Abstract
Protein complexes often represent an ensemble of different assemblies with distinct functions and regulation. This increased complexity is enabled by the variety of protein diversification mechanisms that exist at every step of the protein biosynthesis pathway, such as alternative splicing and post transcriptional and translational modifications. The resulting variation in subunits can generate compositionally distinct protein assemblies. These different forms of a single protein complex may comprise functional variances that enable response and adaptation to varying cellular conditions. Despite the biological importance of this layer of complexity, relatively little is known about the compositional heterogeneity of protein complexes, mostly due to technical barriers of studying such closely related species. Here, we show that native mass spectrometry (MS) offers a way to unravel this inherent heterogeneity of protein assemblies. Our approach relies on the advanced Orbitrap mass spectrometer capable of multi-stage MS analysis across all levels of protein organization. Specifically, we have implemented a two-step fragmentation process in the inject flatapole device, which was converted to a linear ion trap, and can now probe the intact protein complex assembly, through its constituent subunits, to the primary sequence of each protein. We demonstrate our approach on the yeast homotetrameric FBP1 complex, the rate-limiting enzyme in gluconeogenesis. We show that the complex responds differently to changes in growth conditions by tuning phosphorylation dynamics. Our methodology deciphers, on a single instrument and in a single measurement, the stoichiometry, kinetics and exact position of modifications, contributing to the exposure of the multilevel diversity of protein complexes.
Introduction
It is now accepted that multiple layers of regulation contribute to the diversity of individual proteins at every step of the protein biosynthesis pathway1–2. A protein may be encoded by more than one gene3 and a single gene may produce several alternative splice variants4–6. Furthermore, RNA modifications7, multiple translation initiation sites on mRNA transcripts8, or translational slippage sites, can all lead to multiple protein forms9. Moreover, proteins may be cleaved10 or post-translationally modified (PTM) by covalent attachment of more than 400 different types of functional groups, including phosphorylation, glycosylation, methylation and lipidation11–12. These mechanisms of protein diversification generate millions of different protein forms, termed proteoforms13. Recent estimates suggest that the total number of possible human protein variants is somewhere between 10 million14 and 1 billion13, 15.
While this proteoform diversity has been an avenue of intense study on the single protein level, the potential effect on the heterogeneity of protein complexes is relatively unstudied. This layer of complexity is highly relevant considering that most proteins exert their function within protein complexes. Current estimates predict more than 300,000 protein-protein interactions in humans, which correspond to 15 partners per protein16. Although most proteoforms may be extremely rare or have no functional consequence, it is important to elucidate those that impact the protein complex function, sub-cellular localization, and/or interacting partners17. However, studying this inherent heterogeneity of protein complexes is challenging due to the high similarity of the generated species.
Bottom-up proteomic analysis, which is based on the separation of enzymatically digested proteins using liquid chromatography (LC) coupled to tandem mass spectrometry (MS2), is a powerful tool for quantification and identification of proteins and PTMs12, 18–19. Yet, it is constrained in its ability to distinguish between different proteoforms, especially in complex and heterogeneous samples20. This is due to the fact that identical peptides can originate from different proteins or proteoforms, and modifications or sequence variations may occur on distinct peptides, causing their relationship to the precursor protein to be lost following digestion20. This information can be conserved and revealed, however, by introducing intact protein complexes into the mass spectrometer and performing triple-stage MS analysis (MS3), starting from the intact assembly, through its composing subunits, to the primary sequence of each subunit21–22.
Here, we have applied and further refined mass spectrometry (MS) capabilities to investigate the inherent heterogeneity of an endogenous protein complex. We used a modified Q Exactive™ Plus mass spectrometer that enables ion trapping in the inject flatapole of the instrument. During the analysis the intact complex is initially transferred through the mass spectrometer without fragmentation, yielding the MS1 spectrum that elucidates the multiple, coexisting states of the intact assembly. Dissociation of the protein complex into its composing subunits gives the MS2 spectrum, which reveals the heterogeneity that exists within individual subunits. Consequently, selection and fragmentation of an individual subunit enables sequence analysis and PTM mapping through the MS3 spectrum. This approach is demonstrated for the yeast fructose-1,6-bisphosphatase 1 (FBP1) complex, a key regulatory enzyme in the gluconeogenesis pathway that is required for glucose metabolism23. Even in this relatively simple, 156.6 kDa, homotetrameric complex, our approach uncovered the presence of two phosphorylation sites. These modification sites were found to be mutually exclusive and sensitive to different growth conditions. Overall, in addition to uncovering a novel layer of regulation for a key metabolic enzyme, our study lays the groundwork for exposing the hidden heterogeneity of protein complexes by accumulating layers of information from each MS stage.
Experimental Section
Modifications introduced into the Q Exactive Plus instrument
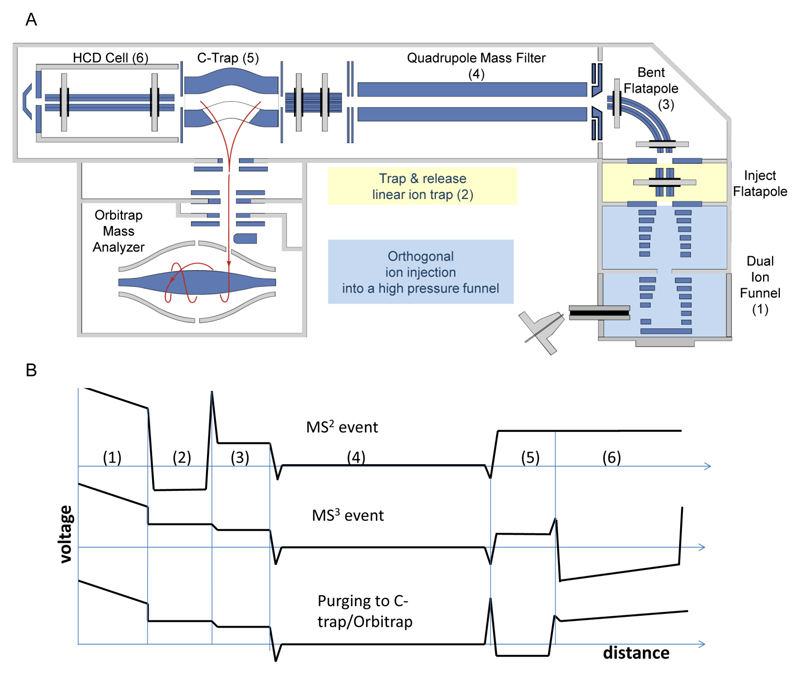
Figure 1 shows the instrument layout and potential profile at different elements of the modified Q Exactive™ Plus mass spectrometer.
Figure 1. Schematic diagram of the modified Q Exactive Plus mass spectrometer, enabling MS3 analysis.
(A) Modifications were introduced in the front section of the instrument (highlighted in colors). Specifically, a pressure-regulated dual-funnel interface was used for improving desolvation and enhancing sensitivity (blue) in the manner described in24. In addition, a trap-and-release mode of analysis was enabled in the inject flatapole by a front-end trap, to allow pseudo-MS2 measurements (yellow). A pressure control valve was coupled to the Edwards forepump to enable fine pressure adjustment in the Lower Pressure Funnel (LPF) region and to optimize ion trapping and activation in the front-end trap. (B) Profiles of experimental voltages applied during three different events. These include: i) ejection of protein subunit from an intact protein complex (MS2); ii) fragmentation of the ejected subunits in the HCD cell (MS3), and iii) purging of fragment ions from the HCD cell into the C-trap for subsequent Orbitrap detection.
A front-end interface encompassing orthogonal ion injection into the radio-frequency (RF) field of a dual-funnel device was implemented as described previously21. The dual-funnel interface (Spectroglyph, Kennewick, USA) was installed in place of the standard outer cage of the IonMax ion source. It was evacuated differentially using the standard rough pump of the instrument (Sogevac, SV65, Leybold, Germany) and an additional rough pump (XDS 35i, Edwards, Surrey, UK). The pressure at the Higher Pressure Funnel, HPF, was 12 mbar as measured with a convection gauge on the dual-funnel interface (Kurt Lesker, Jefferson Hills, PA), while the Lower Pressure Funnel, LPF, was operated at ~1.5 mbar. Both pressures were user-adjustable.
The subsequent inject flatapole device was converted into a linear quadrupole trapand was operated in a “trap and release” mode of analysis as developed in24. Following efficient desolvation in the dual-funnel interface, protein complexes were introduced and activated in the linear ion trap operating at a pressure of 10-2-10-1 mbar. The LPF pressure was adjusted using an inline diaphragm valve (Kurt Lesker, Jefferson Hills, PA) to optimize trapping and fragmentation conditions in the linear ion trap. Since the linear ion trap is located immediately downstream of the LPF, any adjustments in the LPF pressure gave rise to corresponding changes in the linear ion trap pressure. Collisional activation was performed at kinetic energies in the range of 100 to 300 V per elementary charge in the laboratory frame of reference. The linear ion trap is located upstream of the m/z selection device (a quadrupole mass filter). The trap and release events were synchronized with the Orbitrap trigger pulse. A typical injection time into the linear ion trap was 10 ms, while a typical release time was 200 µs, which results in 95% duty cycle. Subsequent to the dissociation event, the ejected subunits of the protein complex were selected by their m/z and were then introduced into another linear ion trap, higher energy collision-induced dissociation (HCD) cell, operating at pressures of 10-1-10-4 mbar. In the HCD cell, the ejected monomer subunits of the protein complex underwent higher energy collisions. The protein ions were activated in the HCD cell at collision energies of 100-200 V per elementary charge in the laboratory frame of reference and then efficiently fragmented to the constituent peptide-level fragments.
Accumulation of molecular ions of large protein complexes inside the linear ion trap at higher translational energy ensures imparting sufficient internal energy into the rotational and vibrational modes of the trapped species. Ejection of subunits from selected protein complexes has previously been reported in the fly-through mode21. In this mode, intact protein complexes undergo collisional activation while traversing the inject flatapole of the instrument, at higher translational energies. The limitations of the fly-through mode are the insufficient number of higher energy collisions per precursor’s residence time in the inject flatapole and translational energy modulation. The former results in reduced efficiency of subunit ejection, thus limiting the approach to weakly bound protein complexes. The latter causes extensive signal loss due to the inability of the downstream RF ion guide (i.e., bent flatapole) to confine higher translational energy species and to force them to a 90° turn. As a result, with an increase in the activation energy (i.e., an increase in the voltage applied to the exit electrode of the ion funnel or S-lens), precursor ions of the more tightly bound protein complexes escape the bent flatapole and yield neither fragmentation nor precursor ion signals. In contrast with the fly-through mode, which limits the interaction time between ions of interest and buffer gas to the time precursor ions take to traverse the collision cell, the trapping capability ensures the required number of higher energy collisions to facilitate protein complex restructuring/unfolding and dissociation.
All other experimental details regarding yeast growth, purification of FBP1, sample preparation, MS data analysis, proteomic experiments and FBP1 modelling are described in the Supporting Information.
Results and Discussion
Multilevel structural analysis in a single experiment
Recently an Exactive™ Extended Mass Range (EMR) mass spectrometer with an Orbitrap™ mass analyzer was introduced, enabling the examination of large protein assemblies in their native state at enhanced resolution and sensitivity25–27. Subsequent developments included the incorporation of high mass selection functionality using a modified quadrupole RF drive21, 28 and the development of MS3 capability for enabling enhanced multi-stage characterization of intact protein complexes21.
Leveraging these instrumentation developments21, we have advanced multi-stage dissociation of large protein complexes by implementing and characterizing a “trap and release” approach. This method allows for a number of capabilities including: (i) high-efficiency desolvation of large precursor species represented by removal of solvents and adduct species and (ii) significantly improved fragmentation into subunits and stripped-complexes. An initial step in this direction was done in a parallel study that demonstrated the improved desolvation capacity provided by the trapping29.
In this work, we have used a dual-funnel interface to enhance protein complex desolvation at higher pressure (12 mbar) and have developed a two-step fragmentation approach to collisionally activate, unfold and fragment larger protein complexes to subunits. The trapping capability was enabled at the front section of the instrument, as described in detail in the Experimental Section. Specifically, to enable the first step of the activation approach, an inject flatapole device was converted to a linear quadrupole trap and was operated in a “trap and release” mode of analysis24 (Fig. 1). Trapping of ions for extended intervals (~ 10 ms) ensured a sufficient number of higher-energy collisions, highly efficient fragmentation, collisional relaxation of the ejected subunits and then purging the fragment ions (or excited precursors) out of the linear ion trap, to be referred to as a release event. In the second step of the activation approach, we elevated the internal energy of the unfragmented precursor ions, which already acquired significant energy (200 V per elementary charge) during the previous activation step. The increase in the translational energy was accomplished by creating a dynamic voltage gradient (100 V/cm) between the rods/exit lens of the linear ion trap and the following RF ion guide, the bent flatapole, during the release event. The combination of high energy collisional activation in the linear ions trap and translational energy modulation during the release event, resulted in drastically improved fragmentation efficiency of the native protein complexes into their composing subunits. Optimization of trapping time, potential well depth, pressure in the linear ion trap and the full energy during the release event, is shown in Figs. S1 and S2. Using these optimized conditions, we have applied the refined MS3 approach to characterize the molecular heterogeneity of an endogenous protein complex.
MS1 captures dynamic responses to growth conditions in an intact complex
To investigate the diversity of protein complexes, we selected the yeast Saccharomyces cerevisiae FBP1 complex. This homotetrameric complex is the rate-limiting enzyme in the yeast gluconeogenesis pathway and is rapidly expressed in the absence of sugar. However, when cells are replenished with glucose, glycolysis takes place and FBP1 is inactivated23. To date, the full repertoire of regulatory events enabling activation and inactivation of FBP1 are yet to be deciphered. To follow FBP1 dynamics, we have grown yeast in the absence of glucose and then shifted the cells to glucose-containing medium. In parallel, we also exposed carbon starved yeast cells to heat shock, reasoning that this treatment may give rise to a distinct response. Then, using a single-step FLAG-affinity purification protocol, FBP1 was isolated from the three different growth conditions (glucose, carbon starved, and carbon starved+heat shock) and the composition of the complex was explored.
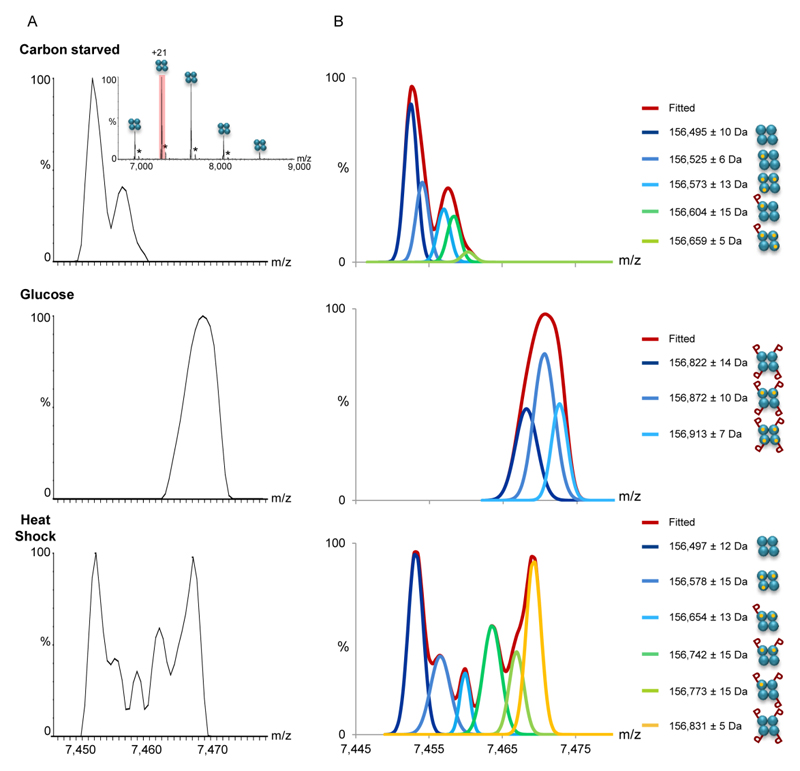
The MS1 spectrum of FBP1 isolated directly from cells grown in the absence of glucose indicates the homotetrameric composition of the complex, as expected (Fig. 2A, Table S1). Detailed assignment of the peaks was assisted by deconvoluting the spectrum (Fig. 2B). The profile of the deconvoluted spectra fitted the observed peaks very closely, as shown in Fig. S3. The analysis indicated that a fraction of the complex is associated with magnesium ions, in accordance with the tendency of this complex to bind three divalent metal ions per subunit30–31. The stoichiometry of the FBP1 complex remained stable despite the exposure of yeast cells to heat shock or to the glucose-containing medium, suggesting that stoichiometry is not used to regulate the complex. However, a distinct pattern of modifications could be identified under each growth condition. A uniform mass shift corresponding to four phosphorylation events was detected for the FBP1 assembly treated with glucose. In contrast, a gradual increase in the number of phosphorylation events from 0 to 4 was detected, following exposure to heat shock. Thus, in addition to determining the assembly state, the MS1 spectrum shows clear differences in the phosphorylation stoichiometry and the rate of occurrence in response to growth conditions.
Figure 2. MS1 analysis indicates that changes in growth conditions are accompanied by differential PTMs.
(A) The FBP1 complex was FLAG-affinity purified from yeast grown in the absence of glucose (top panel), after a shift to glucose (middle panel), or after exposure to heat shock (bottom panel). Mass spectra of purified FBP1 complexes were acquired on the modified Orbitrap™ system, at conditions favoring the preservation of non-covalent interactions, thus preserving the homotetrameric structure (inset). Enlarged view of the +21 charge state reveals differential modifications in response to the different treatments. Black asterisks correspond to homotetrameric FBP1 complexes non-covalently associated with a FLAG peptide adduct. (B) Deconvolution of the measured spectra aided the assignment of the peaks. Assignment was performed by analysis of the deconvoluted spectra and mass measurements. The identified FBP1 species are shown on the right. The results show that the addition of glucose shifted the complex to a uniformly phosphorylated state, at four positions. In contrast, the coexistence of species corresponding in mass to 0 to 4 phosphorylation events was detected, following yeast exposure to heat shock. As expected, association with Mg2+ ions was also obtained. Each FBP1 subunit is graphically depicted as cyan circle. Mg2+ ions are indicated as small orange circles, and phosphorylation is labeled as “P”.
MS2 depicts the heterogeneity that exists at the single subunit level
To determine whether the phosphorylation events observed at the MS1 level are due to mono- or multi-phosphorylation(s) on single or multiple subunit(s), MS2 spectra were collected. Namely, the intact FBP1 complex was collisionally activated and subsequently trapped in the front-end trap to induce complex dissociation into the constituent subunits. This higher-energy dissociation gave rise to the release of individual FBP1 subunits32. Yet, we have also observed the ejection of dimers (Fig. S4). The dissociation into dimers is consistent with the FBP1 structure, which is a dimer of two dimers33. Notably, we have obtained a similar dissociation pattern when FBP1 was fragmented using higher-energy collisional dissociation in the HCD cell of the modified Orbitrap instrument, or when acquiring MS2 spectra with a Synapt G2 mass spectrometer (Waters Corporation, Milford, MA) (Fig. S4). Therefore, we attribute the tendency of FBP1 to dissociate into dimers to its quaternary topology33, and not to the instrument type or dissociation approach.
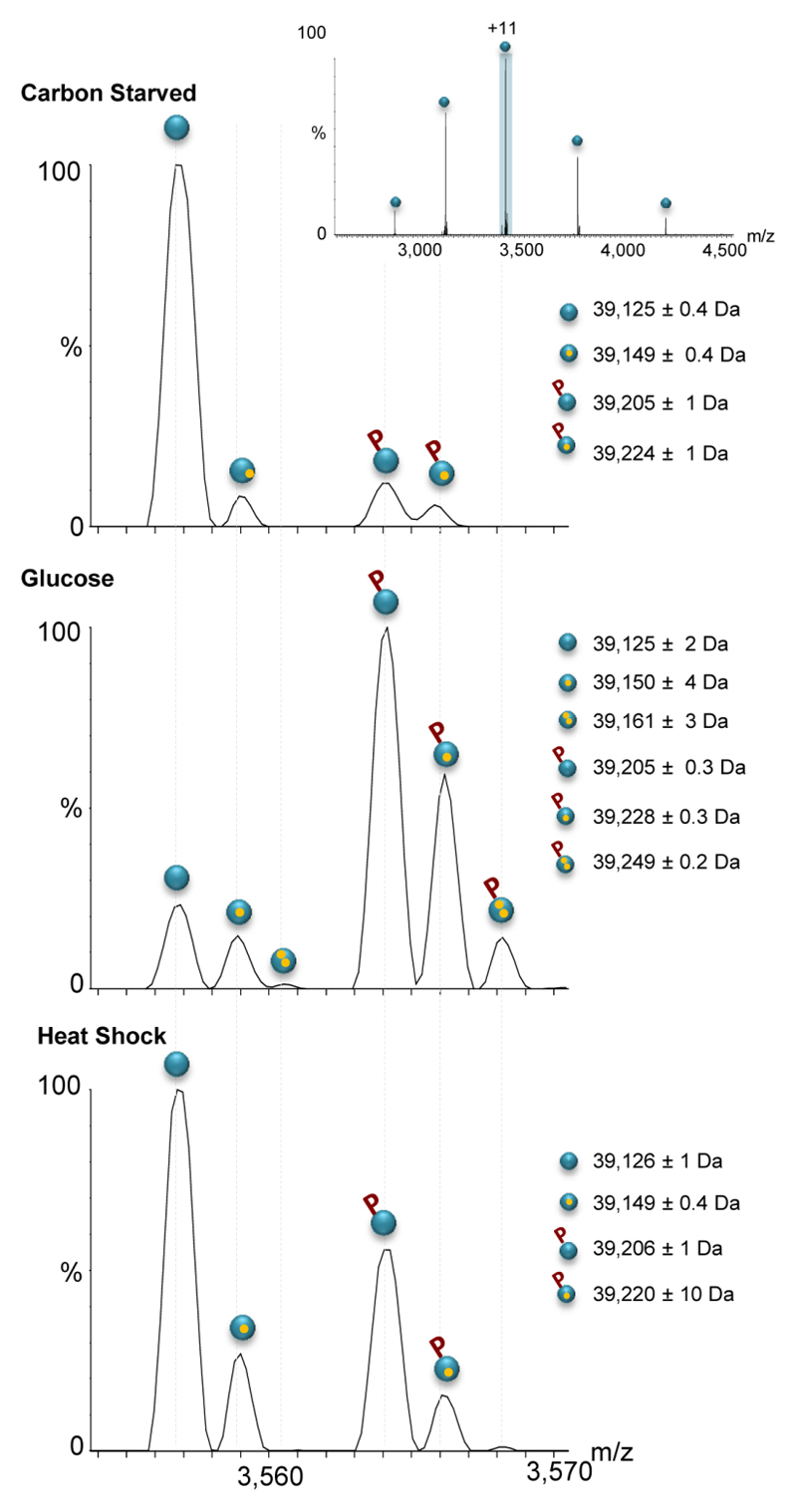
Examining the peaks assigned to the individual FBP1 subunits released during MS2 events revealed differential response to growth conditions. The mass spectrum of FBP1 isolated from yeast grown in the absence of glucose (Fig. 3) indicates that the vast majority of subunits are unmodified with a measured mass corresponding well to the theoretical expected value of the protein after removal of the N-terminal Met34, 39,125 ± 0.4 Da. Additional series of peaks were assigned to lower abundance populations of the FBP1 protein associated with magnesium ions and/or mono-phosphorylation, as indicated by the +24 and +80 mass shifts, respectively. Following the addition of glucose, however, a predominant shift in mass corresponding to mono-phosphorylation was observed. In addition, a minor population of unmodified FBP1 subunits was also detected, and therefore we cannot exclude the possibility that the labile phosphate group is removed during the fragmentation process35. Nevertheless, the results suggest that the measured mass of the intact complex (MS1 spectrum, Fig. 2), corresponds to four mono-phosphorylated subunits.
Figure 3. Changes in growth conditions result in mono-phosphorylation.
The MS2 process leads to dissociation of the FBP1 complex into monomers (inset). Expansion of the +11 charge state shows that carbon starved FBP1 mainly exists in an unmodified form. The addition of glucose leads to mono-phosphorylation of the vast majority of FBP1 subunits (middle panel). In contrast, exposure to heat shock leads to partial mono-phosphorylation events. For an explanation of the labeling, see Figure 1.
In the case of heat shock treatment, two distinct populations were detected: the unmodified and mono-phosphorylated subunits. This bimodal distribution explains the multiple compositions of the tetrameric species observed in the MS1 spectrum. In addition, it implies that the distribution of tetramers in this sample does not follow a binomial distribution, rather the fraction of complexes with four unmodified or four mono-phosphorylated subunits represent the dominant populations, in comparison to the mixed complexes. This result hints to cooperation in either the assembly of mono-phosphorylated subunits with each other, or alternatively that phosphorylation of one subunit favors phosphorylation on the other subunits because of the kinase proximity. Similar results were also obtained by LC-MS analysis of all three samples under denaturing conditions36, confirming the native MS2 data (Fig. S5). Overall, extrapolation of the findings revealed through MS2 analyses, i.e., the existence of different amounts of mono-phosphorylated subunits, elucidated the source of heterogeneity that exists at the intact complex level.
MS3 enables mapping of distinct modifications
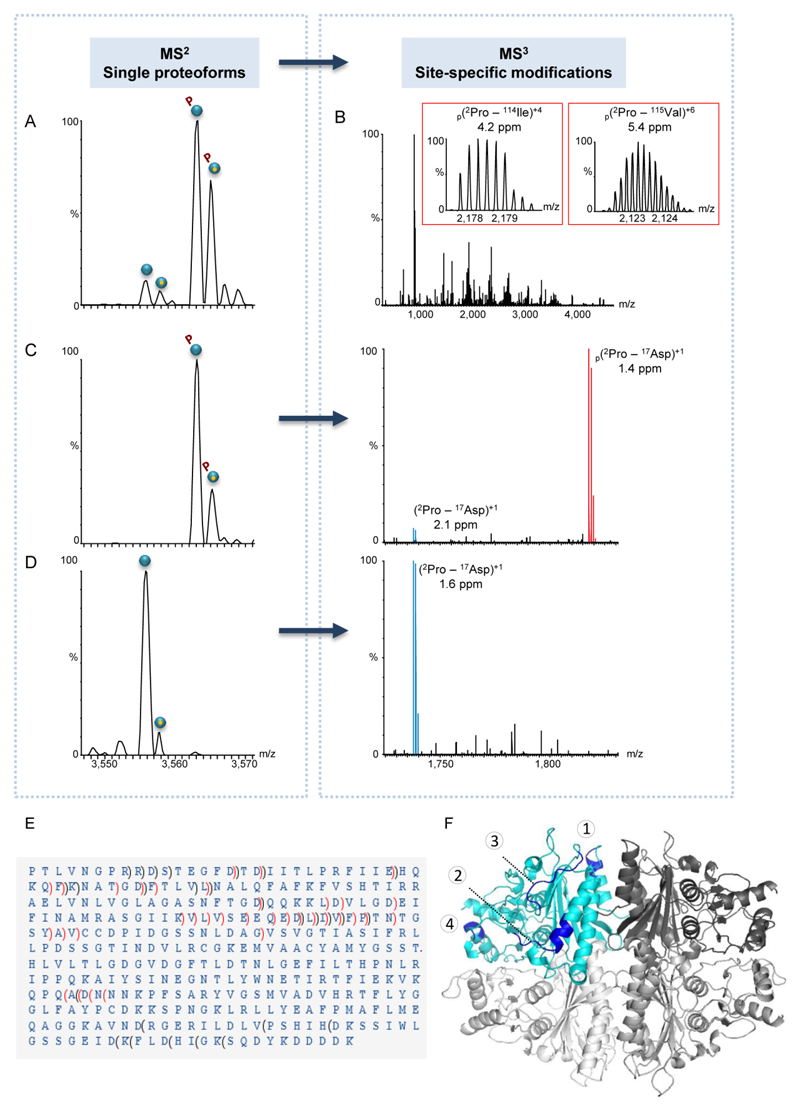
A key question that arises, and cannot be resolved at the level of MS1 or MS2 analyses, is whether the response to glucose supplementation and heat shock treatment leads to mono-phosphorylation at identical sites, or whether it gives rise to distinct modifications. To address this question, we applied the MS3 approach, which enabled selection of a specific FBP1 proteoform within the quadrupole mass analyzer followed by fragmentation in the HCD cell (Figs 4 and S6). The generated multiply-charged fragments were then detected at high mass resolution enabling sequence identification and PTM mapping.
Figure 4. MS3 fragmentation data unravels the existence of two different phosphorylation sites.
(A-D) Representative spectra of FBP1 monomer purified from yeast grown in a glucose-containing medium. In general, individual proteoforms identified in the MS2 spectrum (A) were selected within the quadrupole mass filter and subjected to MS3 fragmentation (B). The insets show fragment ion matches of two b ions, representing identified phosphopeptides. For each fragment, the position, charge and mass accuracy are indicated. Phosphorylated fragments are labeled by p. In all generated MS3 spectra, fully resolved multiply charged fragment ions were detected using a mass resolving power of 140,000. (C-D) Selective MS2 isolation of mono-phosphorylated or unmodified FBP1 proteoforms. For each proteoform selected, the respective MS3 spectrum is shown on the right panel of (C) or (D). Expansion of the 1,700-1,900 m/z region of MS3 data generated upon specific proteoform selection and fragmentation, showing the b16 ion, which matches the 2PTLVNGPRRDSTEGFD17 sequence. The data indicates that the b16 phosphopeptide (labeled in red) is exclusively found in the MS3 spectrum of the mono-phosphorylated FBP1 and not in that of the unmodified proteoform. (E) Analysis of MS2 and MS3 spectra revealed that the FBP1 protein is missing the initial Met, and that it is phosphorylated at two mutually exclusive sites: at either position 12Ser/13Thr or within the stretch of 248Asn-310Asp. In the latter, due to low fragmentation efficiency in this specific region, the exact phosphorylation position could not be mapped. b ions are indicated by brackets pointing towards the N-terminus and y ions are indicated by brackets pointing towards the C-terminus. Non-phosphorylated fragments are indicated in black, phosphorylated fragments are indicated in red. (F) A model of the FBP1 tetramer, demonstrating surface-exposed fragmentation hotspots. One subunit is colored in cyan and the other three are in different shades of gray. Fragmentation hotspot sequences are 30HQKQFK35, 79DQQKKLDVLGD89, 110QEDLIVFPT118 and 229WNETI233 (labeled by numbers 1- 4 respectively). The model highlights the structural organization of the complex, which is assembled from a dimer of dimers, in accordance to our results (Fig. S4).
The peptide fragments obtained in the MS3 experiments were matched against the known FBP1 protein sequence, enabling high confidence identifications of both b- and y-ions, as well as PTM sites (Figs 4 and S6). Mapping the regions prone to fragmentation on the modeled protein structure indicated that they occur predominantly on surface-exposed and loop regions of the tetramer (Fig. 4E), suggesting that these regions are more prone to fragmentation.
Focusing on the PTMs, we have found that all three FBP1 protein complexes encompassed two phosphopeptides corresponding to phosphorylation at either position 12Ser/13Thr or within the stretch of 248Asn-310Asp (Fig. 4B and Fig. S6). Taken together and building on the MS2 data, which clearly show that no subunit has more than a single phosphorylation event, we can now conclude that these two phosphorylation sites are mutually exclusive.
The identification of 12Ser/13Thr phosphorylation site is consistent with previous studies based on mutational analysis, which indicated that FBP1 is deactivated by cAMP-dependent phosphorylation at 12Ser, when cells are exposed to glucose37–38. In addition, a second phosphorylation site was suggested to exist but was not demonstrated experimentally39. Inactivating the enzymatic activity of FBP1 is part of the metabolic rewiring of yeast cells from gluconeogenesis to glycolysis. The finding that 10 minutes after shifting to a glucose containing medium, FBP1 is fully phosphorylated, reflects this efficient adaptation. On the other hand, under heat shock conditions, yeast cells were still deprived of glucose, therefore, the gluconeogenesis pathway needs to be maintained. As a consequence, only partial deactivation of FBP1 takes place, possibly in order to allocate resources to cope with the heat shock.
To validate these findings we performed bottom-up proteomic analysis of tryptic digests of FBP1 from all three samples. In all cases, phosphopeptides were identified at the dominant 12Ser/13Thr site, in accord with our native MS3 results (Fig. S7A). Moreover, in FBP1 from carbon starved cells or from yeast grown on glucose, a 276Tyr phosphorylation site was identified. Intensity label free quantitation40 indicated the dominance of the 12Ser/13Thr site (Fig. S7B). Such resolution in PTM mapping obtained by bottom-up approaches is likely to be provided by MS3 fragmentation analysis, given improvements in fragmentation efficiency and assignment procedures. Taken together, we show that in a single experiment the native FBP1 protein complex was ionized, injected into a mass spectrometer (MS1), and further fragmented to the constituent subunits (MS2), before being subjected to higher-energy fragmentation to map phosphorylation sites at high confidence (MS3).
Conclusions
In this study a linear ion trap operated in a “trap and release” mode was introduced into an Orbitrap Q Exactive™ Plus instrument to enable enhanced desolvation and efficient fragmentation of protein complexes. This set-up facilitates MS3 analysis, enabling characterization across all levels of protein organization, from the quaternary structure through single subunits and up to peptide fragments and PTM mapping. We applied this MS3 approach to investigate the yeast homotetrameric FBP1 complex. The MS1 data revealed that shifting yeast cells from gluconeogenesis to glycolysis is accompanied by uniform phosphorylation of FBP1 at 4 sites. In contrast, heat shock exposure led to progressive phosphorylation events, displaying coexistence of different populations of FBP1 with 0, 1, 2, 3, and 4 phosphorylations. The stoichiometry of phosphorylation was unraveled by the MS2 analysis, which indicated the existence of only mono-phosphorylated subunits. Subsequently in MS3 experiments, we have determined the existence of two, mutual-exclusive modification sites that were common to all three FBP1 forms. Thus, the accumulated information gained at each level of analysis, made possible by the triple-stage MS3 platform, was critical for a holistic view of complex adaptation to different growth conditions.
Not only access to multiple layers of information, but also low sample consumption is an important characteristic of the native MS3 approach. This is extremely relevant in the study of endogenous protein complexes, wherein biochemical preparation of highly purified samples constitutes a major bottleneck for all structural biology approaches. Therefore, given the inherently high sensitivity of the instrument, in-depth multilevel analysis can be performed with reduced amounts of sample on endogenously expressed proteins in a single run.
Considering the significant heterogeneity that was unraveled even for a simple complex like FBP1, it is likely that unexplored diversity exists widely. We anticipate that the MS3 approach will promote the investigation of more complex systems, towards addressing major questions in protein-complex biology such as: How responsive are specific protein complexes to variations in cellular conditions? How many combinations of PTMs can occur on a single subunit? What is the crosstalk between these modifications? How similar are protein complexes isolated from different tissues, cellular compartments, cell-cycle phases, or following various stimuli? Overall, by addressing the many fundamental questions in this field, our perception of protein complexes is likely to change, no longer as uniform entities in terms of form and action, but rather as versatile and dynamic units, capable of modulating their functionality, localization and interacting protein partners to adapt rapidly and accurately to the ever changing needs of the cell.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
Supporting Information: yeast strains, media and transformation, FLAG-affinity purification of FBP1 from yeast, MS3 analysis, data assignment of MS3 spectra, MS measurements using the Synapt G2 instrument, intact protein LC-MS analysis, standard proteomic analysis, deconvolution of the measured MS1 spectra and modeling of the FBP1 structure.
Acknowledgments
We thank Prof. Neil Kelleher’s team (Northwestern University) for helpful discussions. We are grateful for Prof. Vicki Wysocki and Dr. Royston S. Quintyn for helping us implement the SID approach. In addition, we are grateful for the support of a Starting Grant from the European Research Council (ERC) (Horizon 2020)/ERC Grant Agreement no. 636752. MS is an incumbent of the Aharon and Ephraim Katzir Memorial Professorial Chair.
References
- 1.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Nat Genet. 2002;30(1):29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 2.Jensen ON. Nat Rev Mol Cell Biol. 2006;7(6):391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JS, Raes J. Annu Rev Genet. 2004;38:615–43. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 4.Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, Willaime-Morawek S, et al. Genome Biol. 2007;8(6):R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Nat Genet. 2008;40(12):1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456(7221):470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dezi V, Ivanov C, Haussmann IU, Soller M. Biochemical Society transactions. 2016;44(5):1385–1393. doi: 10.1042/BST20160110. [DOI] [PubMed] [Google Scholar]
- 8.Bazykin GA, Kochetov AV. Nucleic acids research. 2011;39(2):567–77. doi: 10.1093/nar/gkq806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagida H, Gispan A, Kadouri N, Rozen S, Sharon M, Barkai N, Tawfik DS. PLoS Genet. 2015;11(8):e1005445. doi: 10.1371/journal.pgen.1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baugh JM, Pilipenko EV. Mol Cell. 2004;16(4):575–86. doi: 10.1016/j.molcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Griffin TJ, Goodlett DR, Aebersold R. Curr Opin Biotechnol. 2001;12(6):607–12. doi: 10.1016/s0958-1669(01)00268-3. [DOI] [PubMed] [Google Scholar]
- 12.Mann M, Jensen ON. Nat Biotechnol. 2003;21(3):255–61. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 13.Smith LM, Kelleher NL, proteomics, C. f. t. d. Nature methods. 2013;10(3):186–7. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz J, Heck AJ. Angew Chem Int Ed Engl. 2014;53(41):10864–10866. doi: 10.1002/anie.201406545. [DOI] [PubMed] [Google Scholar]
- 15.Kelleher NL. Journal of the American Society for Mass Spectrometry. 2012;23(10):1617–24. doi: 10.1007/s13361-012-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, et al. Nature. 2012;490(7421):556–60. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copley SD. Bioessays. 2012;34(7):578–88. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- 18.Jungblut PR, Holzhutter HG, Apweiler R, Schluter H. Chemistry Central journal. 2008;2:16–25. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiolica A, Junger MA, Ezkurdia I, Aebersold R. Journal of proteomics. 2012;75(12):3495–513. doi: 10.1016/j.jprot.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 20.Catherman AD, Skinner OS, Kelleher NL. Biochem Biophys Res Commun. 2014;445(4):683–93. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, Kelleher NL. Analytical chemistry. 2013;85(23):11163–73. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- 22.Skinner OS, Havugimana PC, Haverland NA, Fornelli L, Early BP, Greer JB, Fellers RT, Durbin KR, Do Vale LH, Melani RD, Seckler HS, et al. Nat Methods. 2016;13(3):237–40. doi: 10.1038/nmeth.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holzer H. Revis Biol Celular. 1989;21:305–19. [PubMed] [Google Scholar]
- 24.Belov M. US Patent. 2014 2015/0340213 A1.
- 25.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJ. Journal of the American Chemical Society. 2014;136(20):7295–9. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. Nature methods. 2012;9(11):1084–6. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 27.Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, et al. Nat Methods. 2016;13(4):333–6. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyachenko A, Wang G, Belov M, Makarov A, de Jong RN, van den Bremer ET, Parren PW, Heck AJ. Analytical chemistry. 2015;87(12):6095–102. doi: 10.1021/acs.analchem.5b00788. [DOI] [PubMed] [Google Scholar]
- 29.van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, Heck AJ. Nature methods. 2017 doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 30.Benkovic SJ, deMaine MM. Advances in enzymology and related areas of molecular biology. 1982;53:45–82. doi: 10.1002/9780470122983.ch2. [DOI] [PubMed] [Google Scholar]
- 31.Ruf A, Tetaz T, Schott B, Joseph C, Rudolph MG. Acta crystallographica. Section D, Structural biology. 2016;72(Pt 11):1212–1224. doi: 10.1107/S2059798316016715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benesch JL. Journal of the American Society for Mass Spectrometry. 2009;20(3):341–8. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Barciszewski J, Wisniewski J, Kolodziejczyk R, Jaskolski M, Rakus D, Dzugaj A. Acta crystallographica. Section D, Structural biology. 2016;72(Pt 4):536–50. doi: 10.1107/S2059798316001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dummitt B, Micka WS, Chang YH. J Cell Biochem. 2003;89(5):964–74. doi: 10.1002/jcb.10566. [DOI] [PubMed] [Google Scholar]
- 35.Boersema PJ, Mohammed S, Heck AJ. Journal of mass spectrometry : JMS. 2009;44(6):861–78. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 36.Rozen S, Tieri A, Ridner G, Stark AK, Schmaler T, Ben-Nissan G, Dubiel W, Sharon M. Methods. 2013;59(3):270–7. doi: 10.1016/j.ymeth.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Rittenhouse J, Moberly L, Marcus F. The Journal of biological chemistry. 1987;262(21):10114–9. [PubMed] [Google Scholar]
- 38.Hammerle M, Bauer J, Rose M, Szallies A, Thumm M, Dusterhus S, Mecke D, Entian KD, Wolf DH. The Journal of biological chemistry. 1998;273(39):25000–5. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- 39.Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. The Journal of biological chemistry. 2004;279(47):49138–50. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 40.Shalit T, Elinger D, Savidor A, Gabashvili A, Levin Y. Journal of proteome research. 2015;14(4):1979–86. doi: 10.1021/pr501045t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.