Abstract
Multiple sclerosis (MS) is a common autoimmune disease that targets myelin in the central nervous system (CNS). Multiple GWAS in the last 10 years have uncovered more than 200 loci that independently contribute to disease pathogenesis. As with many other complex diseases, risk to MS is driven by multiple common variants whose biological effects are not immediately clear. This review will present a historical perspective on the progress made on MS genetics and discuss current work geared towards creating a more complete model that accurately represents the genetic landscape of MS susceptibility. Such a model necessarily includes a better understanding of the individual contributions of each common variant to the cellular phenotypes, and interactions with other genes and with the environment. Future genetic studies in MS will likely focus on the role of rare variants and endophenotypes.
Keywords: Multiple sclerosis, Genetic pathways, Gene regulatory networks, GWAS
MS is a genetic disease
Multiple sclerosis (MS) is a chronic disease of the central nervous system (CNS) that affects more than two million individuals worldwide [1, 2]. MS is typically diagnosed between the third and forth decade of life, occurs more frequently in women than men, and is considered the most common cause of non-traumatic neurological disability in young adults. Although the trigger(s) of MS remains unknown, its pathogenesis is best explained by a multifactorial model that incorporates interactions between genetic, epigenetic, and infectious, nutritional, climatic, or other environmental influences including Epstein Barr virus (EBV) infection, sun light exposure and smoking [3–7]. This array of factors results in the loss of immune homeostasis and self-tolerance manifested in brain and spinal cord infiltration by activated peripheral mononuclear cells, and the development of unregulated pathologic inflammatory responses against structural components of the CNS. Myelin loss, gliosis, and the resulting axonal pathology culminate in progressive, often severe neurological dysfunction. Emerging data show that the neuroinflammation and neurodegeneration that occur in MS are overlapping and have a complex dependence [8–11] but the autoimmune model of pathogenesis has set early the tone for immunotherapy as the primary clinical management strategy, first by global immune-suppression using aggressive anti-inflammatory drugs, and more recently by selectively targeting specific elements of the immune response [12–15]. The recognition that axonal damage unresponsive to immunotherapy is the main driver of disability in MS has brought the need to emphasize the genetics of grey and white matter cell death and repair as pressing research frontiers in this disease.
Heritable contributions to MS risk are undisputable. Pivotal epidemiological data consistent with a prominent role of genetic factors in MS include the high disease prevalence in distinct ancestral groups (particularly those of northern European origin) compared with others (e.g. Africans and Asians), in some cases irrespective of geographic location. Remarkably, the prevalence of MS appears to have steadily increased over the past century; this increase has apparently occurred primarily in women [16–18] and in regions previously considered low-incidence [19, 20]. The higher prevalence rates (140–250 per 100,000) are still found in northern Europeans and in whites living in the northern US and Canada [2, 21]. In contrast, low prevalence rates are found in Asian countries (e.g. 6 per 100,000 in Japan) and native populations across the Americas and Oceania [22]. Notwithstanding difficulties in surveillance, MS is almost non-existent in black Africans and early estimates in the US suggested that the disease is significantly less prevalent in admixed African Americans than in European Americans (relative risk of 0.64 [23]). However, more recent studies are challenging this long-held belief, suggesting that MS incidence in African Americans may be equal to, or potentially even higher than in European Americans [24] [25]. Furthermore, compared with Europeans, African Americans also carry a greater risk of ambulatory disability that may be at least in part, genetically determined [26, 27].
Analogous to other autoimmune diseases, individuals with MS tend to cluster in families. Multiple studies have shown for example that monozygotic twins have a higher concordance rate (20% – 30%) compared to dizygotic twins (2% – 5%), providing strong support for a significant but complex genetic etiology in MS [28–30]. Equally important, siblings of an affected individual are at least seven times more likely to develop MS than the general population [31, 32]. Second and third degree relatives, but not spouses, carry a modest increased risk [33, 34], also in agreement with genetic factors causing the familial clustering. Altogether, the population and family data lead to a broad acceptance that the MS-prone genotype results from multiple independent DNA variants relatively frequent in the population [35]. Recognizing the influence of specific HLA variants within the MHC gene complex (chro 6p21) in MS susceptibility in the early 1970’s represented the first empirical demonstration linking disease risk with common genetic variation, but it was the polygenic model of MS heritability that provided the theoretical justification for assembling multi-center large DNA datasets to pursue genome-wide association studies (GWAS).
Ten years of GWAS in MS
The first concerted efforts to identify susceptibility genome-wide were done through linkage analysis in multicase-families. These studies failed to identify any consistent statistically significant signal [36–38]. Yet, a suggestive LOD score found in the HLA region was later confirmed in larger studies, providing a statistical road map to take full advantage of the emerging microarray technology [39, 40]. The first GWAS in MS was completed by the International Multiple Sclerosis Genetics Consortium (IMSGC) on subjects from the US and UK using an unorthodox family-based (MS patients and both parents) study design for discovery coupled with case-control replication [41]. A number of GWAS and large-scale targeted studies followed in the ensuing 10 years [42–48], culminating in the collation of a genetic dataset representing 47,351 MS subjects and 68,284 controls that showed unequivocal statistical evidence for the association of 200 autosomal susceptibility variants outside the MHC, one chromosome X variant, and 32 independent associations within the extended MHC [49]. Altogether, different measures of the heritability explained by all associations are still in the 20–30% range, of which a substantial proportion can be assigned to the MHC region. While the explanation for this missing heritability may be rooted in gene-gene, gene-environment interactions, and epigenetic factors, it is also conceivable that the early measures of heritability were overestimated.
The power of genetic association studies mostly depends on 3 variables: effect size, sample size, and allele frequency. Initial power calculations for MS estimated that to detect an association with OR>1.2 investigating common variants (those with minor allele frequency > 5%) 10,000 samples would be required to ensure that a p-value of 10−7 is a hundred times more likely to be true than false (in contrast, the posterior odds of the same p-value with only 2000 patients would be in favor of a false discovery) [35]. While in later years a p-value threshold of 5 10−8 for GWAS emerged as a consensus [50], a systematic analysis of published studies suggested a further relaxation of this p-value would result in the discovery of additional genuine associations [51].
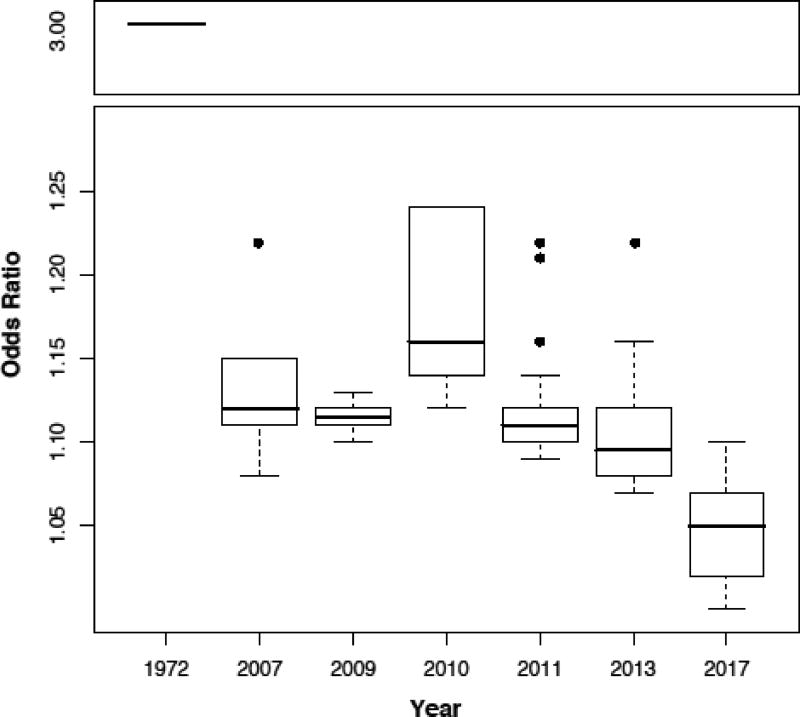
When all MS associations are analyzed by year of discovery (a proxy for study size), they show an ever-decreasing median odds ratio (OR; a measure of effect size) (Fig 1). For example, the association with HLA-DRB1*15:01 has an OR∼3.5 and was first reported in 1972 with just a few hundred cases, while the first discovered non-HLA loci (in 2007) have OR∼1.15 and nearly 1000 cases were needed. The most recent studies (2011, 2013 and 2017) analyzed tens of thousands of cases (10,000, 15,000 and 40,000 respectively), which enabled the reporting of statistically replicated signals down to OR=1.05. This asymptotic trend toward OR∼1 also means that the cost:benefit incentive for performing additional GWAS searching for additional common susceptibility signals in Europeans may no longer be justified or realistic. Associations in that low effect size-range mean that those variants are almost as frequent in cases as they are in controls thus raising the argument of whether they are biologically relevant. Quantitative risk scores have been developed to assess the significance of the “genetic load” of individual patients and whether all associations count equally towards risk [52, 53]. However, at least in these early versions, these algorithms are not able to accurately predict risk of individual subjects, and thus are not yet an option to be used in the clinic for precision medicine purposes.
Fig 1. Decreasing OR as study n increases.
Box plots represent the median and quartiles of the odds ratio (OR) of studies performed at the indicated years. With exception of 2010, each new study revealed associations with smaller OR.
However, this does not mean the GWAS era is over in MS. Due to inherently different LD structures, studies of other populations (African-American/European, Hispanics, or Asians) is a meritorious proposal for fine-mapping and identification of population-specific effects, notwithstanding the difficulties in assembling the required large DNA sample sets [54, 55]. Likewise, the study of disease heterogeneity (clinical subtypes or endophenotypes among cases) by GWAS is a valid approach with high potential for discoveries of disease modifiers, particularly with the large cohorts that have been already assembled. Here, accurate and quantitative description of the phenotypes, beyond whether a subject has the disease or not, is crucial to success. Finally, the search for rare(r) variants by exome sequencing or similar approaches is an active field of investigation.
Nearly all the identified associations map to non-coding regions of the genome, either in intragenic or intergenic regions. Naturally, those mapping to intragenic regions were explored first (IL7RA, TNFSF10, etc.) as their relationship to function is thought to relate to splicing, mRNA stability or promoter activity of the gene in question. However, the vast majority of associations lie in genomic regions distant from any known gene. In those cases, the most likely explanation is that they modify regulatory sites and indirectly influence gene activity. Examples of regulatory regions include open chromatin, histone modifications, enhancers, and repressors. Efforts to integrate statistical evidence of association with their potentially regulatory effect are underway. In the most recent MS GWAS, an analysis of open chromatin regions was performed using DNase I hypersensitivity data collated from 56 tissues from the NIH Roadmap Epigenomics project (REP) [56]. For each tissue, the proportion of SNPs in the list that were located on DHS regions to the proportion of all SNPs located on DHS regions active in that tissue were compared. Notably, statistically significant enrichment was observed only in 9 cells from the immune system, including CD3, CD4, CD8 (T cells), CD14 (monocytes) and CD19 (B cells) [49]. This finding highlights the immune nature of genetic MS susceptibility.
Sharing with other autoimmune diseases
One salient characteristic of the MS susceptibility map is the large proportion of the associations (either the exact same variant, or within the same gene or locus) that are shared with other autoimmune diseases, such as type 1 diabetes mellitus (T1D), rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and Crohn’s disease (CD), among others. Interestingly, some of these shared associations confer risk for one disease but protection for another. For example, allelic heterogeneity was reported in the IL2RA locus, where rs11594656-A was shown to be associated with susceptibility to one disease (MS) and protection to another (T1D). Furthermore, rs41295061-A only conferred susceptibility to T1D but not MS. Finally, rs2104286-G was only associated with T1D [57]. While somewhat surprising at first, genetic and allelic heterogeneity across autoimmune diseases is now recognized to be a distinctive feature of this group of disorders.
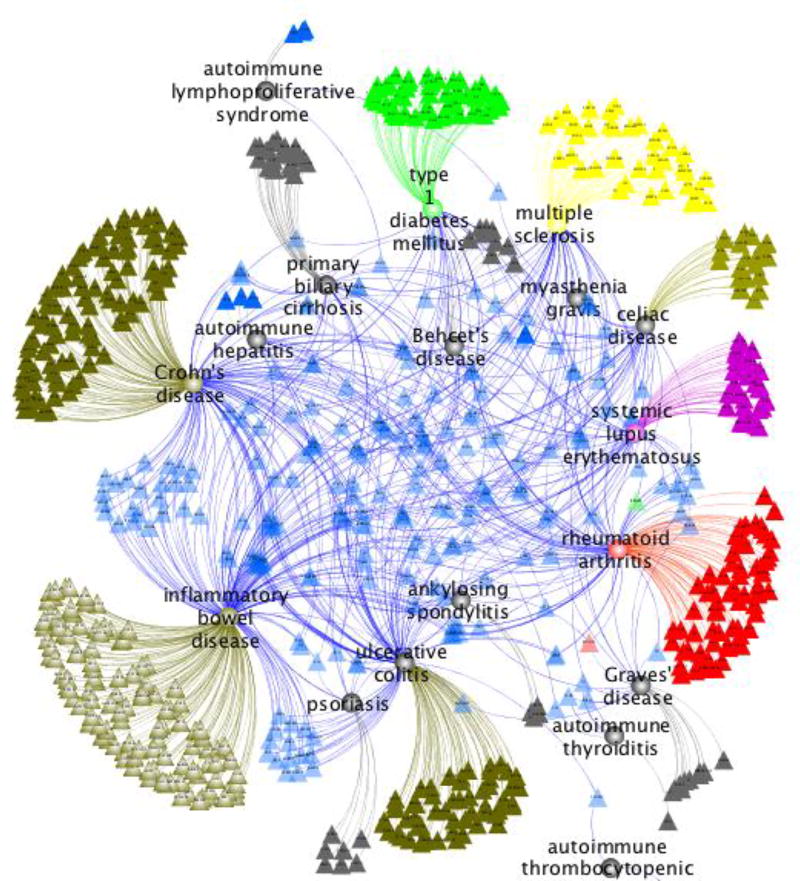
Figure 2 shows associated genes for most autoimmune diseases with GWAS data (obtained from the GWAS Catalog). The figure clearly shows a core of shared loci (blue triangles) and a number of disease-specific ones represented by different colors (MS in yellow). This remarkable sharing is the highest among the complex diseases, including cancers, neurological and metabolic (data not shown). This particular architecture confirms that MS is at its core an autoimmune disease and strongly suggests that susceptibility to autoimmunity is an inherited risk which later in life may be compounded and shaped by additional genetic (and epigenetic) determinants and environmental exposures to ultimately define the predominant effector mechanism (cellular vs. humoral) and target organ of the autodestructive process.
Fig 2. An autoimmunity gene network.
All known associations to common autoimmune diseases were obtained from the GWAS Catalog [92] and mapped to the closest or most likely affected gene, as reported by the authors of each publication. Then, data was displayed graphically as a network using the software Cytoscape [93]. Nodes represent either diseases (circles) or genes (triangles), while edges indicate genetic associations. Genes associated with more than one disease are shown in blue and lie at the center of the network. Genes associated with only one disease are displayed with the same color as their corresponding disease and organized in outward fashion.
From mapping to function
One of the most difficult steps in any GWAS is to infer the biologically relevant consequences of each statistical association. Given that the majority of associations map to non-coding regions, in the absence of obvious functional signals, earlier studies simply reported as candidate causative the closest gene(s) to the most significant independent effect. However, with continuous improvement in our understanding of the molecular machinery of gene regulation it has now become clear that this crude approach needs further refinement. One way to confirm the functional effect of genetic associations is to conduct in-vitro or in-vivo experiments in which the presence of the variant is studied in the context of expression of the putative regulated gene target(s). The first such demonstration in MS was the discovery that rs6897932, a polymorphism located in chromosome 5p13.2 and previously associated with disease susceptibility (OR= 1.18) [41] influenced the function of interleukin 7 receptor alpha gene (IL7RA). Specifically, inheritance of the risk allele (C) disrupts a splicing acceptor site and results in transcriptional skipping of exon 6 of the gene, thus altering the relative amounts of soluble and membrane bound isoforms of the gene [58]. More recently, several SNPs within the RNA helicase DEAD box polypeptide 39B (DDX39B) gene were shown to be associated with MS risk in a meta-analysis [59]. In particular, rs2523506-A --located within the 5’UTR of this gene-- was shown to reduce mRNA translation of DDX39B, which is in turn, a potent activator of IL7R exon 6 transcription. This example provides functional evidence that two associated loci can work together to confer susceptibility to MS. Given the large number of independent associations, and the networked molecular pathways that underlie cellular behavior, this example may only be the tip of the iceberg in unraveling the complex genetic architecture of MS.
Another well-characterized example is the intronic SNP rs1800693 in the tumor necrosis factor receptor super family 1A (TNFRSF1A) gene. In this case, the risk allele also results in skipping of exon 6 and the production of a novel soluble form of the TNF receptor. This soluble protein is able to inhibit TNF signaling inside the cells, mirroring somehow, the exacerbating effects of TNF-blocking drugs on MS course [60].
One of the most complex regions in the MS susceptibility map is 1p22.1, which has been the subject of several reports with seemingly contradictory results, i.e. different genes in the locus were linked to the association signal [61–64]. In this locus, GWAS meta-analysis identified multiple statistically independent genome-wide effects, three of which were found under the same association peak. In 2011 the disease-associated SNP rs11804321, which is located in the last intron of the ecotropic viral integration site 5 (EVI5) gene, was shown to overlap with an insulator element that modulates the expression of the neighboring GFI1 via the transcriptional repressor CCCTC-binding factor (CTCF) [64]. More recently, it was reported that the non-synonymous, strongly associated exonic SNP rs11808092 in the EVI5 gene itself induces changes in superficial hydrophobicity patterns of the coiled-coil domain of EVI5 protein, which, in turns, affects the EVI5 interactome. In particular, this work showed that EVI5 protein bearing the risk allele selectively interacts with sphingosine 1-phosphate lyase (SGPL1)–an enzyme important for the creation of the S1P gradient, which is relevant to adaptive immune response and the therapeutic management of MS [62].Thus, it is conceivable that the association peak in this region truly represents multiple independent functional effects.
A specific DNA polymorphism in TNFSF113B, insertion-deletion GCTGT->A (BAFF-var), was recently reported to be associated with MS risk in Sardinians [65]. Whereas the frequency in Sardinians is approaching 30%, presumably as a result of environmental pressures, BAFF-var frequency in other ancestral groups, including Europeans, ranges from less than 1% to 6%. TNFSF113B encodes the cytokine BAFF, which belongs to the TNF ligand super-family. BAFF is expressed in B lineage cells and acts as a potent B cell differentiator and activator. Noteworthy, BAFF-var results in a shorter transcript that escapes microRNA inhibition, resulting in increased production of soluble BAFF. In addition to disease risk, BAFF-var was shown to be significantly associated with several endophenotypic traits, such as B cell and monocyte counts and total IgG/IgA/IGM levels. The authors hypothesize that BAFF-var carriers would have a weaker response to B-cell depleting therapies due to an accelerated resurgence of memory B cells.
Additional studies with individual risk variants have been performed but our current understanding of the function of each bonafide susceptibility variant in the molecular pathogenesis of MS is merely superficial. Moreover, since DNA variants are likely to act in concert to confer risk, studies on their interaction and on how biological pathways are affected by them are needed.
Gene environment interactions in MS
A reasonable interpretation of the incidence increase and distinctive geo-prevalence distribution of MS implicates precipitating environmental or lifestyle triggers and/or the disappearance of protective environmental factors. The biological interactions between these environmental factors and genetic variance could also explain the inherent complexity of MS as a genetic trait. In recent years, several non-genetic risk factors for MS have been identified. These include vitamin D deficiency, exposure to the Epstein Barr virus (EBV) after early childhood and manifestations of infectious mononucleosis, and cigarette smoking [66, 67]. Although the causality of these associations remains somehow controversial, there is a justifiable interest in formally assessing the additive interactions between exposure and the emerging MS-susceptibility DNA map [68]. These studies have been limited to HLA genes so far, with positive results reported for active and passive smoking, EBV infection and infectious mononucleosis, and childhood/adolescent obesity [reviewed in [5]]. For example, in a pooled analysis of six datasets, smokers carrying HLA-DRB1*15 and lacking HLA-A*02 had a 13-fold increased disease risk compared with never smokers without these genetic risk factors (OR 12.7, 95% CI 10.8–14.9) [69]. Novel modeling approaches will afford the analysis of gene-environment interactions genome-wide [70], potentially addressing an important knowledge gap in the understanding of MS heritability.
From individual loci to pathways and beyond
The multiple efforts in mapping MS susceptibility loci have succeeded in identifying more than 200 independent associations. Without an elaborated analysis, the overall flavor of the associations can be assumed to be heavily immunological. Furthermore, several associated SNPs have been shown to be eQTL in different cells of the immune system. For example, associated variants that are eQTLs for genes like IFITM3, CD37, and CD6 in CD4+ T cells strongly suggest that they may have an effect in modulating adaptive immune responses. Further, eQTL variants in or near CLECL1, RGS1, and MERTK in CD14+ monocytes suggest that the innate immune system might also be involved. Notably, variants in genes expressed in the central nervous system (CNS) are far less common in these studies, thus supporting a model in which genetic susceptibility to MS is mediated primarily by sustained dysregulation of immune responses over several decades before clinical symptoms appear.
However, as described above, the functional consequences have been described for only a handful of loci. Another way to contribute to the interpretability of genetic associations is to conduct pathway analysis. In its simplest form, pathway analysis is the search for enrichment of associated signals at or near genes with similar or complementary functions. Analogous to how large gene sets from transcriptomics data are analyzed, associated polymorphisms can be mined for their effects on genes from common predefined processes like gene ontologies or biological pathways. The main challenge with GWAS signals is that the affected gene(s) is not always obvious. While some analyses focus on the closest gene to the risk-associated SNP, it has now become clear that conserved motifs from promoters, enhancers and regulatory elements can be quite far away from the target gene(s). Modern approaches will likely incorporate this type of information from projects like REP [56], Encyclopedia of DNA elements (ENCODE) [71, 72] and the International Human Epigenome Consortium (IHEC) [73]. Once the SNP-to-gene mappings are performed in a biologically meaningful way, enrichment of the putative genes in specific pathways can be evaluated [74, 75].
Instead of performing enrichment analysis with canonical pathways (an approach that could lead to biased results based on our current knowledge) we and others proposed to merge the statistical evidence of association with the biological evidence of interaction at the protein level by virtue of a protein interaction network (PIN) [76–79]. The main question posed by this approach is whether genes known to be regulated by risk SNPs are more likely to interact among them than what would be expected by chance. A significantly higher number of interactions would indicate that those genes are likely participating in a common biological process (independently of whether that pathway was described in a database or not). We have termed this approach PIN-based pathway analysis (PINBPA) and have created a user-friendly analysis tool (as an App for the software Cytoscape, http://www.cytoscape.org), which has more than 4,000 users worldwide [80, 81].
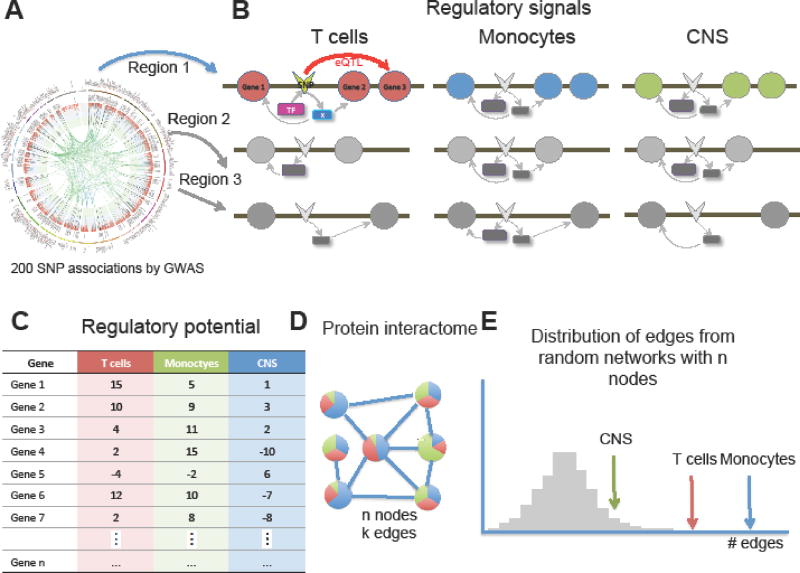
To provide a more accurate representation of the overall risk posed by MS susceptibility loci, we propose an approach that integrates PINBPA with cell specific regulatory signals (Figure 3, Key Figure). Furthermore this approach can also be employed to compute personalized individual’s risk profile, an area of much interest within the precision medicine community.
Figure 3. Strategy for cell specific PINBPA.
A. Each of the 200 MS-associated SNPs and those in LD (regions) is queried for regulatory features such as eQTL, enhancer, DNase hypersensitivity region, histone modification, etc. from ENCODE, REP and IHEC. B. All signals are integrated in a cell specific manner. C. Regulatory potentials are computed for each gene in proximity to associated SNPs for different cell types. D. The regulatory potentials are incorporated as node attributes into a protein interaction network and the number of edges among genes with score > 0 is computed. E. The number of edges among genes for each cell type is displayed along with the distribution of edges from random networks of similar size.
Concluding remarks
It is hard to argue against the contribution of common genetic variation to the susceptibility of MS. While discovery of non HLA-related loci lagged for some time, MS is one of the great successes of the GWAS era in human genetics. The impressive pace of discovery in the last 10 years highlights the need for collaboration, as the number of cases necessary to achieve statistical power is only possible in the context of international consortia.
While additional susceptibility GWAS focusing on common variants will likely be more sporadic in the future, there is still much to be done (See Outstanding Questions Box). As efforts on precision medicine evolve, we anticipate that the study of non-European populations will take a more prominent role. Also, studies of rare variants will likely be undertaken, either by whole genome or exome sequencing, or targeted re-sequencing efforts. Similarly the contribution of genetic variation to endophenotypes has not yet been fully explored and with the advent of wearables and other digital technology that can precisely sense and record internal variables (gait, vision, MRI, micro-movement detection, etc.), we anticipate these studies will see much development.
Outstanding questions (BOX).
Rare variants: The role of rare variation in MS susceptibility has not yet been systematically explored. However, exome sequencing in other common diseases (T2diabetes, etc) has met with only moderate success.
Endophenotypes: In addition to identification of susceptibility (risk) alleles, genetic association studies can also focus on the presence of modifiers of disease severity, response to therapy or other traits that alter disease expressivity. For these studies, availability of quantitative, reproducible and measurements is critical. Such endophenotypes include imaging metrics (MRI/OCT), electrophysiological parameters, biomarkers, and high-definition, computerized gait analysis, among others.
Gene-gene and gene-environment interactions: More sophisticated models of interacting molecular pathways and cellular networks and their interaction with the environment will need to be developed.
Metagenomics: The cross-talk between the genomes of the host and those of human symbionts (bacteria, viruses, archea, etc) is started to be studied in MS. Initial findings suggest the presence of certain gut bacteria can influence susceptibility.
Non-whites: Mounting on the success in Europeans, the next round of studies will likely focus on non-white populations such as African, Asian, Native Americans, Hispanics, etc. Emerging evidence suggests that while some risk loci are conserved, ethnic-specific variants may contribute to shaping disease susceptibility in different populations (50, 69–72).
Risk prediction/precision medicine: Efforts to use risk variants either alone or in combination with other clinical or environmental variables to define an individual-specific susceptibility profile are underway.
Finally, the development of affordable and massively parallel sequencing technologies has enabled the exploration of genomes other than our own, giving birth to the field of metagenomics. It is expected that in the near future large studies will be carried out to characterize the genomes of all microscopic entities that co-exist with humans (e.g. bacteria, archea, viruses, etc.) and their potential relationship to disease triggering or perpetuation.
In summary, the genetics architecture of MS has been largely unraveled by international, collaborative efforts. The next frontier seems to be the identification of their functional consequence, the pathways affected by these variants, and finally, the integration of multi-modal data sources into a subject’s medical record, a process that could bring the promise of precision medicine in MS one step closer.
MHC Box
GWAS have confirmed that the main MS susceptibility association signal genome-wide maps to the HLA-DRB1 locus in the Major Histocompatibility Complex (MHC). This region is located on the short arm of chromosome 6 at p21.3, spans almost 4000 KB of DNA, and contains ∼165 closely linked genes, about half having pivotal roles in the immune system [82]. These include the HLA genes, which have been extensively studied as the principal determinants of allogeneic transplantation outcomes [83]. The polymorphism of many of the HLA genes is extraordinary, with more than 14,000 alleles identified to date; more than 100 infectious, autoimmune and inflammatory disease phenotypes, including MS, as well as drug reactions and cancers are associated with HLA genes variation [84, 85]. The association of the HLA locus with MS risk was first described several decades ago [86] and since then has been observed across all populations studied [87]. HLA-DRB1 allelic heterogeneity vis-a-vis MS risk and copy number effects have been described [88]. Individuals who are HLA-DRB1*15:01 homozygotes carry a high-risk genotype with ORs exceeding 7.0, compared to a range between 3.5 and 5.0 for heterozygotes HLA-DRB1*15:01/X. In addition to risk, HLA-DRB1*15:01 has been associated with phenotypic markers of disease severity [89]. A decade long effort of genomic screens provided statistical evidence for over 30 independent allelic and genetic associations within the extended MHC region. Hierarchical allelic lineages and epistatic effects affecting risk are described but poorly understood [90, 91]. It is sobering to recognize that despite the fact that examination of HLA variation at this level has been performed in MS cohorts comprised of many thousands of individuals, a complete understanding of the role of the MHC in disease pathogenesis remains elusive.
Trends Box.
More than 200 loci have been associated with MS susceptibility to date (half of them in the last 4 years alone).
There is extensive sharing of genetic risk variants between MS and other autoimmune diseases. This suggests a model in which a general risk for autoimmunity is inherited. Additional genetic (and epigenetic) determinants and environmental exposures are compounded to ultimately define the target organ of the autodestructive process.
Efforts to characterize the biological consequences of reported associations are undergoing.
Cell specific pathways are being developed to understand disease heterogeneity and individualized risk assessment.
Acknowledgments
We thank the National Institute of Health’s (NIH) Institute for Neurological diseases and stroke (NINDS) (grants R01NS088155 and RO1NS26799). SEB is the Heidrich Family and Friends Endowed Chair in Neurology at UCSF. JRO is the G. Zimmerman Endowed Chair in Neurology at UCSF.
Glossary
- self-tolerance
Immunological tolerance is the failure to mount an immune response to an antigen. Self-tolerace refers to the (normal) failure to respond to one own’s antigens, thus avoiding avoid autoimmunity
- Gliosis
A process leading to scars in the CNS that involves the production of a dense fibrous network of neuroglia (supporting cells) in areas of damage. In MS gliosis ensues after axonal loss
- GWAS
Genome wide association study. This strategy is an examination of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWASs typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other organism
- HLA
The human leukocyte antigen (HLA) system is a genomic locus encoding the major histocompatibility complex (MHC) proteins in humans
- LOD
Logarithm of the odds ratio (OR). In genetics, the OR is one way to quantify how strongly the presence or absence of a given phenotype is associated with the frequency of a particular allele in a specific population
- MHC
The major histocompatibility complex (MHC) is a set of cell surface proteins essential for the acquired immune system to recognize foreign molecules in vertebrates, which in turn determines histocompatibility. Failure to match HLA types between donor and recipient in an organ or tissue transplant can result in rejection
- Heritability
Is a statistical term to represent the proportion of phenotypic variance attributable to genetic variance
- Splicing acceptor site
In the splicing of RNA, the site at the 3' end of an intron.
- Interactome (protein)
The entire set of molecular interactions of a cell or target protein.
- LD
Linkage disequilibrium. In population genetics, LD is the non-random association of alleles at different loci in a given population
- eQTL (expression quantitative trait loci)
Regions of the genome containing DNA sequence variants that influence the expression level of one or more genes
- Metagenomics
The study of genetic material recovered directly from environmental samples. In this work, it refers to the genomes of bacteria and other microorganisms inhabiting in or around the human body
- REP
Roadmap Epigenomics Project (http://www.roadmapepigenomics.org). An NIH sponsored initiative to produce a public resource of human epigenomic data to catalyze basic biology and disease-oriented research
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hauser SL, Goodin DS. Multiple Sclerosis and other demyelinating diseases. In: Longo DI, editor. Harrison’s principles of internal medicine. 19. McGraw-Hill: 2015. pp. 3395–3409. [Google Scholar]
- 2.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–17. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 3.Hedstrom AK, et al. Environmental factors and their interactions with risk genotypes in MS susceptibility. Curr Opin Neurol. 2016;29(3):293–8. doi: 10.1097/WCO.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 4.Hedstrom AK, et al. Smoking is a major preventable risk factor for multiple sclerosis. Mult Scler. 2016;22(8):1021–6. doi: 10.1177/1352458515609794. [DOI] [PubMed] [Google Scholar]
- 5.Olsson T, et al. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13(1):25–36. doi: 10.1038/nrneurol.2016.187. [DOI] [PubMed] [Google Scholar]
- 6.Rhead B, et al. Mendelian randomization shows a causal effect of low vitamin D on multiple sclerosis risk. Neurol Genet. 2016;2(5):e97. doi: 10.1212/NXG.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundqvist E, et al. Cytomegalovirus seropositivity is negatively associated with multiple sclerosis. Mult Scler. 2014;20(2):165–73. doi: 10.1177/1352458513494489. [DOI] [PubMed] [Google Scholar]
- 8.Baxi EG, et al. Transfer of myelin-reactive th17 cells impairs endogenous remyelination in the central nervous system of cuprizone-fed mice. J Neurosci. 2015;35(22):8626–39. doi: 10.1523/JNEUROSCI.3817-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dendrou CA, et al. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15(9):545–58. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 10.Devier DJ, et al. Increase in NF-kappaB-sensitive miRNA-146a and miRNA-155 in multiple sclerosis (MS) and pro-inflammatory neurodegeneration. Front Mol Neurosci. 2015;8:5. doi: 10.3389/fnmol.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yshii L, et al. Neurons and T cells: Understanding this interaction for inflammatory neurological diseases. Eur J Immunol. 2015;45(10):2712–20. doi: 10.1002/eji.201545759. [DOI] [PubMed] [Google Scholar]
- 12.Kappos L, et al. The 11-year long-term follow-up study from the randomized BENEFIT CIS trial. Neurology. 2016;87(10):978–87. doi: 10.1212/WNL.0000000000003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannoni G, et al. Alemtuzumab improves preexisting disability in active relapsing-remitting MS patients. Neurology. 2016;87(19):1985–1992. doi: 10.1212/WNL.0000000000003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauser SL, et al. Ocrelizumab versus Interferon Beta-1a in Relapsing Multiple Sclerosis. N Engl J Med. 2017;376(3):221–234. doi: 10.1056/NEJMoa1601277. [DOI] [PubMed] [Google Scholar]
- 15.Blinkenberg M, Soelberg Sorensen P. Monoclonal Antibodies for Relapsing Multiple Sclerosis: A Review of Recently Marketed and Late-Stage Agents. CNS Drugs. 2017;31(5):357–371. doi: 10.1007/s40263-017-0414-3. [DOI] [PubMed] [Google Scholar]
- 16.Orton SM, et al. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006;5(11):932–6. doi: 10.1016/S1474-4422(06)70581-6. [DOI] [PubMed] [Google Scholar]
- 17.Sellner J, et al. The increasing incidence and prevalence of female multiple sclerosis--a critical analysis of potential environmental factors. Autoimmun Rev. 2011;10(8):495–502. doi: 10.1016/j.autrev.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Koch-Henriksen N, Sorensen PS. The changing demographic pattern of multiple sclerosis epidemiology. Lancet Neurol. 2010;9(5):520–32. doi: 10.1016/S1474-4422(10)70064-8. [DOI] [PubMed] [Google Scholar]
- 19.Eskandarieh S, et al. Multiple Sclerosis Epidemiology in East Asia, South East Asia and South Asia: A Systematic Review. Neuroepidemiology. 2016;46(3):209–21. doi: 10.1159/000444019. [DOI] [PubMed] [Google Scholar]
- 20.Benito-Leon J. Are the prevalence and incidence of multiple sclerosis changing? Neuroepidemiology. 2011;36(3):148–9. doi: 10.1159/000325368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dilokthornsakul P, et al. Multiple sclerosis prevalence in the United States commercially insured population. Neurology. 2016;86(11):1014–21. doi: 10.1212/WNL.0000000000002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosati G. The prevalence of multiple sclerosis in the world: an update. Neurol Sci. 2001;22(2):117–39. doi: 10.1007/s100720170011. [DOI] [PubMed] [Google Scholar]
- 23.Wallin MT, et al. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann Neurol. 2004;55(1):65–71. doi: 10.1002/ana.10788. [DOI] [PubMed] [Google Scholar]
- 24.Wallin MT, et al. The Gulf War era multiple sclerosis cohort: age and incidence rates by race, sex and service. Brain. 2012;135(Pt 6):1778–85. doi: 10.1093/brain/aws099. [DOI] [PubMed] [Google Scholar]
- 25.Langer-Gould A, et al. Incidence of multiple sclerosis in multiple racial and ethnic groups. Neurology. 2013;80(19):1734–9. doi: 10.1212/WNL.0b013e3182918cc2. [DOI] [PubMed] [Google Scholar]
- 26.Weinstock-Guttman B, et al. Multiple sclerosis characteristics in African American patients in the New York State Multiple Sclerosis Consortium. Mult Scler. 2003;9(3):293–8. doi: 10.1191/1352458503ms909oa. [DOI] [PubMed] [Google Scholar]
- 27.Cree BA, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63(11):2039–45. doi: 10.1212/01.wnl.0000145762.60562.5d. [DOI] [PubMed] [Google Scholar]
- 28.Willer CJ, et al. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100(22):12877–82. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen T, et al. Concordance for multiple sclerosis in Danish twins: an update of a nationwide study. Mult Scler. 2005;11(5):504–10. doi: 10.1191/1352458505ms1220oa. [DOI] [PubMed] [Google Scholar]
- 30.Kuusisto H, et al. Concordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twins. Eur J Neurol. 2008;15(10):1106–10. doi: 10.1111/j.1468-1331.2008.02262.x. [DOI] [PubMed] [Google Scholar]
- 31.Robertson NP, et al. Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain. 1996;119(Pt 2):449–55. doi: 10.1093/brain/119.2.449. [DOI] [PubMed] [Google Scholar]
- 32.Westerlind H, et al. Modest familial risks for multiple sclerosis: a registry-based study of the population of Sweden. Brain. 2014;137(Pt 3):770–8. doi: 10.1093/brain/awt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebers GC. Genetic factors in multiple sclerosis. Neurol Clin. 1983;1(3):645–54. [PubMed] [Google Scholar]
- 34.Ebers GC, et al. A genetic basis for familial aggregation in multiple sclerosis. Canadian Collaborative Study Group. Nature. 1995;377(6545):150–1. doi: 10.1038/377150a0. [DOI] [PubMed] [Google Scholar]
- 35.Sawcer S. The complex genetics of multiple sclerosis: pitfalls and prospects. Brain. 2008;131(Pt 12):3118–31. doi: 10.1093/brain/awn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haines JL, et al. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. The Multiple Sclerosis Genetics Group. Nat Genet. 1996;13(4):469–71. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- 37.Ebers GC, et al. A full genome search in multiple sclerosis. Nat Genet. 1996;13(4):472–6. doi: 10.1038/ng0896-472. [DOI] [PubMed] [Google Scholar]
- 38.Sawcer S, et al. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet. 1996;13(4):464–8. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- 39.Ligers A, et al. Evidence of linkage with HLA-DR in DRB1*15-negative families with multiple sclerosis. Am J Hum Genet. 2001;69(4):900–3. doi: 10.1086/323480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawcer S, et al. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet. 2005;77(3):454–67. doi: 10.1086/444547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.(IMSGC), I.M.S.G.C. et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 42.Jakkula E, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86(2):285–91. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comabella M, et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS ONE. 2008;3(10):e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.(ANZgene), T.A.a.N.Z.M.S.G.C Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–8. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 45.Baranzini SE, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Human molecular genetics. 2009;18(4):767–78. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41(7):776–82. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium IMSG, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–9. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature genetics. 2013;45(11):1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patsopoulos N, et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. 2017 doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jannot AS, et al. P < 5 × 10(−8) has emerged as a standard of statistical significance for genome-wide association studies. J Clin Epidemiol. 2015;68(4):460–5. doi: 10.1016/j.jclinepi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Panagiotou OA, et al. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41(1):273–86. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 52.Gourraud PA, et al. Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann Neurol. 2011;69(1):65–74. doi: 10.1002/ana.22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Jager PL, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8(12):1111–9. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isobe N, et al. An ImmunoChip study of multiple sclerosis risk in African Americans. Brain. 2015;138(Pt 6):1518–30. doi: 10.1093/brain/awv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langefeld CD, et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun. 2017;8:16021. doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maier LM, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5(1):e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregory SG, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39(9):1083–91. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 59.Galarza-Munoz G, et al. Human Epistatic Interaction Controls IL7R Splicing and Increases Multiple Sclerosis Risk. Cell. 2017;169(1):72–84. doi: 10.1016/j.cell.2017.03.007. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488(7412):508–11. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alcina A, et al. Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis. Eur J Hum Genet. 2010;18(7):827–31. doi: 10.1038/ejhg.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Didonna A, et al. A non-synonymous single-nucleotide polymorphism associated with multiple sclerosis risk affects the EVI5 interactome. Hum Mol Genet. 2015;24(24):7151–8. doi: 10.1093/hmg/ddv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoppenbrouwers IA, et al. EVI5 is a risk gene for multiple sclerosis. Genes Immun. 2008;9(4):334–7. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- 64.Martin D, et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes. Nat Struct Mol Biol. 2011;18(6):708–14. doi: 10.1038/nsmb.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steri M, et al. Overexpression of the Cytokine BAFF and Autoimmunity Risk. N Engl J Med. 2017;376(17):1615–1626. doi: 10.1056/NEJMoa1610528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ascherio A, Munger KL. Epidemiology of Multiple Sclerosis: From Risk Factors to Prevention-An Update. Semin Neurol. 2016;36(2):103–14. doi: 10.1055/s-0036-1579693. [DOI] [PubMed] [Google Scholar]
- 67.Amato MP, et al. Environmental modifiable risk factors for multiple sclerosis: Report from the 2016 ECTRIMS focused workshop. Mult Scler. 2017 doi: 10.1177/1352458516686847. 1352458516686847. [DOI] [PubMed] [Google Scholar]
- 68.Ebers G. Interactions of environment and genes in multiple sclerosis. J Neurol Sci. 2013;334(1–2):161–3. doi: 10.1016/j.jns.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 69.Hedstrom AK, et al. The interaction between smoking and HLA genes in multiple sclerosis: replication and refinement. Eur J Epidemiol. 2017 doi: 10.1007/s10654-017-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu M, et al. Logistic Principal Component Analysis for Rare Variants in Gene-Environment Interaction Analysis. IEEE/ACM Trans Comput Biol Bioinform. 2014;11(6):1020–8. doi: 10.1109/TCBB.2014.2322371. [DOI] [PubMed] [Google Scholar]
- 71.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 72.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stunnenberg HG, et al. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell. 2016;167(5):1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Horikoshi M, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538(7624):248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamparter D, et al. Fast and Rigorous Computation of Gene and Pathway Scores from SNP-Based Summary Statistics. PLoS Comput Biol. 2016;12(1):e1004714. doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baranzini SE, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18(11):2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baranzini SE. The genetics of autoimmune diseases: a networked perspective. Curr Opin Immunol. 2009;21(6):596–605. doi: 10.1016/j.coi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 78.Gustafsson M, et al. A validated gene regulatory network and GWAS identifies early regulators of T cell-associated diseases. Sci Transl Med. 2015;7(313):313ra178. doi: 10.1126/scitranslmed.aad2722. [DOI] [PubMed] [Google Scholar]
- 79.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang L, et al. PINBPA: cytoscape app for network analysis of GWAS data. Bioinformatics. 2015;31(2):262–4. doi: 10.1093/bioinformatics/btu644. [DOI] [PubMed] [Google Scholar]
- 81.Wang L, et al. iPINBPA: an integrative network-based functional module discovery tool for genome-wide association studies. Pac Symp Biocomput. 2015:255–66. [PubMed] [Google Scholar]
- 82.Campbell RD, Trowsdale J. Map of the human MHC. Immunol Today. 1993;14(7):349–52. doi: 10.1016/0167-5699(93)90234-C. [DOI] [PubMed] [Google Scholar]
- 83.Opelz G, Lenhard V. Immunological factors influencing renal graft survival. Annu Rev Med. 1983;34:133–44. doi: 10.1146/annurev.me.34.020183.001025. [DOI] [PubMed] [Google Scholar]
- 84.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13(3):175–88. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 85.Tsai S, Santamaria P. MHC Class II Polymorphisms, Autoreactive T-Cells, and Autoimmunity. Front Immunol. 2013;4:321. doi: 10.3389/fimmu.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bertrams J, et al. HL-A antigens and multiple sclerosis. Tissue Antigens. 1972;2(5):405–8. doi: 10.1111/j.1399-0039.1972.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 87.Hollenbach JA, Oksenberg JR. The immunogenetics of multiple sclerosis: A comprehensive review. J Autoimmun. 2015;64:13–25. doi: 10.1016/j.jaut.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barcellos LF, et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet. 2003;72(3):710–6. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Isobe N, et al. Association of HLA Genetic Risk Burden With Disease Phenotypes in Multiple Sclerosis. JAMA Neurol. 2016;73(7):795–802. doi: 10.1001/jamaneurol.2016.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patsopoulos NA, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013;9(11):e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moutsianas L, et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet. 2015;47(10):1107–13. doi: 10.1038/ng.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]