Abstract
As the teleost specific immunoglobulin, IgT plays important roles in systemic and mucosal immunity. In the current study, in rainbow trout, we have cloned the heavy chain (Igτ) genes of a secretory form of IgT2 as well as the membrane and secretory forms of a third IgT subclass, termed IgT3. Conserved cysteine and tryptophan residues that are crucial for the folding of the immunoglobulin domain as well as hydrophobic and hydrophilic residues within CART motif were identified in all IgT subclasses. Through analysis of the rainbow trout genome assembly, Igτ3 gene was found localized upstream of Igτ1 gene, while Igτ2 gene situated on another scaffold. At the transcriptional level, Igτ1 was mainly expressed in both systemic and mucosal lymphoid tissues, while Igτ2 was largely expressed in systemic lymphoid organs. After LPS and poly (I:C) treatment, Igτ1 and Igτ2 genes exhibited different expression profiles. Interestingly the transcriptional level of Igτ3 was negligible, although its protein product could be identified in trout serum. Importantly, a previously reported monoclonal antibody directed against trout IgT1 was able to recognize IgT2 and IgT3. These data demonstrate that there exist three subclasses of IgT in rainbow trout, and that their heavy chain genes display different expression patterns during stimulation. Overall, our data reflect the diversity and complexity of immunoglobulin in trout, thus provide a better understanding of the IgT system in the immune response of teleost fish.
Keywords: IgT, Subclass, Immunoglobulin, Rainbow trout
1. Introduction
The adaptive immune system is based on diverse antigen receptors such as the variable lymphocyte receptor (VLR), which is generated via the recombination of leucine-rich repeat (LRR) modular units in jawless vertebrates, as well as B and T cell receptors (BCRs and TCRs), which are generated via somatic hypermutation and variable-diversity-joining rearrangement (VDJ rearrangement) in jawed vertebrates (Alder et al., 2005; Deng et al., 2013). As key molecules in the adaptive immune system, immunoglobulins (Igs) play a pivotal role in the elimination of extracellular and intracellular pathogens. Functional Igs are primarily expressed on the surface of B lymphocytes as BCRs or are secreted into body fluids as antibodies (Abs) to agglutinate pathogens (Parra et al., 2013). The typical structure of an Ig molecule consists of two heavy (H) chains and two light (L) chains. IgH chains usually contain one variable (V) domain and two to four constant (C) domains, and IgL chains are composed of one V domain and one C domain. The classification of an Ig isotype is based on the C domains of its H chain (Danilova and Amemiya, 2009; Flajnik and Kasahara, 2010; Schroeder and Cavacini, 2010).
To date, five Ig isotypes have been well characterized in mammals: IgM (H chain: μ), IgD (δ), IgG (γ), IgE (ε) and IgA (α) (Schroeder and Cavacini, 2010). Human and mouse also possess certain subclasses of IgG (French and Harrison, 1984) and IgA (Kett et al., 1986), which enable more subtle immune responses. Among those Igs, IgM and IgD are evolutionarily conserved and are thought to be the primordial Ig isotypes (Edholm et al., 2011; Han et al., 2016). However in teleosts, IgM (Bradshaw et al., 1971; Hordvik et al., 2013), IgD (Edholm et al., 2010; Ramirez-Gomez et al., 2012; Wilson et al., 1997) and IgT or IgZ (H chain: τ or ζ) (Hansen et al., 2005; Hu et al., 2010) have been identified in various fish species. The teleost specific IgT has first been reported in zebrafish (Danio rerio) (Danilova et al., 2005) and rainbow trout (Oncorhynchus mykiss) (Hansen et al., 2005). After that, IgT has been cloned in various teleost species except for catfish and medaka (Fillatreau et al., 2013). IgT subclasses are also reported in some teleosts such as Atlantic salmon (Salmo salar) (Yasuike et al., 2010), stickleback (Gasterosteus aculeatus) (Bao et al., 2010; Gambon-Deza et al., 2010) and carp (Cyprinus carpio) (including a chimeric IgM-IgT) (Ryo et al., 2010). Thus, Igs in teleost fish may be more complex than previously expected (Sunyer, 2013). IgT plays important roles in mucosal immunity in the gut (Zhang et al., 2010), skin (Xu et al., 2013) and gill (Xu et al., 2016) in rainbow trout (Oncorhynchus mykiss). In addition, recent studies indicated that IgT+ B cells also participate in the responses against viral infections (Castro et al., 2013) and DNA vaccination (Castro et al., 2014). These evidences indicate that teleost IgT is also involved in systemic immunity (Castro and Tafalla, 2015). In trout, IgT was found in serum as a monomer while in mucus it was characterized as a polymer where the monomers were associated via noncovalent interactions (Zhang et al., 2011).
Two types of IgH locus organization have been reported to date: translocon and cluster (Hikima et al., 2011). IgH genes in tetrapods and teleosts are organized as translocon, with multiple variable (V), diversity (D) and joining (J) segments arrayed upstream of constant (C) gene segments in the following order: (Vn-Dn-Jn-Cn) (Danilova et al., 2005; Mashoof et al., 2014; Sun et al., 2013). The IgH loci of cartilaginous fish exhibit a cluster organization as follows: (V-D-J-C)n (Das et al., 2012; Flajnik, 2002). The V, D and J segments are assembled by RAG-1 and RAG-2 through the recognition of recombination signal sequences (RSSs) at the borders of the V, D and J segments (Hikima et al., 2011). In teleosts, Igτ exons are either located in the 5′ regions of the D and J gene segments of Igμ and Igδ in zebrafish (Danilova et al., 2005) and stickleback (Gambon-Deza et al., 2010) or inserted within the V segments of Igμ and Igδ in rainbow trout (Hansen et al., 2005) and Atlantic salmon (Yasuike et al., 2010), which has led to the evolution of a distinct B cell lineage in addition to IgM (Hansen et al., 2005). Furthermore, the heavy chains of teleost IgT subclasses are encoded by duplicated Igτ genes in the same IgH locus or encoded by different IgH loci (Zhang et al., 2011), potentially resulting from teleost-specific and/or salmonid-specific whole-genome duplication events (Berthelot et al., 2014).
So far in rainbow trout, two IgT subclasses have been reported, the membrane and secretory forms of IgT1 (Hansen et al., 2005) and the membrane form of IgT2 (Danilova et al., 2005). In this study, a third Igτ gene, encoding the H chain of IgT3, was identified in rainbow trout in addition to the secretory form of IgT2. The existence of three IgT subclasses was verified also at the protein level. The Igτ gene loci were also characterized based on the trout genome database. All three Igτ genes could be transcribed and translated into both secretory and membrane forms. Furthermore, to investigate potential functional differences in these IgT subclasses, as well as IgM and IgD, the phylogenetic relationships and expression patterns of trout IgH genes were analyzed.
2. Materials and methods
2.1. Experimental fish and tissue preparation
Rainbow trout (70–100 g) were obtained from Zhanghe Reservoir Rainbow Trout Farm (Jingmen, China) and maintained in aquarium tanks using a water recirculation system involving extensive filtration and temperature control (14–16 °C). The fish were acclimated to the aquarium tanks for 2 weeks before being used in experiments.
Fifty trout were divided into two groups, and intraperitoneally (i.p.) injected with LPS (from Escherichia coli 0111:B4; Sigma-Aldrich) and poly (I:C) (a synthetic analogue of viral dsRNA; Sigma-Aldrich), separately, at a dose of 100 μg per 50 g fish body weight. At 0, 1, 3, 7, 14 and 21 d after injection, three fish from each group were euthanized with tricaine (MS-222, Sigma-Aldrich), and the spleen and gut were collected from euthanized fish under sterile environment. These samples were flash frozen in liquid nitrogen and then transferred to −80 °C freezer for RNA extraction. All animal experiments were approved by the Committee on the Ethics of Animal Experiments of the Chinese Academy of Sciences.
2.2. Total RNA isolation and cDNA synthesis
Total RNA was extracted using the Trizol reagent (Invitrogen) and digested with an RNase-free DNase (Qiagen) according to the manufacturer's instruction. In order to normalize the gene expression level for each sample, same amounts (500 ng) of the total RNA were used for cDNA synthesis in a 25 μl reaction volume with M-MLV reverse transcriptase (Promega). The synthesized cDNA was diluted with 75 μl of RNase-free water and used as template for quantitative real-time PCR (qPCR).
2.3. Cloning of trout Igτ subclasses
The constant regions of trout Igτ1 (GenBank accession No. AY870265, membrane form; AY870263, secretory form) and Igτ2 (GenBank accession No. AY773715, membrane form) were used as queries to search trout expressed sequence tag (EST) database at NCBI (http://www.ncbi.nlm.nih.gov). To obtain the C region cDNAs of the secretory form of Igτ2 and both of the secretory and membrane forms of the currently discovered Igτ subclass (Igτ3), firstly 3′ RACE was carried out with the specific forward primers, designed based on the reported Igτ2 and the identified Igτ3 EST (GenBank accession No. CX150523) sequences, as well as the adaptor primers (Supplemental Table 1). Then the cDNAs encoding a partial VH region and the whole CH region of each Igτ subclass were amplified with the degenerate forward and specific reverse primers (Supplemental Table 1). Ex Taq HS (Takara) was used in all PCR and the PCR products were cloned into the pMD19-T vector (Takara) for sequencing.
2.4. Sequence and phylogenetic analyses
Sequenced trout Igτ subclasses were analyzed using DNAMAN software (Lynnon). To translate the nucleotide sequences to amino acid sequences, the ExPASy online tools (http://web.expasy.org/) were used. Signal peptides were predicted using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Multiple sequence alignment was performed using CLUSTAL X version 2.1 with default parameters and then manually adjusted. On the basis of the alignment, phylogenetic tree was constructed with the MEGA 4.0 package, using a neighbor-joining algorithm and pariwise deletion. Bootstrap values supporting tree nodes were derived from 1000 replications. Trout β2-microglobulin was used as outgroup.
2.5. Extension and annotation of trout IgH locus
Using the published rainbow trout genome database (Berthelot et al., 2014), we extended the trout IgH locus (Hansen et al., 2005) by assembling scaffold_2931 together with the previously reported trout IgH locus (GenBank accession No. AH014877). The software GENSCAN and pairwise TBLASTN alignments were employed for the assembled sequence annotation as described previously (Zhang et al., 2016). According to conserved recombination signal sequence (RSS) motif specificity, D and J segments were identified by pattern search with the program FUZZNUC, an online software package (http://bioinfo.nhri.org.tw/cgi-bin/emboss/fuzznuc). The pattern of RSS heptamer and nonamer sequences with an 11- to 13-nucleotide spacer (CACAGTG-N11-13-ACAAAAACC) allowing a maximum of four mismatches and then manually adjusted.
2.6. qPCR analysis of IgH expression
In order to determine the expression levels of trout IgH genes in different tissues and sorted cells, qPCR was performed with CFX Connect™ (BioRad). The efficiencies for all qPCR primers were analyzed as previously described (Zhang et al., 2009). The primer pairs for IgM, IgD, β-actin and EF-1α were reported previously (Chang et al., 2013; Zhang et al., 2010). All the primer pairs used in this study were qualified, with the efficiencies between 90% and 105%. The specificity of each primer pair was verified by dissociation curve and sequence analyses of the qPCR products. The qPCRs were conducted as follow: 4 μl diluted cDNA template, 5 μl SsoAdvanced™ SYBR Green Supermix (BioRad), and 250 nM forward and reverse primers in 10 μl reaction volumes. The amplification reaction condition consisted of 95 °C for 3 min, 45 cycles of 95 °C for 20 s and 60 °C for 20 s, and then 72 °C for 20 s. Melt curve from 65 °C to 95 °C was added in the final. Three independent qPCR experiments were performed.
2.7. Leukocyte isolation and fractionation
Trout leukocytes were isolated from the peripheral blood (PBLs) and head kidney (HKLs) as described previously (Boshra et al., 2004, 2005). Briefly, trout blood was obtained from the caudal vein and diluted (1/5) immediately with DMEM (Invitrogen) supplemented with 100 μg/ml streptomycin, 100 U/ml penicillin, and 25 U/ml heparin (Invitrogen). Trout head kidney was removed aseptically and passed slowly through a 100 mm nylon mesh (BD Biosciences), then suspended in the same culture medium as described above. The diluted cell suspensions from blood and head kidney were carefully layered onto a 51/34% discontinuous Percoll (GE Healthcare) density gradient and then centrifuged, with zero accelerate and brake, at 400 g for 30 min. The cells laying at the interfaces were carefully collected, and immediately washed with DMEM supplemented with 100 μg/ml streptomycin and 100 U/ml penicillin.
Cell sorting was carried out using the conditions described previously (Zhang et al., 2010). Briefly, isolated trout PBLs and HKLs were suspended in sterility PBS (Invitrogen) supplemented with 2% fetal bovine serum (FBS; Invitrogen), and then stained with mouse anti-trout IgT (Clone: 38.5; isotype: IgG2b) (1 μg/ml) monoclonal antibody (mAb) at 4 °C for 60 min. As a control, cells were incubated with normal mouse IgG. FITC-goat anti-mouse IgG2b (Jackson Scientific) was used as secondary antibody to detect the IgT+ cells. After two times of washing with PBS supplemented with 2% FBS, the IgT+ cells from trout PBLs and HKLs were sorted by flow cytometry using BD FACSAria III (BD Biosciences). The sorted cells were washed and analyzed by qPCR.
2.8. Prokaryotic expression of the recombinant heavy chains of trout IgT subclasses
The subcloning and expression of the heavy chians of trout IgT subclasses in Escherichia coli were performed as described previously (Zhang et al., 2009) with minor modification. The pET-23b(+) expression vector (Novagen) was used for constructing the expression plasmids. The DNA fragments encoding the Cτ2∼Cτ4 of trout IgT1, IgT2 and IgT3 were amplified and digested with EcoR I and Xho I and ligated to pET-23b(+) vector, which was digested with the same restricted enzymes. Then the recombinant plasmids were verified by sequencing. The expression plasmids were subsequently transformed in BL21(DE3)-CodonPlus-RIL expression host competent cells (Stratagene). Bacterial cells were cultured in Luria-Bertani broth containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol and grown at 37 °C with shaking. The OD600 of the culture was allowed to reach 0.6 before induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and allowed to grow for additional 15 h at 15 °C. Cells were harvested by centrifugation and resuspended in PBS (containing the protease inhibitor mixture, Sigma-Aldrich). Cell suspensions were disrupted using sonication and then centrifuged at 12000 g for 20 min to pellet the precipitate. Finally, the supernatants of cell lysates were quantified and subjected to SDS-PAGE for coomassie blue staining and immunoblot analysis.
2.9. Immunoblot analysis
The supernatants (containing the soluble recombinant proteins) of cell lysates were separated by 12% SDS-PAGE under reducing and non-reducing conditions and then transferred to polyvinylidene difluoride membranes (BioRad). The membranes were blocked in TBST buffer (25 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20 [pH 7.5]) containing 5% nonfat dry milk overnight at 4 °C, and then probed with diluted mouse anti-trout IgT mAb (Clone: 38.5; isotype: IgG2b) for 2 h at room temperature. The membranes were washed three times with TBST and incubated with peroxidase-conjugated anti-mouse IgG (NA931; GE Healthcare) for 1 h at room temperature. After three additional washes with TBST, the membranes were stained with Immobilon TM Western Chemiluminescent HRP Substrate (Millipore) and detected using an ImageQuant LAS 4000 system (GE Healthcare).
2.10. Serum IgT purification and LC-MS/MS
Purification and LC-MS/MS analysis of trout serum IgT were performed as described previously (Zhang et al., 2010). Approximately 2 μg of affinity purified trout IgT was resolved on a 4–15% SDS-PAGE under non-reducing conditions and stained with Coomassie blue. The band corresponding to IgT was subjected to in-gel digestion and LC-MS/MS analysis, respectively. The resulting masses and MS/MS spectra were searched against the trout Igτ subclass sequences.
2.11. Statistical analysis
The statistic p values were calculated by Student's t-test (SPSS Statistics, version 19, IBM) between different groups. The p value < 0.05 was considered statistically significant.
3. Results
3.1. Sequence analysis of trout Igτ subclasses
Based on the reported cDNA sequences of trout Igτ1 (membrane and secretory forms) and Igτ2 (membrane form), the cDNAs encoding a partial VH region and the whole CH region of the secretory form of Igτ2 and both of the secretory and membrane forms of the currently discovered Igτ3 were cloned.
To characterize the H chains of trout IgT subclasses, alignment and similarity analyses of the deduced amino acid sequences of the CH domains, secretory tail and transmembrane (TM) region were performed between trout IgM and IgT as well as zebrafish IgM and IgZ. As shown in Fig. 1, trout IgT3 is composed of four putative CH domains, same as IgT1 and IgT2. Within CH1 domain, the first cysteine residue that is considered to be used to form interchain disulfide bond is conserved in all aligned Ig sequences. In addition, conserved cysteine residues (crucial for the forming of intrachain disulfide bonds) and tryptophan residues (crucial for the folding of IgSF domain) (Lesk and Chothia, 1982) are also present within each CH domain of trout IgT3. Furthermore, the TM region of trout IgT3 contains a conserved antigen receptor transmembrane (CART) motif, which is present in the TM regions of all antigen receptors of jawed vertebrates.
Fig. 1.
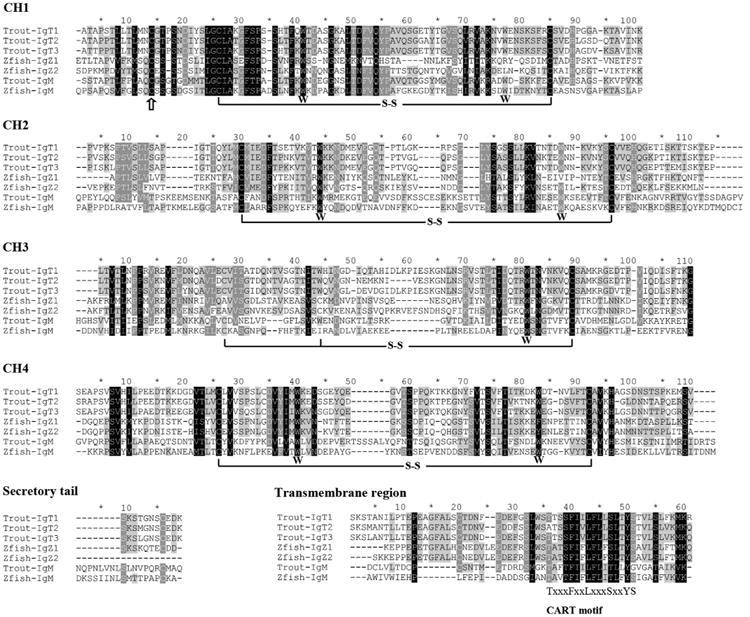
Amino acid sequence alignment of the CH regions of IgM and IgT/Z in trout and zebrafish. Multiple sequence alignment was performed using CLUSTAL X version 2.1 with default parameters. Conserved residues are shaded and gaps are indicated by “-”. The conserved cysteine (C) residues that form the intrachain disulfide bonds are marked with black lines, and the conserved tryptophan (W) residues are shown below the sequences. The conserved cysteine (C) residues that form the interchain disulfide bond with IgL chain are marked by “↑”. the conserved CART motif within transmembrane region is shown below the sequences. The GenBank accession numbers of the IgH sequences used for alignment are: rainbow trout μ (Trout-IgM, AAA56662), rainbow trout τ1 (Trout-IgT1, AAW66978), rainbow trout τ2 (Trout-IgT2, AAV48553), rainbow trout τ3 (Trout-IgT3, ANW11927), zebrafish μ (Zfish-IgM, AAT67447), zebrafish ζ1 (Zfish-IgZ, AAT67446), and zebrafish ζ2 (Zfish-IgZ2, ACH92959).
A similarity comparison of the CH domains within the IgM and IgT subclasses in trout and zebrafish was carried out (Fig. 2). The deduced amino acid sequences of CH1 share the highest similarity among trout IgT subclasses, with IgT1 even reaching 90% of identity with IgT3. However, the CH4 domains, whose identities are less than 80%, exhibited the lowest similarity among trout IgT subclasses. The similarities of the CH2 and CH3 domains are intermediate to those of CH1 and CH4. When the whole CH regions of IgT subclasses in trout are compared, the sequence identity between IgT1 and IgT3 is slightly higher than the identities between IgT2 and IgT3 as well as between IgT2 and IgT1.
Fig. 2.
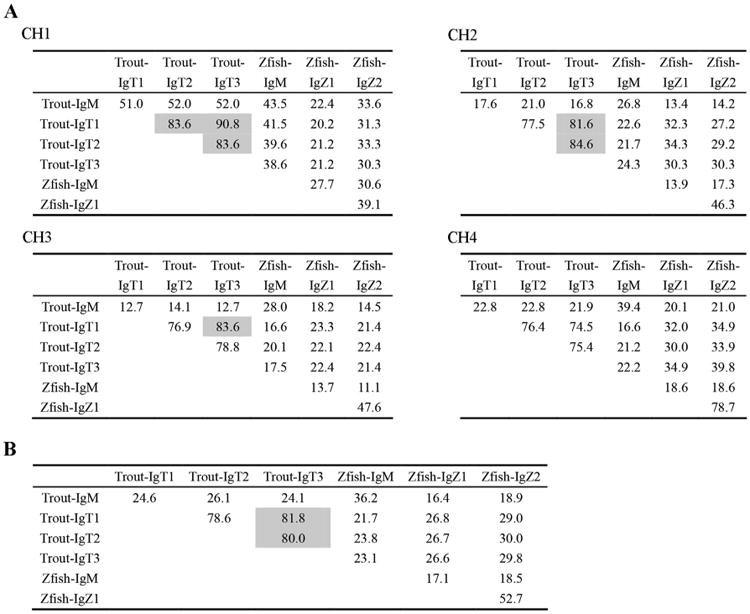
Percentage identity of the amino acid sequences of the CH domains of IgM and IgT/Z in zebrafish and trout. Multiple sequence alignments were performed using CLUSTAL X version 2.1 with default parameters, then the percentage identity (%) was calculated by BioEdit software at default settings (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html). The identity that greater than 80% is shaded. (A) The percentage identity within each CH domain; (B) The percentage identity of the whole CH region.
To explore the relationships between trout Igτ subclasses and other teleost IgH, a phylogenetic analysis was carried out based on the CH regions of diverse teleost species (Fig. 3). The resulting phylogenetic tree indicated that trout Igτ subclasses are closely related to salmon Igτ subclasses and cluster with the Igτ clade of other teleosts. The currently cloned trout Igτ3 is more closely related to trout Igτ1 than to Igτ2.
Fig. 3.
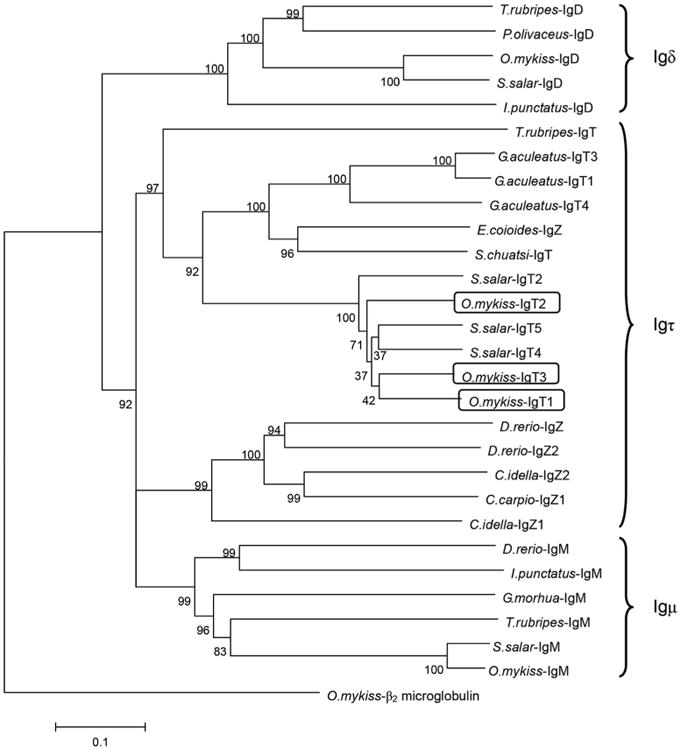
Phylogenetic analysis of teleost IgH based on the CH sequences. Neighbor-joining tree with pairwise gap deletions was drawn using MEGA 4.0 with 1000 bootstrap replications. Trout β2-microglobulin was used as an outgroup. The scale represents the genetic distance. Clusters of sequences are marked and the GenBank accession numbers are: O.mykiss β2-microglobulin (AAB04661); T.rubripes IgD (BAD34542.1); P.olivaceus IgD (BAB41204.1); O.mykiss IgD (AAW66976.1); S.salar IgD (ADD59896.1); I.punctatus IgD (ADF56020.1); D.rerio IgM (AAT67447.1); I.punctatus IgM (A45804); G.morhua IgM (AKL81191.1); T.rubripes IgM (BAD26619.1); S.salar IgM (ACN10415.1); O.mykiss IgM (AAW66972.1); T.rubripes IgT (BAD69712); E.coioides IgZ (ACZ54909); S.chuatsi IgT (AAY42141); S.salar IgT2 (ADD59873); S.salar IgT4 (ADD59858); S.salar IgT5 (ADD59859); D.rerio IgZ (AAT67446); D.rerio IgZ2 (ACH92959); C.idella IgZ (ADD82655); C.idella IgZ2 (ABF19723); C.carpio IgZ1(BAJ41037); O.mykiss IgT1 (AAW66978); O.mykiss IgT2 (AAV48553); O.mykiss IgT3 (ANW11927).
3.2. Genomic organization of trout IgH locus
To identify the Igτ2 and Igτ3 gene loci in trout genome, local blast searching was performed against rainbow trout genome sequences and scaffold database. According to the blast results, the CH regions of the secretory and membrane forms of trout IgT3 are encoded by the same Cτ3 gene through alternative splicing at the 3′ end of the CH4 exon. The secretory Igτ3 transcripts are produced through the strategy that the splice site at the end of CH4 exon is ignored and the transcription continues until reaching the stop codon, whereas the transcripts of the membrane form of the Igτ3 are produced via splicing of the CH4 exon to the TM exon.
In addition, exons of both Cτ1 and Cτ3 were identified in the same scaffold_2931 (GenBank accession No. FR907203), whereas the Igτ2 locus is located in trout scaffold_3178 (GenBank accession No. FR907450) (Fig. 4). Thus, the Igτ locus within trout scaffold_2931 can be assembled with the previously reported trout IgH locus (AH014877). Based on the extended trout IgH locus, the Igτ3 gene is located upstream of the Igτ1 gene and possesses its own V, D and J gene segments, while the relative location of Igτ2 gene with Igτ1 and Igτ3 is still unclear.
Fig. 4.
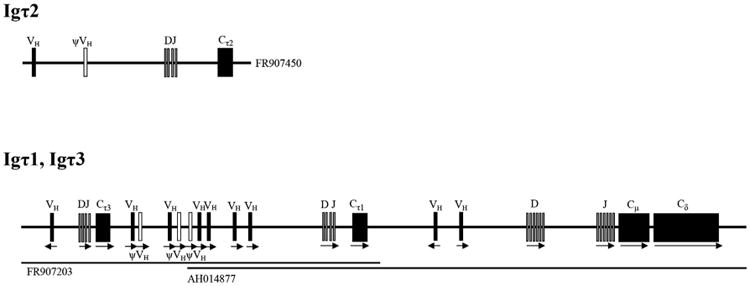
Genomic organization of rainbow trout IgH loci to scale. Arrow represents the transcriptional orientation. Functional gene segments are marked by filled symbols and pseudogenes (ψ) by open symbols.
3.3. Tissue distribution of trout IgH
The expression patterns of the IgH genes were explored using qPCR. As shown in Fig. 5, trout IgH genes were primarily expressed in lymphoid tissues, such as the head kidney, spleen, thymus, gill and blood. Specifically, Igμ transcripts were largely detected in the head kidney, spleen and gill, and moderately in the thymus, blood and gut. Igδ was expressed at considerably high levels in the head kidney, spleen and blood. However, Igτ1 and Igτ2 demonstrated different expression patterns. Igτ1 transcripts were found not only in the systemic lymphoid tissues (such as head kidney, spleen, thymus and blood) but also in the mucosal lymphoid tissues (such as gut, gill and skin). However, Igτ2 transcripts were primarily detected in the systemic lymphoid tissues such as spleen, thymus and blood. In contrast, Igτ3 expression levels were too low to be detected except for a slight transcription in the spleen (data not shown).
Fig. 5.
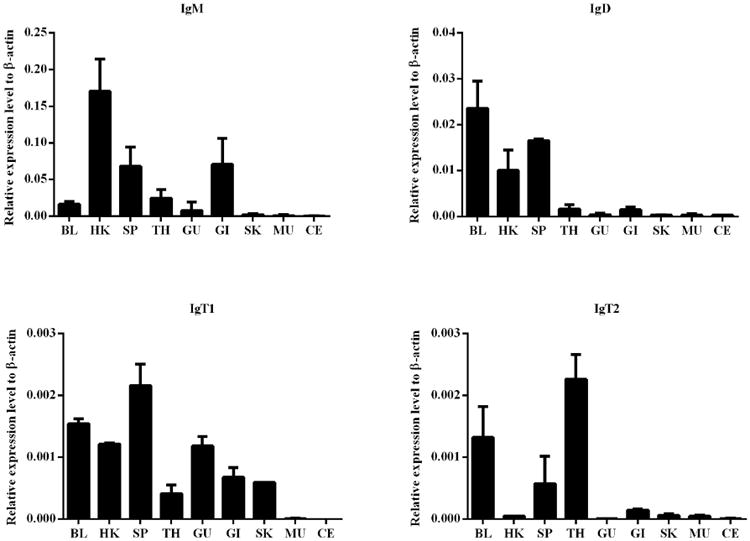
Expression patterns of trout IgH genes in various tissues. The relative expression levels were detected by qPCR, and β-actin was used as an internal control. Abbreviations for tissues are as follows: BL, blood; HK, head kidney; SP, spleen; TH, thymus; GU, gut; GI, gill; SK, skin; MU, muscle; CE, cerebrum. The values represent the mean ± SD of three individual fish.
3.4. Regulation of trout IgH gene expression by LPS and poly (I:C)
Expression levels of the IgH genes in trout gut and spleen were examined after i.p. injection of LPS or poly (I:C) at different time points (Fig. 6). Trout Igμ transcripts in the gut were substantially increased at 1 d after stimulation with LPS and reached a peak at 3 d, whereas its expression in the spleen was obviously increased at 7 d. When stimulated with poly (I:C)), trout Igμ was also up-regulated in the gut at 3 d, but almost no change was observed in the spleen. In the LPS treatment group, the expression of Igδ in the gut and spleen exhibited a similar pattern and slowly increased to a peak at 7 d and 14 d, followed by a decrease at 21 d. After stimulation with poly (I:C), the expression levels of Igδ fluctuated regularly in the spleen but obviously increased in the gut at 14 d. Igτ1 transcripts reached a peak in the gut at 3 d after stimulation with LPS and exhibited a slight increase in the spleen, whereas Igτ1 was slowly up-regulated and reached a peak at 7 d in the spleen and at 14 d in the gut after treatment with poly (I:C). Furthermore, after LPS stimulation, Igτ2 exhibited an obvious increase at 3 d in the gut. However, when stimulated with poly (I:C), Igτ2 increased slightly and reached a peak at 7 d in both the spleen and gut. Taken together, trout IgH responses were much stronger in the gut than in the spleen following i.p. injection of LPS and poly (I:C). In addition, Igτ1 and Igτ2 exhibited differential response patterns.
Fig. 6.
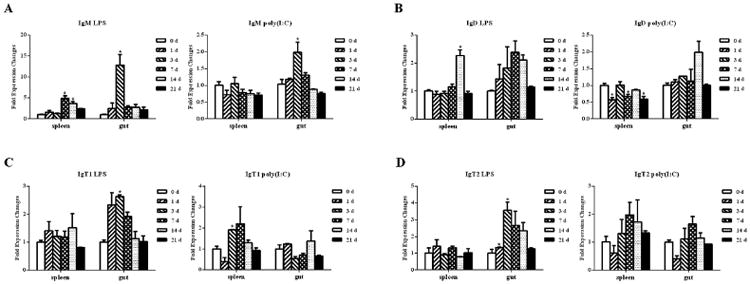
Expression analyses of the IgH genes in the spleen and gut of the trout stimulated with LPS or poly (I:C). The relative expression levels were detected by qPCR and normalized against the expression at 0 d (set as 1). EF-1α and β-actin were used as internal controls. The values represent the mean ± SD of each individual fish. Asterisks indicate significant differences between each time point and control time point (0 d), the p values were calculated by Student's t-test (*, p < 0.05).
3.5. Reactivity of the anti-trout IgT1 mAb to IgT2 and IgT3
A mAb directed to trout IgT1 was developed in our previous study (Zhang et al., 2010). With the discovery and classification of three trout IgT subclasses, we were eager to determine the reactivity of this anti-IgT1 mAb with IgT2 and IgT3. First, IgT+ B cells were sorted from trout PBLs and HKLs using the mouse anti-trout IgT1 mAb. Thereafter, the expression level of each Igτ subclass in the sorted IgT+ B cells was determined using qPCR. As shown in Fig. 7, Igτ1 and Igτ2 transcripts were both detected in the IgT+ B cells from both the HKLs and PBLs. Igτ1 expression levels were higher than that of Igτ2 in HKLs, whereas both Igτ1 and Igτ2 demonstrated equivalent transcriptional levels in PBLs. Igτ3 transcripts were too low to be detected using qPCR.
Fig. 7.
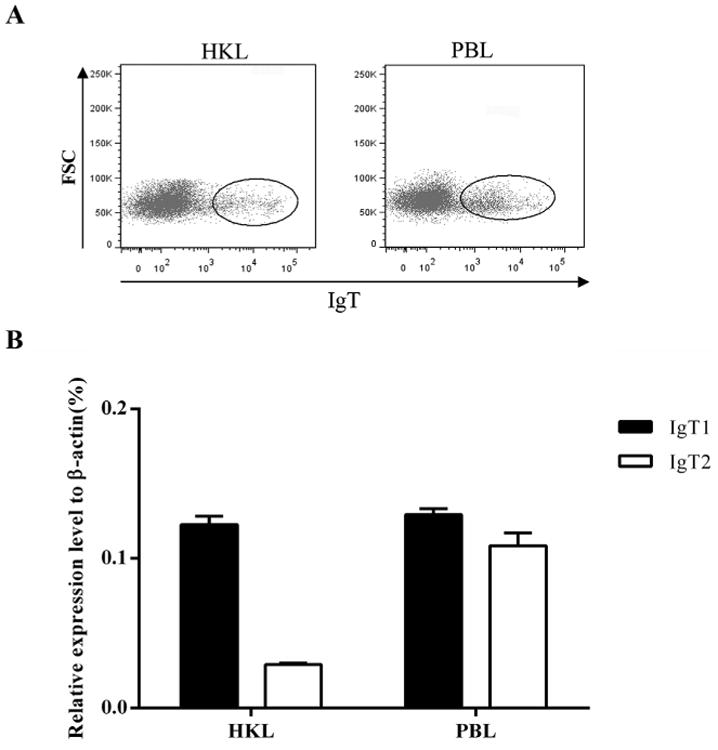
Expression analyses of trout Igτ genes in the sorted IgT+ B cells. (A) Anti-trout IgT1 mAb was used to sort the IgT+ B cells from HKLs and PBLs. (B) The relative expression levels of trout Igτ1 and Igτ2 were detected by qPCR, and β-actin was used as an internal control. The values represent the mean ± SD of three individual fish.
To further investigate the reactivity of the anti-trout IgT1 mAb against the three IgT subclasses, the CH2–CH4 domains of IgT1, IgT2 and IgT3 were subcloned into a prokaryotic expression plasmid, and the soluble expression products were analyzed by Western blotting. As shown in Fig. 8, the recombinant proteins were expressed in soluble form and had a molecular mass of ∼40 kDa, and Western blotting revealed an obvious band in the target area corresponding to Igτ1, Igτ2 and Igτ3 recombinant proteins. Therefore, the mAb that we developed to target IgT1 also recognized the recombinant Igτ2 and Igτ3.
Fig. 8.
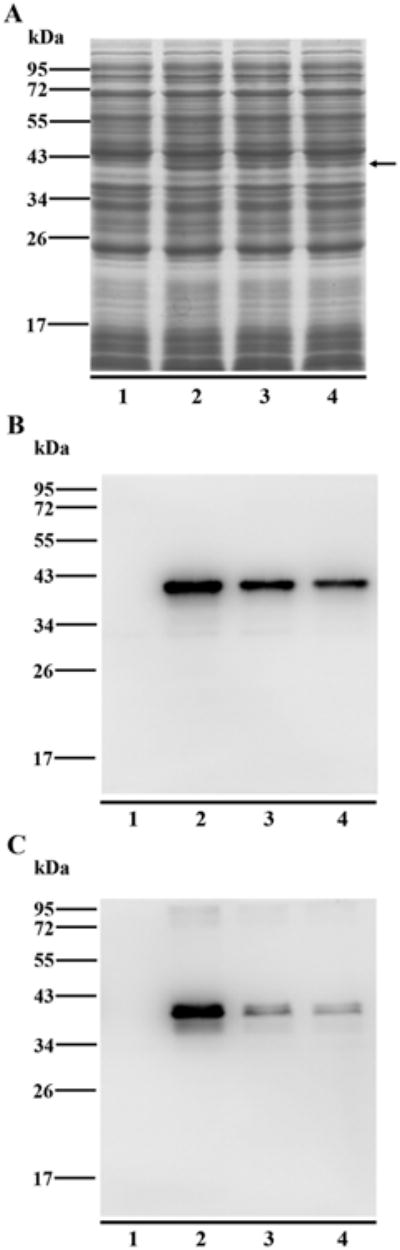
Western blotting analysis of the reactivity of anti-trout IgT1 mAb to other IgT subclasses. The CH2–CH4 regions of trout IgT1, IgT2 and IgT3 were recombinantly expressed in E.coli. Supernatants of the cell lysates (containing the soluble recombinant proteins) were subjected to SDS-PAGE under reducing condition for coomassie blue staining (A) and under both reducing (B) and non-reducing (C) conditions for immunoblotting with mouse anti-trout IgT1 mAb. The total proteins loaded on SDS-PAGE were 15 μg for gel staining and 5 μg for immunoblotting. The expressed recombinant proteins were directed by a black arrow. Lane 1: blank control (trout Igτ1 without IPTG induction); Lane 2: trout Igτ1; Lane 3: trout Igτ2; Lane 4: trout Igτ3.
3.6. Identification of trout IgT subclasses at the protein level
Three IgT subclasses were identified at the protein level in trout serum via LC-MS/MS using affinity purified trout serum IgT by the anti-IgT1 mAb. Mass spectrometry analysis identified a total of 10, 1, and 7 unique peptides belonging to the CH domains of Igτ1, Igτ2 and Igτ3, respectively (Table 1), Additionally, 2 peptides were identified to be shared by both Igτ1 and Igτ3, 1 peptide shared by both Igτ2 and Igτ3, and 2 peptides shared by all three Igτ subclasses. These results supported the existence of three IgT subclasses in rainbow trout and provided complementary evidence that the reactivity of the anti-trout IgT1 mAb with trout IgT2 and IgT3.
Table 1.
LC-MS/MS analyses of the IgT subclasses in trout serum.
| Peptide | Subclass | Position (aa in CH) | Frequency |
|---|---|---|---|
| KGFSPSSHTFQWTDASGK | τ1 | 28–45 | 60 |
| KTAVINKPVPK | τ1 | 92–102 | 35 |
| KRPSGLYSGSSLLK | τ1 | 152–164 | 88 |
| RPSGLYSGSSLLK | τ1 | 152–164 | 2 |
| KTTSKTEPLTVTLNPPR | τ1 | 189–205 | 42 |
| KTEPLTVTLNPPR | τ1 | 193–205 | 59 |
| KVQCSAMKRGEDTPVIQDISFTK | τ1 | 277–299 | 11 |
| RGEDTPVIQDISFTK | τ1 | 285–299 | 30 |
| KEDSGEYQEGVTSPPQK | τ1 | 341–357 | 4 |
| KGNYFVTSVFTITK | τ1 | 360–373 | 12 |
| KSGEDTPVIQDLSFTK | τ2 | 280–295 | 77 |
| KGFSPSSHTFK | τ3 | 28–38 | 5 |
| KGFSPSSHTFKWTDASGK | τ3 | 28–45 | 61 |
| KRGDVTPVIQDLSFNK | τ3 | 284–299 | 1 |
| RGDVTPVIQDLSFNK | τ3 | 285–299 | 13 |
| KVNSGDYQEGVTSPPQK | τ3 | 341–357 | 1 |
| KGNYSVTSVFTTTK | τ3 | 360–373 | 8 |
| KHLGSDNNTTPQGR | τ3 | 387–400 | 9 |
| KALTDFVQYPAVQSGETYTGVSQLR | τ1, τ3 | 45–69a,c | 39 |
| KVTNTDWNNK | τ1, τ3 | 164–173a,c | 3 |
| KTEPLTVTLNPPSVK | τ2, τ3 | 193–207b,c | 2 |
| KNVWENSK | τ1, τ2, τ3 | 72–79a,b,c | 4 |
| RVSTLTIDQTR | τ1, τ2, τ3 | 261–271a,c257–267b | 21 |
Position in the CH region of Igτ1.
Position in the CH region of Igτ2.
Position in the CH region of Igτ3.
4. Discussion
IgT is the most recent immunoglobulin class to be identified in a vertebrate species (Flajnik, 2005), and its genomic organization, protein structure and biochemical functions have been characterized in various teleosts. The mucosal immune function of IgT has been well-characterized in trout gut (Zhang et al., 2010), skin (Xu et al., 2013) and gill (Xu et al., 2016), whereas IgM plays a prevalent role in the systemic immunity. More than one subclass of IgT has been identified in different teleosts, which may be attributable to whole genome duplications. The H chains of IgT subclasses are encoded by duplicated Igτ genes either in the same IgH locus, as in stickleback fish (Bao et al., 2010; Gambon-Deza et al., 2010), or in different IgH loci, as in Atlantic salmon (Yasuike et al., 2010). In the current study, we cloned the H chain genes of the secretory IgT2 as well as the secretory and membrane forms of IgT3 in trout. The H chains of both the secretory and membrane forms of each IgT subclass are encoded by the same Igτ gene through differential splicing site. The transcript of the H chain of the membrane form of each IgT subclass is produced by splicing the Cτ4 exon to the TM exon, similar to mammalian membrane IgM but different from teleost membrane IgM; however, the generation of secretory IgT involves the same mechanism as teleost secretory IgM. Based on the alignment of the CH domains of IgM and IgT subclasses, conserved cysteine residues that participate in the formation of intrachain disulfide bonds were found within the CH1, CH2 and CH4 domains of both IgM and IgT. However, in the CH3 domains of zebrafish IgZ1 and IgZ2, the positions of the first cysteine residues are different with other sequences, this situation may indicate that non-classical cysteine residues are not important for the formation of the three-dimensional structure of IgSF (Danilova and Amemiya, 2009). A conserved cysteine residue responsible for the interchain disulfide bond with IgL was also found in the CH1 domains of all IgT subclasses. The CART motifs identified in the TM regions of trout IgT subclasses are quite similar to those found in mammalian Igμ chains. Thus, the membrane forms of trout IgT subclasses may have the capacity to interact with Igα/Igβ to form BCRs (Campbell et al., 1994).
Alignment of the CH sequences of IgM and IgT in trout and zebrafish indicates that the CH1 is the most highly conserved domain between different IgH isotypes. However, the CH4 domain exhibits the lowest level of sequence identity, not only between different IgT subclasses but also between IgM and IgT, implying that Cτ4 provides the specificity to IgT and its subclasses (Gambon-Deza et al., 2010). The sequence identities of the whole CH regions of IgM and IgT indicate that among the three IgT subclasses in trout, the relationship between IgT1 and IgT3 is much closer than the relationships between either of them to IgT2. In addition, IgT1 and IgT3 are grouped together in the phylogenetic tree, supporting the sequence identity results.
Knowledge of trout IgH loci was extended with the discovery of the Igτ3 gene using local blast. The Igτ3 gene contains its own V, D and J segments and is located upstream of Igτ1 gene. The Igτ2 gene identified in another scaffold indicates that at least two IgH loci may exist in the trout genome (Hansen et al., 2005). This phenomenon was also observed in Atlantic salmon, whose two IgH loci contain 3 functional Igτ genes and additional 5 Igτ pseudogenes (Yasuike et al., 2010). The presence of multiple Igτ genes in trout and salmon might be attributable to teleost-specific and salmonid-specific whole genome duplication events (Berthelot et al., 2014; Hoegg et al., 2004), resulting in a positive and significant increase in the diversity of antibody repertoire. Given the limited information available in terms of trout genome data, further investigation is required to determine if there is any other Ig gene located upstream and downstream of the Igτ2 gene.
The heavy chain genes of zebrafish IgZ1 and IgZ2 were reported to exhibit different expression patterns. IgZ1 was predominantly expressed in the head kidney and thymus, whereas IgZ2 was expressed in both the systemic and mucosal lymphoid organs, similar to IgM (Hu et al., 2010). In the current study, the heavy chain genes of trout IgT1 and IgM demonstrated a similar expression pattern with zebrafish IgZ2 and IgM. Trout IgT2 was primarily expressed in the systemic lymphoid organs, like zebrafish IgZ1. This phenomenon may suggest the functional difference between these two IgT subclasses as well as the non-mucosal role for IgT (Castro et al., 2013, 2014; Castro and Tafalla, 2015). In addition, combined with the previous studies about trout IgT+ B cells in various mucosal tissues during homeostatic conditions (Xu et al., 2013, 2016; Zhang et al., 2011), it seems that there are more IgT+ B cells and IgT transcripts in gill than gut and skin. This may be due to the circulation of B cells in gill (Gomez et al., 2013). Furthermore, IgT3 is slightly expressed only in the spleen. However, the identification of IgT3 peptides in trout serum indicates that its function can not be ignored. Beyond this, transcripts of IgM and IgT could also be detected in trout thymus, where T cells mainly develop. This was also observed in other teleost, like zebrafish (Danilova et al., 2005) and Atlantic salmon (Tadiso et al., 2011). IgM+ B lymphocytes could also be detected in the thymus of zebrafish (Danilova and Steiner, 2002), trout (Korytar et al., 2013) and New Zealand groper (Ployprion oxygeneios) (Parker et al., 2012). Thus, whether IgT+ B cells exist in fish thymus needs further investigation.
When stimulated with E. coli LPS and poly (I:C), the expression patterns of trout IgH genes were notably different, suggesting that all IgH genes are involved in the immune responses in rainbow trout. After stimulated with LPS, the expression levels of IgT1 and IgT2 in trout gut were obviously up-regulated, but the regulation of poly (I:C) to IgT1 and IgT2 was weaker. Compared with the previously reported immune responses to parasites in gut (Zhang et al., 2011), skin (Xu et al., 2013) and gill (Xu et al., 2016), it seems that IgT1 and IgT2 play active roles in the immune response of trout to bacteria and parasites in mucosal tissues. In addition, the responses of trout IgT subclasses to LPS stimuation were similar to zebrafish IgZ1 and IgZ2, which could also be obviously stimulated by LPS with different patterns (Hu et al., 2010). This might be due to that LPS is a typical polyclonal activator which could induce the activation of B cells in gut and skin (Hu et al., 2010; Moller, 1965).
In our previous studies we developed a mAb against trout IgT1, and a hybridoma clone that produced a specific antibody with a high affinity for IgT1 was selected (Zhang et al., 2010). In the current study, IgT+ B cells stained with the anti-IgT1 mAb were sorted from trout HKLs and PBLs and found to express Igτ1 and Igτ2. More interestingly, the anti-IgT1 mAb could recognize all the three recombinant Igτ and trout serum IgT purified with the anti-IgT1 mAb also contained all the three Igτ. Thus, the reactivity of anti-IgT1 mAb with all three IgT subclasses is potentially attributed to the same epitope shared by all of the three Igτ subclasses, which may locate at the CH3 domains.
In summary, a total of three IgT subclasses were characterized in rainbow trout at the nucleotide and protein levels, thus expanding our knowledge of the trout IgH loci. In addition, expression analyses of trout IgH genes in immune tissues under normal and stimulated conditions indicated differential participation of the IgH genes during immune responses. However, further questions remain to be addressed to understand the regulation of expression and functional differences between various IgT subclasses.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31172431) and the National Basic Research Program of China (2014CB138601) to Y.-A. Z and the U.S. Department of Agriculture Grant (USDA-NRI-2013-01107), the National Science Foundation Grant (NSF-IOS-1457282), and the National Institutes of Health Grant (2R01GM085207-05) to J.O.S.
The nucleotide sequences of the secretory form of Igτ2 and the membrane and secretory forms of Igτ3 in trout have been submitted to GenBank with accession numbers KX185935 to KX185937, respectively.
Abbreviations
- BCR
B cell receptor
- C domain
constant domain
- CH
heavy chain constant domain
- CL
light chain constant domain
- D segment
diversity segment
- Ig
immunoglobulin
- IgH
immunoglobulin heavy chain
- IgL
immunoglobulin light chain
- J segment
joining segment
- LC
liquid chromatography
- LLR
leucine-rich repeat
- mAb
monoclonal antibody
- MS
mass spectrometry
- qPCR
quantitative real-time PCR
- RACE
rapid-amplification of cDNA end
- RAG
recombination-activing gene
- RSS
recombination signal sequence
- V domain
variable domain
- VH
heavy chain variable domain
- VL
light chain variable domain
- VLR
variable lymphocyte receptor
- V segment
variable segment
Footnotes
Conflicts of interest statement: The authors have no conflicting commercial or financial interest in publishing this paper.
Appendix A. Supplementary data: Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dci.2017.01.001.
References
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- Bao Y, Wang T, Guo Y, Zhao Z, Li N, Zhao Y. The immunoglobulin gene loci in the teleost Gasterosteus aculeatus. Fish Shellfish Immunol. 2010;28:40–48. doi: 10.1016/j.fsi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noel B, Bento P, Da Silva C, Labadie K, Alberti A, Aury JM, Louis A, Dehais P, Bardou P, Montfort J, Klopp C, Cabau C, Gaspin C, Thorgaard GH, Boussaha M, Quillet E, Guyomard R, Galiana D, Bobe J, Volff JN, Genet C, Wincker P, Jaillon O, Roest Crollius H, Guiguen Y. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshra H, Li J, Peters R, Hansen J, Matlapudi A, Sunyer JO. Cloning, expression, cellular distribution, and role in chemotaxis of a C5a receptor in rainbow trout: the first identification of a C5a receptor in a nonmammalian species. J Immunol. 2004;172:4381–4390. doi: 10.4049/jimmunol.172.7.4381. [DOI] [PubMed] [Google Scholar]
- Boshra H, Wang T, Hove-Madsen L, Hansen J, Li J, Matlapudi A, Secombes CJ, Tort L, Sunyer JO. Characterization of a C3a receptor in rainbow trout and Xenopus: the first identification of C3a receptors in nonmammalian species. J Immunol. 2005;175:2427–2437. doi: 10.4049/jimmunol.175.4.2427. [DOI] [PubMed] [Google Scholar]
- Bradshaw CM, Richard AS, Sigel MM. IgM antibodies in fish mucus. Proc Soc Exp Biol Med. 1971;136:1122. doi: 10.3181/00379727-136-35443. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Backstrom BT, Tiefenthaler G, Palmer E. CART: a conserved antigen receptor transmembrane motif. Semin Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- Castro R, Jouneau L, Pham HP, Bouchez O, Giudicelli V, Lefranc MP, Quillet E, Benmansour A, Cazals F, Six A, Fillatreau S, Sunyer O, Boudinot P. Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013;9:e1003098. doi: 10.1371/journal.ppat.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R, Martinez-Alonso S, Fischer U, Haro NA, Soto-Lampe V, Wang T, Secombes CJ, Lorenzen N, Lorenzen E, Tafalla C. DNA vaccination against a fish rhabdovirus promotes an early chemokine-related recruitment of B cells to the muscle. Vaccine. 2014;32:1160–116. doi: 10.1016/j.vaccine.2013.11.062. 8. [DOI] [PubMed] [Google Scholar]
- Castro R, Tafalla C. Overview of fish immunity. In: Beck BH, Peatman E, editors. Mucosal Health in Aquaculture. Academic Press; London: 2015. pp. 3–54. [Google Scholar]
- Chang MX, Zou J, Nie P, Huang B, Yu Z, Collet B, Secombes CJ. Intracellular interferons in fish: a unique means to combat viral infection. PLoS Pathog. 2013;9:e1003736. doi: 10.1371/journal.ppat.1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Amemiya CT. Going adaptive: the saga of antibodies. Ann N Y Acad Sci. 2009;1168:130–155. doi: 10.1111/j.1749-6632.2009.04881.x. [DOI] [PubMed] [Google Scholar]
- Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- Danilova N, Steiner LA. B cells develop in the zebrafish pancreas. Proc Natl Acad Sci U S A. 2002;99:13711–13716. doi: 10.1073/pnas.212515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Hirano M, Tako R, McCallister C, Nikolaidis N. Evolutionary genomics of immunoglobulin-encoding loci in vertebrates. Curr Genomics. 2012;13:95–102. doi: 10.2174/138920212799860652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Luo M, Velikovsky A, Mariuzza RA. Structural insights into the evolution of the adaptive immune system. Annu Rev Biophys. 2013;42:191–215. doi: 10.1146/annurev-biophys-083012-130422. [DOI] [PubMed] [Google Scholar]
- Edholm ES, Bengten E, Stafford JL, Sahoo M, Taylor EB, Miller NW, Wilson M. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J Immunol. 2010;185:4082–4094. doi: 10.4049/jimmunol.1000631. [DOI] [PubMed] [Google Scholar]
- Edholm ES, Bengten E, Wilson M. Insights into the function of IgD. Dev Comp Immunol. 2011;35:1309–1316. doi: 10.1016/j.dci.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, Boudinot P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front Immunol. 2013;4:28. doi: 10.3389/fimmu.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Flajnik MF. The last flag unfurled? A new immunoglobulin isotype in fish expressed in early development. Nat Immunol. 2005;6:229–230. doi: 10.1038/ni0305-229. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French MAH, Harrison G. Serum IgG subclass concentrations in healthy adults: a study using monoclonal antisera. Clin Exp Immunol. 1984;56:473–475. [PMC free article] [PubMed] [Google Scholar]
- Gambon-Deza F, Sanchez-Espinel C, Magadan-Mompo S. Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev Comp Immunol. 2010;34:114–122. doi: 10.1016/j.dci.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Gomez D, Sunyer JO, Salinas I. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013;35:1729–1739. doi: 10.1016/j.fsi.2013.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Yuan H, Wang T, Li B, Ma L, Yu S, Huang T, Li Y, Fang D, Chen X, Wang Y, Qiu S, Guo Y, Fei J, Ren L, Pan-Hammarstrom Q, Hammarstrom L, Wang J, Wang J, Hou Y, Pan Q, Xu X, Zhao Y. Multiple IgH isotypes including IgD, subclasses of IgM, and IgY are expressed in the common ancestors of modern birds. J Immunol. 2016;196:5138–5147. doi: 10.4049/jimmunol.1600307. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci U S A. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima JI, Jung TS, Aoki T. Immunoglobulin genes and their transcriptional control in teleosts. Dev Comp Immunol. 2011;35:924–936. doi: 10.1016/j.dci.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Biol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hordvik I, Kamil A, Bilal S, Raae A, Fjelldal PG, Koppang EO. Characterization of serum IgM in teleost fish, with emphasis on salmonids. Fish Shellfish Immunol. 2013;34:1656. [Google Scholar]
- Hu YL, Xiang LX, Shao JZ. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: implications for a distinct B cell receptor in lower vertebrates. Mol Immunol. 2010;47:738–746. doi: 10.1016/j.molimm.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Kett K, Brandtzaeg P, Radl J, Haaijman JJ. Different subclass distribution of IgA-producing cells in human lymphoid organs and various secretory tissues. J Immunol. 1986;136:3631–3635. [PubMed] [Google Scholar]
- Korytar T, Jaros J, Verleih M, Rebl A, Kotterba G, Kuhn C, Goldammer T, Kollner B. Novel insights into the peritoneal inflammation of rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2013;35:1192–1199. doi: 10.1016/j.fsi.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Lesk AM, Chothia C. Evolution of proteins formed by beta-sheets. II. The core of the immunoglobulin domains. J Mol Biol. 1982;160:325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- Mashoof S, Pohlenz C, Chen PL, Deiss TC, Gatlin D, Buentello A, Criscitiello MF. Expressed IgH μ and τ transcripts share diversity segment in ranched Thunnus orientalis. Dev Comp Immunol. 2014;43:76–86. doi: 10.1016/j.dci.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller G. 19S antibody production against soluble lipopolysaccharide antigens by individual lymphoid cells in vitro. Nature. 1965;207:1166–1168. doi: 10.1038/2071166a0. [DOI] [PubMed] [Google Scholar]
- Parker S, La Flamme A, Salinas I. The ontogeny of New Zealand groper (Polyprion oxygeneios) lymphoid organs and IgM. Dev Comp Immunol. 2012;38:215–223. doi: 10.1016/j.dci.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Parra D, Takizawa F, Sunyer JO. Evolution of B cell immunity. Annu Rev Anim Biosci. 2013;1:65–97. doi: 10.1146/annurev-animal-031412-103651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, Bromage ES. Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J Immunol. 2012;188:1341–1349. doi: 10.4049/jimmunol.1101938. [DOI] [PubMed] [Google Scholar]
- Ryo S, Wijdeven RHM, Tyagi A, Hermsen T, Kono T, Karunasagar I, Rombout JHWM, Sakai M, Kemenade BMLVv, Savan R. Common carp have two subclasses of bonyfish specific antibody IgZ showing differential expression in response to infection. Dev Comp Immunol. 2010;34:1183–1190. doi: 10.1016/j.dci.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125:S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wei Z, Li N, Zhao Y. A comparative overview of immunoglobulin genes and the generation of their diversity in tetrapods. Dev Comp Immunol. 2013;39:103–109. doi: 10.1016/j.dci.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat Immunol. 2013;14:320–326. doi: 10.1038/ni.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiso TM, Lie KK, Hordvik I. Molecular cloning of IgT from Atlantic salmon, and analysis of the relative expression of tau, mu, and delta in different tissues. Vet Immunol Immunopathol. 2011;139:17–26. doi: 10.1016/j.vetimm.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Wilson M, Bengten E, Miller NW, Clem LW, DuPasquier L, Warr GW. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci U S A. 1997;94:4593–4597. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Parra D, Gomez D, Salinas I, Zhang YA, von Gersdorff Jorgensen L, Heinecke RD, Buchmann K, LaPatra S, Sunyer JO. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci U S A. 2013;110:13097–13102. doi: 10.1073/pnas.1304319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Takizawa F, Parra D, Gomez D, von Gersdorff Jorgensen L, LaPatra SE, Sunyer JO. Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat Commun. 2016;7:10728. doi: 10.1038/ncomms10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuike M, de Boer J, von Schalburg KR, Cooper GA, McKinnel L, Messmer A, So S, Davidson WS, Koop BF. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC genomics. 2010;11:486. doi: 10.1186/1471-2164-11-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang XJ, Song YL, Lu XB, Chen DD, Xia XQ, Sunyer JO, Zhang YA. Preferential combination between the light and heavy chain isotypes of fish immunoglobulins. Dev Comp Immunol. 2016;61:169–179. doi: 10.1016/j.dci.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang YA, Hikima J, Li J, LaPatra SE, Luo YP, Sunyer JO. Conservation of structural and functional features in a primordial CD80/86 molecule from rainbow trout (Oncorhynchus mykiss), a primitive teleost fish. J Immunol. 2009;183:83–96. doi: 10.4049/jimmunol.0900605. [DOI] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Oriol Sunyer J. Recent findings on the structure and function of teleost IgT. Fish Shellfish Immunol. 2011;31:627–634. doi: 10.1016/j.fsi.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


