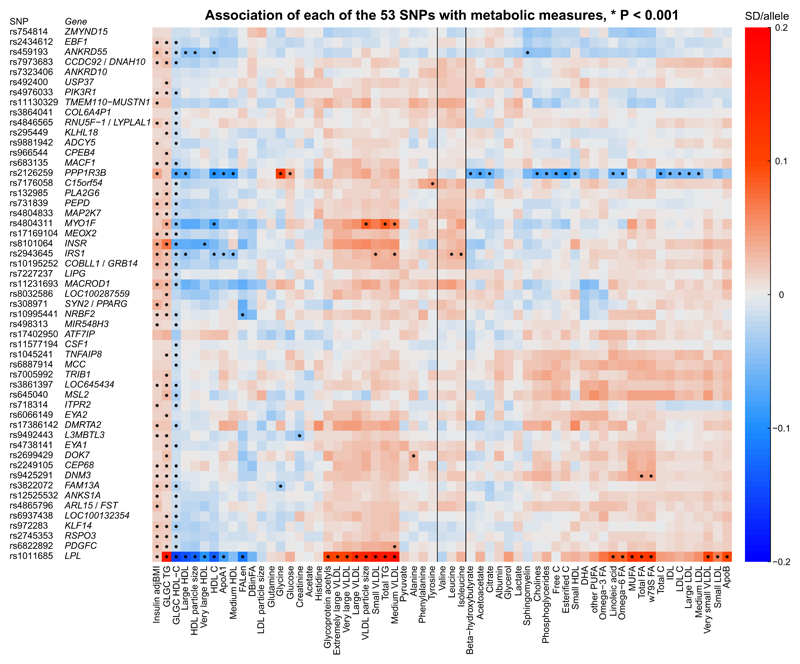
The associations of the 53 SNPs with each of the metabolic measures are shown in Figure 1. As expected, all the SNPs associated with higher BMI-adjusted insulin and triglycerides and lower HDL-C. There were also general trends of the SNPs to associate with higher VLDL and lower HDL traits.
Figure 1. Heat map of the 53 SNPs and their associations with 58 circulating biomarkers.
The units are reported as an SD-difference in metabolic measure per insulin resistance increasing allele. Abbreviations: VLDL, very-low-density lipoprotein; IDL, intermediate-density lipoprotein; LDL, low-density lipoprotein; HDL, high-density lipoprotein; lipoprotein measures without further specification refer to total lipid concentrations; apoA1, apolipoprotein A-I; apoB, apolipoprotein B; w79s FA, omega 7, 9 and saturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; DHA, docosahexaenoic acid; DBinFA, the average number of double bonds in fatty acids; FALen, the average fatty acid chain length. Insulin resistance was defined as a triad of higher fasting insulin (BMI adjusted), higher triglycerides and lower HDL-C. Metabolic measures were quantified by the high-throughput NMR metabolomics platform using primarily fasting serum samples.
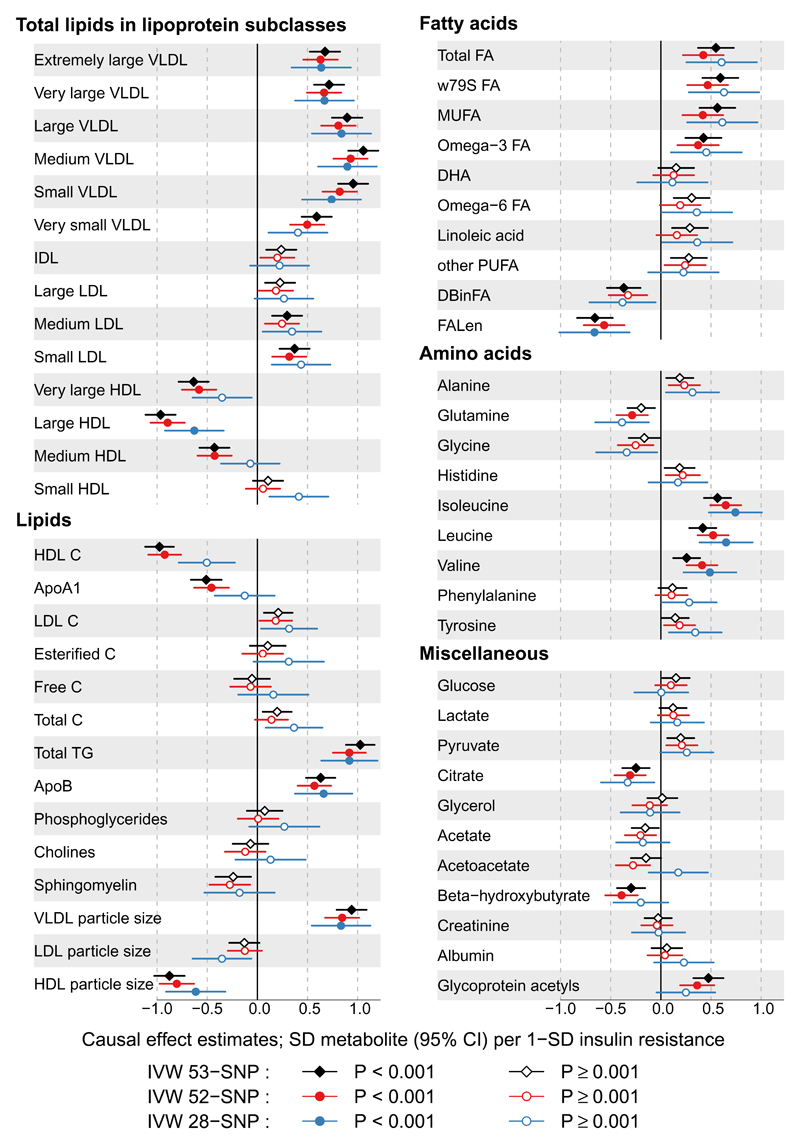
Causal effect estimates of insulin resistance, proxied by the 53-SNP instrument, on the individual metabolic traits are illustrated in Figure 2. The association magnitudes (betas), standard errors and corresponding P-values are reported in Supplemental Table 6. A one-SD genetically higher insulin resistance was associated with 55% (95%CI: 50%, 60%) higher fasting insulin adjusted for BMI, 0.89 mmol/l (95%CI: 0.85, 0.93) higher triglycerides and 0.46 mmol/l (95%CI: 0.44, 0.48) lower HDL-C. In addition, there were clear associations of the genetic instrument for insulin resistance with higher concentrations of all VLDL subclasses with more moderate associations with IDL and LDL subclasses. In contrast, the associations were inverse for most HDL subclasses. The genetic instrument was positively associated with VLDL and negatively with HDL particle size. These findings corroborate the characteristics of the instrument as devised by Lotta et al (7). Similarly, we identified positive associations of the genetic instrument with circulating fatty acids, including monounsaturated and omega-3 fatty acids. Interestingly, the genetic instrument only weakly associated with an increase in NMR-quantified glucose, a finding in keeping with the observation by Lotta et al. using the MAGIC consortium data (7). As reported in the original study (7), the genetic instruments were negatively associated with BMI (Supplemental Table 7).
Figure 2. Forrest plot of the causal effect estimates of insulin resistance on circulating metabolic measures.
Estimates are derived from inverse variance weighted (IVW) Mendelian randomization analyses. The three instruments are: 53 SNPs identified from Lotta et al. (7) (black diamonds); 52 SNPs removing an outlier variant rs1011685 (near LPL) (red circles); 28 SNPs in loci not previously associated with high-density lipoprotein cholesterol and triglycerides at genome-wide significance (blue circles). Open and close symbols indicate P ≥ 0.001 and P < 0.001, respectively. Units are given as SD difference in metabolic measures per 1-SD genetically higher insulin resistance. Abbreviations are as listed in the caption for Figure 1.
We identified strong positive associations of genetically higher insulin resistance with the BCAAs, isoleucine, leucine and valine. These estimates correspond to a 0.56 SD (95%CI: 0.43, 0.70) higher isoleucine, 0.42 SD (95%CI: 0.28, 0.55) higher leucine and 0.26 SD (95%CI: 0.12, 0.39) higher valine per 1-SD higher insulin resistance. Weaker associations were noticed with the other amino acids. In addition, genetically higher insulin resistance was positively associated with glycoprotein acetyls, an inflammation marker (0.47 SD; 95%CI: 0.32, 0.62).
Sensitivity Analyses
Most of the associations identified for the 53-SNP instrument were replicated with the 28-SNP instrument (limited to those SNPs that were not in loci of prior GWAS hits for triglycerides or HDL-C; Figure 2) as well as the 12-SNP instrument (identified in a GWAS of fasting insulin adjusted for BMI; Supplemental Fig. 2). The associations were also consistent when rs1011685 near LPL was removed from the 53-SNP instrument (Figure 2). Removal of 6 SNPs associated with BMI (P<0.001) had no material effect on the MR estimates (data not shown).
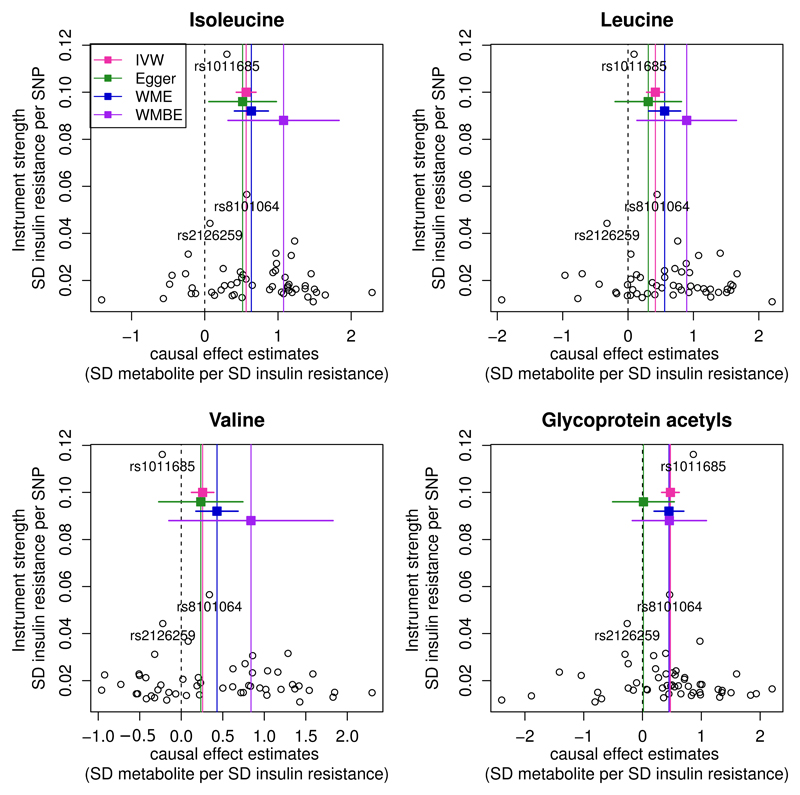
To investigate the robustness of these MR estimates to potential confounding by genetic pleiotropy, we also investigated the association of the 53-SNP instrument with the BCAAs and glycoprotein acetyls (GlycA) using MR-Egger, weighted median and weighted mode-based estimators. Discordance of the point estimates was noticed across the methods, predominantly due to the inclusion of the rs1011685 variant that had minimal effects on insulin adjusted for BMI (Supplemental Table 8). Since MR approaches can be vulnerable to the inclusion of such outliers, we repeated the sensitivity analyses excluding rs1011685, which led to estimates across all MR methods that were comparable to the IVW approach (Figure 3 and Supplemental Table 8).
Figure 3. Funnel plots for the three branched-chain amino acids and glycoprotein acetyls showing the causal effect estimates.
IVW refers to the conventional inverse variance weighted method (using 53 SNPs; red vertical lines), Egger to the MR-Egger (using 52 SNPs; green lines), WME to the weighted median estimator (using 52 SNPs; blue lines) and WMBE for the weighted mode-based estimator (using 52 SNPs; purple line). For the results shown for MR-Egger, WME and WMBE, the outlier SNP rs1011685 near LPL was removed. The 95% CIs for each method are shown as the colourful horizontal lines. Each individual black circle describes the causal effect estimate using the individual SNP as the instrument.
The intercepts of MR Egger were of generally small magnitude (absolute values ≤ 0.01, far smaller than the corresponding beta coefficients) with little or no evidence that they departed from zero, providing little evidence for the presence of genetic pleiotropy (Supplemental Table 9). Because the MR-Egger estimate of the causal effect (obtained from the slope of the regression line) can be underestimated when the assumption of no measurement error of the exposure (NOME) is violated, the heterogeneity index (I2) was used to detect the extent of this potential violation (22). Results remained consistent when SIMEX-adjusted MR-Egger was used to correct potential errors of the SNP to exposure estimates (Supplemental Table 8).
Pathways
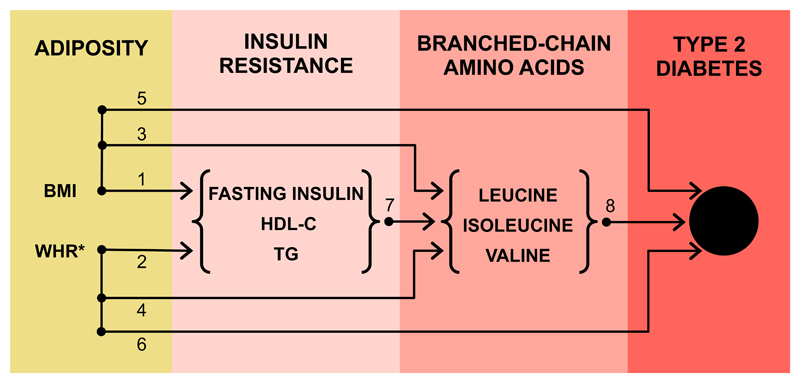
Figure 4 and Supplemental Table 10 illustrates the current evidence base for various pathways leading from adiposity to T2DM. Prior MR studies have shown that general adiposity (measured by BMI) and central adiposity (measured by waist-to-hip ratio adjusted for BMI) causally influence fasting insulin, HDL-C and triglycerides (4). BMI has been previously shown to influence BCAAs (23) and in this study we show that both BMI and waist-to-hip ratio (adjusted for BMI) impact these traits (Supplemental Fig. 3 & 4). Both BMI and waist-to-hip ratio causally affect diabetes (4). Our study shows that insulin resistance impacts on BCAAs, and together with a prior study providing genetic support of BCAAs metabolism in T2DM (11), the various sources of data support a causal pathway that is from adiposity to insulin resistance to BCAAs to diabetes.
Figure 4. Strands of evidence from multiple genetic studies supporting a causal pathway from adiposity, through insulin resistance and branched-chain amino acids, to diabetes.
Sources of evidence: 1. Holmes et al. (3), Dale et al. (4); 2. Dale et al. (4), Emdin et al. (6); 3. Würtz et al. (23), this study (Supplemental Fig. 3); 4. This study (Supplemental Fig. 4); 5. Holmes et al. (3), Dale et al. (4), Lyall et al. (5); 6. Emdin et al. (6), Dale et al. (4); 7. This study (Figure 2), Mahendran et al. (15); 8. Lotta et al. (11). For details of these studies and the MR estimates provided, please see Supplemental Table 10. *refers to adjustment with BMI.
Conclusions
Our study provides genetic evidence in support of higher levels of insulin resistance leading to an elevation in circulating branched-chain amino acids. Within the context of other studies, our findings support the hypothesis that the metabolism of BCAAs may be a mediator that is downstream of adiposity and insulin resistance on the causal pathway to T2DM. If true, then this not only has important aetiological relevance, but also could point towards potential novel opportunities for disease treatment and prevention.
These findings for insulin resistance and BCAAs are consistent with a recent paper by Mahendran et al. (15) in which ten IR associated SNPs were used to quantify the association with a composite measure of BCAAs in a one-sample MR setting of ~1300 individuals. However, the selection of SNPs into the 10-SNP instrument may induce bias as the instrument was enriched for GWAS hits of fasting insulin that were also associated with triglycerides and HDL-C (18). In this study, a more robust approach was taken to instrument derivation by selecting >50 SNPs across the genome, which have recently been identified using a hypothesis-free approach to show directionally consistent associations with a triad of phenotypes that mark insulin resistance; this 53-SNP instrument was used to infer the causality of insulin resistance using a two-sample MR design with little overlap between datasets and with data on ~180,000 individuals for the SNP to exposure (IR) estimates and data on ~25,000 individuals for the SNP to outcome (metabolic markers) estimates. The consistent results that we report derived from multiple genetic instruments and multiple MR sensitivity analyses provide robust evidence that insulin resistance impacts on BCAAs in a cause and effect manner. Particularly, as insulin resistance can be measured by various metrics (e.g. a triad of the phenotypes as defined here and also by fasting insulin alone), the consistent results of the 53-SNP instrument (a genetic proxy for the insulin resistance triad) and 12-SNP instrument (a genetic proxy for fasting insulin alone) across the metabolic profile strengthens the evidence base for a causal role of insulin resistance and potentially validates the biological meaning of insulin resistance as defined by a complex phenotype characterised by higher insulin, higher triglycerides and lower HDL-C.
Interventional studies provide orthogonal support for our findings that obesity and insulin resistance causally impact on circulating BCAAs. Multiple longitudinal studies have shown that BCAA levels were reduced after various insulin-sensitising interventions, including weight loss surgery through gastric bypass, pioglitazone therapy or physical exercise (24–26). Also, a reduction in BCAA concentrations was observed following the secretion of insulin during oral glucose tolerance test (OGTT), with individuals with insulin resistance showing less BCAA suppression (i.e. higher BCAA concentrations) following OGTT (27). Prospective studies have identified circulating BCAAs to be predictive of incident T2DM and a recent genetic study found that the metabolism of BCAAs is likely causally linked to T2DM (11). Triangulating these sources of evidence provides support for the hypothesis that circulating BCAAs may mediate the relation from adiposity and insulin resistance to T2DM. On the other hand, observational studies have reported that higher dietary intake of BCAAs is associated with an improved cardiometabolic risk profile including a lower risk of T2DM (28,29). However, dietary BCAAs, both measured in absolute terms or as a percentage of total protein, are only weakly correlated with circulating concentrations of BCAAs (28,29). There is also evidence that the expression of enzymes involved in BCAA catabolism (e.g. branched-chain α-ketoacid dehydrogenase, BCKD) is reduced in obese and diabetic individuals (11,30). BCKD is responsible for the rate-limiting step of BCAA catabolism and BCKD can be activated by its regulatory phosphatase encoded by PPM1K. Individuals with T2DM have reduced up-regulation of PPM1K in skeletal muscle during OGTT (11). Consistent with this, after weight loss surgery, BCKD concentrations are increased leading to a commensurate reduction in BCAAs (24,30). Thus, elevated circulating BCAA levels observed in obese and diabetic individuals could arise from impaired BCAA catabolism (11). Putting these strands of evidence together, it is plausible that pharmacotherapies to improve or restore the function of BCAA catabolism may represent a means to prevent T2DM. However, further studies are required to understand the exact role of BCAA metabolism in the aetiology of T2DM.
In contrast to the strong effects of IR on the BCAAs, we noticed a generally weaker effect of IR on alanine, glutamine, and aromatic amino acids (phenylalanine and tyrosine). Each of these biomarkers has been associated with the risk of insulin resistance, hyperglycaemia, type 2 diabetes and cardiovascular diseases (8,10,31). Although imprecise estimates were observed for these measures in the MR analyses reported herein, the consistent results from different instruments on these traits merits further investigation in larger datasets to clarify whether these represent causal relationships.
The association that we identify of insulin resistance with GlycA is novel. GlycA is a marker of both acute phase and chronic inflammation and has been linked to neutrophil activity (32). GlycA reflects circulating levels of various inflammatory glycoproteins (primarily alpha-1-acid glycoprotein and haptoglobin), and is also associated with a wide range of inflammatory cytokines (32). Prospective observational studies have identified positive associations of GlycA with cardiovascular disease, T2DM and premature mortality (33,34). A role for inflammation in the development of T2DM has been proposed for many years on account of the observational associations between higher concentrations of biomarkers of inflammation, such as C-reactive protein, interleukin-1, interleukin-6, and the risk of T2DM (35,36). Although recent MR studies have so far failed to provide evidence in support of this hypothesis (36), it remains plausible that such causal pathways (from inflammation to T2DM) exist, and that larger studies and/or investigations of other inflammatory markers and pathways may identify a causal role of inflammation in T2DM. Therefore, GlycA could represent a biomarker either involved in, or correlated to an inflammation pathway involved in the aetiology of T2DM. A causal role of inflammation in vascular disease is gaining traction given recent findings from genetic studies in humans of the interleukin-6 receptor and CHD (37), and more recently in phase III clinical trials of anti-inflammatory drugs for the treatment of CHD (CANTOS trial of canakinumab, a monoclonal antibody to interleukin-1 beta) (38). Of note, a previous study has suggested that BMI has a causal impact on circulating concentrations of GlycA (23) (as we also report in Supplemental Fig. 3 for BMI and Fig.4 for WHR) and our study here provides clarification on the potential causal pathway, showing that insulin resistance is also causal for GlycA. However, elucidating the causal role of GlycA in cardiometabolic disease remains challenging using an MR approach since at present the identified genetic variants associated with GlycA are limited in number (16), thus hindering our ability to answer this important question. Larger GWAS of GlycA may facilitate this endeavor.
Strengths of this study include: (i) a comprehensive genetic instrument for an insulin resistance phenotype using findings from a recent GWAS (7); (ii) characterizing and validating the genetic instrument for IR with a repertoire of biomarkers of triglyceride and HDL-C metabolism; (iii) use of multiple sensitivity analyses (both in the derivation of the genetic instruments and their application to state of the art MR methodologies) which provided robust and consistent evidence; (iv) quantifying the causal effects of insulin resistance on each of the three BCAAs individually; (iv) adding important new information on the effect of insulin resistance on an inflammation marker; and, (v) a data summation that provides evidence of a causal pathway from adiposity through IR and BCAA to T2DM.
Limitations include: (i) analyses were conducted at the summary level and we could not investigate associations by subgroups, e.g., of age or sex, meaning that it is not possible to test whether these associations are modified by age; (ii) our analyses were conducted using European datasets which may hamper their translational relevance to non-Europeans, however risk factors for disease tend to show similar relationships across geographical regions (39) and emerging studies are providing evidence that shows the genetic architecture for common diseases is likely similar across ethnic groups (40) (iii) a meta-GWAS of 3 traits was used to proxy IR, which may not include other traits related to IR and may have limited clinical relevance, although in the original paper by Lotta et al. (7), associations were identified for diabetes and heart disease; (iv) meta-GWAS may select SNPs on the basis of pleiotropy (i.e., by their very selection, they associate with higher fasting insulin, higher triglycerides and lower HDL-C) and thus SNPs may tag heterogeneous pathways, some of which may result in unbalanced horizontal pleiotropy (18). Against this are the consistent associations across the different genetic instruments, their stability to various MR sensitivity analyses (with each MR approach having its own assumptions on the amount and type of genetic pleiotropy, see Supplemental Table 5), and the general consistency with a prior study that used a weaker instrument in a much smaller dataset (15). Finally, the instruments were derived from a meta-GWAS that included fasting insulin adjusted for BMI; conditioning on a trait in discovery GWAS can induce collider bias, as evidenced by the negative association of the instruments with BMI. However, this negative association with BMI would be expected to diminish the association of the genetic instruments with BCAA that we report (and also diminish the association with T2DM and CHD reported by Lotta et al. (7) rather than augment it, and therefore is unlikely to result in major bias in the MR estimates we report. Further, removal of six SNPs that were associated with BMI (at P<0.001 using GIANT summary statistics) had no material impact on the causal estimates derived from MR.
In conclusion, our findings provide new information in support of a causal role of insulin resistance on branched chain amino acids and inflammation. Taken together with recent findings from complimentary studies, these data suggest BCAA metabolism may lie on a causal pathway from adiposity and insulin resistance to type 2 diabetes.
Supplementary Material
Funding
MVH works in a Unit that receives funding from the UK Medical Research Council and is supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre. MAK was supported by the Sigrid Juselius Foundation, Finland. GDS and MAK work in a Unit that is supported by the University of Bristol and UK Medical Research Council (MC_UU_12013/1).
Non-standard Abbreviations and Acronyms
- BCAA
branched-chain amino acid
- IR
insulin resistance
- MR
Mendelian randomization
- IVW
inverse variance weighted
- GlycA
glycoprotein acetyls
- BCKD
branched-chain α-ketoacid dehydrogenase
Footnotes
Duality of Interest
None of the authors report any conflicts of interest.
Author Contributions
Q.W., M.V.H., and M.A.K. designed the study. Q.W. conducted the statistical analyses. Q.W. and M.V.H. wrote the manuscript. All authors interpreted the data and contributed to the discussion, reviewed and edited the manuscript. M.A.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes the responsibility for the integrity of the data and the accuracy of the data analyses.
References
- 1.The GBD 2015 Obesity Collaborators. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N Engl J Med. 2017;377:13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy OT, Czech MP, Corvera S. What causes the insulin resistance underlying obesity? Current opinion in endocrinology, diabetes, and obesity. Curr Opin Endocrinol Diabetes Obes. 2012;19:81–7. doi: 10.1097/MED.0b013e3283514e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes MV, Lange LA, Palmer T, et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet. 2014;94:198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale CE, Fatemifar G, Palmer TM, et al. Causal Associations of Adiposity and Body Fat Distribution With Coronary Heart Disease, Stroke Subtypes, and Type 2 Diabetes Mellitus: A Mendelian Randomization Analysis. Circulation. 2017;135:2373–88. doi: 10.1161/CIRCULATIONAHA.116.026560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyall DM, Celis-Morales C, Ward J, et al. Association of Body Mass Index With Cardiometabolic Disease in the UK Biobank: A Mendelian Randomization Study. JAMA Cardiol. 2017;2:882–9. doi: 10.1001/jamacardio.2016.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emdin CA, Khera AV, Natarajan P, et al. Genetic Association of Waist-to-Hip Ratio With Cardiometabolic Traits, Type 2 Diabetes, and Coronary Heart Disease. JAMA. 2017;317:626–34. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lotta LA, Gulati P, Day FR, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Würtz P, Soininen P, Kangas AJ, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648–55. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guasch-Ferré M, Hruby A, Toledo E, et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;39:833–46. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lotta LA, Scott RA, Sharp SJ, et al. Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med. 2016;13:e1002179. doi: 10.1371/journal.pmed.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles JW, Xie W, Zhang Z, et al. Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J Clin Invest. 2015;125:1739–51. doi: 10.1172/JCI74692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott RA, Fall T, Pasko D, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes. 2014;63:4378–87. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahendran Y, Jonsson A, Have CT, et al. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia. 2017;60:873–8. doi: 10.1007/s00125-017-4222-6. [DOI] [PubMed] [Google Scholar]
- 16.Kettunen J, Demirkan A, Würtz P, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 18.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017 doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017 doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden J, Del Greco MF, Minelli C, et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45:1961–74. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Würtz P, Wang Q, Kangas AJ, et al. Metabolic Signatures of Adiposity in Young Adults: Mendelian Randomization Analysis and Effects of Weight Change. PLoS Med. 2014;11:e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laferrère B, Reilly D, Arias S, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3:80re2. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakazu E, Kondo Y, Ninomiya M, et al. The influence of pioglitazone on the plasma amino acid profile in patients with nonalcoholic steatohepatitis (NASH) Hepatol Int. 2013;7:577–85. doi: 10.1007/s12072-012-9395-y. [DOI] [PubMed] [Google Scholar]
- 26.Kujala UM, Mäkinen V-P, Heinonen I, et al. Long-term leisure-time physical activity and serum metabolome. Circulation. 2013;127:340–8. doi: 10.1161/CIRCULATIONAHA.112.105551. [DOI] [PubMed] [Google Scholar]
- 27.Shaham O, Wei R, Wang TJ, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata C, Nakamura K, Wada K, Tsuji M, Tamai Y, Kawachi T. Branched-chain amino acid intake and the risk of diabetes in a Japanese community: the Takayama study. Am J Epidemiol. 2013;178:1226–32. doi: 10.1093/aje/kwt112. [DOI] [PubMed] [Google Scholar]
- 29.Jennings A, MacGregor A, Pallister T, Spector T, Cassidy A. Associations between branched chain amino acid intake and biomarkers of adiposity and cardiometabolic health independent of genetic factors: A twin study. Int J Cardiol. 2016;223:992–8. doi: 10.1016/j.ijcard.2016.08.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab. 2007;293:E1552–63. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Würtz P, Havulinna AS, Soininen P, et al. Metabolite Profiling and Cardiovascular Event Risk: A Prospective Study of Three Population-Based Cohorts. Circulation. 2015;131:774–85. doi: 10.1161/CIRCULATIONAHA.114.013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie SC, Würtz P, Nath AP, et al. The Biomarker GlycA Is Associated with Chronic Inflammation and Predicts Long-Term Risk of Severe Infection. Cell Systems. 2015;1:293–301. doi: 10.1016/j.cels.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Connelly MA, Gruppen EG, Wolak-Dinsmore J, et al. GlycA, a marker of acute phase glycoproteins, and the risk of incident type 2 diabetes mellitus: PREVEND study. Clinica Chimica Acta. 2016;452:10–7. doi: 10.1016/j.cca.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Fischer K, Kettunen J, Würtz P, et al. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: an observational study of 17,345 persons. PLoS Med. 2014;11:e1001606. doi: 10.1371/journal.pmed.1001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Bao W, Liu J, et al. Inflammatory Markers and Risk of Type 2 Diabetes. Diabetes Care. 2013;36:166–75. doi: 10.2337/dc12-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swerdlow DI. Mendelian Randomization and Type 2 Diabetes. Cardiovasc Drugs Ther. 2016;30:51–7. doi: 10.1007/s10557-016-6638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swerdlow DI, Holmes MV, Kuchenbaecker KB, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379:1214–24. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ridker PM, Everett BM, Thuren T, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017 doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 39.Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 40.Gan W, Walters RG, Holmes MV, et al. Evaluation of type 2 diabetes genetic risk variants in Chinese adults: findings from 93,000 individuals from the China Kadoorie Biobank. Diabetologia. 2016;59:1446–57. doi: 10.1007/s00125-016-3920-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.