Abstract
Diabetes is a risk factor for periodontitis, an inflammatory bone disorder and the greatest cause of tooth loss in adults. Diabetes has a significant impact on the gut microbiota, however studies in the oral cavity have been inconclusive. By 16s rRNA sequencing we show here that diabetes causes a shift in oral bacterial composition and, by transfer to germ free mice, that the oral microbiota of diabetic mice is more pathogenic. Furthermore, treatment with IL-17 antibody decreases the pathogenicity of the oral microbiota in diabetic mice; when transferred to recipient germ-free mice oral microbiota from IL-17-treated donors induced reduced neutrophil recruitment, reduced IL-6 and RANKL, and less bone resorption. Thus, diabetes-enhanced IL-17 alters the oral microbiota and renders it more pathogenic. Our findings provide a mechanistic basis to better understand how diabetes can increase the risk and severity of tooth loss.
eTOC
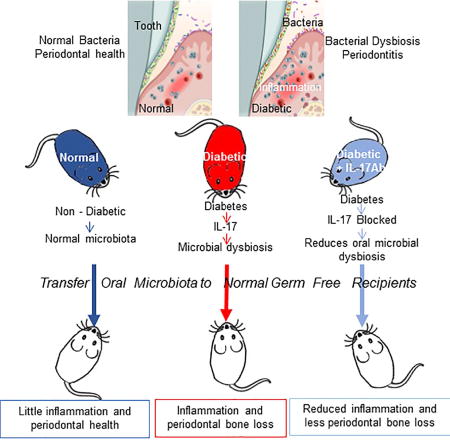
Diabetes increases periodontal disease, a major cause of tooth loss. Xiao et al. demonstrated, by transfer to germ-free mice, that oral microbiota from diabetic mice induced more periodontal inflammation and bone loss than microbiota from normal mice. Diabetes increased the pathogenicity of the oral microbiota through an IL-17 mediated mechanism.
INTRODUCTION
Health is dependent on the homeostasis of both inner and external microenvironments. Diabetes mellitus is characterized by hyperglycemia, inflammation, and high oxidative stress, which leads to systemic complications (Wu et al., 2015). Type II diabetes is caused by insulin resistance frequently associated with obesity and a failure to produce enough insulin. The microbiome, an important component of the external microenvironment, plays a key role in homeostasis and affects several pathologic processes including diabetes (Ussar et al., 2016), hematopoiesis (Khosravi et al., 2014), skeletal health (Hernandez et al., 2016), obesity (Sweeney and Morton, 2013) and carcinogenesis (Li et al., 2016). Thus, dysbiosis of the microbiome has a significant effect on health and disease (Sekirov et al., 2010).
The oral cavity is the initial point of entry to the digestive and respiratory tract. Over 700 bacterial species may be found in the oral cavity of humans (Paster et al., 2006). Oral microbial dysbiosis is linked to oral inflammation, is thought to trigger periodontal disease (Curtis et al., 2011; Hajishengallis et al., 2011) and may contribute to systemic conditions through bacteremia (Han and Wang, 2013). The oral microbiome may impact bacteria which colonize the gut microbiome (Nakajima et al., 2015). There is evidence that some species are more pathogenic than others; the dynamic balance of various bacteria that make up the oral microbiome has been proposed to determine periodontal disease activity (Colombo et al., 2012; Griffen et al., 2012; Hong et al., 2015).
Diabetes is a risk factor for periodontitis and increases disease severity (Emrich et al., 1991; Loe, 1993). Host factors are modified by diabetes and may play a causative role in bacteria-induced tissue damage (Andriankaja et al., 2012; Lalla and Papapanou, 2011; Pacios et al., 2012). Although some microbial studies show changes in the abundance of oral microorganisms in diabetes mellitus, there is no direct evidence that diabetes increases the pathogenicity of the oral microbiome (Ohlrich et al., 2010). A consensus report from the European Federation of Periodontology and the American Academy of Periodontology found that there is no compelling evidence that diabetes has a significant impact on the oral microbiota (Chapple et al., 2013). To investigate this important issue, we examined a mouse model and examined the impact of diabetes on the oral microbial composition and its pathogenicity.
RESULTS
Diabetes causes a shift in the oral microbiome with increased inflammation and bone loss
To characterize the oral microbiome in diabetic mice, we carried out 16S rRNA gene tag sequencing in mice that were homozygous for a mutation in the leptin receptor gene (db/db mice, hereafter). Prior to the experiment, db/db mice and heterozygous control mice were co-housed for 2 weeks and randomly switched between cages at the outset to increase consistency of the oral microbiomes. Oral bacteria were collected before db/db mice became hyperglycemic and again when mice were 12–13 weeks old, an age when db/db mice had blood glucose levels more than 225 mg/dl for 4–5 weeks.
At the outset, db/db mice and their lean littermates had a similar oral microbiome and similar bacterial diversity (Figure 1). The development of hyperglycemia led to a significant difference in the oral microbiota in normoglycemic and diabetic mice determined by alpha diversity; the number of taxonomic units in the normoglycemic group was higher than the number in the diabetic group, reflecting less bacterial diversity in the diabetic animals (Figure 1A, P < 0.05). Similar results were obtained with the Shannon index (Figure 1A, P < 0.05). The diabetic and normoglycemic groups were also distinguished by beta diversity (Figure 1B). Prior to hyperglycemia the diabetes prone group clustered similarly to the normoglycemic. After the onset of hyperglycemia, bacterial community composition in the diabetic and normoglycemic were significantly different (P<0.05). It is known that changes occur in normoglycemic mice over time (Wu et al., 2016). However, diabetes has a distinct effect as shown by the comparison of normoglycemic and diabetic groups that were age matched. Thus, the development of hyperglycemia in diabetic mice had a clear effect on the oral microbial composition.
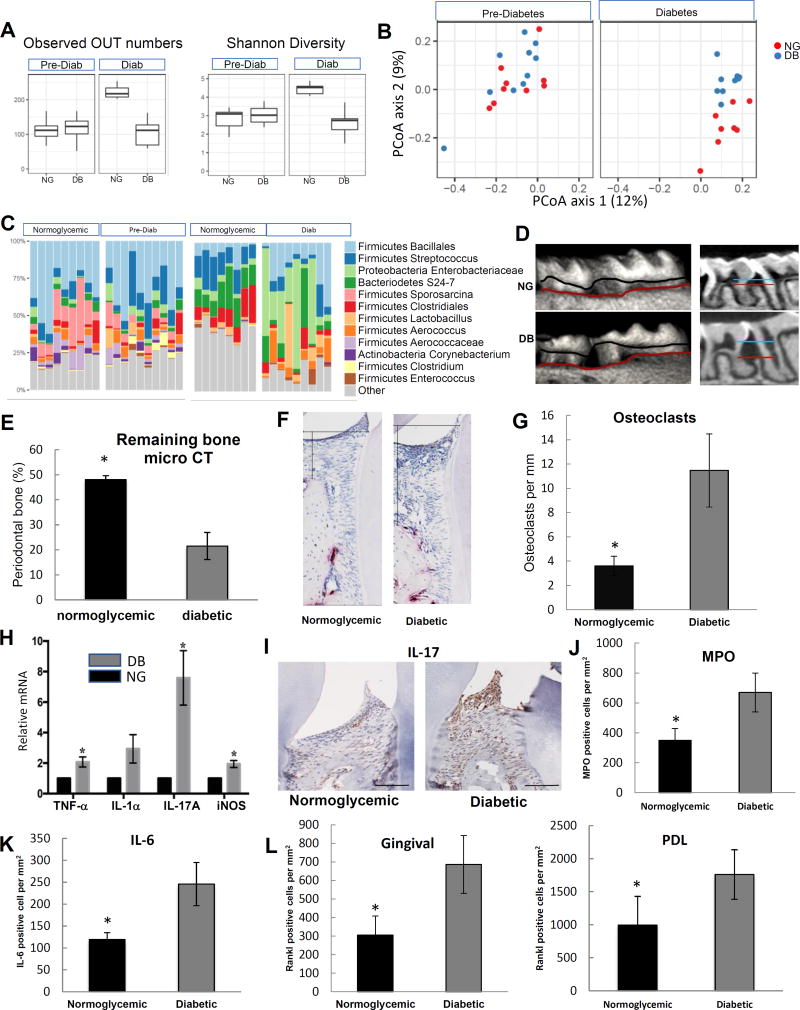
Figure 1. Diabetes alters the composition of the oral microbiome and increases periodontal inflammation and bone loss.
Db/db type 2 diabetes prone and lean normoglycemic control littermates (db/+) were examined. A: Alpha diversity was assessed in the microbiota of 9 normoglycemic and 10 diabetes-prone mice before hyperglycemia and 8 normoglycemic and 9 diabetic mice after the latter developed hyperglycemia. B: Beta (Unweighted Unifrac) diversity was assessed in 9 normoglycemic and 10 diabetes-prone mice before and 8 normoglycemic and 9 diabetic mice after the development of hyperglycemia. C: Prominent bacterial taxa in 9 normoglycemic and 10 diabetes-prone mice before and 8 normoglycemic and 9 diabetic mice after the development of hyperglycemia. D: Micro-CT 3D reconstructions and sagittal slice views of normoglycemic and diabetic mice. The distance between horizontal lines represents the distance between a fixed landmark on tooth surface (cemental-enamel junction) and bone crest. E: Percent periodontal bone remaining in normoglycemic and diabetic mice measured by micro-CT. A minimum of six maxillary samples from different mice were examined per group with the analysis performed 3 times with similar results. F: TRAP stained histologic sections of periodontal tissue from diabetic and normoglycemic mice. The horizontal line represents the position of cemental enamel junction and the vertical line extends from cemental enamel junction to bone height. Osteoclasts are stained red. G: Osteoclast numbers in TRAP stained histologic sections per mm bone length. Two slides from a minimum of 6 maxillary samples were analyzed per group from different animals and the analysis was carried out twice with similar results. H: mRNA was extracted from the gingiva of molar teeth, RNA extracted and mRNA levels of TNF, IL-1α, IL-17A and iNOS were measured by real-time PCR in normoglycemic (NG) and diabetic (DB) groups. A minimum of six maxillary gingival tissue samples per group were examined from different mice by real-time PCR, which was performed three times with similar results. I: Representative immunostaining of histologic sections of normoglycemic and diabetic mice with anti-IL-17A antibody. Horizontal line = 100 µm. J: Immunofluorescence with antibody to myeloperoxidase. Myeloperoxidase-positive cells were counted in gingival connective tissue. K: Immunofluorescence with IL-6 antibody. IL-6 immunopositive cells were counted in gingival connective tissue. L: Immunofluorescence with antibody specific for RANKL. RANKL immunopositive cells were counted in the gingival connective tissue and periodontal ligament. Nuclei were detected with DAPI counterstain. Data are expressed as number of positive cells per mm2 gingival connective tissue or periodontal ligament. For immunofluorescence analysis (I–K) two to three slides were analyzed from at least eight maxillary samples per group, each from a different animal. Immunofluorescence with each control antibody was negative (not shown). Original magnification of fluorescent images 400×. * indicates statistical difference (p<0.05) between diabetic and matched normoglycemic mice.
Analysis of prevalent bacterial taxa revealed genus- and family-level differences between bacterial communities from normoglycemic and hyperglycemic mice (Figure 1C). Specifically, after the onset of hyperglycemia, the diabetic oral cavity had increased levels of Enterobacteriaceae, Aerococcus, Enterococcus, and Staphylococcus, which are often associated with periodontitis or are considered to be associated with impaired healing in diabetic animals (Grice et al., 2010b; Kumar et al., 2006; Vieira Colombo et al., 2016) (Table S1).
A characteristic feature of periodontitis is loss of bone around the teeth. Like diabetic humans, diabetic mice had naturally occurring spontaneous periodontal bone loss (Figure 1D). Quantitative analysis of the microCT results demonstrated that the diabetic mice had less than half the amount of bone present compared to the normoglycemic controls (P<0.05) (Figure 1E). Histologic analysis of tissue sections gave similar results with 57% less interdental bone in the diabetic mice compared to normoglycemic mice (P<0.05) (Figure S1A),
Periodontal bone loss typically reflects the degree of inflammation present. Diabetic mice had increased inflammatory cytokine mRNA levels (Figure 1H). IL-17 has the largest increase of mediators examined, which was confirmed at the protein level (Figure 1I). Immunofluorescence also was carried out to establish the number of neutrophils and the number of cells that express IL-6 and RANKL, cytokines that induce bone-resorption. Diabetes caused a 4-fold increase in the number of neutrophils (P<0.05) that express myeloperoxidase (MPO) (Figures 1J and S1B). In the diabetic periodontium, there was a 2-fold increase in cells expressing IL-6 and RANKL (Figures 1K, 1L, S1C and S1D).
The Diabetic microenvironment increases the pathogenicity of the oral microbiome
Results above indicate that the transition to hyperglycemia in diabetic mice induces a change in periodontal status, periodontal inflammation, and alterations in the microbial composition. These changes are indicative of a more pronounced pathogenic environment in the diabetic group. To determine whether this could be accounted by the microbiota alone, microorganisms were transferred from the oral cavity of diabetic and matched normoglycemic mice to normal germ-free recipient animals with a ligature placed between the left maxillary 1st and 2nd molars. Germ-free mice were chosen as recipients to reduce the influence of the commensal bacteria.
Transfer of bacteria from diabetic mice to germ-free recipients induced a larger amount of bone loss than transfer from normoglycemic donors (Figure 2A). By micro-CT analysis there was 42% less bone in mice that received bacteria from the diabetic group compared to bacteria from the normoglycemic group (Figure 2B, P<0.05). By histologic analysis the recipients of bacteria from the diabetic group had 57% less bone than recipients of bacteria from normoglycemic controls (data not shown). Bacteria from the normal group also had some pathogenic potential, as it induced bone loss compared to germ-free mice without bacterial transfer (P<0.05, Figure 2C), although significantly less than bacteria from diabetic mice. Thus, bacteria from diabetic mice stimulated the most bone loss.
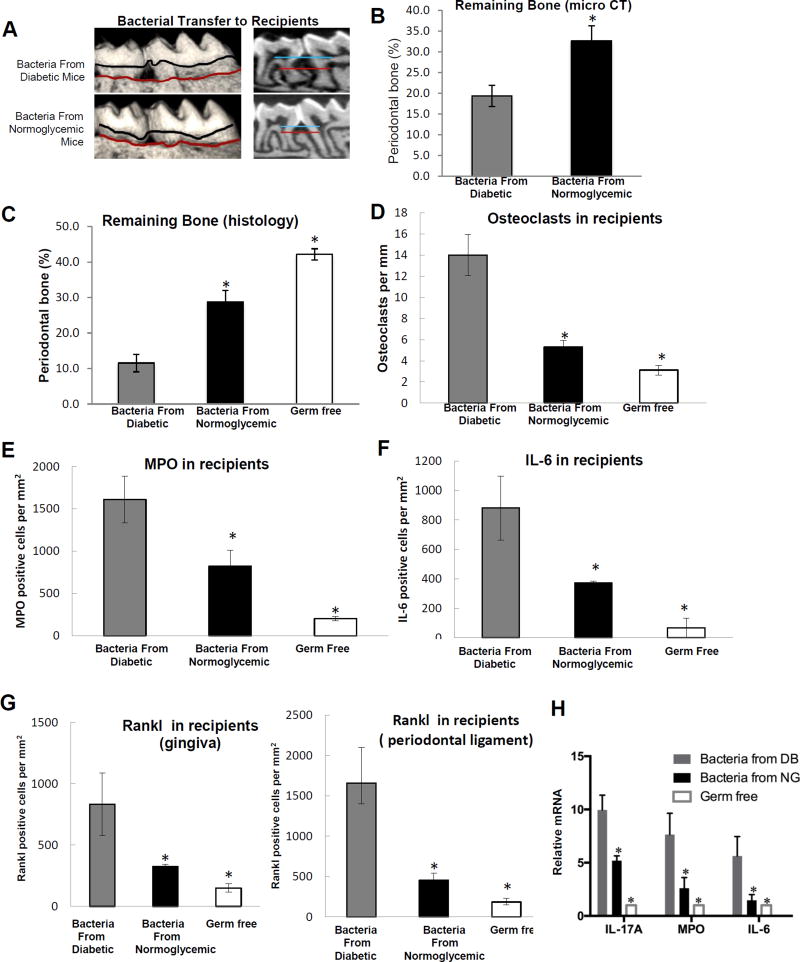
Figure 2. Diabetes increases the pathogenicity of bacteria transferred to normal germ-free mice.
Bacteria were collected by swabbing the teeth and adjacent periodontal tissue of 5 diabetic (db/db) mice or 5 normoglycemic controls (db/+). Collected bacteria were transferred twice with one day in between to normal germ-free mice that had ligatures placed between the left maxillary 1st and 2nd molars. Mice were euthanized and periodontal tissues from recipient normal germ-free mice were examined one week after the first bacterial transfer. A: Representative images of 3D micro-CT reconstructions of periodontal bone following bacterial transfer. B: Periodontal bone was measured by micro-CT following bacterial transfer. Five maxillary samples from 5 different mice per group were analyzed and the analysis was performed 3 times. C: Representative histologic sections of germ-free mice that received bacteria from normoglycemic and diabetic donor mice were examined by TRAP staining. D: The number of TRAP-stained bone-lining multinucleated osteoclasts was counted per mm bone length. E: Immunofluorescence with an antibody specific for myeloperoxidase to quantify neutrophils per mm2 gingival connective tissue after bacterial transfer to normal germ-free mice. Immunofluorescence with control antibody was negative. F: Immunofluorescence with anti-IL-6 antibody following transfer of bacteria to normal germ-free mice. Positive cells were counted per mm2 gingival connective tissue or periodontal ligament. G: Immunofluorescence with antibody specific for RANKL. RANKL immunopositive cells were counted in the gingival connective tissue and in the periodontal ligament. Immunofluorescence with control antibody was negative. Original magnification of fluorescent images 400×. H. mRNA levels in gingiva of recipient mice after transfer of bacteria from diabetic (DB) mice or normoglycemic mice (NG) or germ-free mice without bacterial transfer. Five gingival samples from different mice were examined per group and real-time PCR was performed. The results are representative of three analyses. For histomorphometric analysis (C & D) two to three slides were examined per maxilla with maxillary samples obtained from different animals. Sections were examined twice with similar results. For immunofluorescence studies (E–G) two to three slides were examined per maxilla from five different mice per group and the analysis was carried out twice with similar results. Nuclei were detected by DAPI counterstain. * indicates significantly different from recipients that received bacteria from diabetic mice (p<0.05).
Transfer of bacteria from diabetic mice to germ-free recipients induced 3-fold more osteoclasts than bacteria from normoglycemic mice (P<0.05, Figure 2D), consistent with bone measurements. We previously showed that inoculation of bacteria in diabetic mice induces greater inflammation than an equal inoculation in normoglycemic mice (Naguib et al., 2004a). Here we took the opposite approach to determine whether bacteria from diabetic mice stimulate a greater inflammatory response. The number of neutrophils was higher in the periodontium of germ-free mice exposed to bacteria from diabetic mice compared to normoglycemic animals (Figure 2E). Bacteria from diabetic mice increased MPO-positive neutrophils 2-fold compared to bacteria from normoglycemic controls, which was significantly higher than germ-free mice not exposed to any bacteria (Figure 2E and Figure S2A). Bacteria from diabetic mice increased the number of cells expressing IL-6 and RANKL 2–3 fold higher compared to bacteria from normoglycemic animals (Figure 2F&G and Figures S2B&C). At the mRNA level, there was a significantly greater 2- to 4-fold greater induction of IL-17, MPO, and IL-6 in the gingiva of recipients inoculated with bacteria from diabetic mice compared to bacteria from normoglycemic animals or germ-free mice that did not receive bacteria (Figure 2H). Furthermore, these results were not simply due to higher bacterial loads in the diabetic mice as the amount bacteria obtained from both groups were similar (Figure S3A). That bacteria were successfully transferred from each donor group to germ-free recipients is supported by PCoA data showing distinct bacterial colonization in the recipients (Figure S3B). These findings indicate that the diabetic oral microbiome is considerably more pathogenic than the normal oral microbiome.
IL-17 inhibition reduces the pathogenicity of the oral microbiome in diabetic mice
To test whether diabetes-enhanced inflammation increased the pathogenicity of the oral microbiota, we applied by local injection into the gingiva anti-IL-17 antibody or matched control antibody and transferred bacteria to germ-free recipients after placing a ligature between the first and second molar. A heat map of the bacterial composition of the normoglycemic, diabetic and IL-17 antibody-treated mice shows that the antibody treatment had a significant impact on a number of bacteria (Figure 3A). Several of the Firmicutes and Proteobacteria were elevated in the diabetic group and reduced by treatment with the IL-17 antibody. Moreover, the Unifrac distance between the normoglycemic and diabetic groups was greater than the distance between the normoglycemic and diabetic-IL-17 antibody-treated group (Figure 3B). IL-17 antibody treatment altered the bacterial community composition in germ-free recipients relative to recipients inoculated with bacteria from diabetic control mice (Figure S3B, P<0.05). When the composition of the donor and the recipient mice were compared, they exhibited similar but not identical bacterial communities, consistent with a successful transfer from donors to recipients (Figure 3C). We also examined the diabetes-associated taxa identified in our initial study (Figure 1) to see if their abundance was reduced by IL-17 antibody treatment. Three of the four taxa (Aerococcus, Enterobacteriaceae, Staphylococcus) were not significantly different between the three groups of recipient mice examined and Clostridiales, which was reduced in diabetic mice also had the lowest median abundance in germ-free recipients from diabetic donors but was not statistically significant. However, Enterococcus was significantly different and was elevated in recipients that received bacteria from diabetic animals (Figure 3D, P<0.05) and was similar in recipients that received bacteria from normoglycemic mice and IL-17 antibody treated diabetic donors.
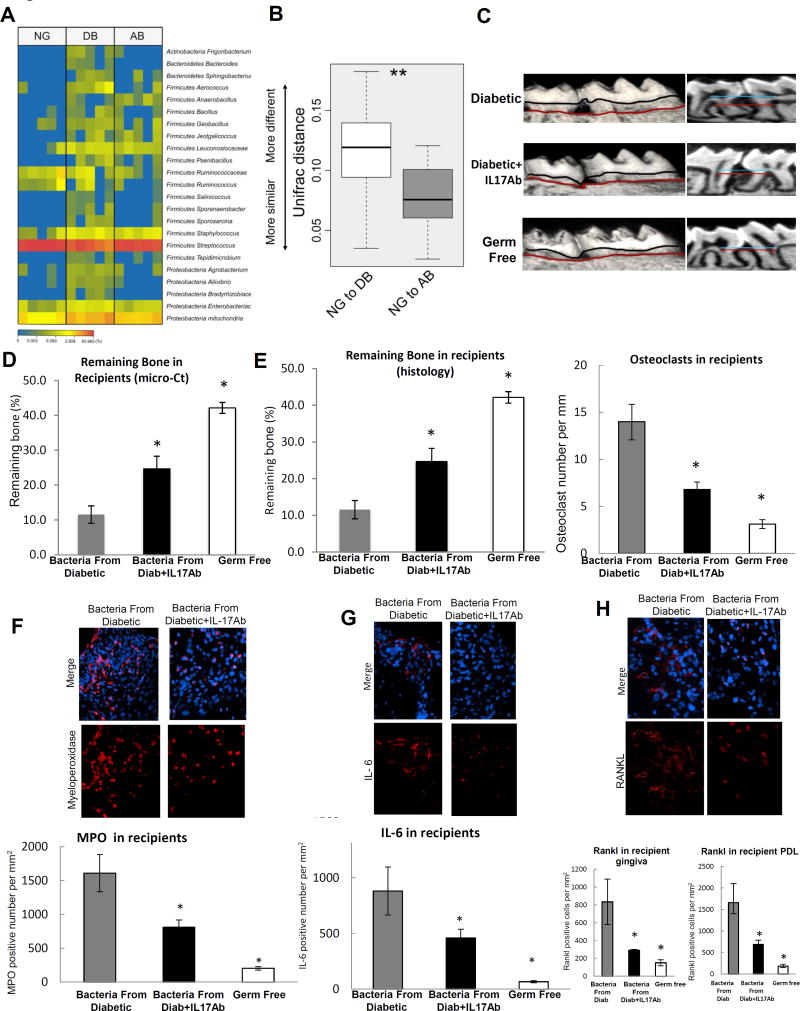
Figure 3. Local injection of IL-17 antibody reduces microbial pathogenicity induced by diabetes.
A & B: The oral bacterial composition from normoglycemic (NG), diabetic (DB) and diabetic IL-17A antibody treated mice (AB) was examined. A. Heat map of oral bacteria from 5 NG, 5 DB and 5 AB groups. B: Unifrac distance of oral bacterial communities from NG, DB and AB groups. C–H: Bacteria from db/db diabetic mice treated with local injection of IL-17A antibody or matched control antibody were transferred to normal germ-free mice that had ligatures placed between the left maxillary 1st and 2nd molars. Mice were euthanized 1 week after bacterial transfer and periodontal tissues were examined. C: Three-dimensional micro-CT reconstruction and sagittal images. D: Periodontal bone remaining after bacterial transfer to normal germ-free mice. Five maxillary samples were examined from 5 different mice per group and the analysis was performed 3 times with similar results. E: Histologic sections were examined to quantify the interdental bone between the first and second molars. Bone lining multi-nucleated TRAP stained osteoclasts were counted per mm bone length. For each measurement two slides were examined from five maxillary samples per group and each maxilla was obtained from a different mouse. The results are representative of two separate analyses. F: Immunofluorescence with anti-myeloperoxidase antibody to quantify neutrophils per mm2 gingival connective tissue following the transfer of bacteria to normal germ-free mice. G: Immunofluorescence with anti-IL-6 antibody. H: Immunofluorescence with antibody specific to RANKL. RANKL positive cells were counted per mm2 in gingival connective tissue or periodontal ligament. For immunofluorescence (F–H) two to three slides were examined from 5 maxillary samples, each from a different mouse. The analysis was carried out twice with similar results. Nuclei were detected by DAPI counterstain. Immunofluorescence with each control antibody was negative (not shown). * Statistical difference (p<0.05) when compared to germ-free normoglycemic mice that received bacteria from diabetic donor mice. Original magnification of fluorescent images 400×.
Treatment with IL-17 antibody had a dramatic effect on the pathogenicity of the oral bacteria. Bacteria from diabetic donors treated with control antibody stimulated twice as much periodontal bone loss in normal germ-free recipients compared to bacteria from diabetic donors treated with IL-17 antibody as measured by micro-CT (P<0.05, Figure 3C&D). Similar results were obtained by histomorphometric analysis of bone sections (Figure 3E). Oral bacteria transferred from diabetic donors induced twice as many osteoclasts as bacteria from diabetic donors treated with IL-17 antibody (P<0.05, Figure 3E). The number of neutrophils (MPO immunopositive cells) recruited in response to bacteria from diabetic mice treated with control antibody was twice that of bacteria from IL-17 antibody treated diabetic mice (P<0.05, Figure 3F). Likewise, oral bacteria from diabetic mice induced a two-fold increase in the infiltration of IL-6 immunopositive cells in germ-free recipients compared to bacteria from diabetic mice treated with the IL-17 antibody (P<0.05, Figure 3G). Rankl positive cells showed similar results as IL-6 and demonstrated that IL-17 antibody had a significant effect on bacteria capable of inducing this osteoclastogenic cytokine (Figure 3H).
DISCUSSION
Diabetes mellitus causes serious complications that are linked to a chronic inflammatory environment (Wu et al., 2015). This includes periodontal inflammation and bone loss (Lalla and Papapanou, 2011) with the source of inflammation assumed to be from alterations in host metabolism caused by hyperglycemia (Wu et al., 2015). Our results showed that diabetes increased inflammation in periodontal tissues and alveolar bone loss in the absence of an exogenous infection in mice. This is unusual since the addition of a periodontal pathogen or placement of a ligature is typically needed to induce a significant periodontal bone loss in rodent models (Graves et al., 2008; Hajishengallis et al., 2015). In humans, it has been demonstrated that the diagnosis of periodontitis is accompanied by a shift in the bacterial community structure and composition (Abusleme et al., 2013; Camelo-Castillo et al., 2015; Griffen et al., 2012; Kistler et al., 2013).
This study demonstrates that the development of hyperglycemia in diabetes-prone animals causes a clear shift in the oral microbiome in animals that were initially similar. This shift increases pathogenicity as shown by changes in the oral cavity of diabetic mice including higher levels of Proteobacteria (Enterobacteriaceae) and Firmicutes (Enterococcus, Staphylococcus and Aerococcus) that are associated with infectious processes, periodontitis and delayed wound healing in diabetic animals (Grice et al., 2010a; Souto and Colombo, 2008; Vieira Colombo et al., 2016). Overall, the Firmicutes phylum is positively associated with periodontitis and insulin resistance (Demmer et al., 2016). Enterococcus was also the predominant taxa in recipient mice which had bacteria transferred from diabetic animals. We found that diabetes led to reduced oral microbial diversity, consistent with reports that diabetic patients and animal models exhibit decreased bacterial diversity in other sites (Hartstra et al., 2015; Patterson et al., 2015; Ussar et al., 2016). The reduced diversity could potentially enhance susceptibility to the formation of a pathogenic microbiome as a highly diverse ecosystems is generally more stable and healthier than communities dominated by fewer taxa (Escalante et al., 2015).
The shift in the microbial composition is consistent with increased pathogenicity but does not prove it. Here we showed that diabetes increases the pathogenicity of oral bacteria by transfer to germ-free mice. Bacteria transferred from the diabetic mice stimulated greater periodontal inflammation, higher expression of the bone resorbing cytokines IL-6 and RANKL, greater osteoclast formation and more periodontal bone loss than bacteria from normoglycemic animals. These results demonstrate unequivocally that the diabetic microbiome has an increased capacity to drive pathologic changes that enhance periodontal bone loss.
IL-17 is a multifaceted cytokine associated with both immune-protection and immunopathology. High levels of IL-17 are found in chronic periodontitis and IL-17 stimulates the production of pro-inflammatory mediators such as IL-6 and RANKL and can indirectly promote osteoclastogenesis (Xiao et al., 2016). Genetic defects that cause leukocyte adhesion deficiency in human patients result in exaggerated IL-17 expression in the periodontium that is accompanied by dysbiotic bacterial communities, which have increased the potential to further drive IL-17 responses, linking our study with observations in humans (Abusleme and Moutsopoulos, 2016; Moutsopoulos et al., 2015). There are several potential mechanisms by which IL-17 inhibition could alter the microbiome such as altering the bacterial growth or colonization by providing substrates generated from increased inflammation or by changes in anti-bacterial defense (Curtis and Way, 2009).
Local injection of IL-17 altered the microbial composition and decreased the capacity of bacteria transferred from diabetic animals to stimulate periodontal inflammation, osteoclast formation and periodontal bone loss in normal germ-free mice. The state of chronic inflammation induced by diabetes could disrupt the homeostasis of the bacterial communities in favor of dysbiotic communities capable of inducing periodontal tissue destruction. Though several diabetes-associated taxa were unaffected by local injection of IL-17, the abundance of Enterococcus was altered between groups and may be a taxon associated with inflammation in the context of diabetes. These results indicate that high levels of IL-17 contribute to diabetes-induced microbial changes that, in turn, increase pathogenicity.
In summary, there is a complex interaction between diabetes, inflammation, the oral microbiome and periodontal disease. We were able to dissect each component separately by inhibiting inflammation through local injection of IL-17 antibody and by examining oral microbial pathogenicity by transfer to normal germ-free recipients. These results suggest that the microbiome and the host response interact with each other and each contributes to diabetes-enhanced periodontal disease. This is based on previous findings that the same bacterial stimulus induces greater inflammation in diabetic animals compared to matched normoglycemic controls (Liu et al., 2006; Naguib et al., 2004b; Wu et al., 2015) and findings here that transfer of microbes from diabetic mice stimulate greater inflammation and bone loss than transfer from normoglycemic donor mice. Moreover, inhibition of IL-17 establishes that the pathogenicity of the diabetic microbiome is affected by the level of inflammation. Thus, we suggest that diabetes enhances periodontal inflammation and periodontal inflammation causes a pathogenic change in the microbiome to significantly increase the susceptibility and severity of periodontal disease.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for the reagents or resources may be directed to and will be fulfilled by the lead contact Dr. Dana Graves (dtgraves@upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Approximately 6-week-old type II diabetic female db/db (BKS.Cg-Dock7m +/+ Leprdb/J) and normoglycemic littermate controls were purchased from Jackson laboratories (Bar Harbor, Maine). For the 1st set of experiments, 10 db/db mice and 9 normoglycemic littermates were used to evaluate the oral microbiota, periodontal inflammation, and bone loss. For the 2nd set, 10 db/db mice were randomly assigned to two groups by using excel “int(rand())” command, one of which group received an oral injection of anti-IL-17A neutralizing antibody (R&D, MAB421, 5ul per site), and the other an equal amount of control IgG. Mice were housed in special cages (Lenderking Caging Products, Millersville, MD) with perforated floors to prevent accumulation of feces and limit coprophagy. Blood glucose was monitored weekly by tail vein bleeds with a digital glucometer (Lifescan Inc., Wayne, PA). Oral swabs were first taken when mice were ~8 weeks and final swabs when they were 12–13 weeks. Hyperglycemia was defined as blood glucose levels greater than 225 mg/dl. The db/db mice with serum glucose concentrations below 225 mg/dl or normoglycemic littermate controls with levels greater than 100 mg/dl at eight weeks were excluded. All mice were housed in the same environment under specific pathogen-free conditions with cycles of 14 hours’ daylight and 10 hours’ dark. Mice were euthanized by an overdose of intra-peritoneal injection of ketamine and xylazine and decapitation. Maxillary and mandibular jaws were harvested for further analysis. The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all procedures and experiments were carried out following their guidelines.
METHOD DETAILS
Oral Microbiome Transplantation
To ensure that recipient mice received similar inoculations, microbiota collected from donor mice of the same group were pooled. Bacteria were eluted and suspended in 1 ml sterile PBS. Colony forming units (CFU) were measured as previously described using a standard curve (Hajishengallis et al., 2011; Jiao et al., 2013; McIntosh and Hajishengallis, 2012; Wu et al., 2016). A standard bacterial reference sample was made consisting of a 1:1 mixture of Pg (ATCC 33277) and Fn (ATCC 25586) serially diluted 1:4 from 109 to 106 CFU. Total DNA was extracted with a DNeasy Blood & Tissue kit. Oligonucleotides primers 5′TCCTACGGGAGGCAGCAGT-3′, 5′-GGACTACCAGGGTATCTAATCCTGTT-3’ for a universal 16S rRNA bacterial sequence was used to identify Ct (cycle threshold) values for each dilution with an ABI 7500 Fast System thermal cycler (Applied Biosystems, Foster City, CA, USA) to generate a standard curve. From the standard curve the CFU was obtained by real-time PCR for each murine bacterial sample. Each recipient mouse was inoculated with 200ul of the bacterial sample in 2% methylcellulose (Sigma, Saint Louis, Missouri). Of the 28 recipient germ-free mice, 10 mice received oral bacteria from normoglycemic donor mice, 9 received samples from diabetic donor mice and 9 received samples from diabetic donor mice treated with IL-17A antibody. Each germ-free mouse was inoculated with bacteria from donor mice twice with a one-day interval between inoculations. Just prior to the first inoculation a ligature was placed between the first and second maxillary molar as previously described to provide conditions that facilitate bacteria-induced periodontal bone loss (Jiao et al., 2013; Maekawa et al., 2014). After 7 days, germ-free recipient mice were euthanized and oral swabs were taken of the teeth and gingiva. Recipient mice in which bacteria were not detected after transfer or mice in which the ligatures were lost were excluded from the study.
Microbial Genomic DNA Extraction and Library Preparation
Bacteria were obtained by oral swab of the teeth and gingival surface with Catch-All™ Sample Collection Swabs (Epicentre Biotechenologies, Madison, WI) and bacteria were eluted in PBS in cell lysis buffer from a DNeasy kit (Qiagen, Valencia, CA, USA) as described by the manufacturer and (Grice et al., 2010a). After a 60 second vortex, DNA present in the buffer was isolated with the DNeasy kit and quantitated with a spectrophotometer (Tecan, Mannedorf, Switzerland). 515F/806R primer with golay barcode in the reverse primer was used to amplify the V4 region of 16S ribosomal DNA (16S rDNA; IDT, Coralville, IA, USA) following the procedures described below and (Grice et al., 2010a).
PCR Full 96 well plate (in duplicate) 25ul total volume
10× Acuprime BufferII (2.5ul)
F Primer (512) 10uM (0.5ul)
R Primer (barcode) 10uM (0.5ul)
PCR clean water (19.3ul)
Accuprime High Fidelity Taq (0.2ul)
DNA (2.0ul)
PCR Run conditions
94°C, 3min; 2) 94°C, 45sec; 3) 50°C, 60sec; 4) 72°C, 90sec; 5) 72°C, 10min; 6) 4°C infinite (Steps 2–4 go through 35 cycles
- Validate PCR
- Pool the duplicates together (50ul total volume)
- Make two gels (60 wells per gel) per each 96 well plate
- 1.2% Agarose gel
-
○100ml TBE
-
○1.2g Agarose
-
○microwave until agarose is fully dissolved (don’t have boil over)
-
○let cool to room temp
-
○add 10ul Gel Red Dye, mix, and pour in mold.
-
○
- 2ul BlueJuice with 10ul PCR product and add 10ul of mixture to each well
- run at 120Volts for 30 min
- PCR Clean-up (with Agencourt XP beads)
- Spin down and then combine reactions to one plate.
- Add 72 ul Agencourt Beads to each well, pipetting up and down 10×.
- Incubate at room temperature for 5 min for DNA to bind to beads.
- Place plate on magnet for 2 min to separate beads from solution.
- Aspirate cleared solution from reaction plate, being careful not to disturb pelleted beads and discard.
- Dispense 200 ul 70% ethanol to each well. Aspirate. Repeat two more times.
- Dry beads 5 min.
- Take plate off of magnet and resuspend beads in 25 ul of TE, pH 8.0.
- Place plate back on magnet and allow beads to separate for 1 minute.
- Transfer eluant to a new plate.
- PCR Product Quantification (Using QuantIT dsDNA High-Sensitivity Assay Kit)
- Bring kit components to room temperature.
- Mix 80 mL of QuantIT buffer with 400 ul of QuantIT reagent (done in duplicates)
- Aliquot 200 ul of buffer-reagent mix to each well of 5–96 well Thermoscan plates.
- Add 10 ul of each standard (8 total standards at 0, 0.5, 1, 2, 4, 6, 8, and 10 ng/ul) to first two columns on each plate. Standards are read in duplicate.
- Add 2 ul of PCR products to wells.
- Measure fluorescence on Thermo plate reader with filter pair 485/538. Also include a 10 second shake step.
- Generate standard curve from standards (subtracting background).
- Calculate PCR product concentration from standard curve and equation.
- PCR product pooling and purification
- Pool equal amounts of each PCR product.
- Purify pool with Qiagen minElute column, following manufacturer’s instructions.
- Elute in 30 ul of TE, pH 8.0.
- Qubit the end product before giving to Sequencing Core
The University of Pennsylvania Genomics Core performed high throughput sequencing with MiSeq-150 paired-end runs. A total of 3,338,666 reads were generated with a mean of 49,098 ± 4421 reads per sample.
Sequencing Data Analysis
Sequence data was analyzed with the QIIME pipeline, version 1.9.1 (Caporaso et al., 2010). The forward and reverse reads were joined with no mismatches permitted. Read quality lower than Q29 or more than 3 consecutive low-quality base calls were discarded. Sequences were clustered into operational taxonomic units at a 97% similarity threshold using the UCLUST method (Edgar, 2010). Taxonomic assignments were generated using the default consensus-based algorithm in QIIME. A phylogenetic tree was inferred using the FastTree method (Price et al., 2010). Diversity, richness and bacterial taxon abundances were compared using the Kruskal-Wallis or Mann-Whitney test, as appropriate. Multiple comparisons were addressed by adjusting for a false discovery rate of 5%. UniFrac distances were compared using the PERMANOVA test (Anderson, 2001). Because non-parametric statistical tests were used, we did not apply additional diagnostic methods to ensure that data conformed to expected parametric distributions. Sequencing was performed in each individual sample once. Raw DNA sequence data was deposited in the NCBI Sequence Read Archive, and is available under accession number SRP108800 (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP108800).
Histologic Study
IL-17A expression in periodontal tissue was detected by immunohistochemistry with decalcified paraffin embedded sections at 4 µm thickness. Primary goat anti-mouse IL-17A (R&D, AF-421-NA) was used and detected by secondary donkey anti-goat IgG conjugated with horseradish peroxidase (PerkinElmer, Waltham, Massachusetts) followed by DAB substrate staining (PerkinElmer, Waltham, Massachusetts). To evaluate bone resorption activity, the number of tartrate-resistant acid phosphatase positive (Sigma-Aldrich, St. Louis, MO, USA) multinucleated cells were counted. Images were captured with a fluorescence microscope and Nikon NIS-Elements software (Nikon, Melville, NY, USA). Immunofluorescence was used to assess RANKL expression, myeloperoxidase-positive cells, and IL-6. Specific primary antibody to RANKL was obtained from Santa Cruz biotechnology, Dallas, TX, USA, to IL-6 from Abcam (Cambridge, MA) and MPO from Abcam. Quantitative analysis of RANKL, IL-6, and MPO was obtained by measuring the number of immunopositive cells by immunofluorescence with specific antibody. In each case a matched control antibody was also used and gave negative results. A histologic section was chosen for each animal so that the analysis was performed in a consistent region defined as the mid interproximal area between the teeth. RANKL was measured at 400× magnification with the region of interest consisting of the entire gingival connective tissue from the alveolar bone crest to the epithelial border bordered laterally by the root surface of each tooth. In addition, the coronal third of the periodontal ligament was assessed. MPO and IL-6 were measured at 400× magnification images of the entire gingival connective tissue as described for RANKL. For histomorphometric analysis a minimum of 6 maxillae were examined per group from different animals and the analysis was carried out twice with similar results. The number of immunopositive cells was counted by a blinded observer per unit area with NIS-Elements software (Nikon) and data are expressed as the number per mm2. For each antibody, a matched control antibody was used with negative results for the immunostaining. TRAP staining was performed by using TRAP staining kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions.
Quantitative Real-time Polymerase Chain Reaction
Total RNA was extracted from gingival tissue by using the RNeasy kit (QIAGEN) followed by revers transcription. 1 µg of total RNA was reverse transcribed using High Capacity RNA-to-cDNA kit (Thermo Fisher Scientific, Waltham, Massachusetts). Primers were obtained from Integrated DNA Technologies (Coralville, Iowa) and probes from Roche Life Science (Indianapolis, IN). Results were normalized with respect to the value obtained for the housekeeping gene RPL32, a ribosomal protein. ΔCt was calculated by subtracting the Ct value of the house-keeping gene (RPL32) from the target gene Ct value. ΔΔCt was calculated by subtracting the experimental group target gene ΔCt with control group target gene ΔCt. Relative mRNA expression was calculated by 2−ΔΔCt. Each experiment was performed two to four times with similar results. All the primers and probe was shown in the key resource table. Details of the primers and probes are given in the Key Resource Table. A minimum of six maxillary gingival tissue samples per group were examined from different mice by real-time PCR, which was performed three times with similar results.
Microscopic Computerized Tomography Analysis
Maxillae were dissected after euthanasia and scanned with the µCT-40 (Scano Medical AG, Bassersdorf, Switzerland). Histomorphometric analysis was performed with the OsirixMD software (version 6.5.2, Pixmeo – Bernex, Switzerland). The amount of alveolar bone present was evaluated as previously described by using Image J and OsirixMD version 6.5.2. The alveolar bone area between the molar teeth was calculated by image J from images obtained with OsirixMD software using the cemental-enamel junction as a coronal landmark to a fixed apical distance as described for histologic sections. The cemental surface of each tooth was used as a lateral boundary and the amount of bone within the space was calculated. A minimum of six maxillary samples from different mice were examined per group with the analysis performed 3 times with similar results.
Statistical Analysis
Statistical analysis of microbial samples is given in the Sequencing Data Analysis section above. Statistical analysis for all other experiments was performed using GraphPad Prism software from GraphPad Software Inc. (La Jolla, CA, USA). All data were expressed as the mean ± SEM. The difference between two groups was established by the Student’s t-test. Multiple group comparisons were performed by one-way ANOVA, with Tukey’s or Bonferroni’s post-hoc test to identify differences between specific groups. A value of P < 0.05 was considered to be statistically significant. No analysis was performed to determine whether the data met assumptions of the statistical approach. Each sample examined was from a different animal and the individual animal was the unit of measurement. The number of animals examined per group and the number of times the experiments were carried out are given in the Methods above and in the Figure Legends. For all experiments a minimum sample size of 5 was required based on our previous experience and published studies.
Supplementary Material
Highlights.
-
-
Diabetes increases periodontal bone resorption and tooth loss in mice.
-
-
Diabetic mice have increased periodontal inflammation and IL-17 levels.
-
-
Diabetic oral microbiota induces periodontitis in wild-type germ-free recipients.
-
-
Blocking IL-17 reduces the pathogenic effect of diabetic oral microbiota.
Acknowledgments
This work was supported by grants R01DE017732 and R01DE021921 from the NIDCR and by assistance from the Center for Host-Bacteria Interactions, University of Pennsylvania School of Veterinary Medicine and the Imaging Core of the Penn Center for Musculoskeletal Diseases. We would like to thank Ana Misic, Dan Beiting, and Aurea Simon-Soro for helpful discussions. We also thank Somreeta Sharma, Lina Jawari and Richa Pande for their assistance with histologic analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
DTG and EX designed experiments; EX, MM, GV, SC, JDC, YW and MLA performed experiments; EX, MM, JDC, KB, and DTG analyzed results; DTG, EX, KB, MM, and JDC wrote the manuscript.
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME journal. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Moutsopoulos NM. IL-17; overview and role in oral immunity and microbiome. Oral Dis. 2016;20:12598. doi: 10.1111/odi.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001:32–46. [Google Scholar]
- Andriankaja OM, Galicia J, Dong G, Xiao W, Alawi F, Graves DT. Gene Expression Dynamics during Diabetic Periodontitis. Journal of dental research. 2012;91:1160–1165. doi: 10.1177/0022034512465292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo-Castillo A, Novoa L, Balsa-Castro C, Blanco J, Mira A, Tomas I. Relationship between periodontitis-associated subgingival microbiota and clinical inflammation by 16S pyrosequencing. J Clin Periodontol. 2015;42:1074–1082. doi: 10.1111/jcpe.12470. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010 May;7(5):335–6. doi: 10.1038/nmeth.f.303. Epub 2010 Apr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple IL, Genco R, working group 2 of the joint, E.F.P.A.A.P.w. Diabetes and periodontal diseases: consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J Periodontol. 2013;84:S106–112. doi: 10.1902/jop.2013.1340011. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst FE, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell host & microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmer RT, Breskin A, Rosenbaum M, Zuk A, LeDuc C, Leibel R, Paster B, Desvarieux M, Jacobs DR, Jr, Papapanou PN. The Subgingival Microbiome, Systemic Inflammation and Insulin Resistance: The Oral Infections, Glucose Intolerance and Insulin Resistance Study (ORIGINS) J Clin Periodontol. 2016 doi: 10.1111/jcpe.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich LJ, Shlossman M, Genco RJ. Periodontal disease in non-insulin-dependent diabetes mellitus. J Periodontol. 1991;62:123–131. doi: 10.1902/jop.1991.62.2.123. [DOI] [PubMed] [Google Scholar]
- Escalante AE, Rebolleda-Gomez M, Benitez M, Travisano M. Ecological perspectives on synthetic biology: insights from microbial population biology. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol. 2008;35:89–105. doi: 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Liechty KW, Segre JA. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010a;107:14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Program NCS, Liechty KW, Segre JA. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010b;107:14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ, Graves DT. The enduring importance of animal models in understanding periodontal disease. Virulence. 2015;6:229–235. doi: 10.4161/21505594.2014.990806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell host & microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92:485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartstra AV, Bouter KE, Backhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes care. 2015;38:159–165. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- Hernandez CJ, Guss JD, Luna M, Goldring SR. Links Between the Microbiome and Bone. J Bone Miner Res. 2016;31:1638–1646. doi: 10.1002/jbmr.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong BY, Furtado Araujo MV, Strausbaugh LD, Terzi E, Ioannidou E, Diaz PI. Microbiome profiles in periodontitis in relation to host and disease characteristics. PLoS One. 2015;10:e0127077. doi: 10.1371/journal.pone.0127077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Nunez G, Giannobile WV, Raes J, et al. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. Gut microbiota promote hematopoiesis to control bacterial infection. Cell host & microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011;7:738–748. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial Biofilms in Colorectal Cancer Initiation and Progression. Trends Mol Med. 2016;13:30170–30178. doi: 10.1016/j.molmed.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Liu R, Bal HS, Desta T, Krothapalli N, Alyassi M, Luan Q, Graves DT. Diabetes enhances periodontal bone loss through enhanced resorption and diminished bone formation. J Dent Res. 2006;85:510–514. doi: 10.1177/154405910608500606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe H. Periodontal disease. The sixth complication of diabetes mellitus. Diabetes Care. 1993;16:329–334. [PubMed] [Google Scholar]
- Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D, Lambris JD, Hajishengallis G. Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. Journal of immunology. 2014;192:6020–6027. doi: 10.4049/jimmunol.1400569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh ML, Hajishengallis G. Inhibition of Porphyromonas gingivalis-induced periodontal bone loss by CXCR4 antagonist treatment. Molecular oral microbiology. 2012;27:449–457. doi: 10.1111/j.2041-1014.2012.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, Chalmers NI, Barb JJ, Abusleme L, Greenwell-Wild T, Dutzan N, Paster BJ, Munson PJ, Fine DH, Uzel G, et al. Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. PLoS pathogens. 2015;11:e1004698. doi: 10.1371/journal.ppat.1004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib G, Al-Mashat H, Desta T, Graves D. Diabetes prolongs the inflammatory response to a bacterial stimulus through cytokine dysregulation. J Invest Dermatol. 2004a;123:87–92. doi: 10.1111/j.0022-202X.2004.22711.x. [DOI] [PubMed] [Google Scholar]
- Naguib G, Al-Mashat H, Desta T, Graves DT. Diabetes prolongs the inflammatory response to a bacterial stimulus through cytokine dysregulation. The Journal of investigative dermatology. 2004b;123:87–92. doi: 10.1111/j.0022-202X.2004.22711.x. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, Ohno H, Yamazaki K. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrich EJ, Cullinan MP, Leichter JW. Diabetes, periodontitis, and the subgingival microbiota. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios S, Kang J, Galicia J, Gluck K, Patel H, Ovaydi-Mandel A, Petrov S, Alawi F, Graves DT. Diabetes aggravates periodontitis by limiting repair through enhanced inflammation. FASEB J. 2012;26:1423–1430. doi: 10.1096/fj.11-196279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios S, Xiao W, Mattos M, Lim J, Tarapore RS, Alsadun S, Yu B, Wang CY, Graves DT. Osteoblast Lineage Cells Play an Essential Role in Periodontal Bone Loss Through Activation of Nuclear Factor-Kappa B. Sci Rep. 2015;5 doi: 10.1038/srep16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Patterson E, Marques TM, O'Sullivan O, Fitzgerald P, Fitzgerald GF, Cotter PD, Dinan TG, Cryan JF, Stanton C, Ross RP. Streptozotocin-induced type-1-diabetes disease onset in Sprague-Dawley rats is associated with an altered intestinal microbiota composition and decreased diversity. Microbiology. 2015;161:182–193. doi: 10.1099/mic.0.082610-0. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Souto R, Colombo AP. Prevalence of Enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Archives of oral biology. 2008;53:155–160. doi: 10.1016/j.archoralbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Sweeney TE, Morton JM. The human gut microbiome: a review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013;148:563–569. doi: 10.1001/jamasurg.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S, Fujisaka S, Kahn CR. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol Metab. 2016;5:795–803. doi: 10.1016/j.molmet.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Colombo AP, Magalhaes CB, Hartenbach FA, Martins do Souto R, Maciel da Silva-Boghossian C. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb Pathog. 2016;94:27–34. doi: 10.1016/j.micpath.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Wu Y, Dong G, Xiao W, Xiao E, Miao F, Syverson A, Missaghian N, Vafa R, Cabrera-Ortega AA, Rossa C, Jr, et al. Effect of Aging on Periodontal Inflammation, Microbial Colonization, and Disease Susceptibility. J Dent Res. 2016;95:460–466. doi: 10.1177/0022034515625962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YY, Xiao E, Graves DT. Diabetes mellitus related bone metabolism and periodontal disease. Int J Oral Sci. 2015;7:63–72. doi: 10.1038/ijos.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Li S, Pacios S, Wang Y, Graves DT. Bone Remodeling Under Pathological Conditions. Front Oral Biol. 2016;18:17–27. doi: 10.1159/000351896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.