Abstract
Cell-cell communication enables bacteria to coordinate their behavior through the production, recognition, and response to chemical signals produced by their microbial neighbors. An important example of coordinated behavior in bacteria is biofilm formation, where individual cells organize into highly complex, matrix-encased communities that differentiate into distinct cell types and divide labor among individual cells. Bacteria rely on environmental cues to influence biofilm development, including chemical cues produced by other microbes. A multitude of recent studies have demonstrated that natural-product antibiotics at subinhibitory concentrations can impact biofilm formation in neighboring microbes, supporting the hypothesis that these compounds may have evolved as signaling molecules that mediate cell-cell interactions. In this review we discuss the role of antibiotics in modulating biofilm formation and interspecies communication in bacteria.
Keywords: Biofilm formation, cell-cell communication, bacterial interactions, natural products, specialized metabolites
Antibiotics as signaling molecules
Antibiotics have long been instrumental in fighting infectious disease. The majority of antibiotics used in clinical settings are derived from small molecule natural products [1] many of which are produced as ‘secondary’ or specialized metabolites by microorganisms [2]. Despite their extensive use as therapeutics, the function of these molecules in the natural environment remains poorly characterized; this disconnect is surprising because such knowledge might accelerate the discovery of additional therapeutic molecules produced by microbes. Although debate remains about the function of these metabolites in their natural environments, the antibiotic concentrations used for killing pathogens in clinical settings are likely higher than those present in nature, where the population of the producing organisms, molecule concentration, and diffusion rate may all fluctuate with environmental variabilities and microbial community composition. In addition, an emerging body of research has highlighted the importance of microbial metabolites as signaling molecules able to modulate gene expression in bacterial populations and affect important physiological functions and cellular processes such as metabolite production, motility, pigmentation, and biofilm formation [3–5]. This suggests that these molecules may not have evolved exclusively for killing, but rather as a means of communication between microorganisms that mediate interspecies and intraspecies interactions. Significant challenges remain for validating this premise, including a need to expand our understanding of the (non-antibiotic) mechanisms of action of these metabolites and to devise experimental capacities that will enable us to monitor their activities at realistic concentrations and spatial scales. Nevertheless, uncovering the potential non-killing biological functions of these compounds is crucial not only for understanding chemical communication within microbial communities but also for ensuring the judicious clinical application of antibiotics, since subinhibitory concentrations could otherwise potentially impact pathogens in a manner that increases their fitness in the host. Here we review recent reports of natural product antibiotics that specifically impact the densely packed, matrix-embedded microbial communities called biofilms, and discuss the potential roles of these natural products in bacterial cell-cell communication (see Table 1 for an overview).
Table 1.
Antibiotics that impact biofilm formation that are mentioned in this paper.
| Compound Source | Compound | Bacterial species effected | Reference |
|---|---|---|---|
|
| |||
| Amphibian skin | Dermaseptin S4 derivative | Pseudomonas fluorescens | 67 |
|
| |||
| Bacteria | 2,4-diacetylphloroglucinol | Bacillus subtilis | 21 |
|
| |||
| Bacteria | Biosurfactants | Acinetobacter baumannii | 53 |
| Escherichia coli | |||
| Staphylococcus aureus | |||
|
| |||
| Bacteria | Biosurfactants putisolvins I and II | Pseudomonas spp. | 52 |
|
| |||
| Bacteria | Polymyxin B | Vibrio cholerae | 59 |
|
| |||
| Bacteria | Subtilosin | Gardnerella vaginalis | 65, 64 |
|
| |||
| Bacteria | Surfactin | B. subtilis | 18 |
| Nystatin | |||
|
| |||
| Bacteria | Tetracycline | Staphylococcus epidermidis | 55 |
|
| |||
| Bacteria | Thiocillin | B. subtilis | 20 |
| Various thiazolyl peptides | |||
|
| |||
| Bacteria | Tobramycin | E. coli | 41 |
| Pseudomonas aeruginosa | |||
|
| |||
| Bacteria/semisynthetic | Carbapenem | Klebsiella pneumoniae | 51 |
|
| |||
| Bacteria/synthetic | Ampicillin | Streptococcus intermedius | 63 |
| Ciprofloxacin | |||
| Tetracycline | |||
|
| |||
| Human | Defence peptide LL-37 | S. aureus | 57 |
| P. aeruginosa | |||
|
| |||
| Lichen | Usnic Acid | P. aeruginosa | 54 |
| S. aureus | |||
|
| |||
| Marine algae | Furanone | E. coli | 48, 62, 60 |
| P. aeruginosa | |||
| S. aureus | |||
| S. epidermidis | |||
|
| |||
| Semisynthetic | Ampicillin | S. aureus | 50 |
| Amoxicillin | |||
| Cloxacillin | |||
| Methicillin | |||
|
| |||
| Semisynthetic | Azithromycin | P. aeruginosa | 44–47 |
|
| |||
| Synthetic | Peptide 1037 | P. aeruginosa | 58 |
| Burkholderia cenocepacia | |||
| Listeria monocytogenes | |||
Biofilm formation
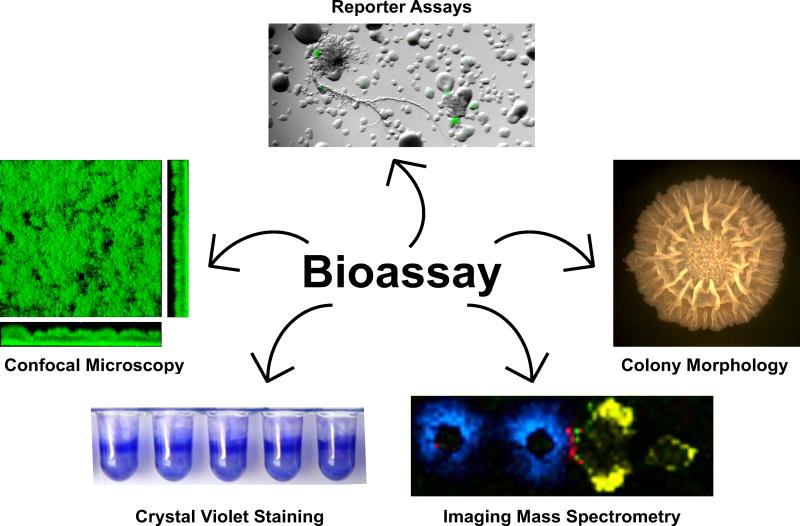
Biofilms are microbial aggregates encased in a self-produced extracellular polymer matrix that can be either surface-attached or free-floating. These highly complex microbial accretions are widely distributed in the natural environment as well as in industrial and medical settings, and are thought to have been a major mode of eubacterial survival for billions of years [6]. Gene expression and metabolic activity is profoundly different between biofilm cells and their planktonic counterparts [7]. Additionally, there is cellular and spatial heterogeneity within biofilms that arises as cells undergo differentiation in response to local conditions and are exposed to different developmental signals [8]. Emergent properties arise in biofilms due to complex social interactions and as the structural and chemical properties of the biofilm matrix develop [8]. Finally, biofilms are highly resistant to environmental stresses, predators, detergents, and antibiotic treatment [6]. These protective effects have been demonstrated to be greatly enhanced when multiple species are present within the biofilm [9], suggesting that multi-species interactions among biofilm cells are beneficial to their fitness. Many natural-product antibiotics affect biofilm formation, suggesting that specialized metabolite production by neighboring microorganisms may act as important environmental signals that regulate biofilm formation and shape multispecies interactions in these communities. A multitude of assays have been used to detect the impacts of these specialized metabolites on biofilm formation, some of which are depicted in Figure 1. We begin by focusing on two bacteria (Bacillus subtilis and Pseudomonas aeruginosa) that are extremely well-characterized in terms of their ability to form biofilms, and thus have also been the focus of many of the studies examining how antibiotics impact the development of biofilms in these species.
Figure 1.
Methods for detecting specialized metabolites that impact biofilm formation. Phenotypic assays that look for effects on biofilm formation include observing colony morphology, biofilm reporter activity, or biofilm architecture and attachment (using confocal microscopy and crystal violet staining) in the presence of purified compound or in co-culture. Imaging mass spectrometry can be used to identify specialized metabolites in respect to their spatial distribution.
Small molecule natural products as a cue for biofilm formation
Bacillus subtilis
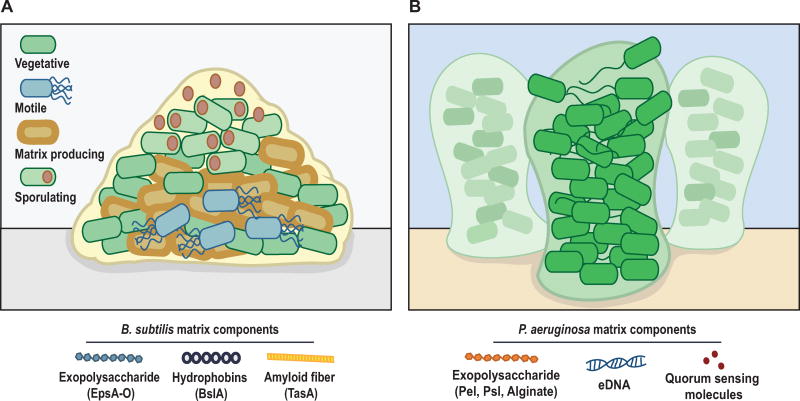
B. subtilis is a Gram-positive soil-dwelling bacterium that has been well-studied for its ability to form highly structured biofilms (Figure 2A). Within a B. subtilis biofilm, subpopulations of matrix-producing, surfactin-producing, sporulating, and motile cells, among other cell types, coexist in discrete regions of the structure [10]. The B. subtilis biofilm matrix is comprised of exopolysaccharides (EPS) and two major matrix proteins: TasA, which provides structural integrity [11] and BslA, which confers hydrophobicity to the biofilm structure [12]. These matrix components are encoded by the epsA-O operon, the tapA operon, and the bslA gene, respectively. The master transcriptional regulator Spo0A controls over 100 genes, depending on its concentration and phosphorylation state, including these three critical structural components of the biofilm matrix, [13,14]. Intermediate levels of phosphorylated Spo0A (Spo0A~P) induce matrix gene expression, while higher levels induce sporulation [13,15]. Levels of Spo0A~P are primarily controlled by the activity of five sensor histidine kinases (KinA, KinB, KinC, KinD, and KinE), which phosphorylate Spo0A both indirectly and directly [16,17]. However, with a few notable exceptions [15] the environmental cues that control the kinase or phosphorylation activities of these kinases are poorly defined.
Figure 2.
Schematic representation of biofilm structure and composition in B. subtilis and P. aeruginosa. (A) B. subtilis and (B) P. aeruginosa.
One of the first studies that indicated that the secretion of small molecules by soil bacteria can influence biofilm development in B. subtilis screened purified compounds for their ability to induce pellicles (floating biofilms that form at the liquid-air interface) [18]. This approach identified nystatin and surfactin as biofilm inducers in B. subtilis [18]. Nystatin is a polyene polyketide produced by Streptomyces noursei and is well-known for its antifungal activity: it forms pores in cell membranes that allow cation efflux. Surfactin is a lipopeptide produced by B. subtilis itself. By characterizing the effect of a variety of structurally and/or functionally related molecules, this study specifically found that cation (potassium) leakage, not simply membrane disruption, was responsible for the induction of biofilm formation in B. subtilis [18]. The authors also showed that kinC was required for B. subtilis to respond to surfactin, suggesting that KinC may be responsible for sensing potassium leakage and mediating biofilm matrix gene expression [18]. Many of the other biofilm-inducing compounds identified in this report were, like nystatin and surfactin, microbially produced metabolites that have characterized antibiotic activity (e.g. gramicidin and valinomycin). Because the concentrations of nystatin and surfactin used did not lead to a decrease in growth rate, this phenomenon does not appear to be a stress response per se, but rather a mechanism B. subtilis might use to sense the presence of specific microbial neighbors by sensing a change in the state of their cell membranes [18].
In a complementary approach, another study used microbial coculture rather than purified compounds to identify soil microbes secreting compounds that stimulated biofilm formation in B. subtilis [19]. This interspecies-interaction screen identified the common soil microbes Bacillus cereus, Bacillus thuringiensis, Bacillus mycoides, Bacillus megaterium, Bacillus luciferensis, and Pseudomonas monteilli all as inducers of biofilm matrix gene expression in B. subtilis [19]. The results from this work demonstrate that B. subtilis responds to the secreted molecules produced by these and other soil bacteria using mechanisms that depend on the phylogenetic relatedness of the interaction partner [19]. A follow-up publication used matrix-assisted laser desorption/ionization time-of-flight imaging mass spectrometry (MALDI-TOF IMS) to identify the thiocillins as biofilm-inducing metabolites produced by B. cereus [20]. Thiocillins are ribosomally encoded, post-translationally modified peptide antibiotics that interfere with the 50S ribosome. In addition to the thiopeptides, this study demonstrated that a range of structurally and functionally diverse thiazolyl peptides (thiostrepton, nosiheptide, berninamycin), as well as two other strains of Bacillus species whose genomes contained putative thiazolyl biosynthetic gene clusters all induced biofilm matrix production in B. subtilis [20]. These results suggest that thiazolyl peptide-induction of biofilm formation may be a widespread signaling mechanism among soil microbes. Indeed, putative thiazolyl-like biosynthesis genes were present in roughly 5 % of the sequenced Bacillus genomes and 40 % of the Streptomyces genomes available at the time this work was published [20]. Although all of these thiazolyl peptides affect B. subtilis biofilm formation under the conditions examined, it remains unknown whether or how these metabolites may influence biofilm formation in the producing species themselves. In addition, although the thiocillins were identified as a biofilm-inducing metabolite secreted by B. cereus, none of the other soil microbes identified in the initial coculture screen appear capable of producing thiazolyl peptides, suggesting that a range of additional metabolites able to alter biofilm formation in B. subtilis await discovery.
The most notable aspect of this work was the discovery that matrix induction by thiocillin is structurally separable from its antibiotic activity – in other words, different parts of the molecule appear to be responsible for its antimicrobial and signalling activities [20]. This was ascertainable because – unlike most antibiotics, which are synthesized using matabolite-specific enzymatic machinery – thiocillin is ribosomally encoded. Therefore, structural variants were easily generated through standard molecular genetics approaches. This allowed the identification of a variant (T4V) that had no antibiotic activity against B. subtilis, yet still induced biofilm matrix gene expression and enhanced colony wrinkling (a phenotype correlated with biofilm matrix production) [20]. This result is important because it provides direct evidence that, at least in the case of thiocillin, biofilm formation in response to antibiotics is not solely a consequence of stress-induced killing, as has frequently been proposed. Instead, this paper establishes that microbial ‘antibiotics’ possess biological activities independent of their killing activity and thus may have evolved as signaling molecules specifically to alter bacterial physiology. Future work will be required to determine how broadly true this finding might be for antibiotics in general. Notably, the identification of a biofilm-inducing, antibiotic-null thiocillin variant (T4V) also provides a unique tool for future studies focused on ascertaining whether the biofilm-enhancing or killing activity (or both) of this metabolite are pertinent at microbially relevant spatial scales and concentrations.
In addition to these antibiotics that enhance biofilm formation in B. subtilis, some bacterial natural products have been identified that inhibit biofilm formation in this bacterium. Pseudomonas protogens is a Gram-negative bacterium found in soil and associated with plants, and thus inhabits the same environments as B. subtilis. P. protegens produces 2,4-diacetylphloroglucinol (DAPG), a broad-spectrum antibiotic whose biosynthesis genes are conserved across many pseudomonads [21], and which has been shown to stimulate biofilm formation in Azospirillum brasilense at subinhibitory concentrations [22]. In contrast, when applied at subinhibitory concentrations to B. subtilis, DAPG was recently shown to inhibit biofilm formation (and sporulation) [21]. One intriquing detail that emerged from this study is that DAPG (a biofilm-inhibitor) appears to be overproduced on the medium MSgg (a B. subtilis-biofilm-inducing medium); this suggests that in the same environmental conditions where B. subtilis is forming biofilms, P. protegens produces large amounts of its biofilm-inhibiting compound DAPG. This raises interesting questions about how these two bacteria may interact and coexist in nature.
These findings highlight a challenge with the effort to understand the biological activities of microbial metabolites: DAPG has been shown in different in vitro contexts to kill fungi, to kill bacteria, to affect the hatching of nematodes, and to modulate B. subtilis biofilm formation. A similarly broad range of roles have been attributed to molecules such as surfactin, and to a variety of other microbal metabolites as well. How then are we to know which (if any) of these activities are relevant to the producing organisms in the natural environment? In some ways it may be irrelevant: these studies have advanced our understanding of diverse biological processes and provided potential chemical tools to modulate interspecies interactions to our benefit. But for those driven by a desire to discern the native functions of microbial metabolites, such reports serve as a vital starting point, providing critical reagents (signaling mutants, purified compounds, etc.) to test the hypothesis of whether the observed in vitro results align with their in vivo or in situ activity. This question will be facilitated by the development of experimental systems that allow us to examine chemically mediated interactions at the microscale: we predict that the behavior of bacteria at the single cell (or tens-of-cells level) will differ dramatically compared to the responses of the massive populations of cells typically examined in in vitro laboratory assays.
Pseudomonas aeruginosa
P. aeruginosa is a Gram-negative model bacterium whose biofilm formation has been extensively studied (Figure 2B). This opportunistic pathogen is capable of colonizing plants and humans and is of considerable medical importance as a colonizer of the lungs of cystic fibrosis patients. The ability of P. aeruginosa to form biofilms is thought to be a major contributor to its ability to cause disease in cystic fibrosis patients, as biofilm cells exhibit increased tolerance to antibiotics and resistance to phagocytosis [23]. Biofilm formation in P. aeruginosa requires flagellar motility (which facilitates localization to the liquid-surface interface) as well as type IV pili-mediated twitching motility (which mediates bacterial movement along surfaces) [24]. P. aeruginosa produces three matrix polysaccharides (alginate, Pel, and Psl) that contribute to the stability and structure of the biofilm [25,26]. In addition, extracellular DNA is important for biofilm structure in P. aeruginosa [27,28]. Cyclic di-guanosine monophosphate (c-di-GMP) is a second messenger molecule that controls polysaccharide production in P. aeruginosa [29,30] and promotes cell surface adhesion [31]. Levels of c-di-GMP are modulated in response to environmental signals by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), which produce and degrade c-di-GMP respectively [32]. Quorum sensing (QS) is also integrated into the biofilm regulatory network in P. aeruginosa [33]. The QS system in P. aeruginosa is comprised of two hierarchical systems, the autoinducer synthetases/regulatory protein pairs LasI/LasR and RhlI/RhlR. LasR binds to the promoter region of the psl operon [34] and a lack of lasI causes a biofilm defect [35]. Both lasI and rhlI enhance Pel polysaccharide biosynthesis in P. aeruginosa [36].
P. aeruginosa has been isolated from numerous environments including plants, animals, soil, natural and man-made aquatic environments, and clinical settings [37–40]; thus there is great potential for interspecies and interkingdom interactions with this organism. Subinhibitory concentrations of tobramycin, an aminoglycoside produced by the soil bacterium Streptomyces tenebrarius, was found to induce biofilm formation in P. aeruginosa [41]. Other aminoglycosides (streptomycin and gentamicin, produced by the soil bacteria Streptomyces griseus and Micromonospora purpurea, respectively) induce biofilm formation in P. aeruginosa as well [41]. Aminoglycosides are protein synthesis inhibitors that are a clinically important for treatment of chronic heart, lung, and urinary tract infections. The fact that these antibiotics provide a signal to P. aeruginosa to form highly recalcitrant biofilms is therefore of significant clinical relevance, since subinhibitory concentrations of these compounds may stimulate a bacterial response that is counter to their desired clinical effect.
The induction of biofilm formation by tobramycin requires a functional arr (aminoglycoside response regulator) gene, which encodes an inner membrane PDE that degrades c-di-GMP [41]. Tobramycin-inducible biofilm formation is inhibited by the addition of exogenous GTP (a c-di-GMP inhibitor), suggesting that tobramycin impacts biofilm formation through the Arr phosphodiesterase via a still undefined mechanism [41]. Analysis of the P. aeruginosa transcriptome upon exposure to antibiotic treatment (at subinhibitory concentrations) revealed that tobramycin differentially regulates 40 genes (out of 555 genes represented in the microarray used), including those involved in transcriptional regulation, secondary metabolite regulation, outer membrane composition, secretion, chemotaxis, and motility [42]. This study also demonstrated that subinhibitory concentration of tobramycin and tetracycline (a protein synthesis inhibitor produced by multiple streptomycete species) both stimulated biofilm gene expression, but otherwise altered expression in unique subsets of genes [42]. Thus, P. aeruginosa may have multiple sensing systems that integrate to influence biofilm formation in response to subinhibitory concentrations of antibiotics. Also notable is the fact that these antibiotics cause major changes to the transcriptome of P. aeruginosa without measurably altering its growth rate or global protein synthesis [41], further suggesting that these antibiotics may have specific biological functions beyond just killing.
Another indication that antibiotics are molecules important for bacterial communication is that several antibiotics impact the QS pathway in P. aeruginosa (as well as in other bacteria as discussed below), which may be an important mechanism for controlling bacterial biofilm formation. Azithromycin (AZM) is a macrolide antibiotic derived from the soil-dwelling bacterium Saccharopolyspora erythraea commonly used for treating chronic lung infections in cystic fibrosis patients. Subinhibitory concentrations of AZM transcriptionally repress both the las and rhl QS systems in P. aeruginosa, and inhibit the expression of the N-acylhomoserine lactone (AHL) synthesis enzymes upstream of lasI and rhlI, leading to lower concentrations of autoinducers being produced [43,44]. Transcriptome and proteome analysis of P. aeruginosa indicates that AZM influences mRNA message abundance of over 10 % of the QS regulon and almost 32 % of QS-dependent proteins, while also reducing the expression of multiple flagellar biosynthesis proteins required for biofilm surface attachment [45]. AZM also delays biofilm formation in P. aeruginosa in flow cell systems [46], and inhibits biofilm formation in static systems [47], presumably through its effect on QS pathways and by impairing motility.
Other natural products also inhibit biofilm formation in P. aeruginosa by impacting QS pathways. Halogenated furanones are natural compounds secreted by the red alga Delisea pulchra. Using a lasB-based AHL reporter assay, a halogenated furanone was found to reduce quorum sensing-controlled gene expression at concentrations that had no effect on growth [48]. Confocal imaging demonstrated that this antibiotic penetrates microcolonies within the P. aeruginosa biofilm and blocks cell signaling, which in turn impacts biofilm architecture and biomass [48]. Several other natural compounds from plants and animals have anti-QS properties in P. aeruginosa [4]. Thus, chemical communication mechanisms that influence biofilm formation in P. aeruginosa appear to be widespread in both the eubacterial and eukaryotic kingdoms.
Antibiotics impacting biofilm formation in other bacteria
Fewer studies have examined the impact of antibiotics on biofilm development in bacterial species other than the model microbes B. subtilis and P. aeruginosa. Nevertheless, such studies, some of which are summarized below, demonstrate that many microoganisms produce specialized metabolites that affect biofilm formation in other bacteria, and suggest that microbes possess a range of possible mechanisms for sensing and responding to subinhibitory concentrations of antibiotics. In many cases, however, there is insufficient data available to discern whether the tested antibiotics impact biofilm formation independently of their killing activity, or whether they are simply acting as antimicrobial and/or antibiofilm molecules, a distinction that should be more thoroughly explored by the field going forward.
Sensing and responding to subinhibitory concentrations of antibiotics
We begin by providing examples of publications that describe the effects of subinhibitory concentrations of antibiotics on biofilms. Subinhibitory concentrations of β-lactams have been shown to disperse established Listeria monocytogenes biofilms; to stimulate biofilm formation in L. monocytogenes [49]; to induce biofilm formation and extracellular DNA release in multiple strains of S. aureus [50]; and to cause Klebsiella pneumoniae biofilm cells to round, bleb, and dimple [51]. These studies demonstrate that functionally related β-lactam molecules have widely divergent impacts on different bacterial species. Similar conclusions can be drawn from studies examining the effects of biosurfactants on biofilms. Surfactin, as mentioned above, is a biosurfactant that activates the biofilm formation pathway in B. subtilis [18]. Meanwhile, the putisolvin lipopeptides inhibit or break down Pseudomonas biofilms [52], while other biosurfactants inhibit adhesion and biofilm formation in Acinetobacter baumannii, Escherichia coli, and Staphylococcus aureus [53]. As a final example of this concept, P. aeruginosa grown on a polymer surface loaded with usnic acid (a specialized metabolite produced by lichens) showed enhanced biofilm structure (but no change in cell number compared to its growth on a non-usnic acid surface), while no such difference was observed with S. aureus grown on the same surfaces [54]. These data all emphasize that the same compound can have significantly different effects on different bacteria.
On the other hand, it is also the case that different molecules have unique impacts on the same organism: one study demonstrated that while two antibiotics (tetracycline and a streptogramin) at subinhibitory concentrations enhanced biofilm gene expression in Staphylococcus epidermidis, numerous others had no impact [55]. Taken together, these studies exemplify a significant challenge in the field: while it would be satisfying to be able to build a coherent framework for predicting the action of subinhibitory antibiotics on bacterial biofilms, the diversity of activities currently observed makes this impossible. Indeed, such a consistent framework may not exist. However, only by achieving a better understanding the molecular targets of the non-killing activities of antibiotics, as well as by constructing reproducible in vitro assay systems that allow direct comparisons to be made across different bacterial systems (and laboratories) will we be able to make a conclusion either way.
In an effort to identify the impact of subinihibitory antibiotics on non-stress-related, biofilm-relevant genes, some studies have used global analyses to examine the effects of subinhibitory antibiotics on the transcriptional responses of bacteria. For instance, subinhibitory concentrations of penicillin results in the differential regulation of 386 genes in Streptococcus pneumoniae, including those involved in competence, quorum sensing, cell envelope, capsular polysaccharide biosynthesis, fatty acid chain elongation, and polyamine transporters [56]. A similar study identified genes involved in twitching motility, flagellar assembly, biofilm formation, and quorum sensing that were differentially regulated in P. aeruginosa in response to subinhibitory concentrations of the human host defense peptide LL-37 [57]. The results from such studies emphasize the breadth of cellular functions that subinhibitory antibiotics affect, but in most cases they provide only tantalizing hints as to the potential mechanisms by which these non-killing functions are mediated.
One way to gain additional insight into this problem is to compare the transcriptional responses of related molecules within the same bacterium. For example, the biofilm-inhibiting peptides LL-37 and 1037 were observed to dysregulate more than 400 genes in S. aureus, but only 14 of them were similarly differentially regulated, nearly all of which were important in biofilm formation; these genes thus represent potential targets of action of these peptides [58]. In other cases, more directed studies have begun to elucidate the connections between antibiotic sensing and a change in biofilm physiology. In Vibrio cholerae, for instance, the two-component signal transduction system CarRS was shown to directly regulate the transcription of the almEFG operon, which both promotes polymyxin B resistance and also represses the major biofilm transcriptional regulators vpsR and vpsT [59]. In spite of these examples, however, in most instances significant future work needs to be applied to follow-up on either transcriptional or biofilm analyses to identify the potential mechanisms by which subinhibitory antibiotics impact bacterial physiology. Only then will it be possible to more accurately assess whether these metabolites have distinct modes of action for their killing and biofilm-modulating activities (as suggested by the thiocillin results obtained in B. subtilis described above).
Antibiotic signals that feed into quorum sensing networks
As described in the P. aeruginosa section above, another clue that antibiotic compounds may be important interspecies signals is the fact that they often impact QS systems, which in many bacteria then go on to impact biofilm formation. One furanone (structurally related both to bacterial acylhomoserine lactones (AHL) and the furanones that impact P. aeruginosa biofilms) was shown to enhance biofilm formation of Staphylococcus epidermidis and Staphylococcus aureus at subinhibitory concentrations in a manner that appears to be via the staphylococcal QS pathway [60]. Other studies have found that different furanones inhibit biofilm formation in S. epidermidis [61], B. subtilis [35–37] and E. coli [62], yet it remains unknown if QS pathways are impacted in these examples. In addition to furanones, subinhibitory concentrations of β-lactams have also been shown to act via QS pathways: penicillin stimulates biofilm formation in Streptococcus pneumoniae [56], while ampicillin induces biofilm formation in Staphylococcus intermedieus [63]. Finally, healthy vaginal lactobacilli produce bacteriocins that work as antibiofilm agents against the Gardnerella vaginalis [64,65] and reduce AI-2 production in this bacterial vaginosis-associated pathogen [66].
Conclusion: challenges and opportunities
Few studies have attempted to address the function of antibiotics in the natural environment, and many questions still remain about their potential roles there (see Outstanding Questions Box). However, subinhibitory concentrations of antibiotic compounds clearly affect biofilm formation in a multitude of bacterial species, and coculture experiments indicate that, at least in vitro, antibiotic production by one bacterium can affect biofilm gene expression and structure in other bacterial species. This indicates that natural-product antibiotics may play an important role as chemical signals that provide cues impacting biofilm development in nature.
Outstanding Questions Box.
How do antibiotics affect biofilm development in non-model bacteria?
What are the molecular mechanisms by which antibiotics impact biofilm formation?
Are there commonalities in the mechanisms used to sense and respond to antibiotics among diverse bacterial species?
Will expanding the breadth of data available (more compounds, bacterial species, conditions), allow us to formulate a coherent framework for predicting the activity of antibiotics on biofilm formation in different species?
How do the effects of purified compounds on biofilm formation compare to those resulting from microbial coculture interactions?
Do the activities of specialized metabolites observed in standard laboratory assays differ from those obtained at realistic (microbial) spatial scales from environmentally relevant (typically small) populations of cells?
How do native environmental conditions impact antibiotic communication between bacteria?
Yet, as mentioned throughout this review, our knowledge of the mechanistic details of these effects and the identification of the (non-killing, biofilm-modulating) targets of subinhibitory antibiotics is still extremely limited-to-nonexistent. This makes it nearly impossible to know whether the observed effects are dependent or independent of the antimicrobial effect of the compound, a distinction critical for drawing conclusions about the potential function of these compounds in the natural environment. To address this challenge, we need experimental approaches that allow us to directly probe how metabolites affect microbial cells, ideally in situ and in real time. One such method is Attenuated Total Reflectance-Fourier Transform InfraRed (ATR-FTIR) spectroscopy, which was recently applied to gain insight into the molecular mechanism of action of an antimicrobial peptide (dermaseptin) against Pseudomonas fluorescens biofilms [67]. This study revealed that dermaseptin led to a loss of membrane lipids and a rapid inhibition of nucleic acid biosynthesis in young P. fluorescens biofilms [67]. ATRFTIR thus can be used to probe and monitor, in situ and in real time, the biochemical changes induced by antibiotics in bacteria cells at the molecular scale during early biofilm formation. The further application of this and other novel spectroscopic methods may provide much-needed insights into how microbial metabolites impact biofilm formation in diverse bacteria.
In addition to obtaining a more mechanistic understanding of these interactions, future studies characterizing bacterial interactions mediated by specialized metabolites in non-model organisms and at small, microbially relevant spatial scales may provide important knowledge about the potential roles antibiotics play in nature. This information will be crucial to understanding how subinhibitory antibiotic concentrations can enhance potentially adaptive characteristics that increase fitness in bacterial pathogens. Furthermore, studies that address the role that antibiotics play in shaping microbial communities would provide important insights into how these compounds affect complex systems such as the human, plant, and soil microbiomes.
Trends Box.
Many antibiotics are derived from natural products produced as specialized metabolites by microorganisms
The function of antibiotics in the natural environment remains poorly characterized
Bacteria produce a variety of natural-product antibiotics that can impact biofilm formation in other bacteria
A single metabolite can have divergent effects on biofilm formation in different bacterial species
The argument that the natural function of antibiotics may be signaling is strengthened by the finding that (at least for one natural product), different parts of the molecule are responsible for its biofilm-enhancing and killing activities.
Specialized metabolites may act as important environmental cues that regulate biofilm formation and shape multispecies interactions in microbial communities
Acknowledgments
Work related to biofilm formation and bacteria interactions in our laboratory is supported by grants from the National Institutes of Health (GM112981) and the Department of Energy (DE-SC0013887) to E.A.S. We kindly thank Kriti Sharma for her input on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Clardy J, et al. The natural history of antibiotics. Curr. Biol. 2009;19:437–441. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero D, et al. Antibiotics as signal molecules. Chem. Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasamiravaka T, et al. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res. Int. 2015;2015:1–17. doi: 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, et al. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Stoodley P, et al. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 8.Flemming H-C, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 9.Burmølle M, et al. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014;22:84–91. doi: 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Vlamakis H, et al. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero D, et al. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol. Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 13.Molle V, et al. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 14.Fujita M, et al. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlamakis H, et al. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013;11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang M, et al. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 17.Ireton K, et al. Integration of Multiple Developmental Signals in Bacillus subtilis through the Spooa Transcription Factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 18.Lopez D, et al. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shank EA, et al. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc. Natl. Acad. Sci. 2011;108:E1236–E1243. doi: 10.1073/pnas.1103630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bleich R, et al. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc. Natl. Acad. Sci. 2015;112:3086–3091. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers MJ, et al. Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens. J. Bacteriol. 2015;197:2129–2138. doi: 10.1128/JB.02535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Combes-Meynet E, et al. The Pseudomonas secondary metabolite 2,4-Diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol. Plant-Microbe Interact. 2011;24:271–284. doi: 10.1094/MPMI-07-10-0148. [DOI] [PubMed] [Google Scholar]
- 23.Høiby N, et al. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–74. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 24.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 25.Ghafoor A, et al. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryder C, et al. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swartjes JJTM, et al. A functional DNase I coating to prevent adhesion of bacteria and the formation of biofilm. Adv. Funct. Mater. 2013;23:2843–2849. [Google Scholar]
- 28.Whitchurch CB, et al. Extracellular DNA required for bacterial biofilm formation. Adv. Sci. 2011;295:1–2. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 29.Merighi M, et al. The second messenger bis-(3’–5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 30.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha D, O’Toole GA. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 2015 doi: 10.1128/microbiolspec.MB-0003-2014.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romling U, et al. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirisits MJ, Parsek MR. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 2006;8:1841–1849. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- 34.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science (80-.) 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert KB, et al. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 2007;189:5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grosso-Becerra M-V, et al. Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genomics. 2014;15:318. doi: 10.1186/1471-2164-15-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selezska K, et al. Pseudomonas aeruginosa population structure revisited under environmental focus: Impact of water quality and phage pressure. Environ. Microbiol. 2012;14:1952–1967. doi: 10.1111/j.1462-2920.2012.02719.x. [DOI] [PubMed] [Google Scholar]
- 39.Nair AV, et al. A comparative study of coastal and clinical isolates of Pseudomonas aeruginosa. Braz. J. Microbiol. 2015;46:725–34. doi: 10.1590/S1517-838246320140502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trust TJ, Bartlett KH. Isolation of Pseudomonas aeruginosa and other bacterial species from ornamental aquarium plants. Appl. Environ. Microbiol. 1976;31:992–994. doi: 10.1128/aem.31.6.992-994.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman LR, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 42.Linares JF, et al. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tateda K, et al. Azithromycin inhibits quorum wensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001;45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kai T, et al. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009;22:483–486. doi: 10.1016/j.pupt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Nalca Y, et al. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 2006;50:1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillis R, Iglewski B. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 2004;42:5842–5845. doi: 10.1128/JCM.42.12.5842-5845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Favre-Bonte S, et al. Biofilm formation by Pseudomonas aeruginosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 2003;52:598–604. doi: 10.1093/jac/dkg397. [DOI] [PubMed] [Google Scholar]
- 48.Hentzer M, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen UT, et al. Role of PBPD1 in stimulation of Listeria monocytogenes biofilm formation by subminimal inhibitory beta-lactam concentrations. Antimicrob. Agents Chemother. 2014;58:6508–6517. doi: 10.1128/AAC.03671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan JB, et al. Low levels of beta-lactam antibiotics Induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3:2–9. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Laar TA, et al. Sublethal concentrations of carbapenems alter cell morphology and genomic expression of Klebsiella pneumoniae biofilm. Antimicrob. Agents Chemother. 2015;59:1707–1717. doi: 10.1128/AAC.04581-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuiper I, et al. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 2004;51:97–113. doi: 10.1046/j.1365-2958.2003.03751.x. [DOI] [PubMed] [Google Scholar]
- 53.Sambanthamoorthy K, et al. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 2014;14:197. doi: 10.1186/1471-2180-14-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francolini I, et al. Usnic acid, a natural antimicrobial agent able to inhibit bacterial biofilm formation on polymer surfaces. Antimicrob. Agents Chemother. 2004;48:4360–4365. doi: 10.1128/AAC.48.11.4360-4365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rachid S, et al. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000;44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers PD, et al. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 2007;59:616–626. doi: 10.1093/jac/dkl560. [DOI] [PubMed] [Google Scholar]
- 57.Overhage J, et al. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008;76:4176–4182. doi: 10.1128/IAI.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De La Fuente-Nunez C, et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012;56:2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bilecen K, et al. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect. Immun. 2015;83:1199–1209. doi: 10.1128/IAI.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuehl R, et al. Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob. Agents Chemother. 2009;53:4159–4166. doi: 10.1128/AAC.01704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lönn-Stensrud J, et al. Furanones, potential agents for preventing Staphylococcus epidermidis biofilm infections? J. Antimicrob. Chemother. 2009;63:309–316. doi: 10.1093/jac/dkn501. [DOI] [PubMed] [Google Scholar]
- 62.Ren D, et al. Inhibition of biofilm formation and swarming of Escherichia coli by ( 5Z )-4-bromo-5-(bromomethylene)-3-butyl-2( 5H )-furanone. Environ. Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 63.Ahmed NA, et al. AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob. Agents Chemother. 2009;53:4258–4263. doi: 10.1128/AAC.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turovskiy Y, et al. Susceptibility of Gardnerella vaginalis biofilms to natural antimicrobials subtilosin, ε-poly-L-lysine, and lauramide arginine ethyl ester. Infect. Dis. Obstet. Gynecol. 2012 doi: 10.1155/2012/284762. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Algburi A, et al. Natural antimicrobials subtilosin and lauramide arginine ethyl ester synergize with conventional antibiotics clindamycin and metronidazole against biofilms of Gardnerella vaginalis but not against biofilms of healthy vaginal lactobacilli. Pathog. Dis. 2015;73:1–12. doi: 10.1093/femspd/ftv018. [DOI] [PubMed] [Google Scholar]
- 66.Algburi A, et al. Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiotics Antimicrob. Proteins. 2016 doi: 10.1007/s12602-016-9242-x. [DOI] [PubMed] [Google Scholar]
- 67.Quiles F, et al. In situ and real time investigation of the evolution of a Pseudomonas fluorescens nascent biofilm in the presence of an antimicrobial peptide. Biochim. Biophys. Acta - Biomembr. 2016;1858:75–84. doi: 10.1016/j.bbamem.2015.10.015. [DOI] [PubMed] [Google Scholar]