ABSTRACT
Congenital disorders of glycosylation (CDG) comprise a family of human multisystemic diseases caused by recessive mutations in genes required for protein N-glycosylation. More than 100 distinct forms of CDGs have been identified and most of them cause severe neurological impairment. The Conserved Oligomeric Golgi (COG) complex mediates tethering of vesicles carrying glycosylation enzymes across the Golgi cisternae. Mutations affecting human COG1, COG2 and COG4–COG8 cause monogenic forms of inherited, autosomal recessive CDGs. We have generated a Drosophila COG7-CDG model that closely parallels the pathological characteristics of COG7-CDG patients, including pronounced neuromotor defects associated with altered N-glycome profiles. Consistent with these alterations, larval neuromuscular junctions of Cog7 mutants exhibit a significant reduction in bouton numbers. We demonstrate that the COG complex cooperates with Rab1 and Golgi phosphoprotein 3 to regulate Golgi trafficking and that overexpression of Rab1 can rescue the cytokinesis and locomotor defects associated with loss of Cog7. Our results suggest that the Drosophila COG7-CDG model can be used to test novel potential therapeutic strategies by modulating trafficking pathways.
KEY WORDS: COG7, Drosophila, GOLPH3, Golgi, Glycosylation
Highlighted Article: A Drosophila model of congenital disorders of glycosylation diseases (mutation of Cog7) suggests that modulating secretory trafficking could provide a novel means of therapy.
INTRODUCTION
Glycosylation, the enzymatic addition of monosaccharides or polysaccharides (glycans) to proteins or lipids, plays crucial roles in the metabolism of eukaryotic cells (Freeze and Ng, 2011; Ohtsubo and Marth, 2006). The synthesis of N-linked or O-linked glycans starts in the endoplasmic reticulum (ER) and it is completed in the Golgi. It requires the action of glycosyltransferases, glycosidases and nucleotide-sugar transporters (Ohtsubo and Marth, 2006). Congenital disorders of glycosylation (CDGs) comprise a family of human diseases caused by mutations in genes required for synthesis of glycoconjugates (Freeze et al., 2015; Jaeken, 2013). More than 100 distinct forms of CDGs have been identified and more than 80% of these diseases display severe neurological impairment (Barone et al., 2014). Because 2% of the translated genome encodes the glycosylation machinery, many other CDGs remain to be discovered (Freeze et al., 2015). CDGs are divided into two large groups (Freeze et al., 2014; Goreta et al., 2012). Type I CDGs affect the synthesis of the dolichol-linked oligosaccharide, the major precursor of N-linked glycoproteins, and its transfer to acceptor proteins (Freeze et al., 2014). Type II CDGs disrupt either the processing of N-linked glycans, the biosynthesis of O-linked oligosaccharides or the addition of glycans to lipids (Freeze et al., 2014; Goreta et al., 2012). The eight-subunit Conserved Oligomeric Golgi (COG) complex is required for tethering of vesicles that recycle Golgi resident proteins and glycosylation enzymes (Climer et al., 2015; Miller and Ungar, 2012). The COG complex has a bi-lobed structure, with eight subunits arranged in two distinct subcomplexes: lobe A (COG1–COG4) and lobe B (COG5–COG8) (Miller and Ungar, 2012; Smith and Lupashin, 2008). In Saccharomyces cerevisiae, only mutants carrying lobe A deletions are not viable, whereas lobe B mutants display defects of glycosylation and internal membrane organization (Miller and Ungar, 2012; Smith and Lupashin, 2008). Mutations affecting human COG1, COG2 and COG4–COG8 cause monogenic forms of inherited, autosomal recessive CDGs-II (Foulquier et al., 2006; Fung et al., 2012; Kranz et al., 2007; Kodera et al., 2015; Lübbehusen et al., 2010; Morava et al., 2007; Ng et al., 2007; Paesold-Burda et al., 2009; Reynders et al., 2009; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009). Most patients carrying mutations in COG complex subunits (COG-CDG), share many developmental deficits, including microcephaly, mental retardation, hypothonia associated with dysmorphic features and feeding problems (Foulquier et al., 2006; Fung et al., 2012; Kranz et al., 2007; Kodera et al., 2015; Lübbehusen et al., 2010; Morava et al., 2007; Ng et al., 2007; Paesold-Burda et al., 2009; Reynders et al., 2009; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009). Among COG mutations, those affecting the COG7 gene cause the highest mortality within the first year of life (Spaapen et al., 2005; Wu et al., 2004). The first COG7-CDG (or CDG-IIe according to a previous nomenclature) patients reported were two siblings carrying a homozygous intronic IVS1+4A>C mutation in a splice motif, resulting in a truncated polypeptide (Wu et al., 2004). Both patients displayed several phenotypic traits including dysmorphic facial features, skeletal anomalies, hypotonia, hepatomegaly, progressive jaundice and cardiac insufficiency. The neurological aspects of the disease (microcephaly and severe epilepsy) were relevant (Spaapen et al., 2005; Wu et al., 2004). Six COG7-CDG patients, from four different families, were reported with the same IVS1+4A>C mutation and similar clinical characteristics, including the effects on the nervous system (Morava et al., 2007; Ng et al., 2007; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009). Another intronic mutation in COG7 was more recently reported (Zeevaert et al., 2009). The patient carrying this mutation lacked dysmorphic features and survived longer; however, he displayed cerebral atrophy.
It has been proposed that the COG complex orchestrates retrograde traffic, acting as a protein interaction hub (Climer et al., 2015; Hong and Lev, 2014; Miller et al., 2013; Willet et al., 2013a). Consistent with this role, COG proteins were shown to interact with intra-Golgi SNAREs, the COPI coat proteins, small Rab GTPases and other tethering proteins and to play an essential role in regulating assembly of functional SNARE complexes (Bailey Blackburn et al., 2016; Hong and Lev, 2014; Willet et al., 2013a). However, full elucidation of the functional interplay between the COG proteins and the vesicle trafficking machinery is lacking. Defects in COG proteins have been linked to glycosylation alterations in several model organisms and in human cells (Kingsley et al., 1986; Struwe and Reinhold, 2012; Suvorova et al., 2002). CHO cells carrying COG1 or COG2 mutations exhibit abnormal synthesis of N-, O- and ceramide-linked oligosaccharides, indicating alterations in multiple Golgi glycosylation pathways (Kingsley et al., 1986). Most studies on COG7-CDG patients clearly document defects in the N-linked and O-linked glycosylation with hyposialylation (Morava et al., 2007; Ng et al., 2007; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009). Typical neurological manifestations of COG7-CDG include psychomotor delay, microcephaly, brain atrophy and epileptic seizures (Freeze et al., 2014, 2015; Morava et al., 2007; Ng et al., 2007; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009).
It is unclear how defective glycosylation causes the neuropathology of COG7-CDG and other CDGs. This issue can only be addressed by developing animal models of the specific CDG (Freeze et al., 2014). However, in vivo models of CDGs are scarce and none has been described for COG7-CDG (Chu et al., 2013; Freeze et al., 2014; Thiel and Körner, 2011). Drosophila melanogaster has emerged as a suitable organism for studying in vivo glycan-dependent functions in the central nervous system (CNS) and the involvement of N-glycosylation in neuropathologies (Katoh and Tiemeyer, 2013; Scott and Panin, 2014). In vertebrates, the complexity of the nervous system, the redundancy of glycosylation enzymes and the ubiquity of sialylation in glycoconjugates make these studies particularly challenging (Freeze et al., 2014; Scott and Panin, 2014). By contrast, Drosophila allows investigation of aspects of neural glycosylation at a simplified level and with the ease of genetic tools and well-established neurobiological approaches (Scott and Panin, 2014).
Drosophila melanogaster genome harbors a single gene encoding a sialyltransferase (DSiaT), which is most homologous to the mammalian ST6GalI enzyme (Repnikova et al., 2010). DSiaT expression is restricted to a subset of cells in the CNS and is required for neuronal excitability (Repnikova et al., 2010). Although α2,6-sialylated N-linked glycans represent a small fraction of the Drosophila N-glycome, they have been unambiguously detected in embryos and adult heads of this organism by mass spectrometry (Aoki et al., 2007; Koles et al., 2007), thus supporting the usefulness of the fruit fly as a model system for CDG. We previously showed the requirement for Cog7 protein for cytokinesis in Drosophila neuroblasts and spermatocytes. In this study, we show that loss-of-function Cog7 mutations reduce life span and cause neuromotor defects similar to those seen in patients suffering with COG7-CDG. We demonstrate that Cog7 mutant flies exhibit N-linked glycome defects with hyposialylation that are consistent with a role for Cog7 protein in N-glycosylation at the Golgi membranes. Additionally, we show that larval neuromuscular junctions of Cog7 mutants exhibit a significant reduction in bouton number. Finally, we dissect the interactions of Cog7 protein with proteins required for Golgi trafficking in Drosophila. Based on our findings, we propose that Cog7 and the COG complex cooperate with Rab1 and Golgi phosphoprotein 3 (GOLPH3) to regulate retrograde COPI-mediated vesicle trafficking.
RESULTS
Cog7 mutants display decreased longevity, uncoordinated movement and temperature-sensitive paralysis
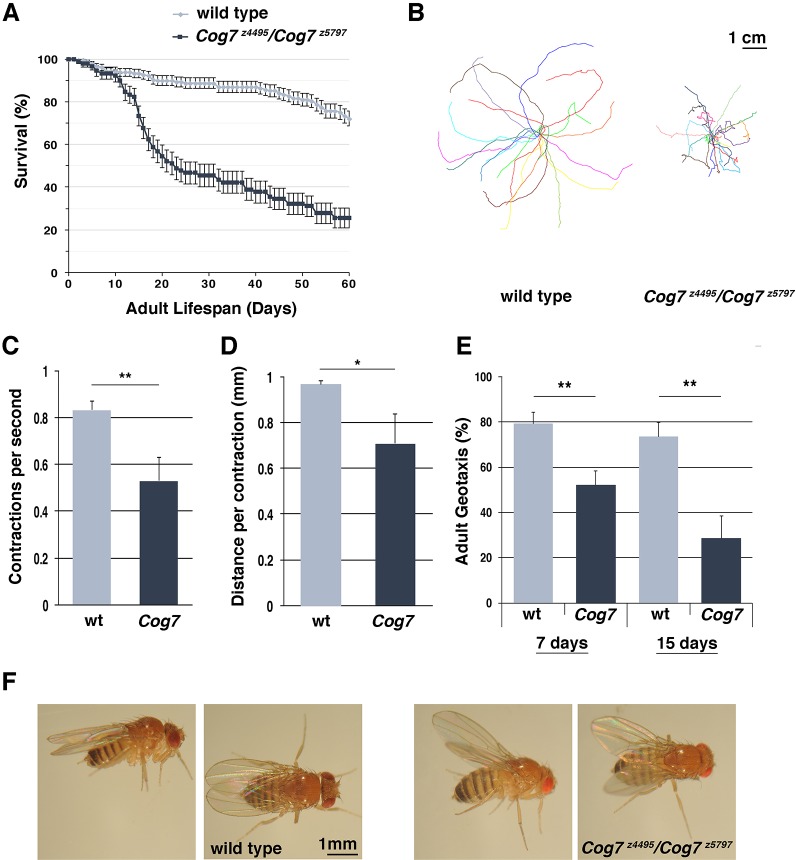
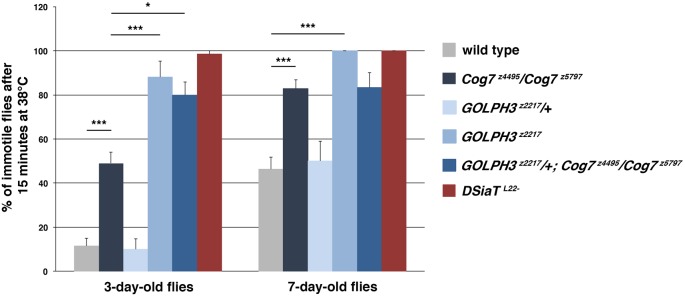
Analysis of wild-type and Cog7 mutant viability revealed decreased adult lifespan in Cog7 mutants (Fig. 1A). In addition, loss of Drosophila Cog7 resulted in pronounced postural and locomotor defects (Fig. 1B–F, Fig. 2). Cog7 mutant flies displayed a highly penetrant ‘held out wings’ posture (Fig. 1F), which indicates altered flight-muscle control of wing positioning (Müller et al., 2010; Parkinson et al., 2016; Zaffran et al., 1997). For locomotion assay of larvae, we used the crawling pattern assay (Fig. 1B–D). The crawling of mutant larvae was irregular, displaying more turns compared with the wild type (Fig. 1B) when subjected to an open field crawling assay and appeared significantly slower, as indicated by the reduction in frequency of body muscle contractions (Fig. 1C,D). The negative geotaxis test was used to assess the animals' ability to climb 2 cm up from the bottom of the vial. Fruit flies are negatively geotropic and rapidly climb to the top of an empty vial after being tapped to the bottom. However, Cog7z4495/Cog7z5797 mutants are significantly sluggish (Fig. 1E). Because neurological mutants, including DSiaT mutants, display temperature-sensitive (TS) paralysis, we tested the behavior of Cog7 at elevated temperatures. Like DSiaT mutants, Cog7 flies exhibit TS paralysis (Repnikova et al., 2010). Flies carrying Cog7 mutation became fully paralyzed at 38°C within a few minutes (Fig. 2). Paralysis is reversible as flies fully recover within 10-15 min when transferred to 25°C. However, compared with DSiaT flies, the TS defect was weaker. The percentage of Cog7 and DSiaT mutant flies that exhibited the sensitivity to elevated temperatures increased with age, suggesting a progressive age-dependent alteration of the nervous system (Fig. 2).
Fig. 1.
Behavioral phenotype of Cog7 mutants. (A) Adult lifespan of Cog7z4495/Cog7z5797 mutants compared with controls (wild type). 150 flies for each genotype, were assayed for 60 days. Note that the percentage survival of Cog7z4495/Cog7z5797 mutant flies halved after 20 days. (B) Crawling pattern assay on third instar larvae of each genotype (wild type Oregon-R and Cog7z4495/Cog7z5797 mutant larvae of the same age). The crawling motion for each larva was recorded on agar plates during 1 min and the digitized tracks analyzed. Graphs show 22 superimposed larval tracks, with the initial positions overlaid in the middle. Mutant larvae exhibit crawling defects. (C) Normalized quantification of 3rd instar larval locomotion (peristaltic waves per second) in Oregon-R (wild type) and Cog7z4495/Cog7z5797 (Cog7) animals. (D) Normalized quantification of third instar larval locomotion (distance per contraction) in Oregon-R (wild type) and Cog7z4495/Cog7z5797 (Cog7) animals. (E) Quantification of adult negative geotaxis as percentage of animals climbing to 2-cm height at 10 s, for Oregon-R (wild type) and Cog7z4495/Cog7z5797 (Cog7) animals; n=60 flies (males) for each genotype. Error bars indicate s.e.m. *P<0.05; **P<0.01 (unpaired Student's t-test). (F) Loss of Cog7 results in a highly penetrant ‘held-out wings’ posture, which indicates defects in flight-muscle control. 200 flies were examined for each genotype.
Fig. 2.
Cog7 and GOLPH3 have synergistic genetic interaction. Quantification of TS-paralysis phenotype in flies of the indicated genotype. 100 flies per genotype (in groups of 10 flies) were assayed at 38°C for 15 min. Note that the severity of locomotor defects increases with age. Error bars represent s.e.m. *P<0.05; ***P<0.0001 (unpaired Student's t-test).
Cog7 mutants exhibit defects in the development of larval neuromuscular junctions
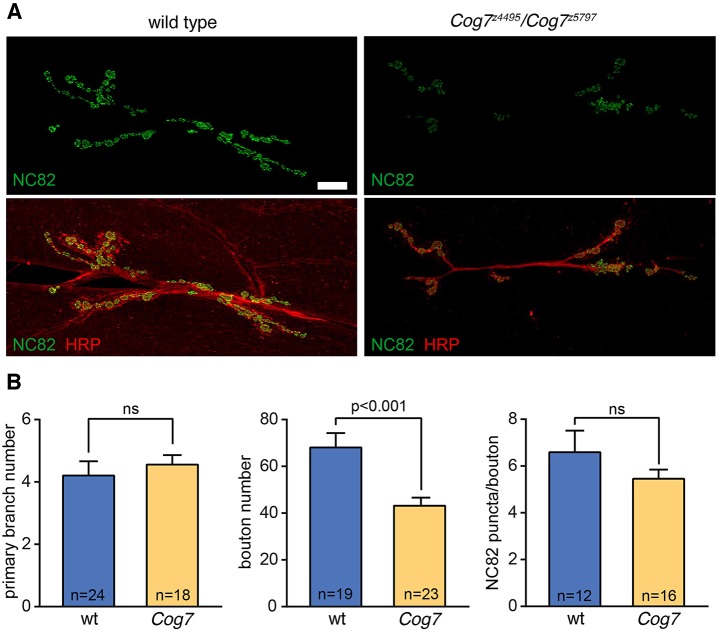
Because many neurological Drosophila mutations, including DSiaT, affect the function and development of neuromuscular junctions (NMJs) (Parkinson et al., 2016; Repnikova et al., 2010), we analyzed the neuroanatomy of NMJs and compared the phenotypes of Cog7 mutant larvae with the wild type. The number of primary nerve branches, boutons and active zones at the NMJ between larval muscles 6 and 7 was quantified by immunofluorescence confocal imaging using monoclonal antibody NC82, anti-Brüchpilot (Wagh et al., 2006) and polyclonal anti-horseradish peroxidase (HRP) antiserum. The number of primary branches at the NMJs of Cog7 mutant larvae was unchanged compared with wild type. However, Cog7 mutant larvae exhibited a statistically significant reduction in bouton number (Fig. 3). The density of active zones, defined as NC82-postive punctae per bouton, was also slightly decreased but not to a level of statistical significance (Fig. 3). Therefore, NMJs from Cog7 mutants contain a slightly reduced number of synaptic release sites distributed across a significantly reduced area of junctional contact.
Fig. 3.
Loss of Cog7 results in decreased bouton number at the larval neuromuscular junction. (A) The NMJs between muscles 6 and 7 were imaged by confocal scanning laser microscopy. NC82 anti-Brüchpilot was used to visualize active zones within synaptic boutons, whereas anti-HRP was used to assess primary branch number and NMJ bouton morphology. (B) Primary branch number, bouton number and total active zones (NC82 puncta) were quantified in the wild type and in Cog7z4495/Cog7z5797mutants. Primary branch number was unchanged in the Cog7z4495/Cog7z5797 mutant larvae, but total bouton number decreased significantly. Total active zone abundance was also decreased, but not to a statistically significant level. The number of NMJs quantified for each parameter is indicated. Statistical significance was determined by unpaired Student's t-test; ns, not significant (P≥0.05). Scale bar: 20 µm.
Loss of Cog7 affects N-glycosylation
To investigate the fidelity of N-linked glycoprotein glycosylation in Cog7 mutants, we used multidimensional ion trap mass spectrometry to profile N-linked glycoprotein glycans. To address neural glycosylation more directly, fly heads were removed from the whole body and used as starting material for glycomic profiling. Three different allelic combinations of Cog7 mutants exhibited increased glycoprotein glycosylation (Fig. 4A). Most of this alteration resulted from increased abundance of high mannose and paucimannose type glycans compared with wild type, because these glycans contribute most to the total glycan profile (Fig. 4B-D). Several of the less abundant complex glycans were also increased in the mutant backgrounds, but to a lesser extent than the less processed high mannose and paucimannose glycans. The most striking fold-change in glycan abundance in the Cog7 mutant heads was detected as a substantial increase in the abundance of a family of neural-specific, difucosylated N-glycans known as HRP epitopes (Fig. 4E). However, not all N-glycans were increased in the mutant background. The most complex N-glycans that bear terminal sialic acid (NeuAc) are among the least abundant structures in Drosophila at any stage of development (Aoki et al., 2007; Koles et al., 2007). A single sialylated N-glycan was detected among the adult head glycans and quantification relative to standard indicated that it was decreased in the two allelic combinations in which it was detected, compared with wild type (Fig. 4F,G). Notably, serum proteins from human COG7-CDG patients also exhibit reduced glycoprotein sialylation (Morava et al., 2007; Ng et al., 2007; Zeevaert et al., 2009).
Fig. 4.
N-linked glycoprotein glycosylation is altered in Cog7-deficient adult heads. (A) Total N-glycan abundance was increased in all three allelic combinations compared with wild type. (B) The major class of N-glycans in Drosophila includes those of the high mannose type and most of the high mannose glycans were increased in the Cog7 alleles. (C) The single most abundant N-glycan in the adult Drosophila head is the paucimannose glycan M3N2F, which was also increased in all Cog7 allelic combinations compared with the wild type. (D) Hybrid and complex type N-glycans were also increased, although not as consistently or as dramatically. (E) Neural-specific N-glycans, known as HRP-epitopes, were profoundly increased in the Cog7-deficient adult heads. (F) The least abundant complex glycans in Drosophila are sialylated with NeuAc; unlike the other classes of N-glycans, the only detected sialylated glycan was reduced in Cog7 mutants. (G) Full MS spectra zoomed to present the naturally occurring isotope distribution of the sialylated glycan quantified in (F) demonstrates reduced abundance of sialylated glycans in the Cog7 mutant. The three most abundant isotopes of the biantennary, disialylated complex glycan depicted in (F) correspond to the all 12C form (13C0) and two other isotopic variants that contain either one or two heavy 13C atoms. All of the isotopic variants were reduced in the mutant backgrounds.
Cog7 protein interacts with vesicle trafficking proteins required for cytokinesis
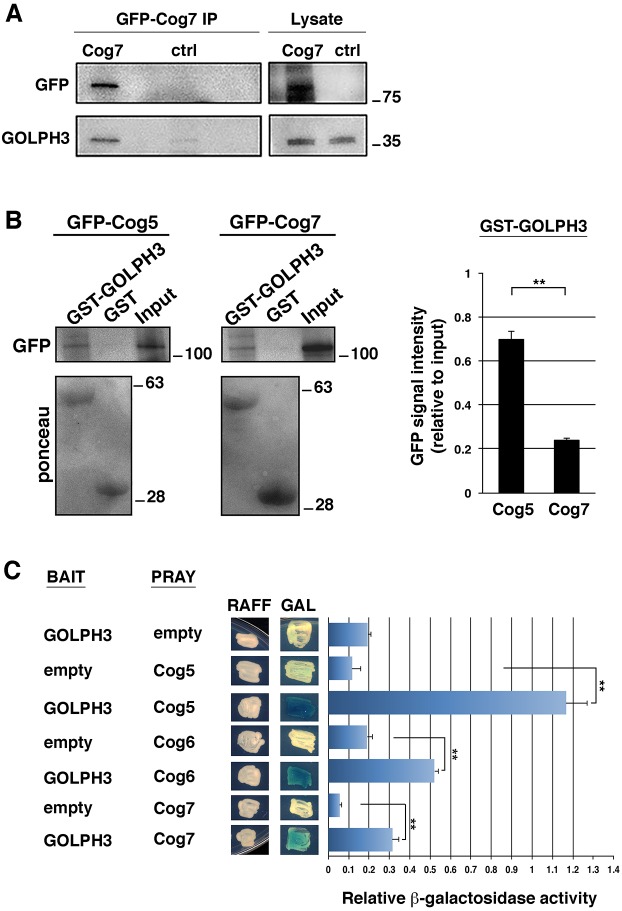
Our previous results (Belloni et al., 2012) showed that the Drosophila phosphatidyl transfer protein (PITP) Giotto (Gio) co-precipitates with both Cog7 and Rab11 in Drosophila testes, suggesting that these proteins might form a complex. Recent work indicates that GOLPH3 proteins are essential for proper localization of several Golgi glycosyltransferases (Eckert et al., 2014; Isaji et al., 2014; Schmitz et al., 2008; Sechi et al., 2015b; Tu et al., 2008). Increasing evidence indicates that GOLPH3 proteins help to retain Golgi enzymes by binding to the coatomer and several Golgi glycosyltransferases, including α2,6-sialyltransferase I (Eckert et al., 2014; Schmitz et al., 2008; Tu et al., 2008). Human GOLPH3 interacts with sialyltransferases and affects α2,6-sialylation of N-glycans (Isaji et al., 2014). Based on these data, we reasoned that GOLPH3 might cooperate with the COG complex in Golgi retention and proper functioning of glycosyltransferases. Drosophila GOLPH3 protein co-localizes with Cog7 to Golgi structures in interphase spermatocytes (Sechi et al., 2014, 2015a), raising the question of whether these proteins could physically interact. To test the interaction of GOLPH3 with Cog7 we used co-immunoprecipitation (Co-IP). GOLPH3 co-immunoprecipitated with GFP–Cog7 in larval brain extracts (Fig. 5A). We next tested the interaction between GOLPH3 and the COG complex proteins Cog5 and Cog7 by using glutathione S-transferase (GST) pull-down analysis and yeast two-hybrid assays (Fig. 5B,C). Our results indicate positive interactions of GOLPH3 with multiple subunits of the COG complex (Fig. 5B,C).
Fig. 5.
GOLPH3 interacts with COG complex proteins. (A) GOLPH3 co-precipitates with Cog7 protein in Drosophila larval brains. Larval brain extracts from either Oregon-R (ctrl) or individuals expressing GFP–Cog7 protein were immunoprecipitated with anti GFP (using GFP-trap) and blotted for either GOLPH3 or GFP. 2% of the total lysate (Lysate) and one-third of the immunoprecipitates were loaded and probed with the indicated antibody. Molecular mass is given in kilodaltons. The Co-IP experiment was performed three times with identical results. (B) GST and recombinant GST–GOLPH3 proteins immobilized on glutathione beads were incubated with larval brain extracts expressing either GFP–Cog5/Fws (Farkas et al., 2003) or GFP–Cog7 (Belloni et al., 2012). GST–GOLPH3 but not GST precipitated both GFP–Cog7 and GFP–Cog5 proteins. Ponceau staining is shown as a loading control. 2% of the input and 25% of the pull-downs were loaded and probed with the indicated antibody. Molecular mass is given in kilodaltons. The graph represents quantification of the amount of GFP–Cog7 and GFP–Cog5 that were pulled down from GST–GOLPH3 in western blotting analysis. Protein band intensities were obtained from three independent experiments. (C) Yeast two-hybrid assay was used to test GOLPH3 interaction with Cog5, Cog6 and Cog7 proteins. In the presence of the GOLPH3 bait, all the indicated COG proteins induce LacZ expression (blue color indicates positive interaction). The graph shows quantification of LacZ reporter expression induced with different combinations of bait and prey plasmids. See Materials and Methods for further details. Error bars indicate s.e.m. **P<0.01 (unpaired Student's t-test).
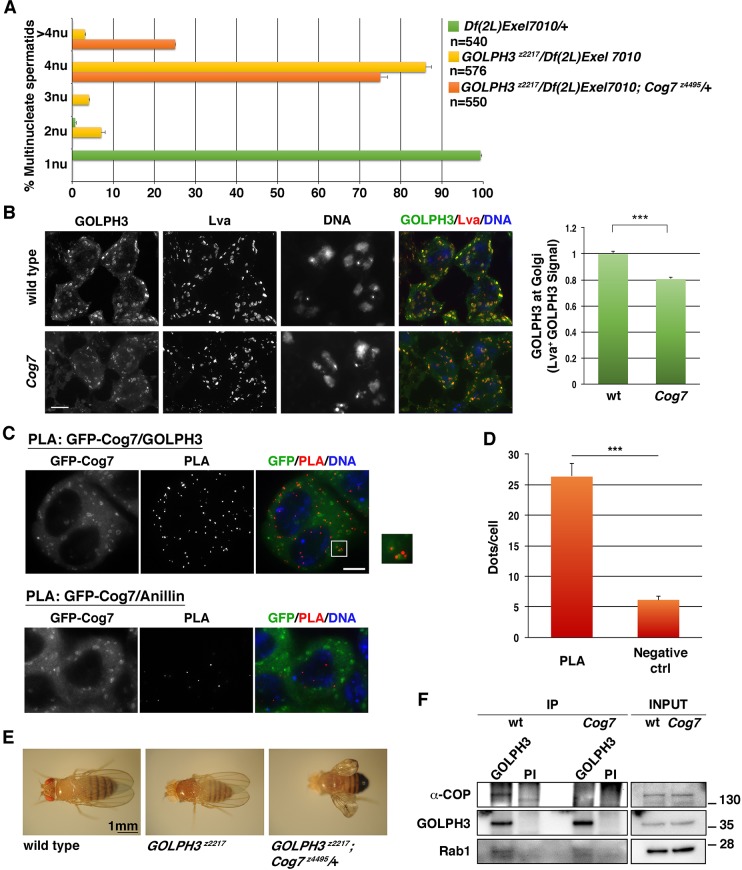
We constructed double mutants carrying Cog7 mutations (Cog7z4495/Cog7z5797) and simultaneously homozygous or hemizygous for GOLPH3z2217. Double mutants were examined for potential synthetic lethality compared with single mutants. Consistent with the idea that Cog7 and GOLPH3 proteins contribute to the same process, double mutants carrying both Cog7 and GOLPH3 mutations were synthetic lethal (Fig. S1). In addition, heterozygosity for Cog7 increased the frequency of cytokinesis failures, resulting in an increased number of multinucleate spermatids in testes of males that were hemizygous or homozygous for GOLPH3 mutations (Fig. 6A). Animals that were heterozygous for Cog7 and homozygous for GOLPH3 (GOLPH3z2217/GOLPH3z2217; Cog7z4495/+ or GOLPH3z2217/GOLPH3z2217; Cog7z5797/+) also displayed the ‘held out wings’ posture (Fig. 6E). In addition, wings of GOLPH3z2217; Cog7z4495/+ appeared severely swollen and crumpled (Fig. 6E). We analyzed genetic interactions between GOLPH3 and Cog7 using the TS-paralysis assay (Fig. 2). GOLPH3z2217/+ flies were not significantly different from the wild type. By contrast, similar to Cog7 mutants, GOLPH3z2217 mutants displayed a TS-paralysis phenotype. A single copy of GOLPH3z2217 allele could also significantly enhance the TS-paralysis phenotype of Cog7z4495/Cog7z5797. Taken together these results indicate a synergistic interaction between Cog7 and GOLPH3 (Fig. 2).
Fig. 6.
Cog7 interacts with GOLPH3 and is required for proper localization of GOLPH3 protein to Golgi membranes. (A) Frequency of spermatids, containing 1, 2, 3, 4 or more than 4 nuclei per mitochondrial derivative, in testes of control males (Df(2L)Exel7010/+) and males that are either hemizygous for GOLPH3 (GOLPH3z2217/Df(2L)Exel7010), or heterozygous for Cog7 and hemizygous for GOLPH3 (GOLPH3z2217/Df(2L)Exel7010; Cog7z4495/+). (B) Cog7 mutants display a decreased amount of GOLPH3 protein at the Golgi membranes. Interphase spermatocytes from wild type and Cog7z4495/Df(3R)BSC861 (Cog7) mutants were stained for GOLPH3, the Golgi marker Lava lamp (Lva) and DNA. GOLPH3 levels in the Golgi were quantified as mean fluorescence intensity of GOLPH3 in Lva-positive regions (Lva+). In total, we examined 112 Golgi from Cog7z4495/Df(3R)BSC861 (Cog7), and 105 Golgi in wild-type interphase spermatocytes. The cells examined for Golgi analysis were randomly selected from three independent experiments. (C) PLA to visualize GFP–Cog7/GOLPH3 interaction in fixed spermatocytes. PLA with monoclonal antibodies against GFP and GOLPH3 (rabbit anti-GOLPH3 G49139/77) was used to test the interaction in interphase spermatocytes stained for DNA. Zoomed panel shows co-localization of the proteins at the Golgi. Negative control experiments were performed with antibodies against GFP and anillin. Scale bars: 10 µm. (D) Average number (±s.e.m.) of PLA dots per cell in interphase spermatocytes. Quantification of the number of PLA signals per cell was obtained as described in the Materials and Methods. (E) Oregon-R GOLPH3z2217 flies display regular wings, but GOLPH3z2217;Cog7z4495/+ exhibit ‘held out wings’ posture. In addition, the wings of GOLPH3z22177; Cog7z4495/+ appear severely swollen and crumpled. (F) Loss of Cog7 impairs the interaction of GOLPH3 with the coatomer protein α-COP. Protein extracts from fly heads were immunoprecipitated with antibodies against Drosophila GOLPH3 (rabbit G49139/77) and blotted with mouse anti-GOLPH3 S11047/1/56, guinea pig anti-α-COP, mouse anti-Rab1 S12085a antibodies. Preimmune serum (PI; G49139/1, from the same animal before the immunization), was used as control. 2% of the total lysate and one-third of the immunoprecipitates were loaded and probed with the indicated antibody. Molecular mass is given in kilodaltons. The Co-IP experiment was performed three times with identical results. Error bars indicate s.e.m. ***P<0.0001 (unpaired Student's t-test).
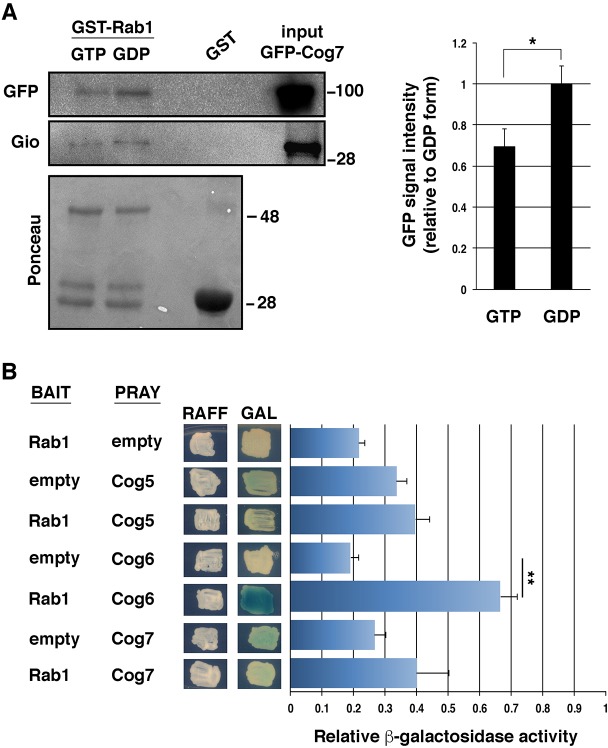
Based on previous work in mammalian cells, another partner of the COG complex might be Rab1 (Miller et al., 2013). Indeed, we recently found that the COG complex might interact with both Rab1 and Rab11 proteins in Drosophila testes (Sechi et al., 2017). Here, we used GST-pull-down experiments to investigate whether Rab1 and Rab11 proteins form a complex with both Cog7 and Gio in larval brains and to test the GTP/GDP dependence of the complexes. As shown in Fig. 7A and in Fig. S2, both Rab1 and Rab11 proteins interact with Cog7 and Gio in larval brains. The lack of strong preferential binding of Cog7 to activated (GTP-bound) Rab11 and Rab1 proteins suggests that the COG complex, rather than being a Rab1 or Rab11 effector, might regulate these proteins in vivo. Consistent with this hypothesis, Cog7 is required for normal recruitment of Rab1 to Golgi membranes of primary spermatocytes (Sechi et al., 2017). By using the yeast two-hybrid approach, we demonstrated that Drosophila Rab1 directly binds the Cog6 subunit but not the Cog5 and Cog7 subunits (Fig. 7B). Yeast two-hybrid experiments also confirmed the interaction of the COG subunit Cog7 with the PITP Gio (Fig. S3).
Fig. 7.
Drosophila Rab1 interacts with the COG complex. (A) Recombinant GST–Rab1 protein, immobilized on glutathione beads and loaded with either GDP-β-S (GDP) or GMP–PNP (GTP), was incubated with larval brain extracts expressing GFP–Cog7. GST–Rab1, but not GST, precipitated both GFP–Cog7 and Gio from brain protein extracts. Ponceau staining is shown as a loading control. 2% of the input and 25% of the pull-downs were loaded and probed with the indicated antibody. Molecular mass is given in kilodaltons. The graph represents quantification of the amount of GFP–Cog7 that was pulled down from each form of GST–Rab1 in western blotting analysis. Protein band intensities were obtained from three independent experiments. (B) Yeast two-hybrid assay was used to test Rab1 interaction with Cog5, Cog6 and Cog7 proteins. In the presence of Rab1 bait, only Cog6 induces LacZ expression (blue color indicates positive interaction). Graph shows quantification of LacZ reporter expression (graph) induced with different combinations of bait and prey plasmids. See Materials and Methods for further details. Error bars indicate s.e.m. *P<0.05; **P<0.01 (unpaired Student's t-test).
Loss of Cog7 affects GOLPH3 localization at the Golgi membranes and GOLPH3 interaction with COPI alpha and Rab1 proteins
We investigated the functional dependence between Cog7 and GOLPH3 at the Golgi membranes using immunofluorescence analysis (as described by Sechi et al., 2017). We performed these experiments in male meiotic cells, which provide a suitable cell system for Golgi analysis (Giansanti and Fuller, 2012). Cog7 mutant spermatocytes displayed a significant reduction in Golgi-localized GOLPH3 protein levels (Fig. 6B). In agreement with this result, the proximity ligation assay demonstrated that Cog7 and GOLPH3 proteins interact at the Golgi membranes (Fig. 6C,D). We then examined whether the marked effect of Cog7 mutation on GOLPH3 could influence the assembly of GOLPH3–COPI and GOLPH3–Rab1 complexes (Fig. 6F). Our data indicate that loss of Cog7 strongly affects the interaction of GOLPH3 with α-COP and reduces the efficiency of GOLPH3–Rab1 complex formation in fly heads.
Effects of overexpression of Rab proteins on Cog7 mutant phenotypic defects
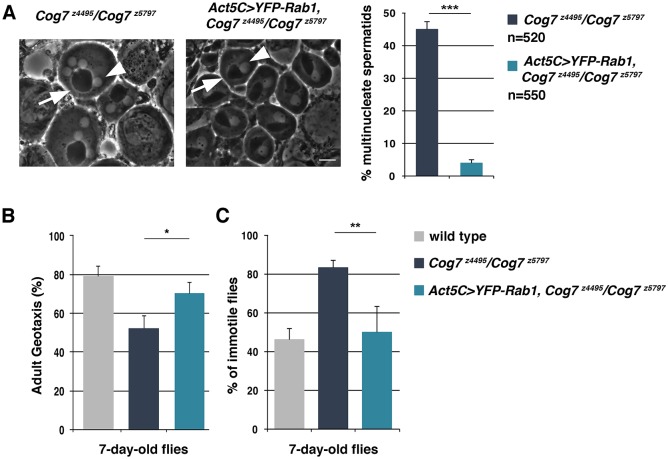
We next tested whether overexpression of Golgi trafficking regulators could rescue the phenotypic defects associated with loss of Cog7. We used the UAS/GAL4 binary system to overexpress Rab1 GTPase in Drosophila tissues using the UAS-YFP-Rab1 transgenic fly line in combination with Act5C-GAL4 (for ubiquitous expression). Animals of genotype Cog7z4495/Cog7z5797 and overexpressing Rab1 were examined for a rescue of the cytokinesis failures in Cog7z4495/Cog7z5797 mutants. This analysis was performed in testes, where defects in spermatocyte cytokinesis result in multinucleate spermatids. Overexpression of Rab1 can suppress the cytokinesis defects associated with Cog7 mutation, as indicated by a tenfold decrease in the percentage of multinucleate spermatids (4% of multinucleate spermatids compared with 45%; P<0.0001) (Fig. 8A). Overexpression of Rab1 also rescued the climbing defects and the TS-paralysis associated with loss of Cog7 (Fig. 8B,C).
Fig. 8.
Overexpression of Rab1 rescues cytokinesis failures and neuromotor defects associated with loss of Cog7. (A) Rescue of Cog7 cytokinesis defects by overexpression of Rab1 (Act5C>YFP-Rab1, Cog7z4495/Cog7z5797). Testes from 3-day-old males of the indicated genotypes were viewed using phase-contrast microscopy. Cog7z4495/Cog7z5797 display irregular spermatids containing multiple nuclei (phase light, arrowhead) associated with enlarged mitochondrial derivatives (phase dark, arrow). Spermatids from Act5C>YFP-Rab1, Cog7z4495/Cog7z5797 males display a single nucleus (arrowhead) associated with a mitochondrial derivative (arrow) of similar size. Scale bar: 10 µm. The graph shows the percentage of multinucleate spermatids in the indicated genotypes. Overexpression of Rab1 can suppress the cytokinesis defects associated with Cog7 mutation, as indicated by the significant decrease in multinucleate spermatids. (B) Quantification of adult negative geotaxis as percentage of animals climbing to 2-cm height at 10 s, for Cog7z4495/Cog7z5797 and Act5C>YFP-Rab1, Cog7z4495/Cog7z5797 animals aged 7 days; n=60 flies for each genotype. (C) Quantification of TS-paralysis phenotype in flies of the indicated genotypes. 100 flies for each genotype were assayed at 38°C for 15 min. Error bars represent s.e.m. *P<0.05; **P<0.01; ***P<0.0001 (unpaired Student's t-test).
DISCUSSION
One of the most challenging issues in studying CDG is to understand the relationship between glycosylation and neurological defects (Freeze et al., 2015). Although the defective glycosylation of proteins associated with COG7-CDG and other COG-CDGs has been characterized, it is still unknown how these alterations cause microcephaly and malfunction of the CNS, which represent the most widespread symptoms of CDGs. Animal models for the specific CDG are necessary to address the impact of glycosylation impairment on CNS function.
Common clinical manifestations of COG-CDG patients include hypothonia, brain atrophy, dysmorphia, episodes of hyperthermia and seizures (Morava et al., 2007; Ng et al., 2007; Spaapen et al., 2005; Wu et al., 2004; Zeevaert et al., 2009). Here, we have addressed the developmental consequences of loss of Cog7 in Drosophila melanogaster. Like COG7-CDG patients, Cog7 mutants display reduced life span and severe psychomotor alterations, including severe postural and movement deficits. We have demonstrated that the neurological alterations in Cog7 mutants are associated with distinctive changes in the N-glycome of adult heads. Our analysis of Cog7 mutants revealed an increased abundance of major glycan types; high mannose, paucimannose, non-sialylated hybrid/complex and difucosylated glycans are all increased compared with the wild type. Increased glycan abundance (detected as more moles of N-linked oligosaccharide per milligram of protein) can only be accomplished by enhancing the efficiency of the oligosaccharyltransferase complex (OST) in the endoplasmic reticulum (ER), where OST co-translationally transfers a high mannose N-glycan (Glc3Man9GlcNAc2) en bloc from a dolichol-linked precursor to appropriate asparagine residues. Although COG proteins have not been implicated directly in co-translational protein glycosylation, the results presented here are consistent with previous observations that Golgi disruptions, such as those induced by COG deficiencies, can also impact ER function (Steet and Kornfeld, 2006; Xiang et al., 2013). Despite detecting enhanced abundance of N-glycans on glycoproteins in Cog7 mutant heads, not all N-glycan structures were increased. The least abundant but most complex N-glycans in Drosophila are capped with sialic acid; these N-glycans were decreased in Cog7 mutants, similar to the hyposialylation that has been reported in COG7 patients (Morava et al., 2007; Zeevaert et al., 2009). In agreement with the involvement of Cog7 function in sialylation, Cog7 mutants exhibited TS-paralysis, a phenotype that was previously observed in DSiaT mutants.
The growing list of Drosophila mutants that affect protein glycosylation also impacts the development or stability of the larval NMJ (Baas et al., 2011; Dani et al., 2012, 2014; Itoh et al., 2016; Johnson et al., 2006; Jumbo-Lucioni et al., 2014, 2016; Parkinson et al., 2013, 2016; Repnikova et al., 2010). Among the proteins encoded by these mutated genes are glycosaminoglycan core proteins, glycan degradative and biosynthetic enzymes, as well as cell signaling molecules and cell adhesion molecules. Thus, NMJ architecture appears to provide a sensitive readout for the morphological consequences of altered protein glycosylation. Cog7 can now be added to this list, emphasizing the underlying importance of Golgi trafficking for appropriate protein glycosylation in the regulation of cell–cell interactions. Cog1 deficiency was also recently demonstrated to alter synaptic architecture at the larval NMJ, not by impacting branching but by increasing total junctional length without affecting bouton density (Comstra et al., 2017). Comparison of glycomic changes in Cog1 mutants (although currently unknown) with glycomic changes in Cog7 mutants might highlight different functional requirements for specific glycan classes in various stages of NMJ elaboration.
It has been proposed that the COG complex functions as a vesicle-tethering factor in intra-Golgi retrograde trafficking, thereby helping to regulate the recycling of resident Golgi proteins, including glycosylation enzymes (Smith and Lupashin, 2008; Miller and Ungar, 2012; Reynders et al., 2009). Consistent with this hypothesis, the interactome of the COG complex comprises several components of the trafficking machinery, including Rab and SNARE proteins (Miller et al., 2013; Willett et al., 2013b). We previously demonstrated that Drosophila Cog7 interacts with both Rab11 and the PITP Gio in testis extracts (Belloni et al., 2012). Here, we confirm the interaction of the COG complex with Rab1 and identify GOLPH3 as a novel molecular partner. Evidence indicates that GOLPH3 function is required for recruitment to the Golgi cisternae of several glycosylation enzymes, including Exostosins in Drosophila and α2,6-sialyltransferase I in human cells (Chang et al., 2013; Eckert et al., 2014; Isaji et al., 2014). Because the cytoplasmic tails of Golgi glycosyltransferases lack COPI sorting motifs, GOLPH3 was shown to mediate the intra-Golgi distribution of these proteins by dual interaction with the cytosolic portions of these enzymes and the COPI coatomer (Isaji et al., 2014). Remarkably, human GOLPH3 interacts with both Core 2 N-acetylglucosaminyltransferase 1 (C2GnT1) and α2,6-sialyltransferase I (Isaji et al., 2014). Recent data suggest that GOLPH3-dependent sialylation of N-glycans is required for integrin-mediated cell migration and Akt phosphorylation, both of which are affected in GOLPH3-depleted HeLa cells (Isaji et al., 2014). Our data suggest that the COG complex is required for proper localization of GOLPH3 at the Golgi and cooperates with GOLPH3 in controlling retrograde trafficking of Golgi glycosyltransferases, at least in part by facilitating GOLPH3 binding with the coatomer and Rab1-GTPase.
The phenotypic characteristics of our Drosophila COG7-CDG model closely parallel the pathological characteristics of COG7-CDG patients. We are confident that our studies can provide a novel means of dissecting the molecular circuits underlying the neurological impairments associated with COG7-CDG and related diseases. The Drosophila disease model that we have developed, with its powerful genetic tools, offers unique opportunities to identify novel targets and new strategies for therapeutic intervention. In this regard, we have shown that overexpression of Rab1 can rescue the cytokinesis and neuromotor defects associated with loss of Cog7, providing a proof of principle that genetic modifier screens in Drosophila could be used to identify potential targets for CDG clinical therapy. Thus, the Drosophila Cog7 disease model can be used to investigate the pathophysiology of COG-CDG and to perform therapeutic screening based on modulators of vesicle trafficking pathways.
MATERIALS AND METHODS
Fly stocks and transgenes
Flies were raised at 25°C by standard procedures. Oregon-R flies were used as wild-type controls. Cog7z4495, Cog75797 and GOLPH3z2217 mutant strains were described previously (Belloni et al., 2012; Sechi et al., 2014) and were from the C. Zuker collection (Giansanti et al., 2004). The chromosomal deficiencies Df(3R)BSC861, Df(2L)Exel7010, the fly strains UAS-YFP-Rab1 (Zhang et al., 2007) and Act5C-GAL4 were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN). The DSiaTL22− mutant allele, described by Repnikova et al. (2010), was kindly provided by Dr V. Panin (Texas A&M University). Flies expressing GFP–Cog7 and flies expressing GFP–Cog5/Fws were described by Belloni et al. (2012) and by Farkas et al. (2003), respectively. To verify whether overexpression of Rab1 could rescue the cytokinesis failures and the neuromotor defects associated with loss of Cog7, females of genotype Act5C-GAL4/CyO; Cog7z5797/TM6B were crossed with males of genotype UAS-YFP-Rab1, Cog7z4495/TM6B. Males of genotype Act5CGAL4/+; Cog7z5797/UAS-YFP-Rab1, Cog7z4495 were examined for rescue of cytokinesis failures and tested in the locomotor and TS assays.
Behavioral assays
Larvae
Larval locomotion assays were performed following the procedures described by Repnikova et al. (2010). Third instar larvae were rinsed in 25% sucrose and in distilled water before being transferred to the center of a 3% agar plate. The plate was placed on a 1 mm×1 mm grid paper and larval movement recorded using a Canon HD digital camera. Speed and number of waves of body contraction were measured from straight-path crawling intervals. Distance per contraction was evaluated as the ratio between larval speed and number of waves of body contraction. For crawling pattern assays, the digital tracks were obtained by following the larval head movements for 60 s.
Adult flies
Analysis of lifespan was performed as described by Repnikova et al. (2010). For negative geotaxis assay, adult males aged 7 days or 15 days were placed in empty fly vials and left for 15 min to acclimate. Fly tubes were then sharply tapped to let the flies fall to the bottom, as described by Parkinson et al. (2016). Movement was recorded with a Canon HD digital camera. The percentage of animals that climbed above 2 cm was evaluated after 10 s in the movies. The TS-assays were performed according to the procedures described by Repnikova et al. (2010). Briefly, adult males were transferred to empty vials that were submerged in a controlled-temperature water bath at 38°C. Paralysis was evaluated in groups of ten flies by calculating the number of individuals lying on the bottom of the vial and unable to stand and walk during the first 30 s.
Western blotting and co-immunoprecipitation
Immunoblotting analysis was performed on protein extracts of adult testes, larval brains or adult heads. Briefly, 200 adult testes, 100 larval brains or 150 heads from males of each genotype were homogenized on ice in 500 µl of lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1 mM PMSF, 1× Protease Inhibitor Cocktail) using a Dounce homogenizer. Samples were separated on Mini-protean TGX precast gels (Bio-Rad) and blotted to PVDF membranes (Bio-Rad). Membranes were blocked in Tris-buffered saline (Sigma-Aldrich) with 0.05% Tween-20 (TBST) containing 5% nonfat dry milk (Bio-Rad; Blotting GradeBlocker) for 3–4 h at room temperature, followed by incubation with primary and secondary antibodies diluted in TBST. Co-IP experiments on larval brains expressing GFP–Cog7 were performed using the GFP trap-A kits purchased from ChromoTek (Planegg-Martinsried), following the previously described protocol (Sechi et al., 2014). To immunoprecipitate Drosophila GOLPH3, we used the protocol described by Sechi et al. (2017). 150 adult heads were homogenized in 500 µl of lysis buffer (see above) for 40 min on ice. The lysate was precleared and divided into two halves. Fractions were incubated with either 5 µg of rabbit anti-GOLPH3 G49139/77 antibody or 5 µg of rabbit pre-immune serum (G49139/1, from the same animal before the immunization). After antibody incubation, immunoprecipitation was performed using the immunoprecipitation kit-Protein G (Roche) following the manufacturer's instructions. Primary antibodies used for immunoblotting were as follows: mouse anti-GOLPH3 S11047/1/56 (1:2500; Sechi et al., 2017), rabbit anti-GOLPH3, (1:2500; G49139/77; Sechi et al., 2014), mouse anti-Rab1 antibody S12085a (1:750; Sechi et al., 2017), mouse HRP anti-GFP (1:1000; Vector-Lab); guinea pig anti α-COP (1:1000; Kitazawa et al., 2012, gift of Dr Inoue) and rabbit anti-Gio (1:5000; Giansanti et al., 2006). HRP-conjugated secondary antibodies (GE Healthcare) were used at 1:5000. After incubation with antibodies, blots were washed in TBST and imaged using the ECL detection kit (GE Healthcare).
GST pull-down assays
GST, GST-Rab1, GST-Rab11 and GST-GOLPH3 proteins were expressed in bacteria and purified using glutathione–Sepharose 4B beads (GE Healthcare) following the manufacturer's instructions, as described by Giansanti et al. (2015) and Sechi et al. (2017). At least 200 larval brains were homogenized for 40 min on ice in 500 µl of lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% NP40, 1 mM EDTA) with the addition of protease and phosphatase inhibitors cocktails (Roche), using a Dounce homogenizer. After clearing the lysates by centrifugation, protein concentration of the supernatants was determined by Bradford Assay (Bio-Rad). GST pull-down was performed by incubating larval brain lysates with either GST, GST–Rab1 or GST–Rab11 (at the appropriate concentration) bound to glutathione–Sepharose 4B beads, with gentle rotation at 4°C for 2 h. After rinsing in ‘wash buffer’ (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP40, 1 mM EDTA, protease and phosphatase inhibitors) for three times, the beads were boiled in SDS sample buffer and separated by SDS-PAGE. The bound proteins were analyzed by western blotting (see above). Before immunoblotting, PVDF membranes were stained with Ponceau (Sigma-Aldrich). Guanosine 5′-[β-thio]diphosphate trilithium salt (GDP-β-S) and guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate (GMP-PNP) were purchased from Sigma-Aldrich.
Immunofluorescence staining of testis preparations
Cytological preparations were made with testes from third instar larvae. To visualize Cog7–GFP, larval testes were fixed in 4% methanol-free formaldehyde (Polysciences, Warrington, PA), as previously described (Frappaolo et al., 2017; Sechi et al., 2017). To visualize GOLPH3 and Lava lamp (Lva), testes were fixed using 3.7% formaldehyde in PBS and then squashed in 60% acetic acid as previously described (Frappaolo et al., 2017). Monoclonal antibodies were used to stain GFP (see above). Polyclonal antibodies were rabbit anti-Lva (1:500; Sisson et al., 2000), which was a gift from Chris Field (Harvard University), and mouse anti-GOLPH3 S11047/1/56 (1:1000; Sechi et al., 2014, 2017). Secondary antibodies were Alexa 555-conjugated anti-rabbit IgG (1:300; Life Technology), and FITC-conjugated anti-mouse IgG (1:30; Jackson ImmunoResearch). All incubations with primary antibodies (diluted in PBT containing 3% BSA) were performed overnight at 4°C. Incubations with secondary antibodies were performed at room temperature for 1 h. After immunostaining, samples were rinsed in PBS and mounted in Vectashield mounting medium with DAPI (H-1200, Vector Laboratories). Images were captured with a charged-coupled device (CCD camera, Qimaging QICAM Mono Fast 1394 Cooled) connected to a Nikon Axioplan epifluorescence microscope equipped with an HBO 100-W mercury lamp and 40× and 100× objectives. Images from spermatocytes treated for proximity ligation assay (PLA) were captured with a charged-coupled device (Axiocam 503 mono CCD camera) and ZEN2 software, connected to a Zeiss Cell Observer Z1 microscope equipped with an HXP 120 V inclusive built-in power supply, lamp module and 63× (N.A. 1.4) objective. Images were analyzed with ImageJ and processed in Photoshop. The protein content of GOLPH3 in the Golgi membranes (Fig. 6) was measured using the ImageJ software, as described by Sechi et al. (2017). The Golgi compartment was demarcated using the freehand selection tool, and mean signal intensity of the protein was measured in the selected compartment.
Analysis of neuromuscular junction morphology
NMJ morphology was assessed in wandering third instar larvae raised at 18°C. Larval dissections were performed as previously described (Kaufmann et al., 2002). Larval body wall preparations were stained with anti-HRP antibody (Jackson ImmunoResearch) to visualize the motor nerve and associated boutons (Jan and Jan, 1982; Baas et al., 2011). Active zones within NMJ boutons were visualized with mAb NC82 (anti-Brüchpilot, Developmental Studies Hybridoma Bank) diluted 1:100 (Wagh et al., 2006). The NMJ at muscles 6 and 7 (abdominal segments 3 and 4) were imaged by laser scanning confocal microscopy (LSC, Olympus FV1000) with a 40× (N.A. 1.30) oil objective. Stacks of optical sections were collected in the z-dimension. Laser intensity, slice thickness and all other acquisition parameters were identical for imaging wild-type and mutant embryos. Image analysis was accomplished off-line with the Slidebook software package (Intelligent Imaging Innovations, Denver) on non-compressed z-stacks.
N-linked glycan preparation
Fly heads were harvested from adults within 2 days of eclosion and stored frozen at −80°C until enough heads were accumulated for analysis, generally between 200 and 500 heads for each genotype. Adult heads were pulverized in liquid nitrogen and then delipidated by organic extraction to produce a protein-enriched powder that was used as starting material for N-linked glycan analysis, as described previously (Aoki, et al., 2007). Briefly, pulverized heads were homogenized on ice in cold 50% methanol. The lysate was then adjusted to a chloroform–methanol–water ratio of 4:8:3 (v/v/v) and incubated at room temperature. Following centrifugation at 2000×g for 15 min at 4°C, the resulting pellet was dried under nitrogen stream to produce a protein powder that was stored at −20°C until use. N-glycans were released from the protein powder as previously described (Aoki et al., 2007). Briefly, 2–5 mg of protein powder was digested with trypsin, adjusted to 5% acetic acid and loaded onto a Sep-Pak C18 cartridge pre-conditioned with acetonitrile and 5% acetic acid. After washing the column with 5% acetic acid, glycopeptides were eluted with 0.5 ml of 20% isopropanol in 5% acetic acid and 0.5 ml of 40% isopropanol in 5% acetic acid. The isopropanol eluates were combined and dried by vacuum centrifugation. The dried glycopeptides were subjected to digestion with either PNGaseF (Prozyme, Hayward, CA) or PNGaseA (Sigma, St. Louis, MO). For PNGaseF, dried glycopeptides were resuspended in 0.1 ml of 20 mM sodium phosphate buffer (pH 7.5) and the N-glycans were released by incubation with 2 μl of the enzyme (10 U/ml) for 18 h at 37°C. For PNGaseA, dried glycopeptides were resuspended in 50 μl of citrate-phosphate buffer (pH 5.0) and the N-glycans were released by incubation with 0.5 μl of the enzyme (88 mU/ml) for 18 h at 37°C. Following either PNGaseF or PNGaseA digestion, released N-glycans were separated from peptide and enzyme by passage through a Sep-Pak C18 cartridge. Released glycans were permethylated with methyliodide prior to mass spectrometry (MS) analysis according to the method of Anumula and Taylor (1992). Known amounts of malto-series oligosaccharide standards (dp3 and dp4) were permethylated with isotopically heavy methyliodide (CD3I) for use as reference standards for quantification (Mehta et al., 2016).
Glycan mass spectrometry
Glycan samples were permethylated and analyzed by nanospray ionization (NSI)-MS. Briefly, permethylated glycans were reconstituted in 50% methanol containing 1 mM NaOH for infusion and injected into a linear ion trap mass spectrometer with an orbital trap mass analyzer (LTQ-Orbi; Thermo Fisher) using a nanoelectrospray source at a syringe flow rate of 0.50 μl/min and capillary temperature set to 210°C (Aoki, et al., 2007). The instrument was tuned with a permethylated oligosaccharide standard in positive ion mode. For fragmentation by collision-induced dissociation (CID in MS/MS and MSn), a normalized collision energy of 45% was used. Most permethylated glycan components were identified as singly, doubly and triply charged sodiated species [M+Na] in positive mode. The intensities of oligosaccharide standards were used for quantification of individual glycans. Peaks for all charge states were deconvoluted by the charge state and summed for quantification. All spectra were manually interpreted and annotated. Permethylated glycans released from three separate preparations of wild-type and mutant protein powder were analyzed. Replicate analyses of each genotype gave similar profiles for that genotype when the abundance of each glycan was calculated as its percent of the total profile (i.e. when the abundance of each individual glycan was normalized to the total signal derived for all glycans). The reproducibility of the normalized quantification indicated the robustness of the analytic approach, but this method of quantification cannot reveal changes in absolute glycan abundance (i.e. moles of glycan per milligram of protein). Therefore, one preparation of N-linked glycans harvested from wild-type and mutant heads was supplemented with quantitation standard before analysis to allow calculation of the amount of glycan per milligram of protein. The MS-based glycomics data generated in these analyses, and the associated annotations, are presented in accordance with the MIRAGE standards and the Athens Guidelines (Wells and Hart, 2013; York et al., 2014). Graphical representation of monosaccharide residues are as shown in Fig. 4 and are consistent with the symbol nomenclature for glycans (SNFG), which has been broadly adopted by the glycomics community (Varki et al., 2015).
Proximity ligation assay
Proximity ligation assay (PLA) was performed on testis preparations that were fixed using 4% methanol-free formaldehyde, as described above. Slides were blocked by immersing in the blocking solution contained in the kit (Duolink In Situ PLA Probes, Sigma-Aldrich), following the instructions provided by the supplier. After blocking, samples were incubated in a humid chamber overnight at 4°C with the following primary antibodies: rabbit anti-GOLPH3 G49139/77, diluted 1:1000; mouse monoclonal anti-GFP (3E6, Thermofisher Scientific) diluted 1:750; and anti-Anillin (Giansanti et al., 2015) diluted 1:2000 in the Duolink In Situ Antibody Diluent, equipped with the kit (Duolink In Situ PLA Probes, Sigma-Aldrich). The PLA probe incubation and detection protocols were performed in accordance with the procedures described in the Duolink In Situ-Fluorescence User Guide, using the provided probes and reagents. Following the detection steps, specimens were mounted using Vectashield Mounting Medium containing DAPI. Quantification of the number of PLA signals per cell was obtained using the Analyze Particles tools of the ImageJ software. In detail, for each genotype, 20 cells were randomly selected in the images collected from three experiments and manually demarcated using the freehand selection tool. The ‘analyze particles’ command in ImageJ was then used to count the number of dots in the selected area.
Yeast two-hybrid assay
The assay was performed using the B42/lexA system with strain EGY48 (Mata his3 ura3 trp1 6lexAOP-LEU2; lexAOP-lacZ reporter on plasmid pSH18-34) as the host strain (Cassani et al., 2013). This strain was co-transformed with various combinations of bait (pEG202) and prey (pJG4-5) plasmids carrying GOLPH3, Rab1, Gio cDNAs or Cog5, Cog6 and Cog7 cDNAs. To assess two-hybrid interaction, the strains were spotted on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-GAL) selective synthetic plates containing either raffinose (RAFF, prey not induced) or 2% galactose (GAL, prey expressed), as described by Cassani et al. (2013). To quantify the yeast two-hybrid results, a β-galactosidase assay was performed. Strains were first grown in a selective medium containing galactose, then cells were collected and protein extracts prepared in order to test β-galactosidase enzyme activity using o-nitrophenyl-d-galactoside (ONPG) as a substrate, as described by Polevoy et al. (2009). For each sample, five independent transformants were used and the assays were performed in duplicate to calculate average β-galactosidase units and standard error.
Statistical analysis
For all the immunofluorescence assays, the differences between wild-type and mutant cells were examined for statistical significance using unpaired Student's t-test with Prism 6 (Graphpad). In western blotting analysis, the band intensities were quantified using Image Lab software (version 4.0.1; Bio-Rad Laboratories, Hercules, CA, USA). The representative results from at least three independent experiments were analyzed using the unpaired Student's t-test with Prism 6 (Graphpad). For yeast two-hybrid experiments, differences between each group were examined for statistical significance using the unpaired Student's t-test using Prism 6 (Graphpad). P<0.05 was considered statistically significant. Data are expressed as mean±s.e.m. For N-glycan analysis, the data represent a single biological replicate of the wild type and three different Cog7 allelic combinations.
Supplementary Material
Acknowledgements
We thank Dr K. Field for anti-Lva and Dr Y.H. Inoue for α-COP antibodies. We are grateful to Prof. M. T. Fuller for flies expressing GFP-Fws. We also thank Dr V. Panin for DSiaTL22− alleles and for helpful suggestions and discussion. We are grateful to the Bloomington Drosophila Stock Center (Indiana University) for essential genetic stocks and the Developmental Studies Hybridoma Bank (University of Iowa) for essential antibodies used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.F., S.S., S.R., R.F., M.T., M.G.G.; Methodology: A.F., S.S., T.K., S.R., M.G.G.; Software: T.K.; Validation: A.F., S.S., T.K., S.R., R.F., R.P., M.T., M.G.G.; Formal analysis: A.F., T.K., S.R., R.F., M.G.G.; Investigation: A.F., S.S., T.K., R.F., A.K., G.B., R.P., K.H.T., M.G.G.; Data curation: M.T., M.G.G.; Writing - original draft: M.T., M.G.G.; Writing - review & editing: R.F., R.P., M.T., M.G.G.; Visualization: A.F., S.S., T.K., S.R., A.K., G.B., K.H.T., M.G.G.; Supervision: M.T., M.G.G.; Funding acquisition: M.T., M.G.G.
Funding
This work was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) (grant number IG14671), a grant from Fondazione Telethon Italy (grant number GEP 14076) to M.G.G. and a grant to M.T. from the National Institute of General Medicine of the National Institutes of Health (P41 GM103490). A.F. was supported by a fellowship from the AIRC (19686). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.209049.supplemental
References
- Anumula K. R. and Taylor P. B. (1992). A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 203, 101-108. 10.1016/0003-2697(92)90048-C [DOI] [PubMed] [Google Scholar]
- Aoki K., Perlman M., Lim J.-M., Cantu R., Wells L. and Tiemeyer M. (2007). Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 282, 9127-9142. 10.1074/jbc.M606711200 [DOI] [PubMed] [Google Scholar]
- Baas S., Sharrow M., Kotu V., Middleton M., Nguyen K., Flanagan-Steet H., Aoki K. and Tiemeyer M. (2011). Sugar-free frosting, a homolog of SAD kinase, drives neural-specific glycan expression in the Drosophila embryo. Development 138, 553-563. 10.1242/dev.055376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey Blackburn J., Pokrovskaya I., Fisher P., Ungar D. and Lupashin V. V. (2016). COG complex complexities: detailed characterization of a complete set of HEK293T cells lacking individual COG subunits. Front. Cell Dev. Biol. 4, 23 10.3389/fcell.2016.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone R., Fiumara A. and Jaeken J. (2014). Congenital disorders of glycosylation with emphasis on cerebellar involvement. Semin. Neurol. 34, 357-366. 10.1055/s-0034-1387197 [DOI] [PubMed] [Google Scholar]
- Belloni G., Sechi S., Riparbelli M. G., Fuller M. T., Callaini G. and Giansanti M. G. (2012). Mutations in Cog7 affect Golgi structure, meiotic cytokinesis and sperm development during Drosophila spermatogenesis. J. Cell Sci. 125, 5441-5452. 10.1242/jcs.108878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani C., Raspelli E., Santo N., Chiroli E., Lucchini G. and Fraschini R. (2013). Saccharomyces cerevisiae Dma proteins participate in cytokinesis by controlling two different pathways. Cell Cycle 12, 2794-2808. 10.4161/cc.25869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.-L., Chang C.-W., Chang Y.-Y., Sung H.-H., Lin M.-D., Chang S.-C., Chen C.-H., Huang C.-W., Tung K.-S. and Chou T.-B. (2013). The Drosophila GOLPH3 homolog regulates the biosynthesis of heparan sulfate proteoglycans by modulating the retrograde trafficking of exostosins. Development 140, 2798-2807. 10.1242/dev.087171 [DOI] [PubMed] [Google Scholar]
- Chu J., Mir A., Gao N., Rosa S., Monson C., Sharma V., Steet R., Freeze H. H., Lehrman M. A. and Sadler K. C. (2013). A zebrafish model of congenital disorders of glycosylation with phosphomannose isomerase deficiency reveals an early opportunity for corrective mannose supplementation. Dis. Model. Mech. 6, 95-105. 10.1242/dmm.010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climer L. K., Dobretsov M. and Lupashin V. (2015). Defects in the COG complex and COG-related trafficking regulators affect neuronal Golgi function. Front. Neurosci. 9, 405 10.3389/fnins.2015.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstra H. S., McArthy J., Rudin-Rush S., Hartwig C., Gokhale A., Zlatic S. A., Blackburn J. B., Werner E., Petris M., D'Souza P. et al. (2017). The interactome of the copper transporter ATP7A belongs to a network of neurodevelopmental and neurodegeneration factors. Elife 6, e24722 10.7554/eLife.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N., Nahm M., Lee S. and Broadie K. (2012). A targeted glycan-related gene screen reveals heparan sulfate proteoglycan sulfation regulates WNT and BMP trans-synaptic signaling. PLoS Genet. 8, e1003031 10.1371/journal.pgen.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N., Zhu H. and Broadie K. (2014). Two protein N-acetylgalactosaminyl transferases regulate synaptic plasticity by activity-dependent regulation of integrin signaling. J. Neurosci. 34, 13047-13065. 10.1523/JNEUROSCI.1484-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert E. S. P., Reckmann I., Hellwig A., Röhling S., El-Battari A., Wieland F. T. and Popoff V. (2014). Golgi phosphoprotein 3 triggers signal-mediated incorporation of glycosyltransferases into coatomer-coated (COPI) vesicles. J. Biol. Chem. 289, 31319-31329. 10.1074/jbc.M114.608182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas R. M., Giansanti M. G., Gatti M. and Fuller M. T. (2003). The Drosophila Cog5 homologue is required for cytokinesis, cell elongation, and assembly of specialized Golgi architecture during spermatogenesis. Mol. Biol. Cell 14, 190-200. 10.1091/mbc.E02-06-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier F., Vasile E., Schollen E., Callewaert N., Raemaekers T., Quelhas D., Jaeken J., Mills P., Winchester B., Krieger M. et al. (2006). Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc. Natl. Acad. Sci. USA 103, 3764-3769. 10.1073/pnas.0507685103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappaolo A., Sechi S., Belloni G., Piergentili R. and Giansanti M. G. (2017). Visualization of cleavage furrow proteins in fixed dividing spermatocytes. Methods Cell Biol. 137, 85-103. 10.1016/bs.mcb.2016.03.035 [DOI] [PubMed] [Google Scholar]
- Freeze H. H. and Ng B. G. (2011). Golgi glycosylation and human inherited diseases. Cold Spring Harb. Perspect. Biol. 3, a005371 10.1101/cshperspect.a005371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze H. H., Chong J. X., Bamshad M. J. and Ng B. G. (2014). Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 94, 161-175. 10.1016/j.ajhg.2013.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze H. H., Eklund E. A., Ng B. G. and Patterson M. C. (2015). Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 38, 105-125. 10.1146/annurev-neuro-071714-034019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C. W., Matthijs G., Sturiale L., Garozzo D., Wong K. Y., Wong R., Wong V. and Jaeken J. (2012). COG5-CDG with a mild neurohepatic presentation. JIMD Rep. 3, 67-70. 10.1007/8904_2011_61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G. and Fuller M. T. (2012). What Drosophila spermatocytes tell us about the mechanisms underlying cytokinesis. Cytoskeleton 69, 869-881. 10.1002/cm.21063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Farkas R. M., Bonaccorsi S., Lindsley D. L., Wakimoto B. T., Fuller M. T. and Gatti M. (2004). Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell. 15, 2509-2522. 10.1091/mbc.E03-08-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Bonaccorsi S., Kurek R., Farkas R. M., Dimitri P., Fuller M. T. and Gatti M. (2006). The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 16, 195-201. 10.1016/j.cub.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Vanderleest T. E., Jewett C. E., Sechi S., Frappaolo A., Fabian L., Robinett C. C., Brill J. A., Loerke D., Fuller M. T. et al. (2015). Exocyst-dependent membrane addition is required for anaphase cell elongation and cytokinesis in Drosophila. PLoS Genet. 11, e1005632 10.1371/journal.pgen.1005632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goreta S. S., Dabelic S. and Dumic J. (2012). Insights into complexity of congenital disorders of glycosylation. Biochem. Med. 22, 156-170. 10.11613/BM.2012.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. J. and Lev S. (2014). Tethering the assembly of SNARE complexes. Trends Cell. Biol. 24, 35-43. 10.1016/j.tcb.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Isaji T., Im S., Gu W., Wang Y., Hang Q., Lu J., Fukuda T., Hashii N., Takakura D., Kawasaki N. et al. (2014). An oncogenic protein Golgi phosphoprotein 3 up-regulates cell migration via sialylation. J. Biol. Chem. 289, 20694-20705. 10.1074/jbc.M113.542688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Akimoto Y., Fuwa T. J., Sato C., Komatsu A. and Nishihara S. (2016). Mucin-type core 1 glycans regulate the localization of neuromuscular junctions and establishment of muscle cell architecture in Drosophila. Dev. Biol. 412, 114-127. 10.1016/j.ydbio.2016.01.032 [DOI] [PubMed] [Google Scholar]
- Jaeken J. (2013). Congenital disorders of glycosylation. Handb. Clin. Neurol. 113, 1737-1743. 10.1016/B978-0-444-59565-2.00044-7 [DOI] [PubMed] [Google Scholar]
- Jan L. Y. and Jan Y. N. (1982). Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA 79, 2700-2704. 10.1073/pnas.79.8.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. G., Tenney A. P., Ghose A., Duckworth A. M., Higashi M. E., Parfitt K., Marcu O., Heslip T. R., Marsh J. L., Schwarz T. L. et al. (2006). The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 49, 517-531. 10.1016/j.neuron.2006.01.026 [DOI] [PubMed] [Google Scholar]
- Jumbo-Lucioni P., Parkinson W. and Broadie K. (2014). Overelaborated synaptic architecture and reduced synaptomatrix glycosylation in a Drosophila classic galactosemia disease model. Dis. Model. Mech. 7, 1365-1378. 10.1242/dmm.017137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumbo-Lucioni P. P., Parkinson W. M., Kopke D. L. and Broadie K. (2016). Coordinated movement, neuromuscular synaptogenesis and trans-synaptic signaling defects in Drosophila galactosemia models. Hum. Mol. Genet. 25, 3699-3714. 10.1093/hmg/ddw217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T. and Tiemeyer M. (2013). The N's and O's of Drosophila glycoprotein glycobiology. Glycoconj. J. 30, 57-66. 10.1007/s10719-012-9442-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann N., DeProto J., Ranjan R., Wan H. and Van Vactor D. (2002). Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron 28, 27-38. 10.1016/S0896-6273(02)00643-8 [DOI] [PubMed] [Google Scholar]
- Kingsley D. M., Kozarsky K. F., Segal M. and Krieger M. (1986). Three types of low density lipoprotein receptor-deficient mutant have pleiotropic defects in the synthesis of N-linked, O-linked, and lipid-linked carbohydrate chains. J. Cell Biol. 102, 1576-1585. 10.1083/jcb.102.5.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa D., Yamaguchi M., Mori H. and Inoue Y. H. (2012). COPI-mediated membrane trafficking is required for cytokinesis in Drosophila male meiotic divisions. J. Cell Sci. 125, 3649-3660. 10.1242/jcs.103317 [DOI] [PubMed] [Google Scholar]
- Kodera H., Ando N., Yuasa I., Wada Y., Tsurusaki Y., Nakashima M., Miyake N., Saitoh S., Matsumoto N. and Saitsu H. (2015). Mutations in COG2 encoding a subunit of the conserved oligomeric Golgi complex cause a congenital disorder of glycosylation. Clin. Genet. 87, 455-460. 10.1111/cge.12417 [DOI] [PubMed] [Google Scholar]
- Koles K., Lim J.-M., Aoki K., Porterfield M., Tiemeyer M., Wells L. and Panin V. (2007). Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology 17, 1388-1403. 10.1093/glycob/cwm097 [DOI] [PubMed] [Google Scholar]
- Kranz C., Ng B. G., Sun L., Sharma V., Eklund E. A., Miura Y., Ungar D., Lupashin V., Winkel R. D., Cipollo J. F. et al. (2007). COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum. Mol. Genet. 16, 731-741. 10.1093/hmg/ddm028 [DOI] [PubMed] [Google Scholar]
- Lübbehusen J., Thiel C., Rind N., Ungar D., Prinsen B. H., de Koning T. J., van Hasselt P. M. and Körner C. (2010). Fatal outcome due to deficiency of subunit 6 of the conserved oligomeric Golgi complex leading to a new type of congenital disorders of glycosylation. Hum. Mol. Genet. 19, 3623-3633. 10.1093/hmg/ddq278 [DOI] [PubMed] [Google Scholar]
- Mehta N., Porterfield M., Struwe W. B., Heiss C., Azadi P., Rudd P. M., Tiemeyer M. and Aoki K. (2016). Mass spectrometric quantification of N-linked glycans by reference to exogenous standards. J. Proteome. Res. 15, 2969-2980. 10.1021/acs.jproteome.6b00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. J. and Ungar D. (2012). Re‘COG'nition at the Golgi. Traffic 13, 891-897. 10.1111/j.1600-0854.2012.01338.x [DOI] [PubMed] [Google Scholar]
- Miller V. J., Sharma P., Kudlyk T. A., Frost L., Rofe A. P., Watson I. J., Duden R., Lowe M., Lupashin V. V. and Ungar D. (2013). Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the Golgin TATA element modulatory factor (TMF). J. Biol. Chem. 288, 4229-4240. 10.1074/jbc.M112.426767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morava E., Zeevaert R., Korsch E., Huijben K., Wopereis S., Matthijs G., Keymolen K., Lefeber D. J., De Meirleir L. and Wevers R. A. (2007). A common mutation in the COG7 gene with a consistent phenotype including microcephaly, adducted thumbs, growth retardation, VSD and episodes of hyperthermia. Eur. J. Hum. Genet. 156, 638-645. 10.1038/sj.ejhg.5201813 [DOI] [PubMed] [Google Scholar]
- Müller D., Jagla T., Bodart L. M., Jährling N., Dodt H.-U., Jagla K. and Frasch M. (2010). Regulation and functions of the lms homeobox gene during development of embryonic lateral transverse muscles and direct flight muscles in Drosophila. PLoS ONE 5, e14323 10.1371/journal.pone.0014323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B. G., Kranz C., Hagebeuk E. E., Duran M., Abeling N. G., Wuyts B., Ungar D., Lupashin V., Hartdorff C. M., Poll-The B. T. et al. (2007). Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol. Genet. Metab. 91, 201-204. 10.1016/j.ymgme.2007.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K. and Marth J. D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126, 855-867. 10.1016/j.cell.2006.08.019 [DOI] [PubMed] [Google Scholar]
- Paesold-Burda P., Maag C., Troxler H., Foulquier F., Kleinert P., Schnabel S., Baumgartner M. and Hennet T. (2009). Deficiency in COG5 causes a moderate form of congenital disorders of glycosylation. Hum. Mol. Genet. 18, 4350-4356. 10.1093/hmg/ddp389 [DOI] [PubMed] [Google Scholar]
- Parkinson W., Dear M. L., Rushton E. and Broadie K. (2013). N-glycosylation requirements in neuromuscular synaptogenesis. Development 140, 4970-4981. 10.1242/dev.099192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson W. M., Dookwah M., Dear M. L., Gatto C. L., Aoki K., Tiemeyer M. and Broadie K. (2016). Synaptic roles for phosphomannomutase type 2 in a new Drosophila congenital disorder of glycosylation disease model. Dis. Model. Mech. 9, 513-527. 10.1242/dmm.022939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoy G., Wei H.-C., Wong R., Szentpetery Z., Kim Y. J., Goldbach P., Steinbach S. K., Balla T. and Brill J. A. (2009). Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J. Cell Biol. 187, 847-858. 10.1083/jcb.200908107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnikova E., Koles K., Nakamura M., Pitts J., Li H., Ambavane A., Zoran M. J. and Panin V. M. (2010). Sialyltransferase regulates nervous system function in Drosophila. J. Neurosci. 30, 6466-6476. 10.1523/JNEUROSCI.5253-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders E., Foulquier F., Leão Teles E., Quelhas D., Morelle W., Rabouille C., Annaert W. and Matthijs G. (2009). Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum. Mol. Genet. 18, 3244-3256. 10.1093/hmg/ddp262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K. R., Liu J., Li S., Setty T. G., Wood C. S., Burd C. G. and Ferguson K. M. (2008). Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell 14, 523-534. 10.1016/j.devcel.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott H. and Panin V. M. (2014). N-glycosylation in regulation of the nervous system. Adv. Neurobiol. 9, 367-394. 10.1007/978-1-4939-1154-7_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi S., Colotti G., Belloni G., Mattei V., Frappaolo A., Raffa G. D., Fuller M. T. and Giansanti M. G. (2014). GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster. PLoS Genet. 10, e1004305 10.1371/journal.pgen.1004305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi S., Frappaolo A., Belloni G. and Giansanti M. G. (2015a). The roles of the oncoprotein GOLPH3 in contractile ring assembly and membrane trafficking during cytokinesis. Biochem. Soc. Trans. 43, 117-121. 10.1042/BST20140264 [DOI] [PubMed] [Google Scholar]
- Sechi S., Frappaolo A., Belloni G., Colotti G. and Giansanti M. G. (2015b). The multiple cellular functions of the oncoprotein Golgi phosphoprotein 3. Oncotarget 6, 3493-3506. 10.18632/oncotarget.3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi S., Frappaolo A., Fraschini R., Capalbo L., Gottardo M., Belloni G., Glover D. M., Wainman A. and Giansanti M. G. (2017). Rab1 interacts with GOLPH3 and controls Golgi structure and contractile ring constriction during cytokinesis in Drosophila melanogaster. Open Biol. 7, 160257 10.1098/rsob.160257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson J. C., Field C., Ventura R., Royou A. and Sullivan W. (2000). Lava lamp, a novel peripheral Golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151, 905-918. 10.1083/jcb.151.4.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. D. and Lupashin V. V. (2008). Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 343, 2024-2031. 10.1016/j.carres.2008.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaapen L. J. M., Bakker J. A., van der Meer S. B., Sijstermans H. J., Steet R. A., Wevers R. A. and Jaeken J. (2005). Clinical and biochemical presentation of siblings with COG-7 deficiency, a lethal multiple O- and N-glycosylation disorder. J. Inherit. Metab. Dis. 28, 707-714. 10.1007/s10545-005-0015-z [DOI] [PubMed] [Google Scholar]
- Steet R. and Kornfeld S. (2006). COG-7-deficient human fibroblasts exhibit altered recycling of Golgi proteins. Mol. Biol. Cell 17, 2312-2321. 10.1091/mbc.E05-08-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W. B. and Reinhold V. N. (2012). The conserved oligomeric Golgi complex is required for fucosylation of N-glycans in Caenorhabditis elegans. Glycobiology 22, 863-875. 10.1093/glycob/cws053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E. S., Duden R. and Lupashin V. V. (2002). The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157, 631-643. 10.1083/jcb.200111081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C. and Körner C. (2011). Mouse models for congenital disorders of glycosylation. J. Inherit. Metab. Dis. 34, 879-889. 10.1007/s10545-011-9295-7 [DOI] [PubMed] [Google Scholar]
- Tu L., Tai W. C. S., Chen L. and Banfield D. K. (2008). Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321, 404-407. 10.1126/science.1159411 [DOI] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T. et al. (2015). Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323-1324. 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh D. A., Rasse T. M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.-C., Schmidt M., Qin G. et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833-844. 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Wells L. and Hart G. W. (2013). Glycomics: building upon proteomics to advance glycosciences. Mol. Cell. Proteomics 12, 833-835. 10.1074/mcp.E113.027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett R., Ungar D. and Lupashin V. (2013a). The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem. Cell Biol. 140, 271-283. 10.1007/s00418-013-1117-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett R., Kudlyk T., Pokrovskaya I., Schönherr R., Ungar D., Duden R. and Lupashin V. (2013b). COG complexes form spatial landmarks for distinct SNARE complexes. Nat. Commun. 4, 1553 10.1038/ncomms2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Steet R. A., Bohorov O., Bakker J., Newell J., Krieger M., Spaapen L., Kornfeld S. and Freeze H. H. (2004). Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat. Med. 10, 518-523. 10.1038/nm1041 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhang X., Nix D. B., Katoh T., Aoki K., Tiemeyer M. and Wang Y. (2013). Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat. Commun. 4, 1659 10.1038/ncomms2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W. S., Agravat S., Aoki-Kinoshita K. F., McBride R., Campbell M. P., Costello C. E., Dell A., Feizi T., Haslam S. M., Karlsson N. et al. (2014). MIRAGE: the minimum information required for a glycomics experiment. Glycobiology 24, 402-406. 10.1093/glycob/cwu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Astier M., Gratecos D. and Sémériva M. (1997). The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 124, 2087-2098. [DOI] [PubMed] [Google Scholar]
- Zeevaert R., Foulquier F., Cheillan D., Cloix I., Guffon N., Sturiale L., Garozzo D., Matthijs G. and Jaeken J. (2009). A new mutation in COG7 extends the spectrum of COG subunit deficiencies. Eur. J. Med. Genet. 52, 303-305. 10.1016/j.ejmg.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., Fish M., Acar M., Hoskins R. A., Bellen H. J. and Scott M. P. (2007). Thirty-one flavors of Drosophila rab proteins. Genetics 176, 1307-1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.