Abstract
Background
Subtribe Orchidinae (Orchidaceae, Orchidoideae) are a nearly cosmopolitan taxon of terrestrial orchids, comprising about 1800 species in 47 to 60 genera. Although much progress has been made in recent years of phylogenetics of Orchidinae, considerable problems remain to be addressed. Based on molecular phylogenetics, we attempt to illustrate the phylogenetic relationships and discuss generic delimitation within Orchidinae. Seven DNA markers (five plastid and two nuclear), a broad sampling of Orchidinae (400 species in 52 genera) and three methods of phylogenetic analysis (maximum likelihood, maximum parsimony and Bayesian inference) were used.
Results
Orchidinae s.l. are monophyletic. Satyrium is sister to the rest of Orchidinae s.l. Brachycorythis and Schizochilus are successive sister to Asian-European Orchidinae s.s. Sirindhornia and Shizhenia are successive sister to clade formed by Tsaiorchis-Hemipilia-Ponerorchis alliance. Stenoglottis is sister to the Habenaria-Herminium-Peristylus alliance. Habenaria, currently the largest genus in Orchidinae, is polyphyletic and split into two distant clades: one Asian-Australian and the other African–American–Asian. Diplomeris is sister to Herminium s.l. plus Asian-Australian Habenaria.
Conclusions
We propose to recognize five genera in the Ponerorchis alliance: Hemipilia, Ponerorchis s.l., Sirindhornia, Shizhenia and Tsaiorchis. Splitting Habenaria into two genera based on morphological characters and geographical distribution may be the least disruptive approach, and it is reasonable to keep Satyrium in Orchidinae.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1160-x) contains supplementary material, which is available to authorized users.
Keywords: Gennaria, Habenaria, Orchidinae s.l., Orchid generic delimitation, Ponerorchis alliance, Platanthera
Background
Subtribe Orchidinae are a nearly cosmopolitan clade of mostly terrestrial orchids, comprising about 1800 species in 47 to 60 genera [1, 2]. Morphologically, Orchidinae are characterized by: fleshy roots or tubers; non-plicate, convolute leaves that are basal or spirally arranged along the stem (Fig. 1); terminal, racemose inflorescences carrying mostly resupinate, small flowers (predominantly white, green or purple; Fig. 1); a single, usually erect anther lacking an operculum; two sectile pollinia; a column with two lateral appendages; stigma appearing 2–3-lobed (Fig. 1) [2–4]. Various genera within Orchidinae have been the subject of recent systematic studies, including Dactylorhiza [5, 6]; Gymnadenia [7, 8]; Habenaria [9]; Herminium [10]; Himantoglossum [11]; Ophrys [12–14]; Orchis [15–17]; Platanthera [18–22]; and Ponerorchis [23, 24]. These studies inevitably addressed only part of the diversity of the subtribe as a whole.
Fig. 1.
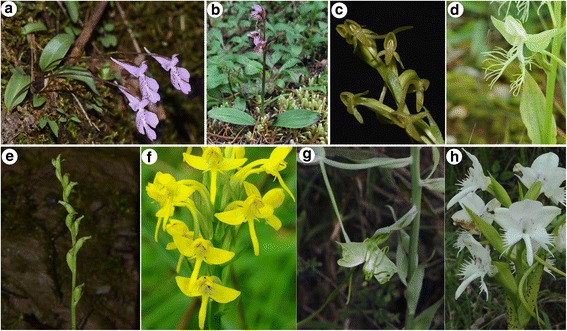
Representative species of Orchidinae. a, Shizhenia pinguicula; b, Galearis spathulata; c, Platanthera bakeriana; d, Habenaria davidii; e, Gennaria griffthii; f, Habenaria linguella; g, Bonatea steudneri; h. Pecteilis susannae. (a by Weitao Jin, g by Bing Liu, others by Xiaohua Jin)
More comprehensive systematic investigations of Orchidinae have elucidated certain parts of the clade and also indicated areas where our knowledge is still inadequate [25–27]. Using nuclear ribosomal ITS (nrITS) from 190 species, Bateman et al. [26] identified 12 well-resolved clades within Orchidinae, confirming triphyly of Orchis as traditionally delimited, polyphyly of Habenaria, and considerable levels of homoplasy of various morphological characters. Most of these findings have been confirmed by subsequent results [9, 19, 23–25]. The first study with a good selection of Asian taxa [23], based on three markers (plastid matK/rbcL and nrITS) from 146 species, found that several genera as traditionally delimited were not monophyletic. These authors combined Hemipiliopsis Y.B.Luo & S.C.Chen with Hemipilia Lindl., Amitostigma Schltr. and Neottianthe (Rchb.) Schltr. with Ponerorchis, Smithorchis Tang & F.T.Wang with Platanthera, and Aceratorchis Schltr. and Neolindleya Kraenzl. with Galearis Raf. (Galearis, Fig. 1b), and proposed the new genus Hsenhsua X.H.Jin, Schuit. & W.T.Jin to accommodate Ponerorchis chrysea (W.W.Sm.) Sóo. Tang et al. analysed the Amitostigma/Ponerorchis/Hemipilia alliance using a wider sampling of this group than previous studies and concluded that still broader generic limits in this alliance were appropriate. They combined Ponerorchis, Hemipilia and related genera into a single genus, applying the oldest available name, Hemipilia.
These results, together with the above-mentioned studies on particular genera of Orchidinae, have greatly improved our understanding of phylogenetics of the subtribe, although some controversy remains in the Ponerorchis/Hemipilia alliance, as we will demonstrate. With about 835 species [1], Habenaria is one of the largest genera in the Orchidaceae; it is widespread throughout the tropical and subtropical regions of the world [2]. Currently, it undoubtedly presents the biggest issue in Orchidinae phylogenetics [1]. Generic delimitation and infrageneric classification of Habenaria have been much debated [2, 28–30]. Recent molecular studies found Habenaria to be para- or polyphyletic [9, 19, 23, 25, 26]. The most extensive phylogenetic analysis of Habenaria published so far, Batista et al. [9], focused on the South American representatives and found these to form a clade nested within a larger group of African taxa.
In the present study, phylogenetic relationships were inferred using seven DNA markers (five plastid and two nuclear regions) based on a much broader sampling (approximately 52 genera and 400 species) across Orchidinae, especially for those undersampled or unrepresented in previous studies, with the aims of: (1) reconstructing the phylogenetic interrelationships within Orchidinae; and (2) rationalizing generic delimitation within Orchidinae in accordance with a phylogenetic framework. Of the genera recognized as members of Orchidinae [1], we were unable to obtain material of the following: Bartholinia, Centrostigma, Dracomonticola, Holothrix, Megalorchis, Neobolusia, Oligophyton, Platycoryne, Roeperocharis, Silvorchis, Thulinia and Veyretella. Shizhenia was proposed after publication of Chase et al. [1], and we have included it.
Results
Sequences and alignment
In total, 2474 sequences were included, of which 1294 were newly produced for this paper. The number of missing sequences in the combined matrix of all markers is 767. The characteristics of maximum parsimony analyses of each nuclear gene, combined nuclear genes, combined plastid genes and combined nuclear and plastid datasets are summarized in Additional file 6: Table S4.
Phylogenetic inference
Based on the combined nuclear nrITS, Xdh and plastid DNA data, our findings are consistent in overall topology with the trees produced using ML, MP and BI methods, except for a few of the terminal nodes.
Phylogeny inferred from plastid and nuclear data
The maximum parsimony analysis of the plastid data produced 375 MPTs with a length of 8819 steps, consistency index (CI) of 0.44, and retention index (RI) of 0.82. The strict consensus tree shows that most of the ingroup clades form a polytomy with Satyrium sister to the rest but with low support (Additional file 1: Figure S1). On the other hand, the ML tree and BI trees inferred from plastid data are highly resolved (Additional file 1: Figure S1). Nodes of the trees with BSML/ BSMP > 50 are considered weakly supported, > 70 as moderately supported, and > 80/85 and Bayesian inference (BI) 0.95 as strongly supported.
The maximum parsimony analysis of nrITS (including nrITS of the conflicting species) retrieved 75 MPTs of 7352 steps, consistency index of 0.20, and retention index of 0.79. The strict consensus tree (Additional file 2: Figure S2) includes ten major clades, the relationships of which are not well resolved: (1) 110 species of the Platanthera-Galearis-Dactylorhiza-Gymnadenia-Orchis-Ophrys-Himantoglossum alliance (about 20 genera) with high support (BSML = 100, BSMP = 100, PP = 1.0; in this same order in the following results and discussion); (2) 65 species of Ponerorchis-Hemipilia-Tsaiorchis-Shizhenia-Sirindhornia (five genera) with moderate to weak support (69, 55, 0.74), within which six species (including Ponerorchis graminifolia) are sister to Hemipilia plus the remaining species of Ponerorchis; (3) four species of Brachycorythis with high support (100, 100, 1.0); (4) 100 species of the Herminium-Habenaria (p.p.) alliance (often considered to be four genera) with high to moderate support (92, 77, 0.99); (5) 19 species of Peristylus with high support (100, 100, 1.00); (6) a clade of 26 species Habenaria (including the type) with high support (100, 100, 1); (7) seven species of Bonatea-Habenaria (p.p.) with high support (100, 100, 1); (8) nine species of Habenaria (p.p.) with high support (100, 100, 1); (9) two species of Gennaria (100, 100, 1); (10) six species of Satyrium (100, 100, 1). The ML and BI nrITS trees are a better resolved than that from MP (Additional file 2: Figure S2).
The maximum parsimony analysis of nrITS (excluding the species that, exhibit topological incongruence between data sets) retrieved 30 MPTs with a tree length of 7241 steps, CI = 0.20, and RI = 0.79. The strict consensus tree (Additional file 3: Figure S3) shows a similar topology as Additional file 2: Figure S2, with the exception of the six species excluded due to their incongruent positions.
The maximum parsimony analysis of nrITS + Xdh (excluding nrITS of incongruent species) retrieved 240 MPTs with a score of 9082, CI = 0.26, and (RI) = 0.80. The strict consensus tree (Additional file 4: Figure S4) shows a similar topology (Additional file 2: Figure S2) except for the position of the six incongruent species. These six species are here nested within the clade formed by Ponerorchis and Hemipilia (Additional file 4: Figure S4).
Trees inferred from combined molecular data
The MP analysis retrieved 120 MPTs with a tree length of 18,051, CI = 0.34, and RI = 0.81. The strict consensus tree (Figs. 2, 3) shows that Orchidinae can be divided into 32 strongly or moderately supported clades along the backbone. Inter-relationships among most of these clades are well resolved. The ML tree inferred from the combined data has a similar topology to the MP strict consensus tree (Figs. 2, 3; Additional file 4: Figure S4). The BI topology from the combined dataset is presented as the tree for the discussion of phylogenetic relationships (Figs. 2, 3) because it is better resolved and supported.
Fig. 2.

Tree of Orchidinae s.l. from Bayesian inference based on the combined nuclear ITS (excluding ITS of six conflict species), Xdh, and five plastid markers. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. A dash (−) indicates support at a node < 50%, Asterisk (*) indicates BS = 100 or PP = 1.0
Fig. 3.

Continuation of the tree in Fig. 2
Clade I comprises 74 species of Platanthera s.l. with high or weak support (55, < 50, 0.98; Fig. 2). Clade II consists of nine species of Galearis s.l. (in which we including three monotypic genera formerly recognized, Aceratorchis, Amerorchis Hultén and Neolindleya) with high to weak support (67, < 50, 0.98; Fig. 2). Clade III includes two species of Pseudorchis Ség. with high to weak support (81, 65, 1) (Fig. 2). Clade IV consists of 11 species of Dactylorhiza (90, 89, 1; Fig. 2). Clade V includes ten species of Gymnadenia R.Br., and, like its sister clade IV, is strongly supported (95, 96, 0.99). It is subdivided into two weakly support subclades, one of which corresponds to the formerly recognized genus Nigritella Rich. The sister group relationship of clades IV and V is highly to moderately supported (73, 59, 0.99) (Fig. 2). Clade VI consists of two species of Neotinea Rchb.f. sister to the group formed by clades I–V with weak support < 50, < 50, 0.51,) (Fig. 2). Strongly supported clade VII consists of eight species of Anacamptis Rich., and is sister with strong support to the likewise strongly supported clade VIII made up of two species of Serapias (99, 100, 1). Clade IX consists of two species Himantoglossum (L.) W.D.J.Koch with strong support (100, 100, 1); it is sister to the group formed by clades VII and VIII with strong support. Clade X (Ophrys) and clade XI (Steveniella Schltr.) are sister groups with strong (ML + BI) or weak (MP) support (93, 1, 68). Clade XII consists of seven species of Orchis s.s. with strong support (95, 96, 1), subdivided into two well supported subclades. Clade XIII is formed by Chamorchis Rich. and Traunsteinera Rchb. with weak to strong support (64, 50, 1). These 13 clades form the moderately supported superclade I (86, 72, 1).
Clade XIV includes 44 species of Ponerorchis s.l. with strong to weak support (91, < 50, 1) and is subdivided into two moderately to strongly supported subclades: subclade I includes Neottianthe, Ponerorchis s.s. and some species of Amitostigma (79, 67, 1), including the type species of all three genera; subclade II includes most of the species formerly assigned to Amitostigma (64, < 50, 0.99) (Fig. 2). Six taxa from Japan, i.e. Amitostigma keiskei (Finet) Schltr., A. kinoshitae (Makino) Schltr., A. lepidum (Rchb.f.) Schltr., Ponerorchis chidori (Makino) Ohwi, P. graminifolia Rchb.f., and P. [graminifolia var.] suzukiana (Ohwi) J.M.H.Shaw, formed a strongly supported clade in the nrITS phylogram, sister to the clade formed by Hemipilia + Amitostigma + Ponerorchis + Tsaiorchis. However, these same species have widely different phylogenetic positions in the Xdh + plastid trees (Additional file 4: Figure S4). Our analyses suggest that this hard incongruence needs more sampling and research to explain (see discussion below).
Clade XV is also strongly supported and consists only of Hemipilia, including a few species previously assigned to Amitostigma and Ponerorchis (100, 99, 1). Clade XVI includes three species of Tsaiorchis Tang & F.T.Wang with strong support (100, 100, 1). Clade XVII only includes the recently established monotypic genus Shizhenia X.H.Jin, L.Q.Huang, W.T.Jin & X.G.Xiang (Fig. 1a), which is sister to the moderately to strongly supported clades XIV–XVI (83, 71, 1). Clade XVIII only includes Sirindhornia H.A.Pedersen & Suksathan. These last five clades (XIV–XVIII) form the strongly supported superclade II (99, 88, 1). Clade XIX (just Brachycorythis) was strongly supported as sister to superclades I + II with moderate to strong support (70, 66, 1), whereas clade XX, consisting of Schizochilus Sond., is sister to clades I–XIX with weak support (72, < 50, 0.6).
Clades I–XX and the following clades XXI–XXXI are two major, but highly supported sister groups (93, 81, 1), corresponding to the split between Orchidinae s.s. and Habenariinae in other classifications (e.g. [3, 24]).
The weakly supported clade XXI (< 50, 52, 0.81) consists of Herminium s.l., subdivided into six subclades that have moderate to strong support. Clade XXII includes two samples of Hsenhsua (formerly in Ponerorchis) sister to Herminium. These two clades form superclade III with high support (95, 91, 1).
Clade XXIII includes six species of Pecteilis Raf. and 45 species of Habenaria with strong support (100, 99, 1), subdivided into two well-supported groups: (i) four species from tropical Asia (H. longicorniculata–H. rhodocheila) and (ii) species from both tropical and subtropical Asia (Pecteilis and species of Habenaria). Habenaria delavayi Finet is sister to a strongly to moderately supported (97, 55, 1) group of 20 species, within which Pecteilis is deeply nested. Three yellow-flowered species, including H. linguella Lindl., are sister to the white-flowered H. marginata Colebr. Clade XXIV, consisting entirely of Habenaria species, includes a poorly resolved group formed by ten species. Habenaria josephi Rchb.f. is sister to the remainder of this group. Sister clades XXIII and XXIV with strong support (90, 90, 1) form superclade IV.
Strongly supported (100, 99, 1.00) clade XXV is formed by 19 species of Peristylus. Clade XXV (= superclade V) is sister to superclades III + IV with weak to strong support (62, 50, 0.96). Strongly supported clade XXVI (100, 100, 1) consists of two samples of Diplomeris D. Don, which are sister to superclades III–V, with weak support (< 50, < 50, 0.63).
Clade XXVII consists of Bonatea Willd. and 40 species of Habenaria, including H. macroceratitis Willd. (the type of the genus) with strong support (91, 79, 1). It is subdivided into three well-supported groups (Fig. 3). Subclade I is the core Habenaria group comprising Habenaria rolfeana–H. lithophila (99, 97, 1). This group, which includes H. macroceratitis, is characterized by bilobed petals (except for H. petitiana (A.Rich.) T. Durand & Schinz), consists of species from tropical Africa, Asia and South America. Habenaria rolfeana–H. tridens are entirely American, and no other Habenaria species in our dataset are from the Americas. Subclade II comprises H. limprichtii–H. keniensis (100, 96, 1). This is a group of (sub)tropical to subalpine African and Asian species, which is characterized by the pectinate lateral lobes of the lip, and entire petals (Fig. 1d). Weakly to well- supported (76, 63, 0.95) Subclade III includes Bonatea and some African species of Habenaria. Clade XXVII forms superclade VI.
Clade XXVIII includes Benthamia Rich. (one species) and Tylostigma Schltr. (one species) with strong support (100, 100, 1). Clade XXIX includes Gennaria Parl. (two species) with strong support (100, 100, 1). The last three clades form a polytomy in all three analyses.
Clade XXX includes Cynorkis Thouars (three species) and Physoceras Schltr. (one species) with strong support (99, 99, 1). Clade XXXI includes two species of Stenoglottis Lindl. with strong support (89, 86, 1). Clades XXX and XXXI are resolved as successive sisters to the group formed by superclades III–VI and clades XXVIII and XXIX. Clade XXXII includes six species of Satyrium with strong support (100, 99, 1) and is sister to the rest of Orchidinae with strong support (99, 97, 1).
Discussion
Phylogenetics and distribution of Orchidinae s.l.
With broad sampling across Orchidinae and seven DNA markers, many new or previous overlooked phylogenetic relationships were discovered in our analyses. Orchidinae s.l. (comprising Orchidinae s.s., Habenariinae and Satyriinae of other classifications, e.g. [26]) are monophyletic with strong support and divided into three main clades: Satyrium (Africa and Asia), Orchidinae s.s. (mainly Northern Hemisphere), and the formerly recognised Habenariinae’ (Fig. 3; nearly cosmopolitan). Satyrium is sister to the rest of Orchidinae s.l. with strong support.
Orchidinae s.s. include two superclades and two smaller clades sister to these. Superclade I has high to moderate support and includes 12 sampled genera: Anacamptis, Chamorchis, Dactylorhiza, Galearis, Gymnadenia, Himantoglossum, Neotinea, Ophrys, Orchis s.s., Platanthera, Steveniella, and Traunsteinera. Most of these genera with ovoid/ellipsoid/globose tubers have their center of diversity in Europe and the Mediterranean region, but the two genera with palmately lobed tubers, i.e., Dactylorhiza and Gymnadenia, extend into the Far East (Dactylorhiza into North America), whereas two genera with rhizomes or fleshy rootstocks, i.e., Galearis and Platanthera, have two centers of diversity, the Pan-Himalayan region and North America.
Superclade II (97, 88, 1), according to our delimitation (discussed below), includes five genera: Hemipilia, Ponerorchis, Shizhenia, Sirindhornia, and Tsaiorchis. This is a subtropical to temperate clade with ovoid/ellipsoid/globose tubers and basal leaf (leaves) occurring almost exclusively in eastern Asia and the eastern Himalayas, with a single species (Ponerorchis cucullata (L.) X.H. Jin, Schuit. & W.T.Jin) occurring as far west as Poland. In the subtropical zone the members of this superclade is normally found above 1000 m asl., often in alpine regions [4, 31].
Two relatively isolated genera, Schizochilus and Brachycorythis, were resolved as successive sister to these two superclades with weak to strong support (Fig. 2). Schizochilus is exclusively from eastern to southern Africa, whereas Brachycorythis is most diverse in tropical and southern Africa, but also has some species in tropical Asia as far east as Taiwan. Unlike many other Orchidinae, Brachycorythis does not occur at alpine elevations in Asia [32]; it is essentially a tropical montane genus.
The clade formerly referred to as Habenariinae (Clades XXI–XXXI) include four superclades (III–VI) and four isolated clades. Chase et al. (2003; 2015) decided not to recognise Habenariinae because of uncertainty over phylogenetic placements of genera such as Androcorys (synonym of Herminium, see Raskoti (2016)), Brachycorythis and Holothrix, and until all of the missing genera of Orchidinae s.l. are included, it would be premature to resurrect Habenariinae and not necessary regardless of the final phylogenetic outcome. Superclade III comprises the genera Herminium and Hsenhsua and is exclusively Asian, except for a single taxon that extends into Europe (H. monorchis (L.) R.Br. ex Aiton). Most are alpine plants, including the species that probably holds the elevational record in Orchidaceae (H. pugioniforme Lindl. ex Hook.f. at c. 5200 m). Superclade IV includes Pecteilis (often included in Habenaria) and about 50 species of Habenaria mainly distributed in tropical Asia. Superclade V includes only one genus, Peristylus, sister to superclades III + IV with weak support. Peristylus is only found in tropical Asia, from the lowlands to the alpine zone. The small tropical Asian genus Diplomeris is sister to these three superclades with weak support. Superclade VI includes Bonatea and 42 species of Habenaria; this superclade is distributed in the Americas, tropical and southern Africa, and tropical Asia. A number of small clades are well supported, but their relationships with other Habenariinae are still largely unresolved. Benthamia and Tylostylis are two sister genera from Madagascar, comprising clade XXVIII, whereas clade XXIX contains the two species of Gennaria, one from the Canary Islands and the other from the Himalayan region, a remarkable disjunction. Clade XXX contains two genera from Madagascar and the Mascarenes, Cynorkis (also in southern Africa) and Physoceras and is resolved as sister to former Habenariinae with strong support. The African genus Stenoglottis, often included in Habenarrinae, is sister to the remainder of Habenariinae, but with weak support.
Phylogeny and generic delimitation
Usually there are many ways to translate a phylogenetic tree into a Linnean classification. In the interest of nomenclatural stability it is desirable that a classification is broadly accepted. One way to help ensure such acceptance is by using objective criteria for generic delimitation as far as possible. Bateman, as cited in Tang et al. (2015), proposed the following five guidelines:
Recognize only monophyletic groups (clades);
Preferentially divide the tree at branches that are relatively robust;
Preferentially divide the tree at branches that receive similar levels of statistical support (obviously, there exists considerable overlap between Rules 2 and 3);
Minimize the proportion of branches in the tree that represent more than one taxonomic rank (notably monotypic higher taxa);
Preferentially divide the tree in a way that minimizes the need (a) to create new names and/or (b) to create new combinations of existing names.
Although we would agree to a large extent with the utility of these guidelines (always allowing for exceptions), we think that one consideration is missing: there is no reference to morphology. We would suggest that a classification supported by morphology is likely to gain acceptance more readily than one based solely on molecular criteria. Therefore, we propose the following addition:
-
(6)
Preferentially divide the tree in such a way that the points of division correspond to morphological discontinuities (in particular those representing evolutionary innovations).
Ultimately, we are mainly interested in the topology of our phylogenetic tree. Measures of internal support inform us of clarity in the phylogenetic signal of the data analysed. Morphological criteria should also be applied; they not only help us reduce the large number of potential classification schemes based on a given tree, but also make the resulting classification of greater practical value, and thereby easier to accept. However, without a formal cladistics analysis of morphological data, such applications of morphological criteria are ad hoc and likely to be subjective and ignore conflicting characters while emphasising those preferred by one set of authors. Below we discuss a number of troublesome alliances in which consensus has not yet been reached, as well as others in which we are able to present a consensus agreed by most authors.
Bonatea, Diplomeris, Habenaria, Herminium, Pecteilis, and Peristylus
Morphologically, Diplomeris is an isolated genus in Orchidinae, characterized by its large, white flowers with an entire lip, a column with well developed, semi-circular connective about two thirds the length of the column, two elongate, oblong stigmatic lobes extending from near the entrance of the spur, and a large, shield-like midlobe of the rostellum. Ecologically, it is unusual in Orchidinae in that the species grow as lithophytes on cliffs with seasonal running water trickling down the moss-covered rock. This genus of two species is mainly distributed in tropical to subtropical regions in the Himalayas [2, 4, 33] and would be difficult to place without molecular data. It is somewhat unexpected that Diplomeris is sister to the group formed by superclades III–V, but support for this position is as yet weak only.
Peristylus consists of about 70 species mainly distributed in tropical Asia [2, 34]. It is not always easy to distinguish from Peristylus from Habenaria and Herminium [2, 10, 23, 33–35]. Recent studies have demonstrated that most alpine species of Peristylus actually belong to Herminium [10, 23]. With the exclusion of these species, Peristylus is distributed mainly in the lowland and montane zones of tropical and subtropical Asia. Our results indicate that, excluding the misplaced species, Peristylus is monophyletic and characterized by its elliptic tuber, more or less clearly trilobed and spurred lip, narrow connective, and two stigma lobes adnate to the entrance of the spur. The species of Peristylus analysed are distributed over four strongly supported clades: (1) P. chapaensis in subtropical limestone regions; (2) a tropical group, including P. stocksii (Hook.f.) Kraenzl.; (3) a subtropical montane subclade including P. affinis; (4) another subtropical montane subclade including P. densus (Lindl.) Santapau & Kapadia. Each subclade is to a large extent characterized by minor morphological characters, especially of the spur and lip.
Pecteilis consists of about eight species distributed in southern and eastern Asia and the Malay Archipelago [4, 30, 31]. Morphologically, it is similar to Habenaria, but differs in its concave, sessile stigma lobes, unlike the stalked stigmaphores of Habenaria [2, 30]. We find that Pecteilis is not monophyletic and deeply nested within clade XXIII (otherwise containing species currently accepted as members of Habenaria by most authors), and forms a strongly supported subclade interspersed with species of Habenaria. It seems that the stigmaphore has been lost independently at least twice in this subclade. Except for the stigmaphore, several members of Habenaria of clade XXIII share morphological characters with Pecteilis, such as an anther with a broad connective, thecae extending from base of column, and large, subrhombic to flabellate lateral lobes of the lip (Fig. 1h). On the other hand, many other species have linear to filiform lateral lobes, such as H. plurifoliata Tang & F.T.Wang, and H. propinquior Rchb.f. Species in clade XXIV, which most recent authors include in Habenaria, are mostly from alpine habitats in Asia (especially in Himalayan region), and are characterized by two basal, almost opposite leaves appressed to the ground, lateral lobes of the lip linear to filiform, green to white flowers, and petals often more or less distinctly bilobed. Some species in clade XXIII also have leaves appressed to the ground, such as H. roxburghii D.H.Nicolson and H. delavayi (the latter also with linear lateral lobes to the lip, but usually with three or more leaves in a rosette). On the other hand, the petal lobes in clade XXIV are sometimes indistinct, making the two clades ultimately difficult to tell apart. Together they form superclade IV.
Our results indicate that Habenaria is polyphyletic but with low support; it is divided into two well supported but not immediately related groups, superclades IV and VI. Superclade IV, discussed above under Pecteilis, is exclusively Asian, and is sister to superclade III (Herminium and Hsenhsua). A sister-group relationship of superclade VI with any other clade in Habenariinae (Fig. 2) has not been established, since superclade VI consisting of all African and southern American taxa sampled so far as well as some Asian ones is part of a polytomy.
There are at least two divergent options to treat the polyphyly of Habenaria. One option would be to expand the delimitation of Habenaria to include Benthamia, Bonatea, Diplomeris, Gennaria, Herminium, Hsenhsua, Pecteilis, and Tylostigma (Clades XXI–XXIX), many of which have been in the past considered members of Habenaria (e.g., many species of Benthamia, Bonatea, Gennaria, Herminium, Pecteilis, Tyolostigma have combinations in Habenaria). Herminium would be the earliest available name for this assemblage. Therefore, ironically, an attempt to keep all the species of Habenaria within a single genus would lead to the loss of the name Habenaria, unless it was conserved. This option would cause minimal nomenclatural upheaval, and the resulting genus could be diagnosed by the characters used to delimit the formerly recognised Habenariinae. A perhaps more disruptive option would be to split Habenaria into at least two genera, a decision that would likely result in no agreement over the number of segregate genera and how to recognise them.
The main problem with recognising two genera, the two larger clades into species of Habenaria fall, is that there appear to be no consistent morphological differences between the two superclades. However, perhaps it is possible to circumscribe the two groups in a more circuitous way, based on polythetic sets. For example, bilobed petals are, as far as we can tell, only found within certain subclades (in both superclades, unfortunately); similarly, pectinate or laciniate lateral lobes of the lip (Fig. 1d) only occur in some branches of our tree. When the group has been more fully studied it is perhaps possible to identify the two superclades using multiple sets of character states that are unique to each of the two.
Splitting Habenaria into more than two genera is another option to be considered. We note, however, that many character states show much homoplasy, and suspect that a more finely split classification will not be easier to use. Instead of having two difficult-to-define genera, we may end up with five or more difficult-to-define genera. This would only increase the potential for misidentifications, without having any obvious benefits, but some authors D. Szlachetko and collaborations have already started down this route, describing new genera based on only morphological characters, mostly column details, which typically results in polyphyletic circumscriptions (such as [36–38]). At this stage it would be premature to propose new combinations.
Brachycorythis, Hemipilia, Ponerorchis, Tsaiorchis, Sirindhornia (superclade II)
Brachycorythis, consisting of 30–35 species, is mainly distributed in Africa, with a few species in Asia. It is characterized by leaf-like floral bracts and a more or less bipartite, spurred lip. Recent molecular studies indicate that Brachycorythis is sister to the rest of Orchidinae s.s. [23, 25, 26], and that Sirindhornia is sister to the clade formed by Ponerorchis, Hemipilia and related genera [23]. Both findings are also supported by our results, in which Sirindhornia is sister to clades XIV–XVII. Tang et al. [24] inferred (but with only moderate support) that Brachycorythis is sister to Hemipilia s.l. (in which these authors include Ponerorchis, Amitostigma, Neottianthe and Tsaiorchis), and that Sirindhornia is sister to the remaining Orchidinae s.s. We have been unable to replicate this result in our phylogenetic analyses. Morphologically, Brachycorythis is less similar morphologically to the Hemipilia alliance than Sirindhornia.
Tang et al. [24] detected a hard incongruence involving a group of six taxa (Ponerorchis graminifolia, P. [graminifolia var.] suzukiana, P. chidori, Amitostigma lepidum, A. keiskei, and A. kinoshitae) between nrITS and four plastid markers and stated that this most likely reflected plastid capture. They concluded that their ITS results were more likely to reflect the actual phylogenetic relationships. Our results indicate that, based on plastid markers, these six taxa, which are endemic to Japan, are deeply nested within clade XIV, but in different positions in the tree. Amitostigma keiskei and A. kinoshitae are sister to the subclade formed of Ponerorchis alpestris–P. physoceras (Fig. 2). Amitostigma lepidum is sister to a subclade formed exclusively of Amitostigma (s.l.) species: A. parciflora–A.trifurcata. The remaining three species (including Ponerorchis graminifolia) are sister to the subclade formed of Neottianthe species + Amitostigma gracile (Blume) Schltr. Based on nrITS alone, according to the results of Tang et al. [24] and our analyses of nrITS (Additional file 2: Figure S2), these six species form a strongly supported clade sister to our clades XIV–XVI. Since they include the type species of Ponerorchis (P. graminifolia), this would imply, as Tang et al. [24] correctly pointed out, that our clade XIV could not be called Ponerorchis, as it would not include the type species of Ponerorchis.
Morphologically, these six species do not form a uniform group, and the scattered placement of its members in the plastid tree more accurately reflects their diverse morphology than their occurrence in a single clade. We consider it unlikely that six taxa that are all endemic to Japan and morphologically disparate, happen to form a single clade of which the members have somehow integrated the plastid genomes of at least three different, now extinct progenitors that are closely related to geographically isolated, extant Chinese taxa.
Based on our results from nuclear markers (ITS +Xdh) we infer that the six taxa are nested within a polytomy of Ponerorchis s.l., which is sister to Hemipilia + Tsaiorchis with moderate support (BSML = 78, BSMP = 67, PP = 0.72) (Additional file 4: Figure S4). They do not form a clade, and their placement does not render Ponerorchis in the sense of Jin et al. [23] poly- or paraphyletic.
It remains an interesting problem to explain how ITS data could produce such discordant results in the case of these six taxa. It is well known that certain molecular genetic processes involving ITS, such as ancient or recent array duplication events, genomic harbouring of pseudogenes, and incomplete intra- or inter-array homogenization, may distort results of phylogenetic inference and need to be examined [39]. Whatever the cause for the incongruence, on morphological grounds and the plastid and combined nuclear molecular results, we prefer to continue to recognize Hemipilia, Ponerorchis and Tsaiorchis as distinct genera, while adding Shizhenia, which is sister to the three others combined.
Platanthera
Although most species-rich in Asia, Platanthera is also diverse in North America, and a few segregate genera have been recognized there (e.g. Blephariglottis Raf., Limnorchis Rydb., Piperia Rydb., and others). In a recent revision of the Asian taxa [20], the genus was taken in the broad sense, which corresponds with our clade I.
Platanthera is among the largest genera of the tribe Orchideae, consisting of some 130 species, and is mainly distributed in Asia and North America, with a few species in Europe and North Africa [2, 19]. Historically, the generic delimitation and phylogenetics of Platanthera were confusing [19, 21, 28, 40]. Recent results indicate that once a few misplaced species are transferred to the proper genera, Platanthera s.l. is monophyletic and relatively well characterized by morphology, such as fleshy, more or less tapering roots, often forming fusiform tuberoids, a spurred lip, a short and truncate column, and a broad connective [19, 23, 41] (Fig. 1c).
Efimov [20] recognized five subgenera, which were recovered in our tree. The Platanthera minor group comprises about eight species distributed in subtropical regions in East Asia; it is characterized by several cauline leaves attenuating into bracts, a dense inflorescence, and a narrowly elliptic lip narrowed at the base. This group in subgenus Platanthera could not be assigned to a section by Efimov [20]. The Platanthera minutiflora group displays disjunction between East Asia and North America. It was recognized by Efimov [20] as subgen. Platanthera sect. Lysiella (Rydberg) Efimov. The Platanthera carnosilabris group (99, 100, 1) in subgen. Platanthera includes about eight species that are characterized by small flowers, a lip more or less concave at base, and spur shorter than the lip. Efimov [20] could not place these in a section. This group includes the misplaced Herminium orbiculare Hook.f. Moderate support (75, 75, 0.97) is shown for subgen. Platanthera sect. Stigmatosae (Lang) Efimov (P. bakeriana–P. leptocaulon).
Stenoglottis, Schizochilus
Both Stenoglottis and Schizochilus are small genera endemic to southern Africa. The phylogenetic positions of Stenoglottis and Schizochilus have not been fully established in previous studies, and are still uncertain. Bateman et al. [26] resolved Stenoglottis as sister to Habenariinae s.s. Inda et al. [25], on the other hand, found Stenoglottis to be sister to the rest of their tribe Orchideae (our Orchidinae s.l.). Our own results show weak support for the position inferred by Bateman et al. (< 50, 60, 0.64).
Schizochilus is here found to be sister to Orchidinae s.s., but with weak support (72, < 50, 0.6). Morphologically, Schizochilus is similar to Brachycorythis and Sirindhornia, especially in column morphology.
Satyrium (Clade XXXII)
Satyrium comprises about 90 species and is mainly distributed in temperate and montane regions of Sub-Saharan Africa, with a few species occurring in Asia, mainly in the Himalayan region [2]. Based on its distinctive morphology, including non-resupinate flowers, a galeate lip with two spurs, stalked column, and pad-like stigmatic lobes, which sets it apart from other Orchidinae, Satyrium has been treated as a monotypic subtribe Satyriinae [2, 42]. Published molecular studies indicate that Satyrium is monophyletic and sister to Orchidinae s.l. [25, 26, 43, 44], which is also strongly supported by our results 99, 97, 1; Fig. 2). Chase et al. [1] assigned Satyrium to Orchidinae, which was motivated especially by the desire to minimize the number of monogeneric higher taxa. This is also in agreement with guideline 4 above (chapter 4.2). Considering the distinctive morphology of Satyrium, a case could be made to reinstate Satyriinae, but similar arguments could be advanced for many other subtribes. We would prefer not to increase the complexity of our classification if this can be avoided. Limiting the number of monogeneric higher taxa as far as possible is a means towards this end. In addition, it would appear from the work of Inda et al. [25] that the genus Holothrix is sister to Satyrium + the remainder of Orchidinae s.l. Therefore, recognition of a subtribe Satyriinae would also require the recognition of another subtribe for Holothrix (which might include other, related genera that have not been analysed yet). It is certainly simpler to keep them all within Orchidinae.
Gennaria (Clade XXIX)
There are two species in Gennaria, the type species G. diphylla Parl. in the Mediterranean region plus the Canary Islands, the other (the recently transferred G. griffthii (Hook.f.) X.H.Jin., L.Q.Quang, W.T.Jin & X.G.Xiang, Fig. 1e) in the Pan-Himalayan region [2, 45]. Morphologically, Gennaria is characterized by subcampanulate flowers more or less secund along the rachis, a short column with stalked lateral appendages, and convex stigmas [2, 46]. Inda et al. [25] found Gennaria to be deeply nested within Habenaria. Batista et al. [9] had Gennaria as sister to Habenaria + Bonatea, but with low support, which is similar to the analysis of Jin et al. (2014). Our new results show that Gennaria is a strongly supported clade that does not nest within Habenaria. Instead, it is one of the branches of a polytomy formed otherwise by superclade VI, Clade 26, and Clade 25. It may be an outlier in this alliance, which is also suggested by the extremely wide but fragmented distribution area of the clade, with a low number of extant species.
Conclusions
With increased understanding of the phylogenetics of Orchidinae s.l. a stable classification system of generic level is close to being reached. The main outstanding problem is the apparent polyphyly of the largest genus in this alliance, Habenaria. Another area of contention is in the Ponerorchis-Hemipilia alliance, where a hard conflict exists between results from ITS and those from plastid + Xdh analyses. Satyrium is best included within a broader Orchidinae rather than in its own monotypic subtribe Satyriinae, especially since its exclusion would probably require that a separate subtribe is also needed for Holothrix. Future studies will undoubtedly shed light on the geographical origin of the various clades and the timing of their divergence.
Methods
Taxon sampling, DNA extraction, amplification and sequencing
We sampled 436 accessions representing approximately 400 species in 52 genera of Orchidinae (Additional file 5: Table S1). Five species from tribes Cranichideae and Diurideae were used as outgroups based on previous results [1, 47, 48].
Total genomic DNA was isolated from silica-gel-dried tissue using a Plant Genomic DNA Kit (Tiangen Biotech, LTD, Beijing, China) following the manufacturer’s protocols and a modified CTAB method [49]. For this study, seven markers including two nuclear markers (nrITS and Xdh) and five plastid markers (matK, psaB, psbA-trnH, rbcL, and trnL-F) were used. Amplification of all DNA regions was performed in 25 μL reactions containing 10–50 ng DNA, 12.5 μL 2× Taq PCR Master Mix (Biomed, Beijing, China), 0.4 μM of each primer, and ddH2O to reach a total volume of 25 μL. The PCR profiles used in the current study are presented in Additional file 6: Table S2. The sequencing reactions were performed with the ABI Prism Bigdye Terminator Cycle Sequencing Kit (Applied Biosystems, ABI). Amplification and sequencing primers are listed in Additional file 6: Table S3.
Phylogenetic analyses
Sequences were edited independently and assembled using the ContigExpress program in Vector NTI Suite v. 6.0 (Informax, North Bethesda, Maryland, USA). The edited sequences were aligned using MAFFT v. 7.221 [50] with auto strategy and default settings. The aligned sequences were then manually adjusted using BioEdit v. 7.25 [51]. Ambiguously aligned characters in the psbA-trnH and trnL-F datasets were excluded before phylogenetic analyses.
Congruence between nuclear (nrITS and Xdh) and plastid datasets (matK, psaB, psbA-trnH, rbcL, and trnL-F) was tested using the incongruence length difference (ILD) [52] implemented in PAUP v4.0b10 [53]. The ILD test used an analysis of 100 replicates, with random addition sequence, TBR swapping, and no more than 15 trees of score (length) greater than or equal to 5 were saved in each replicate. The P-value was 0.01 in this study, and regarded as significant incongruence following Cunningham (1997). In the present study, the incongruence between data sets was also assessed by a comparing the topologies and support of the trees produced by the data partitions. The thresholds of hard incongruence here followed previous work [54]: (1) ILD P < 0.01; (2) bootstrap percentages ≥ 80 and/or posterior probabilities (PP) ≥ 95. These tests indicated a hard incongruence between nrITS topologies on the one hand and plastid, Xdh, plastid + Xdh topologies on the other. When the nrITS dataset of these six conflicting taxa was removed from the combined matrix, the results (see results) did not indicate hard incongruence. Therefore, we combined the nuclear and plastid datasets in SequenceMatrix v. 1.7.8 [55] to perform further phylogenetic analyses, but with the nrITS dataset of these six taxa excluded from the combined matrix.
Phylogenetic analyses were performed for each matrix using maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI). ML inference relied on RAxML-HPC2 v. 8.2.4 [56] on XSEDE via the CIPRES Science Gateway online web server [57]. The ML analysis used the GTRCAT nucleotide substitution model following other default settings and ran 1000 rapid bootstrap replicates to obtain the best-scoring ML tree with support percentages. The MP analyses were performed in PAUP v. 4.0b10 (Swofford, 2002). All characters were equally weighted, and gaps were coded as missing data. Heuristic searches of 1000 random addition replicates with tree bisection reconnection (TBR) branch swapping were conducted to obtain the most parsimonious trees; 10 trees were held in each iteration step of the TBR-swapping procedure. To evaluate the node support, bootstrap analyses [58] were performed using 500 replicates with 10 random taxon additions and heuristic search. For BI analysis, posterior probabilities (PP) for individual clades were inferred with MrBayes v. 3.2.3 [59] on XSEDE via the CIPRES Science Gateway online web server [57]. The best-fitting DNA substitution model for each matrix was determined using MrModeltest v. 2.3 [60] under the Akaike information criterion (AIC). Two separate four-Markov-chain Monte Carlo (MCMC) analyses were performed, starting with a random tree, proceeding for 10,000,000 generations, and sampling every 1000 generations. The tree or posterior probabilities was constructed after discarding the burn-in phase samples (the first 25% of sampled trees).
Additional files
Tree of Orchidinae s.l. from Bayesian inference based on plastid markers. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. The dash (−) indicates support at a node < 50%, Asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 427 kb)
Tree of Orchidinae s.l. from Bayesian inference based on nrITS, including ITS of six species for which position conflict between the plastid and nrITS results. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. The dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 438 kb)
Tree of Orchidinae s.l. from Bayesian inference based on ITS, excluding nrITS of the six species with incongruent positions. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. A dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 435 kb)
Tree of Orchidinae s.l. from Bayesian inference based on nrITS and Xdh, excluding the nrITS sequences of the six species with incongruent positions. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. A dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 439 kb)
Voucher information and GenBank accession numbers for the sequences analysed in this study, *indicates newly produced sequences for this paper. The dash (−) indicates the missing sequences. (XLSX 46 kb)
The PCR programs used for amplifying the DNA regions. Table S3. List of primers used for PCR and sequencing in this study. Table S4. Statistics from analyses of nuclear and plastid datasets. (DOCX 21 kb)
Acknowledgements
We are sincerely grateful to Forest Department of Yunnan Province and Nature Reserve of Gaoligongshan Mountains for their invaluable assistance.
Funding
This research was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31,470,299, 31,670,194, J1310002), Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Grant No. Y4ZK111B01). The funders played no role in research design, data collection and analysis, preparation of the manuscript, or decision to publish.
Availability of data and materials
All of the phylogenetic sequence data in this study was deposited in GenBank (National Center for Biotechnology Information) under accession numbers: ITS, MF 944259-MF944417; matK, MF945394-MF945550; psaB, MF 944418-MF944626; psbA-trnH: MF 944627–944,832; rbcL, MF944833–944986; trnL-F, MF945188–945393; Xdh, MF944987–945187 (Additional file 5: Table S1). All additional materials supporting the results of the article are included as additional files.
Abbreviations
- AIC
Akaike information criterion
- BI
Bayesian inference
- ILD
Incongruence length difference
- MCMC
Markov-chain Monte Carlo
- ML
Maximum likelihood
- MLBS
Bootstrap values of maximum likelihood
- MP
Maximum parsimony
- MPBS
Bootstrap values of maximum parsimony
- PP
Posterior probabilities
Authors’ contributions
XHJ conceived the study. WTJ performed the experiments and conducted statistical analysis. XHJ and WTJ wrote the manuscript. AS and MWC revised the manuscript greatly. XHJ, JWL, SWC and TCH provided important materials and revised manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
Plant materials used in this study were collected in field with necessary permissions from local authorities of Forest Department. All voucher specimens have been deposited in publicly available herbaria (Additional file 5: Table S1).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1160-x) contains supplementary material, which is available to authorized users.
Contributor Information
Wei-Tao Jin, Email: jinweitao@ibcas.ac.cn.
André Schuiteman, Email: A.Schuiteman@kew.org.
Mark W. Chase, Email: M.Chase@kew.org
Jian-Wu Li, Email: ljw@xtbg.org.cn.
Shih-Wen Chung, Email: biflora@gmail.com.
Tian-Chuan Hsu, Email: lecanorchis@gmail.com.
Xiao-Hua Jin, Phone: 86-10-62836582, Email: xiaohuajin@ibcas.ac.cn.
References
- 1.Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Van den Berg C, Schuiteman A: An updated classification of Orchidaceae. Bot J Linn Soc 2015, 177(2):151-174.
- 2.Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. Genera Orchidacearum. Oxford: Oxford University Press; 2001. [Google Scholar]
- 3.Dressler RL. Phylogeny and classification of the orchid family. Cambridge: Cambridge University Press; 1993. [Google Scholar]
- 4.Chen X-Q, Liu Z-J, Zhu G-H, Lang K-Y, Ji Z-H, Luo Y-B, Jin X-H, Cribb PJ, Wood JJ, Gale SW, et al. Flora of China. Beijing: Science Press; 2009. [Google Scholar]
- 5.Shipunov AB, Fay MF, Pillon Y, Bateman RM, Chase MW. Dactylorhiza (Orchidaceae) in European Russia: combined molecular and morphological analysis. Am J Bot. 2004;91(9):1419–1426. doi: 10.3732/ajb.91.9.1419. [DOI] [PubMed] [Google Scholar]
- 6.Stahlberg D, Hedren M. Systematics and phylogeography of the Dactylorhiza Maculata Complex (Orchidaceae) in Scandinavia: insights from cytological, morphological and molecular data. Plant Syst Evol. 2008;273(1–2):107–132. doi: 10.1007/s00606-008-0035-x. [DOI] [Google Scholar]
- 7.Efimov PG. Sibling species of fragrant orchids (Gymnadenia: Orchidaceae, Magnoliophyta) in Russia. Russ J Genet. 2013;49(3):299–309. doi: 10.1134/S102279541302004X. [DOI] [PubMed] [Google Scholar]
- 8.Bateman RM, Rudall PJ, James KE. Phylogenetic context, generic affinities and evolutionary origin of the enigmatic Balkan orchid Gymnadenia frivaldii Hampe ex Griseb. Taxon. 2006;55(1):107–118. doi: 10.2307/25065532. [DOI] [Google Scholar]
- 9.Batista JAN, Borges KS, de Faria MWF, Proite K, Ramalho AJ, Salazar GA, van den Berg C. Molecular phylogenetics of the species-rich genus Habenaria (Orchidaceae) in the new world based on nuclear and plastid DNA sequences. Mol Phylogenet Evol. 2013;67(1):95–109. doi: 10.1016/j.ympev.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Raskoti BB, Jin W-T, Xiang X-G, Schuiteman A, Li D-Z, Li J-W, Huang W-C, Jin X-H, Huang L-Q. A phylogenetic analysis of molecular and morphological characters of Herminium (Orchidaceae, Orchideae): evolutionary relationships, taxonomy, and patterns of character evolution. Cladistics. 2016;32(2):198–210. doi: 10.1111/cla.12125. [DOI] [PubMed] [Google Scholar]
- 11.Sramko G, Molnar AV, Hawkins JA, Bateman RM. Molecular phylogeny and evolutionary history of the Eurasiatic orchid genus Himantoglossum s.l. (Orchidaceae) Ann Bot. 2014;114(8):1609–1626. doi: 10.1093/aob/mcu179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devey DS, Bateman RM, Fay MF, Hawkins JA. Friends or relatives? Phylogenetics and species delimitation in the controversial european orchid genus Ophrys. Ann Bot. 2008;101(3):385–402. doi: 10.1093/aob/mcm299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devey DS, Bateman RM, Fay MF, Hawkins JA. Genetic structure and systematic relationships within the Ophrys fuciflora aggregate (Orchidaceae: Orchidinae): high diversity in Kent and a wind-induced discontinuity bisecting the Adriatic. Ann Bot. 2009;104(3):483–495. doi: 10.1093/aob/mcp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillon Y, Fay MF, Hedren M, Bateman RM, Devey DS, Shipunov AB, van der Bank M, Chase MW. Evolution and temporal diversification of western European polyploid species complexes in Dactylorhiza (Orchidaceae) Taxon. 2007;56(4):1185–1208. doi: 10.2307/25065911. [DOI] [Google Scholar]
- 15.Aceto S, Caputo P, Cozzolino S, Gaudio L, Moretti A. Phylogeny and evolution of Orchis and allied genera based on ITS DNA variation: morphological gaps and molecular continuity. Mol Phylogenet Evol. 1999;13(1):67–76. doi: 10.1006/mpev.1999.0628. [DOI] [PubMed] [Google Scholar]
- 16.Gamarra R, Ortunez E, Galan Cela P, Guadano V. Anacamptis versus Orchis (Orchidaceae): seed micromorphology and its taxonomic significance. Plant Syst Evol. 2012;298(3):597–607. doi: 10.1007/s00606-011-0569-1. [DOI] [Google Scholar]
- 17.Tyteca D, Ceinos M, Gathoye J-L, Brys R, Jacquemyn H. On the morphological, biological and genetic heterogeneity of the genus Orchis (Orchidaceae, Orchidinae) Phytotaxa. 2012;75:19–32. doi: 10.11646/phytotaxa.75.1.2. [DOI] [Google Scholar]
- 18.Bateman RM, Sramko G, Rudall PJ. Floral miniaturisation and autogamy in boreal-arctic plants are epitomised by Iceland's most frequent orchid, Platanthera Hyperborea. PeerJ. 2015;3 [DOI] [PMC free article] [PubMed]
- 19.Bateman RM, James KE, Luo Y-B, Lauri RK, Fulcher T, Cribb PJ, Chase MW. Molecular phylogenetics and morphological reappraisal of the Platanthera clade (Orchidaceae: Orchidinae) prompts expansion of the generic limits of Galearis and Platanthera. Ann Bot. 2009;104(3):431–445. doi: 10.1093/aob/mcp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efimov PG. A revision of Platanthera (Orchidaceae; Orchidoideae; Orchideae) in Asia. Phytotaxa. 2016;254(1):1–233. doi: 10.11646/phytotaxa.254.1.1. [DOI] [Google Scholar]
- 21.Lang K-Y. A new subgenus of Platanthera (Orchidaceae) Acta Phytotaxonomica Sinica. 1998;65:449–458. [Google Scholar]
- 22.Gamarra R, Galan P, Herrera I, Ortunez E. Seed micromorphology supports the splitting of Limnorchis from Platanthera (Orchidaceae) Nord J Bot. 2008;26(1–2):61–65. doi: 10.1111/j.1756-1051.2008.00135.x. [DOI] [Google Scholar]
- 23.Jin W-T, Jin X-H, Schuiteman A, Li D-Z, Xiang X-G, Huang W-C, Li J-W, Huang L-Q. Molecular systematics of subtribe Orchidinae and Asian taxa of Habenariinae (Orchideae, Orchidaceae) based on plastid matK, rbcL and nuclear ITS. Mol Phylogenet Evol. 2014;77:41–53. doi: 10.1016/j.ympev.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Yukawa T, Bateman RM, Jiang H, Peng H. Phylogeny and classification of the east Asian Amitostigma alliance (Orchidaceae: Orchideae) based on six DNA markers. BMC Evol Biol. 2015;15 [DOI] [PMC free article] [PubMed]
- 25.Inda LA, Pimentel M, Chase MW. Phylogenetics of tribe Orchideae (Orchidaceae: Orchidoideae) based on combined DNA matrices: inferences regarding timing of diversification and evolution of pollination syndromes. Ann Bot. 2012;110(1):71–90. doi: 10.1093/aob/mcs083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman RM, Hollingsworth PM, Preston J, Yi-Bo L, Pridgeon AM, Chase MW. Molecular phylogenetics and evolution of Orchidinae and selected Habenariinae (Orchidaceae) Bot J Linn Soc. 2003;142(1):1–40. doi: 10.1046/j.1095-8339.2003.00157.x. [DOI] [Google Scholar]
- 27.Douzery EJP, Pridgeon AM, Kores P, Linder HP, Kurzweil H, Chase MW. Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. Am J Bot. 1999;86(6):887–899. doi: 10.2307/2656709. [DOI] [PubMed] [Google Scholar]
- 28.Luer CA. The native orchids of the United States and Canada. New York: The New York Botanical Garden; 1975. [Google Scholar]
- 29.Seidenfaden G. The orchids of Indochina. Opera Botanica. 1992;114:1–505. [Google Scholar]
- 30.Pedersen HÆ, Suddee S, Cribb PJ. Flora of Thailand. Vol. 12. Part 1. The Forest Herbarium: Bangkok; 2011. [Google Scholar]
- 31.Pearce NR, Cribb PJ. The orchids of Bhutan. Huddersfield: Charlesworth Group; 2002. [Google Scholar]
- 32.Pedersen HAE. Species delimitation and recognition in the Brachycorythis helferi complex (Orchidaceae) resolved by multivariate morphometric analysis. Bot J Linn Soc. 2010;162(1):64–76. doi: 10.1111/j.1095-8339.2009.01015.x. [DOI] [Google Scholar]
- 33.Lang K-Y. Flora Republicae Popularis Sinicae vol. 17, vol. 17. Beijing: Science Press; 1999. [Google Scholar]
- 34.Lang K-Y. The genus Peristylus Bl. (Orchidaceae) in China. Acta Phytotaxonomica Sinica. 1987;25:442–459. [Google Scholar]
- 35.Seidenfaden G. Orchid genera in Thailand: 5. Orchidoideae. Dansk Botanisk Arkiv. 1977;31(3):1–149. [Google Scholar]
- 36.Szlachetko DL. Matériaux pour la révision des Habenariinae (Orchidaceae, Orchidoideae)-5. Richardiana. 2004;4(3):103–108. [Google Scholar]
- 37.Szlachetko DL. Rhinorchis (Orchidaceae, Orchidoideae), a new neotropical genus highlighted from Habenaria. Richardiana. 2012;XIII:71–79. [Google Scholar]
- 38.Szlachetko DL, Kras M. Notes sur le genre Plantaginorchis (Orchidaceae, Orchidoidae, Habenariinae) Richardiana. 2006;6:31–32. [Google Scholar]
- 39.Alvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol. 2003;29(3):417–434. doi: 10.1016/S1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 40.Pridgeon AM, Bateman RM, Cox AV, Hapeman JR, Chase MW. Phylogenetic of subtribe Orchidinae (Orchidoideae, Orchidaceae) based on nuclear ITS sequences. 1. Intergeneric relationships and polyphyly of Orchis sensu lato. Lindleyana. 1997;12(2):89–109.
- 41.Hapeman JR, Inoue K. Plant-pollinator interactions and floral radiation in Platanthera (Orchidaceae) In: Givnish TJ, Sytsma KJ, editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; 1997. pp. 433–454. [Google Scholar]
- 42.Dressler RL. The orchids: natural history and classification. Cambridge, Massachusetts: Harvard University Press; 1981. [Google Scholar]
- 43.Van Der Niet T, Linder HP, Bytebier B, Bellstedt DU. Molecular markers reject monophyly of the subgenera of Satyrium (Orchidaceae) Syst Bot. 2005;30(2):263–274. doi: 10.1600/0363644054223530. [DOI] [Google Scholar]
- 44.Bytebier B, Antonelli A, Bellstedt DU, Linder HP. Estimating the age of fire in the cape flora of South Africa from an orchid phylogeny. Proc R Soc B-Biol Sci. 2011;278(1703):188–195. doi: 10.1098/rspb.2010.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin W-T, Xiang X-G, Jin X-H. Generic delimitation of Orchidaceae from China: current situation and perspective. Biodivers Sci. 2015;23:237–242. doi: 10.17520/biods.2014268. [DOI] [Google Scholar]
- 46.Jin X-H, Li D-Z, Xiang X-G, Lai Y-J, Shi X-C. Nujiangia (Orchidaceae: Orchideae): a new genus from the Himalayas. J Syst Evol. 2012;50(1):64–71. doi: 10.1111/j.1759-6831.2011.00167.x. [DOI] [Google Scholar]
- 47.Kores PJ, Molvray M, Weston PH, Hopper SD, Brown AP, Cameron KM, Chase MW. A phylogenetic analysis of Diurideae (Orchidaceae) based on plastid DNA sequence data. Am J Bot. 2001;88(10):1903–1914. doi: 10.2307/3558366. [DOI] [PubMed] [Google Scholar]
- 48.Clements MA, Jones DL, Sharma IK, Nightingale ME, Garratt MJ, Fitzgerald KJ, Mackenzie AM, Molloy BPJ. Phylogenetics of Diurideae (Orchidaceae) based on the internal transcribed spacer (ITS) regions of nuclear ribosomal DNA. Lindleyana. 2002;17:135–171. [Google Scholar]
- 49.Li J-L, Wang S, Yu J, Wang L, Zhou S-L. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013;48(1):72–78. doi: 10.3724/SP.J.1259.2013.00072. [DOI] [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 52.Farris JS, Kallersjo M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44(4):570–572. doi: 10.1093/sysbio/44.4.570. [DOI] [Google Scholar]
- 53.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Massachusetts: Sinauer, Sunderland; 2003. [Google Scholar]
- 54.Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit JW, Watson LE. Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Am J Bot. 2010;97(5):856–873. doi: 10.3732/ajb.0900287. [DOI] [PubMed] [Google Scholar]
- 55.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27(2):171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 56.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large Phylogenetic trees. 2010. [Google Scholar]
- 58.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian Phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53(1):47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tree of Orchidinae s.l. from Bayesian inference based on plastid markers. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. The dash (−) indicates support at a node < 50%, Asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 427 kb)
Tree of Orchidinae s.l. from Bayesian inference based on nrITS, including ITS of six species for which position conflict between the plastid and nrITS results. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. The dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 438 kb)
Tree of Orchidinae s.l. from Bayesian inference based on ITS, excluding nrITS of the six species with incongruent positions. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. A dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 435 kb)
Tree of Orchidinae s.l. from Bayesian inference based on nrITS and Xdh, excluding the nrITS sequences of the six species with incongruent positions. Numbers above branches indicate bootstrap percentages (BS) for ML and MP analyses and posterior probabilities (PP) for BI analysis, respectively. A dash (−) indicates support at a node < 50%, and an asterisk (*) indicates BS = 100 or PP = 1.0. (PDF 439 kb)
Voucher information and GenBank accession numbers for the sequences analysed in this study, *indicates newly produced sequences for this paper. The dash (−) indicates the missing sequences. (XLSX 46 kb)
The PCR programs used for amplifying the DNA regions. Table S3. List of primers used for PCR and sequencing in this study. Table S4. Statistics from analyses of nuclear and plastid datasets. (DOCX 21 kb)
Data Availability Statement
All of the phylogenetic sequence data in this study was deposited in GenBank (National Center for Biotechnology Information) under accession numbers: ITS, MF 944259-MF944417; matK, MF945394-MF945550; psaB, MF 944418-MF944626; psbA-trnH: MF 944627–944,832; rbcL, MF944833–944986; trnL-F, MF945188–945393; Xdh, MF944987–945187 (Additional file 5: Table S1). All additional materials supporting the results of the article are included as additional files.


