Abstract
Background
Artemisinin resistance, linked to polymorphisms in the Kelch gene on chromosome 13 of Plasmodium falciparum (k13), has outpaced containment efforts in South East Asia. For national malaria control programmes in the region, it is important to establish a surveillance system which includes monitoring for k13 polymorphisms associated with the clinical phenotype.
Methods
Between February and December 2013, parasite clearance was assessed in 35 patients with uncomplicated P. falciparum treated with artesunate monotherapy followed by 3-day ACT in an isolated area on the Myanmar–Thai border with relatively low artemisinin drug pressure. Molecular testing for k13 mutations was performed on dry blood spots collected on admission.
Results
The proportion of k13 mutations in these patients was 41.7%, and only 5 alleles were detected: C580Y, I205T, M476I, R561H, and F446I. Of these, F446I was the most common, and was associated with a longer parasite clearance half-life (median) 4.1 (min–max 2.3–6.7) hours compared to 2.5 (min–max 1.6–8.7) in wildtype (p = 0·01). The prevalence of k13 mutant parasites was much lower than the proportion of k13 mutants detected 200 km south in a much less remote setting where the prevalence of k13 mutants was 84% with 15 distinct alleles in 2013 of which C580Y predominated.
Conclusions
This study provides evidence of artemisinin resistance in a remote part of eastern Myanmar. The prevalence of k13 mutations as well as allele diversity varies considerably across short distances, presumably because of historical patterns of artemisinin use and population movements.
Electronic supplementary material
The online version of this article (10.1186/s12936-017-2128-x) contains supplementary material, which is available to authorized users.
Keywords: Plasmodium falciparum malaria, Artemisinin, Parasite clearance, k13 mutation, Drug resistance
Background
Resistance to the artemisinin derivatives in Plasmodium falciparum, characterized by delayed parasite clearance in patients treated with artesunate, emerged in Western Cambodia in 2007 [1–3]. This phenotypic trait was also documented in a large study on the Thai–Myanmar border in 2012 and was [1] found to be heritable [4] and associated with strong selective sweeps in the plasmodial genome [5]. In 2014, non-synonymous single nucleotide polymorphisms in the Kelch propeller domain on chromosome 13 (k13) were found to be strongly associated with resistance to artemisinin derivatives [6]. The link between certain k13 mutations and the clinical resistance phenotype (delayed parasite clearance) has been demonstrated by a number of clinical [7–10], in vitro [11] and transfection studies [12]. In the areas where k13 mutations have been found, the distribution of different alleles has been variable. For example, the C580Y polymorphism is the most common mutation in Cambodia [13, 14], Laos [15, 16] and Vietnam [16], while the F446I mutants are most common on the Myanmar–China border [9, 17, 18]. On the Thailand–Myanmar border near Mae Sot, the E252Q allele was the most abundant before 2008, but it has since been replaced by C580Y, an allele now close to fixation in this area [19]. The C580Y mutant parasites have emerged at least twice at different locations [20, 21]. Isolates from Cambodia and Vietnam appear to have a common ancestor and they differ from resistant parasites in Myanmar [13, 21]. The C580Y allele found on the Myanmar–China border appears to have originated from the Thailand–Myanmar border [17]; indicating that genetic mutations (at least C580Y) associated with artemisinin resistance in P. falciparum are not only emerging “de novo” in different locations but also spreading contiguously.
In 2011, the World Health Organization (WHO) proposed a strategy of containment of artemisinin resistant malaria in the region [22], but this has not contained the spread of resistance; parasites carrying k13 mutations have now spread to the China–Myanmar [23] and India–Myanmar borders [7, 21, 23, 24]. The C580Y genotype has increased in frequency replacing other genotypes in most of Southeast Asia, and a single C580Y parasite lineage has swept across the Eastern Mekong subregion spreading from Western Cambodia to Southern Vietnam indicating that a “hard” selective sweep has now replaced initial “soft” selective sweeps [19].
More importantly, k13 alleles under strong selective pressure confer (or are associated with) slow parasite clearance after artemisinin treatment leaving a larger number of surviving (artemisinin resistant) parasites exposed to the partner drugs [25]. The loss in efficacy of DHA-piperaquine in Cambodia and Viet Nam [26, 27] and mefloquine-artesunate on the Thai–Myanmar border [28] have demonstrated how slow parasite clearance due to artemisinin resistance unavoidably leads to the failure of ACT.
This report presents data collected in 2013 from a remote region along the Myanmar–Thai border in Hpa-pun District, Myanmar (Fig. 1). While artemisinin resistance was well established by 2010 [4] in the border district of Mae Sot (Thailand), 200 km further south, which is a well-connected area encompassing the Trans-Asia highway, data from elsewhere in the remote, hilly and forested parts of eastern Myanmar are few [10, 24]. This study provided an opportunity for examining the phenotype and genetic diversity of k13 of parasites from a secluded relatively inaccessible region with limited access to ACT (Fig. 1). Parasite clearance half-life and k13 genotyping results from 35 patients treated with 3 days artesunate monotherapy followed by a full course of ACT are presented.
Fig. 1.
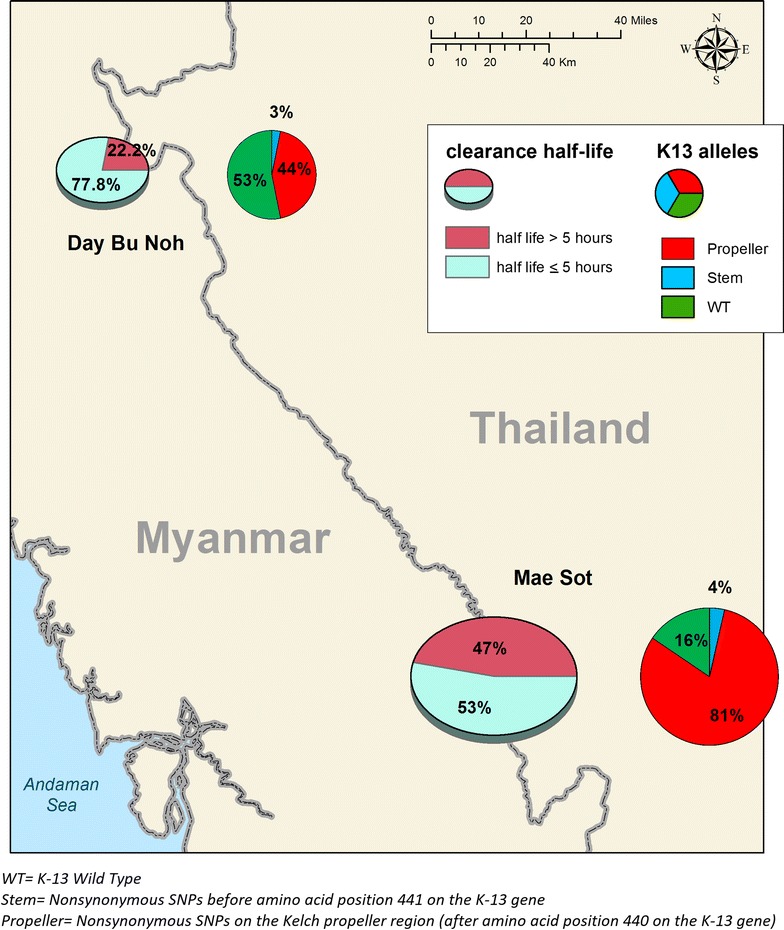
Map showing study site with summary of K-13 mutation and parasite clearance comparison with Maesot
Methods
Study site and study design
This was an open label non-randomized trial designed to evaluate the association between the efficacy of artemisinin against P. falciparum malaria (i.e. parasite clearance) and k13 mutations using the same protocol as a larger multicentre trial [7]. The study was conducted in a remote and isolated region of Hpa-Pun district in Eastern Myanmar in northern Kayin state (Fig. 1). Much of Kayin State has been affected by political instability and conflict for over half a century. The public health infrastructure in this remote district has lagged behind that of other neighbouring areas that have benefited from a relatively more stable situation in recent years. There has been less intense artemisinin usage in the district and malaria remains endemic.
Patient recruitment
Patients who presented at Day Bu Noh clinic with fever (axillary temperature ≥ 37.5 °C) or history of fever within the last 24 h and who were between the ages of 6 months to 65 years were screened for malaria parasites using microscopy of thick and thin Giemsa-stained blood smears. Patients with uncomplicated P. falciparum malaria and asexual parasitaemia above or equal to 10,000/µL were enrolled. Pregnant women and patients with severe malaria [29] or treated with an ACT in the past 7 days were excluded. On admission, capillary blood was collected on Whatman® 903 (W-903) filter paper for parasite genotyping. Haemoglobin colour scale Copack® was used for haemoglobin estimation. A clinical examination was also performed, and vital signs were recorded as well as demographic data.
Drug administration and follow up
All patients received oral artesunate (Guilin Pharmaceutical Co, PRC), 4 mg/kg/day for 3 days. After 3 days of artesunate monotherapy, either
artesunate-mefloquine (Eloquine® Medochemie Ltd., Cyprus) (4 mg/kg artesunate day 3–5 and mefloquine 15 mg/kg day 4 & 10 mg/kg day 5) or
dihydroartemisinin-piperaquine (Duo-Cotecxin® Beijing Holley-Cotec Pharmaceuticals Co., Ltd, China) at a dose of 2.5 mg/kg dihydroartemisinin and 20 mg/kg piperaquine day 3–5 was administered.
All treatments were supervised. Peripheral parasitaemia and temperature were measured at 0, 4, 6, 8, 12 and every 6 h thereafter until two consecutive negative slides were obtained. No further follow up was undertaken after the patient finished the full course of treatment and had a negative malaria slide on two occasions. Other clinical data and vital signs were recorded daily.
Sequencing of the Plasmodium falciparum kelch13 gene
Kelch genotyping was performed at the malaria molecular laboratory of Mahidol Oxford Tropical Medicine Research Unit using the method reported previously [24, 28] (see more detail in Additional file 1).
Data management and statistical analysis
Patient data were recorded on individual Case Report Forms (CRFs) and later entered into a Microsoft Access database and analysed using STATA (Version 13, STATA Corp) and Graphpad prism (version 5). Normally distributed data were compared by Student’s t test and non-normally distributed data by the Wilcoxon rank-sum (Mann–Whitney) test. Parasite clearance half-lives were calculated using the World Wide Antimalarial Resistance Network (WWARN) Parasite Clearance Estimator [2, 30].
Ethical approval
Written informed consent was obtained after the screening process and the study was explained in detail through the use of a local interpreter. Consent was obtained from a parent or guardian for participants below 18 years.
Ethical approval was given by the Oxford Tropical Research Ethics Committee (OXTREC 06-11). Verbal approval was given locally by existing health authorities KDHW (Karen Department of Health and Welfare) in the absence of a local ethics committee. However, a community advisory board composed of members of the local population also approved this study [9].
Results
Patients disposition
Between February and December 2013, a total of 1736 febrile patient presented at the clinic, and 27.4% (475) of them were microscopically confirmed to have malaria (Fig. 2). Of the 291 patients with P. falciparum malaria, 36 that met the inclusion criteria and gave consent were recruited into the study (including 3 mixed infections). The main reason for exclusion was low parasitaemia. The baseline demographic and clinical data of the patients are summarized in Table 1.
Fig. 2.
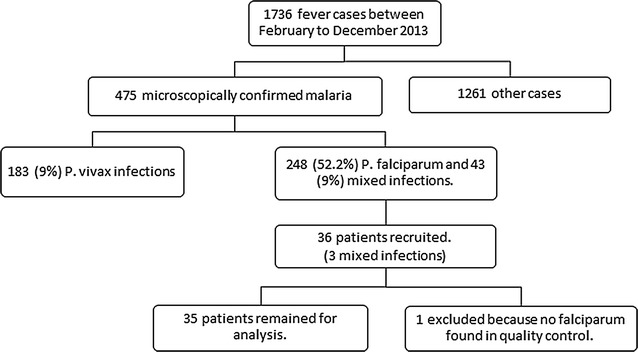
Study flow diagram
Table 1.
Baseline demographic and clinical data of recruited patients
| Screened, n | 291 |
| Recruited, n | 35 |
| Male, n (%) | 20 (57.1) |
| Age, years | |
| Median (IQR) | 14.0 (8.5) |
| < 5, n (%) | 2 (5.6) |
| 5–15, n (%) | 17 (47.2) |
| > 15, n (%) | 17 (47.2) |
| Weight, kg | |
| Median (IQR) | 41.0 (28.0) |
| Clinical profiles (Day 0) | |
| Proportion of patients with fever | |
| n (%) | 23 (65.7%) |
| Temperature | |
| Mean (SD) | 38.5 (0.8) |
| Haemoglobin (g/dL) | |
| Median (IQR) | 12.0 (0.5) |
| Parasite density | |
| Geometric mean (per µL) | 86,063 |
| Min | 22,608 |
| Max | 310,860 |
| Gametocytaemia | |
| n (%) | 3 (8.3%) |
All patients had a history of fever before coming to the clinic and 23/35 (65.7%) had fever on admission. By 48 h 30/35 (85.7%: 95% CI 74.1–97.3) of the patients were afebrile and all had cleared their fever by 72 h. After 3 days of artesunate, 13 (37.1%; 95% CI 21.1–53.1%) patients remained parasitaemic but all were negative by microscopy by day 4. Three (8.6%) patients had gametocytes on admission that were cleared in 2 patients by day 6.
Prevalence of k13 mutations
From the 35 recruited patients, 32 (91.4%) parasite isolates were successfully genotyped for k13 sequence. Five different non-synonymous polymorphisms were detected in 46.9% (15/32; 95% CI 29.6–64.2%) of samples including 4 in the propeller region and 1 in the ‘stem’ region of the kelch protein. No sample had more than one mutation. Among these, the most common allele was F446I (31.3%) (Fig. 3).
Fig. 3.
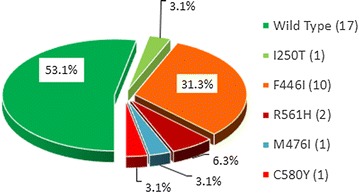
Proportion of K-13 SNPs in recruited patients
Association of k13 genotype and treatment outcomes
There was a statistically significant difference in the parasite clearance half-life (median, IQR) of patients infected with wild type k13 (2.5, 1.3 h) compared to those infected with isolates carrying a mutation in the propeller region (4.1, 3.3 h) (p = 0.04). Again, the patients infected with the most common allele, F446I cleared their parasites more slowly (4.1, 2.0 h) (p = 0.04) compared to those infected with wild type K-13 (Fig. 4; Additional file 2). The longest parasite clearance half-life (9.0 h) was in the patient infected with a parasite carrying the C580Y mutation.
Fig. 4.
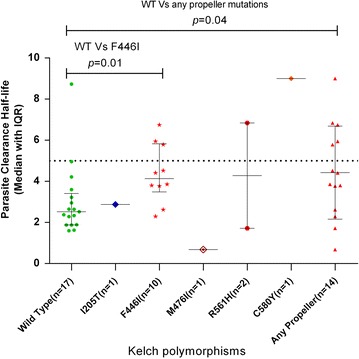
Comparison of parasite clearance half-lives of different K-13 SNP
Safety
The 6-day treatment (3-day artesunate monotherapy follow by 3-day ACT) was well tolerated. None of the patients developed severe malaria. There were no serious adverse events.
Discussion
Throughout South East Asia parasite clearance rates are decreasing among patients with uncomplicated P. falciparum malaria, a pattern that is strongly associated with mutations in the k13 gene of P. falciparum and accompanying partner drug resistance. Until now, 108 non-synonymous polymorphisms of k13 gene have been found in all malarious regions of the world. Most of the mutations in the propeller region of the gene are associated with the slow parasite clearance phenotype, and in Asia show evidence of recent selection [16, 19]. Historical data from the Thai–Myanmar border (Mae Sot area) show that the prevalence of k13 mutations has increased significantly, and among them the C580Y allele has out-competed others in recent years signifying that a “hard” selective sweep has replaced an initially “soft” one [19]. The C580Y mutation also dominates in the Eastern Greater Mekong sub-region although this successful lineage had a different genetic origin to the lineage in Eastern Myanmar. In this study in a relatively inaccessible region 200 km away but days travel from Mae Sot, the most frequent kelch mutant allele was F446I. This is the predominant allele in the North of Myanmar which was significantly associated with slow parasite clearance.
The main limitation of this study is the small sample size, which is related to both the remoteness of the study site, logistical difficulties related to working in this remote area, and the relatively high parasitaemia threshold for inclusion (i.e. ≥ 10,000/µL). The parasitaemia threshold was chosen in order to obtain reliable parasite clearance curves.
However, this small study highlights the complexity of the spatial dynamics of artemisinin resistance in P. falciparum with significant differences in allele frequencies across small distances. Even though the prevailing alleles may be different it is clear that artemisinin resistance has emerged even in geographically secluded areas and ultimately may be unstoppable. In the absence of alternative treatments ready to replace the failing ACT, elimination remains the best practical strategy to slow down or halt progression of artemisinin resistance in this region.
Additional files
Additional file 1. Genetic markers of artemisinin resistance.
Additional file 2. Parasite clearance curves for individual patients.
Authors’ contributions
CAB, EAA, FN and NJW designed the study. CAB implemented the study and collected the clinical data. MI, SP and KS did laboratory work. CAB, EAA, DMP, FN and APP analysed the data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank all participants that took part in this study. Especially to Naw Chit and Aye Thar the two field microscopists who made this study possible.
Competing interests
All authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding or last author on reasonable request.
Conflict of interest
NJW is co-chairman of the WHO antimalarial treatment guidelines committee. All other authors declare that they have no conflicts of interest.
Funding
This document is an output from a project funded by the UK Department for International Development (DFID) for the benefit of developing countries as well as from Wellcome Trust–Mahidol University–Oxford Tropical Medicine Research Programme. However, the views expressed, and information contained in it are not necessarily those of or endorsed by funders, which can accept no responsibility for such views or for any reliance place on them.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12936-017-2128-x) contains supplementary material, which is available to authorized users.
Contributor Information
Craig A. Bonnington, Email: c.a.bonnington@gmail.com
Aung Pyae Phyo, Email: aungpyaephyo@shoklo-unit.com.
Elizabeth A. Ashley, Email: liz@tropmedres.ac
Mallika Imwong, Email: noi@tropmedres.ac.
Kanlaya Sriprawat, Email: poo@shoklo-unit.com.
Daniel M. Parker, Email: dparker1@uci.edu
Stephane Proux, Email: steph@shoklo-unit.com.
Nicholas J. White, Email: nickwdt@tropmedres.ac
Francois Nosten, Email: francois@tropmedres.ac.
References
- 1.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flegg JA, Guerin PJ, Nosten F, Ashley EA, Phyo AP, Dondorp AM, et al. Optimal sampling designs for estimation of Plasmodium falciparum clearance rates in patients treated with artemisinin derivatives. Malar J. 2013;12:411. doi: 10.1186/1475-2875-12-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noedl H, Se Y, Sriwichai S, Schaecher K, Teja-Isavadharm P, Smith B, et al. Artemisinin resistance in Cambodia: a clinical trial designed to address an emerging problem in southeast Asia. Clin Infect Dis. 2010;51:e82–e89. doi: 10.1086/657120. [DOI] [PubMed] [Google Scholar]
- 4.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tun KM, Jeeyapant A, Imwong M, Thein M, Aung SS, Hlaing TM, et al. Parasite clearance rates in Upper Myanmar indicate a distinctive artemisinin resistance phenotype: a therapeutic efficacy study. Malar J. 2016;15:185. doi: 10.1186/s12936-016-1240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, et al. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyunt MH, Hlaing T, Oo HW, Tin-Oo LL, Phway HP, Wang B, et al. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis. 2015;60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 11.Boulle M, Witkowski B, Duru V, Sriprawat K, Nair SK, McDew-White M, et al. Artemisinin-resistant Plasmodium falciparum K13 mutant alleles, Thailand–Myanmar border. Emerg Infect Dis. 2016;22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, et al. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin–piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. Artemisinin and artemisinin based combination therapy resistance; status report 2016. Geneva: World Health Organization; 2016.
- 16.Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye R, Hu D, Zhang Y, Huang Y, Sun X, Wang J, et al. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China–Myanmar border. Sci Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z. Artemisinin resistance at the China–Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson TJ, Nair S, McDew-White M, Cheeseman IH, Nkhoma S, Bilgic F, et al. Population parameters underlying an ongoing soft sweep in southeast Asian malaria parasites. Mol Biol Evolut. 2017;34:131–144. doi: 10.1093/molbev/msw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong Subregion: a molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in southeast Asia. J Infect Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. Global Plan for Artemisinin Resistance Containment (GPARC). Geneva: World Health Organization; 2011.
- 23.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra N, Bharti RS, Mallick P, Singh OP, Srivastava B, Rana R, et al. Emerging polymorphisms in falciparum Kelch 13 gene in northeastern region of India. Malar J. 2016;15:583. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dondorp AM, Fairhurst RM, Slutsker L, Macarthur JR, Breman JG, Guerin PJ, et al. The threat of artemisinin-resistant malaria. N Engl J Med. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thanh NV, Thuy-Nhien N, Tuyen NTK, Tong NT, Nha-Ca NT, Quang HH, et al. Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J. 2017;16:27. doi: 10.1186/s12936-017-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phyo AP, Ashley EA, Anderson TJC, Bozdech Z, Carrara VI, Sriprawat K, et al. Declining efficacy of artemisinin combination therapy against P. falciparum malaria on the Thai–Myanmar border (2003–2013): the role of parasite genetic factors. Clin Infect Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO. Guidelines for the treatment of malaria. Geneva: World Health Organization; 2015. [PubMed]
- 30.Parasite clearance esimator [https://www.wwarn.org/pce]. Accessed 11 Nov 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Genetic markers of artemisinin resistance.
Additional file 2. Parasite clearance curves for individual patients.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding or last author on reasonable request.


