Abstract
Intestinal Paneth cells limit bacterial invasion by secreting antimicrobial proteins including lysozyme. However, invasive pathogens can disrupt the Golgi apparatus, interfering with secretion and compromising intestinal antimicrobial defense. Here we show that during bacterial infection, lysozyme is rerouted through secretory autophagy, an autophagy-based alternative secretion pathway. Secretory autophagy was triggered in Paneth cells by bacteria-induced endoplasmic reticulum (ER) stress, required extrinsic signals from innate lymphoid cells, and limited bacterial dissemination. Secretory autophagy was disrupted in Paneth cells of mice harboring a mutation in autophagy gene Atg16L1 that confers increased risk for Crohn’s disease in humans. Our findings identify a role for secretory autophagy in intestinal defense and suggest why Crohn’s disease is associated with genetic mutations that impact both the ER stress response and autophagy.
The mammalian intestine is home to a diverse population of bacteria, which includes pathogens that can disrupt host cellular functions. The intestinal epithelium defends against bacterial encroachment through multiple mechanisms including antimicrobial protein secretion and destruction of invading bacteria through autophagy (1). Paneth cells are specialized intestinal epithelial cells that secrete abundant antimicrobial proteins, including lysozyme, and thus disrupting Paneth cell secretion can lead to inflammatory disease (2–4). Pathogenic microbes can trigger ER stress that interferes with protein secretion (5, 6) and compromises antimicrobial protein delivery, raising the question of how Paneth cells preserve their antimicrobial function during pathogen-induced stress.
Invasive bacteria, including Salmonella enterica Serovar Typhimurium (S. Typhimurium), trigger autophagy in intestinal enterocytes. This is indicated by abundant epithelial cell autophagosomes, marked by microtubule-associated protein 1 light chain 3 (LC3), that capture and eliminate invading bacteria (7). S. Typhimurium also invaded Paneth cells (fig. S1), and invasion was associated with elevated numbers of LC3+ puncta in Paneth cells (Fig. 1A,B). The puncta numbers were comparable to those in mice subjected to fasting, a trigger of canonical autophagy (8) (Fig. 1A,B). However, many of the LC3+ structures in infected Paneth cells were markedly larger (0.2 to 7 μm in diameter) than the LC3+ puncta in S. Typhimurium-infected enterocytes (~1 μm) or in Paneth cells of fasted (~0.5 μm) mice (Fig. 1C,D).
Figure 1. Large LC3+ vesicles in S. Typhimurium-infected mice contain lysozyme.
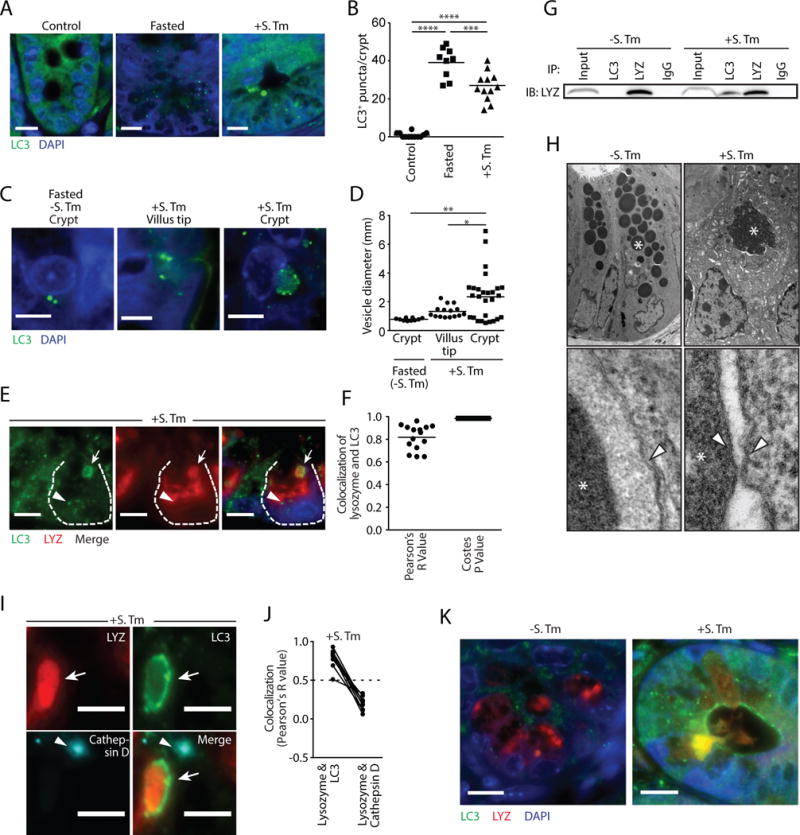
(A) Immunofluorescence of LC3 in intestinal crypts. Nuclei are stained with DAPI. Scale bars=10 μm. (B) Quantification of LC3+ puncta. Each point represents one mouse. (C) Immunofluorescence of LC3 in intestinal crypts. Scale bars=5 μm. (D) LC3+ vesicle diameter measurements (E) Immunofluorescence of LC3 and lysozyme in S. Typhimurium-infected intestinal crypts. A Paneth cell is outlined. Arrows indicate a lysozyme-filled LC3+ vesicle. Arrowhead indicates an autophagosome that does not contain lysozyme. Scale bars=5 μm. (F) Co-localization of LC3 and lysozyme in intestinal crypts from S. Typhimurium-infected mice. Each point represents one lysozyme granule. (G) Co-immunoprecipitation of intestinal lysates using the indicated antibodies. Immunoblot was performed with anti-lysozyme (LYZ) antibody. (H) Transmission electron microscopy (TEM) of Paneth cells from uninfected (-S. Tm) and infected (+S. Tm) mice. Asterisks indicate secretory granules. Arrowheads indicate surrounding membranes. (I) Immunofluorescence of lysosomes (cathepsin D+), LC3 and lysozyme in S. Typhimurium-infected intestinal crypts. Arrows indicate a lysozyme-filled LC3+ vesicle with no lysosome (cathepsin D) signal. Arrowheads indicate lysosomes that are not coincident with lysozyme-filled LC3+ vesicles. Scale bars=5 μm. (J) Quantification of lysosome (cathepsin D), LC3 and lysozyme co-localization in I. Each point represents one lysozyme-containing granule. Two points connected by a line represent the same granule. Dotted line represents limit of strong co-localization. (K) Immunofluorescence of LC3 and lysozyme in intestinal crypts. Scale bars=10 μm. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; One-way ANOVA (B,D); S. Tm, Salmonella Typhimurium; LYZ, lysozyme.
To characterize the contents of the LC3+ vesicles we performed immunofluorescence, transmission electron microscopy (TEM), and co-immunoprecipitation assays. These assays revealed that the large LC3+ vesicles contained lysozyme (Fig. 1E–G) and were absent in Paneth cells of uninfected and fasted mice, where lysozyme was packaged into LC3− vesicles (fig. S2). Ultrastructure analysis showed that the large granules in Paneth cells of infected mice contained lysozyme (fig. S3) and were surrounded by a double membrane (Fig. 1H), a hallmark of autophagosomes (8). Granules from uninfected mice were surrounded by a single membrane (Fig. 1H). The LC3+ vesicles did not contain bacteria (fig. S4A,B) or the antimicrobial proteins REG3γ or cryptdin 5 (fig. S5). Also, cryptdin 5 was not packaged in secretory granules and was excluded from the LC3+ vesicles in infected mice (fig. S6A,B). This suggested that infection interferes with packaging and secretion of Paneth cell antimicrobial proteins and that lysozyme might be rerouted through an alternative secretion pathway involving an LC3+ vesicle.
Canonical autophagy targets the cargo in LC3+ autophagosomes for degradation in lysosomes (8). Immunofluorescence analysis indicated that the lysozyme-filled LC3+ vesicles do not fuse with lysosomes as the vesicles did not colocalize with the lysosomal marker Cathepsin D (Fig. 1I,J), implying that lysozyme is not targeted for degradation. Additionally, the lysozyme-filled LC3+ vesicles also did not colocalize with p62/Sequestesome 1 (SQSTM1), which selects autophagosome cargo for degradation (8). SQSTM1 was associated with the smaller LC3+ puncta in Paneth cells but was not associated with the larger LC3+ lysozyme+ vesicles in infected mice (fig. S7), implying that lysozyme is not selected for degradation in the autolysosome through the canonical selective autophagy pathway. LC3 also accumulated at the apical surface of Paneth cells from infected mice and there were LC3+ vesicles that had fused with the apical surface and discharged lysozyme into the intestinal lumen (Fig. 1K). This was not observed in uninfected mice (fig. S7) and implied that the LC3+ vesicles might be involved in lysozyme secretion.
During conventional protein secretion proteins are transported through the ER-Golgi complex, packaged in secretory granules, and released to the extracellular space. There are various alternative secretory pathways, including one that utilizes components of the autophagy pathway and is known as secretory autophagy (9). In secretory autophagy, cargo is transported in an LC3+ vesicle and discharged at the plasma membrane, thus bypassing the ER-Golgi complex (9). Rab8α, a marker of secretory autophagy vesicles (10), colocalized with the lysozyme-filled LC3+ vesicles and was co-immunoprecipitated with LC3 only in infected mice, but did not colocalize with cryptdin-5 (fig. S8). This suggested that lysozyme might be selectively secreted through the secretory autophagy pathway during infection.
To further test this idea we isolated Paneth cell-containing crypts, infected them in vitro while treating with chemical inhibitors of conventional secretion and autophagy, and analyzed the supernatants for lysozyme secretion. Inhibiting ER-Golgi trafficking with Brefeldin A (BFA) did not affect lysozyme secretion in infected or uninfected crypts (Fig. 2A,B), indicating that lysozyme secretion can bypass the ER-Golgi pathway. Lysozyme secretion was also not altered by treatment with chloroquine (Fig. 2A,B), which prevents lysosome acidification (8), implying that inhibiting autophagic degradation does not affect lysozyme secretion. However, treatment with 3-methyladenine (3-MA), which inhibits autophagosome nucleation (8), impaired lysozyme secretion and caused an accumulation of intracellular lysozyme in infected crypts (Fig. 2A,B). Accordingly, secretions from BFA-treated but not 3-MA-treated crypts killed bacteria (Fig. 2C), indicating that secretory autophagy is essential for antibacterial defense in infected crypts.
Figure 2. Lysozyme is secreted via secretory autophagy during bacterial infection.
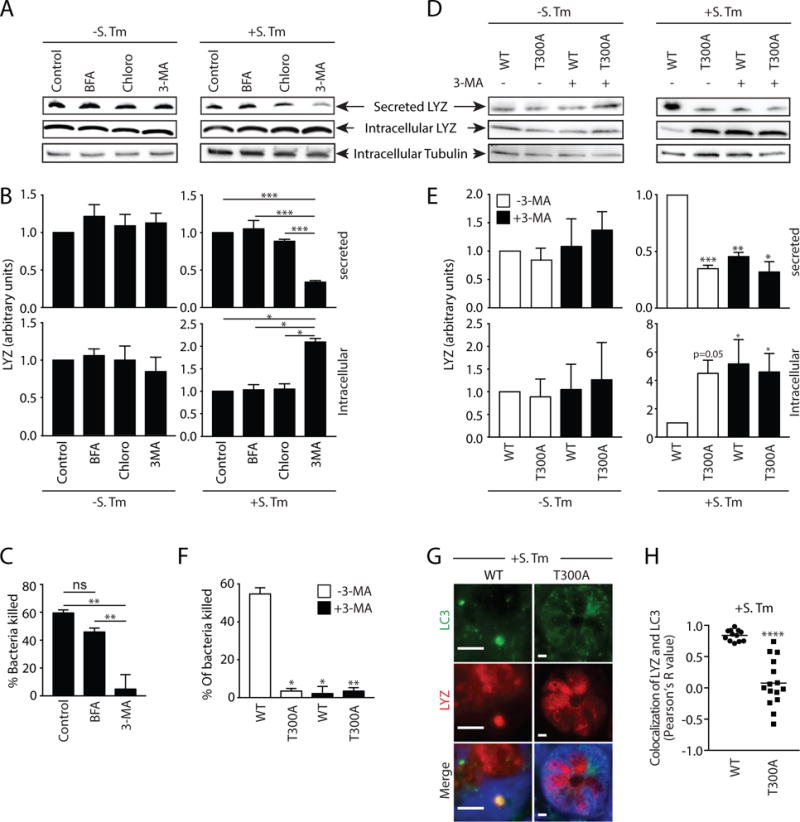
(A) Immunoblot of intracellular and secreted fractions of ex vivo small intestinal crypts. Crypts were treated as indicated and blots were detected with an anti-lysozyme antibody. (B) Quantification of data in A. (C) Bacterial killing assay against S. Typhimurium using secreted fraction from A. (D) Immunoblot of intracellular and secreted fractions of ex vivo small intestinal crypts from wild type and Atg16L1T300A (T300A) mice. Crypts were treated as indicated and blots were detected with an anti-lysozyme antibody. (E) Quantification of data in D. p values are relative to control group. (F) Bacterial killing assay against S. Typhimurium using the secreted fraction from D. (G) Immunofluorescence of LC3 and lysozyme in intestinal crypts of S. Typhimurium-infected wild type and T300A mice. (H) Quantification of LC3 and lysozyme colocalization in G. Each point represents one lysozyme-containing granule. Error bars represent SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; One-way ANOVA (B,C,E and F); Student’s t-test (H). S. Tm, Salmonella Typhimurium; LYZ, lysozyme; BFA, Brefeldin A; Chloro, chloroquine; 3-MA, 3-methyladenine; T300A, Atg16L1T300A mice.
We next studied mice in which autophagy is perturbed by a mutation in the autophagy related 16-like 1 gene (Atg16L1). A mutation in the Atg16L1 gene (T300A) confers an increased risk of developing Crohn’s disease in humans (11). Mice harboring this mutation (Atg16L1T300A) exhibit decreased antibacterial autophagy and abnormal Paneth cell lysozyme distribution (12). While crypts from uninfected wild type and Atg16L1T300A mice secreted similar amounts of lysozyme, lysozyme secretion was impaired in crypts from S. Typhimurium-infected Atg16L1T300A mice (Fig. 2D,E). Treatment of infected crypts from Atg16L1T300A mice with 3-MA did not further hinder lysozyme secretion (Fig. 2D,E), suggesting that 3-MA inhibition of lysozyme secretion in wild-type crypts was not due to off-target effects. Accordingly, crypts from Atg16L1T300A mice showed reduced bacterial killing and 3-MA did not further impact this reduction (Fig. 2F). Furthermore, infection of Atg16L1T300A mice did not produce lysozyme-filled LC3+ vesicles as observed in wild type mice (Fig. 2G,H). These results support a role for autophagy in lysozyme secretion during infection, and suggest how ATG16L1 gene mutations could lead to the aberrant lysozyme packaging and secretion that characterizes Paneth cells from Crohn’s disease patients (2).
We next sought to identify the cellular signals that trigger secretory autophagy. S. Typhimurium disrupts the ER-Golgi complex in infected cells and thus interferes with conventional secretion (13). Accordingly, S. Typhimurium induced Golgi fragmentation in Paneth cells (fig. S9), while non-invasive bacteria, including the commensal B. thetaiotaomicron and the mutant S. Typhimurium ΔinvA, did not cause Golgi breakdown (fig. S9) or trigger secretory autophagy of lysozyme (Fig. 3A,B). Disruption of the ER-Golgi complex activates ER stress pathways that maintain cellular functions (14). This response includes elevated expression of C/EBP homologous protein (CHOP)(15). CHOP levels were elevated in the intestines of S. Typhimurium-infected mice (Fig. 3C), indicating activation of the ER stress response. This suggested that ER stress might trigger secretory autophagy of lysozyme in order to circumvent the secretion block caused by ER-Golgi disruption. Supporting this idea, the ER stress inhibitor tauroursodeoxycholic acid (TUDCA)(6) reduced ER stress in infected mice as indicated by lowered CHOP expression (Fig. 3C), and also reduced secretory autophagy of lysozyme (Fig. 3D,E). Conversely, treatment of uninfected mice with the ER stress inducer thapsigargin (6) elevated CHOP expression (Fig. 3C) and induced secretory autophagy of lysozyme (Fig. 3D,E). Thus, ER stress triggers secretory autophagy of lysozyme.
Figure 3. ER stress caused by invasive bacteria triggers secretory autophagy.
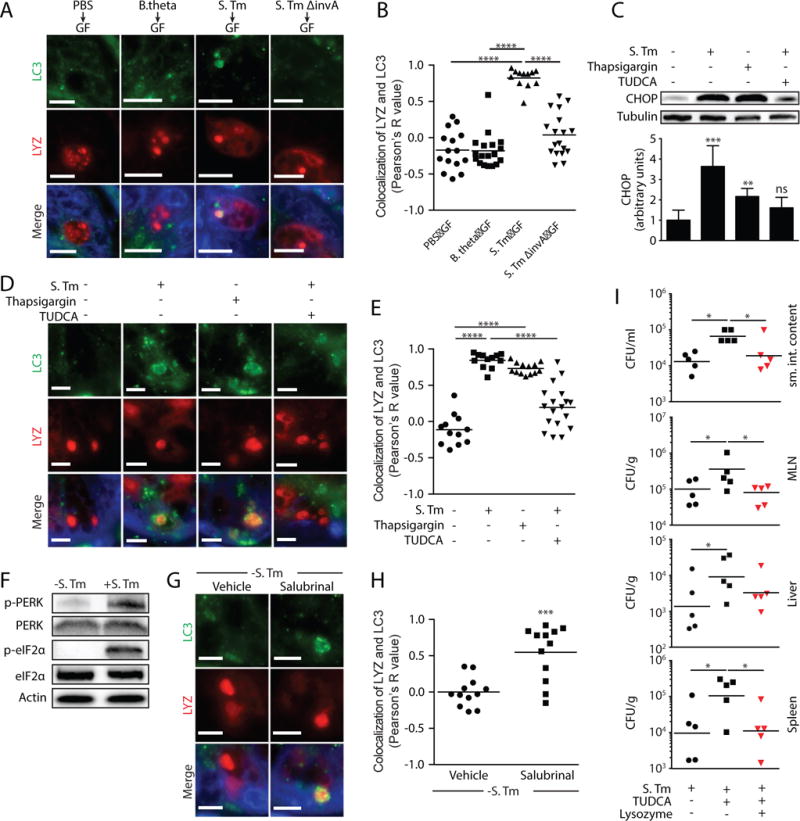
(A) Immunofluorescence of LC3 and lysozyme in intestinal crypts of germ-free (GF) mice inoculated with the indicated bacterial strains. (B) Quantification of LC3 and lysozyme colocalization in A. Each point represents one lysozyme-containing granule. (C) Representative immunoblot of small intestines from mice treated as indicated, with detection of CHOP. (D) Immunofluorescence of LC3 and lysozyme in intestinal crypts of mice treated as indicated. (E) Quantification of LC3 and lysozyme colocalization in D. Each point represents one lysozyme granule. (F) Representative immunoblot of small intestines from infected and uninfected mice, with detection of PERK and eF2α. (G) Immunofluorescence detection of LC3 and lysozyme in crypts of uninfected mice treated with vehicle or salubrinal. (H) Quantification of LC3 and lysozyme co-localization in G. Each point represents one lysozyme granule. (I) Bacterial burdens (CFU) in intestinal contents, MLNs, liver, and spleen of mice infected with S. Typhimurium and treated as indicated. Each point represents one mouse, and geometric means are shown. Error bars represent SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; One-way ANOVA (B,C,E and I); Student’s t-test (H). Scale bars=5 μm. S. Tm, Salmonella Typhimurium; LYZ, lysozyme; TUDCA, tauroursodeoxycholic acid; MLN, mesenteric lymph nodes.
We next investigated the intracellular signaling pathways that link ER stress to secretory autophagy in Paneth cells. When cells sense ER stress, protein kinase RNA-like endoplasmic reticulum kinase (PERK) is activated by phosphorylation. p-PERK then phosphorylates elongation initiation factor 2α (eIF2α) which inactivates it and attenuates translation (16). This pathway links ER stress to compensatory autophagy activation in Paneth cells (17). S. Typhimurium infection increased phosphorylation of intestinal PERK and eIF2α (Fig. 3F and fig. S10), consistent with its activation of ER stress pathways. eIF2α was not activated by infection with the non-invasive bacteria B. thetaiotaomicron and S. Typhimurium ΔinvA (fig. S11), indicating that activation depends upon bacterial invasion. Treatment of uninfected mice with salubrinal, a selective inhibitor of p-eIF2α dephosphorylation (18), promoted secretory autophagy of lysozyme in Paneth cells (Fig. 3G and H), supporting the idea that ER stress triggers secretory autophagy through the PERK-eIF2α pathway.
We next tested whether inhibiting secretory autophagy would compromise intestinal defense against oral S. Typhimurium infection. Inhibiting secretory autophagy by TUDCA treatment of S. Typhimurium-infected mice led to increased numbers of S. Typhimurium in the intestine, mesenteric lymph nodes (MLN), liver, and spleen (Fig. 3I). Lysozyme gavage of TUDCA-treated infected mice rescued the increased bacterial burden (Fig. 3I), suggesting that the increased bacterial numbers were not due to other effects of TUDCA. Thus, secretory autophagy is essential for host defense against invasive bacteria.
Activation of antibacterial autophagy in intestinal enterocytes requires epithelial cell expression of the Toll-like receptor (TLR) signaling adaptor MyD88 (7). Secretory autophagy was inhibited in Paneth cells of Myd88 deficient mice (Fig. 4A,B), producing a diffuse distribution of lysozyme similar to that seen in mice hypomorphic for Atg16L1 (3) (Fig. 4A,C). Secretory autophagy was still evident in infected mice with an epithelial cell-specific deletion of Myd88 (Myd88ΔIEC; Fig. 4A,B), indicating that epithelial cell Myd88 is dispensable. In contrast, infected mice harboring a dendritic cell (DC)-specific Myd88 deletion (Myd88ΔDC) failed to show secretory autophagy and exhibited a diffuse distribution of lysozyme (Fig. 4A–C). Thus, Paneth cell secretory autophagy requires DC MyD88.
Figure 4. A DC-ILC3 circuit controls secretory autophagy in Paneth cells.
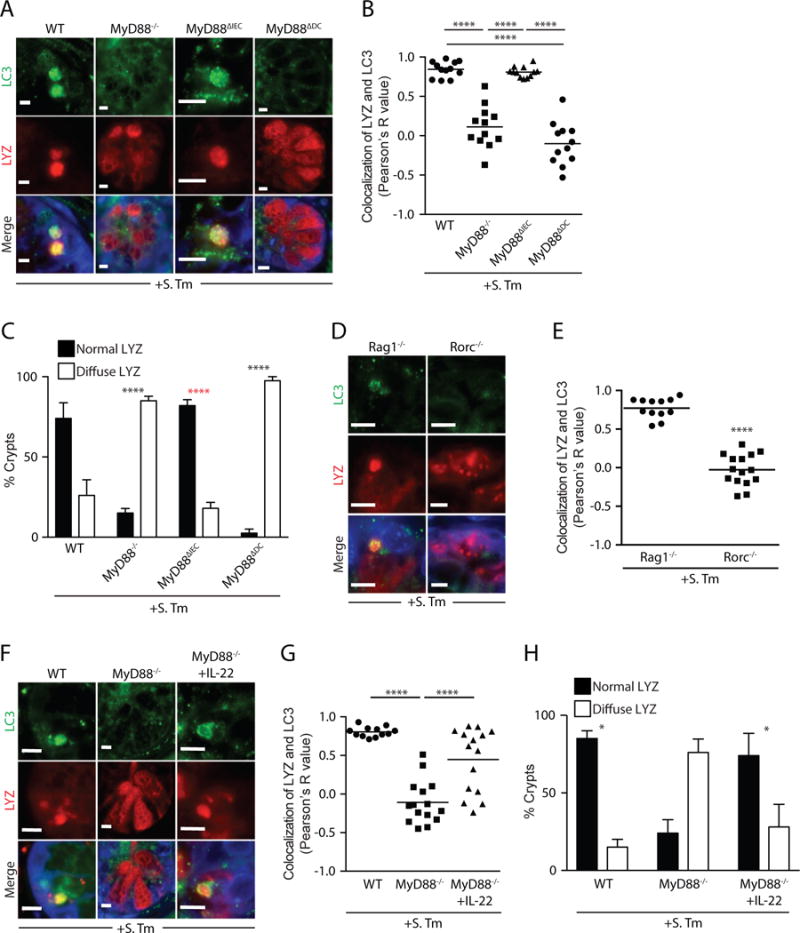
(A) Immunofluorescence of LC3 and lysozyme in intestinal crypts of S. Typhimurium-infected mice. (B) Quantification of LC3 and lysozyme co-localization in A. Each point represents one lysozyme granule. (C) Quantification of intestinal crypts displaying a diffuse lysozyme signal. p values in black are relative to WT group. p value in red is relative to MyD88−/− and Myd88ΔDC mice. (D) Immunofluorescence of LC3 and lysozyme in small intestinal crypts of S. Typhimurium-infected mice. (E) Quantification of LC3 and lysozyme colocalization in D. Each point represents one lysozyme granule. (F) Immunofluorescence of LC3 and lysozyme in intestinal crypts of S. Typhimurium-infected mice. (G) Quantification of LC3 and lysozyme colocalization in F. Each point represents one lysozyme granule. (H) Quantification of small intestinal crypts displaying a diffuse lysozyme signal. Error bars represent SEM. *p<0.05, ****p<0.0001; Student’s t-test (E); One-way ANOVA (B,G) Two-way ANOVA (C,H). Scale bars=5 μm. S. Tm, Salmonella Typhimurium; LYZ, lysozyme.
The requirement for DC Myd88 suggested the involvement of a known cellular relay in which DC TLRs capture bacterial signals and relay them to epithelial cells via type 3 innate lymphoid cells (ILC3) and their secretion of interleukin-22 (IL-22)(19). While secretory autophagy occurred upon infection of mice lacking T cells (Rag1−/−), it was inhibited in infected Rorc−/− mice which lack both T helper 17 (TH17) cells and ILC3 (Fig. 4D,E). This suggests that ILC3 are essential for secretory autophagy of lysozyme. Supporting the requirement for ILC3, treatment of infected Myd88−/− mice with recombinant IL-22 rescued the diffused distribution of lysozyme and restored secretory autophagy of lysozyme in Paneth cells without affecting lysozyme transcript levels (Fig. 4F–H; fig. S12). These results argue that Paneth cell secretory autophagy requires activation of the DC-ILC3 circuit, which may provide a cell-extrinsic licensing signal that allows secretory autophagy to be rapidly activated upon detection of Paneth cell-intrinsic ER stress.
Our results illuminate how the intestine preserves antimicrobial function in the face of a pathogenic bacterial infection (fig. S13), and suggest how simultaneous disruption of both ER stress and autophagy pathways leads to severe inflammation in mice (17). Our findings also provide potential clues about how inflammation can arise in human inflammatory bowel disease. Genes that govern both the ER stress response and autophagy are frequently mutated in people with Crohn’s disease (2, 4). Since disruption of either pathway precludes secretory autophagy, our results suggest how infection of intestinal epithelial cells could trigger chronic inflammation in people with these genetic abnormalities.
Supplementary Material
One sentence summary.
During pathogen-induced cellular stress Paneth cells reroute a key antimicrobial protein through an autophagy-based secretion pathway.
Acknowledgments
We thank Beth Levine for helpful discussions and C.L. Behrendt-Boyd for assistance with mouse experiments. This work was supported by the NIH (DK070855 to L.V.H.; AI118807 and AI128151 to S.E.W.), the Burroughs Wellcome Foundation (Investigators in the Pathogenesis of Infectious Diseases Award to L.V.H.), the Welch Foundation (I-1874 to L.V.H.; I-1858 to S.E.W.), and the Howard Hughes Medical Institute (L.V.H.). S.B. was supported by a Gruss-Lipper postdoctoral fellowship.
Footnotes
References
- 1.Ramanan D, Cadwell K. Cell Host Microbe. 2016;19:434–441. doi: 10.1016/j.chom.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadwell K, et al. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadwell K, et al. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaser A, et al. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celli J, Tsolis RM. Nat Rev Microbiol. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keestra-Gounder AM, et al. Nature. 2016;532:394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamin JL, Sumpter R, Levine B, Hooper LV. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Yoshimori T, Levine B. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponpuak M, et al. Curr Opin Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupont N, et al. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khor B, Gardet A, Xavier RJ. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lassen KG, et al. Proc Natl Acad Sci U S A. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escoll P, Mondino S, Rolando M, Buchrieser C. Nat Rev Microbiol. 2015;14:5–19. doi: 10.1038/nrmicro.2015.1. [DOI] [PubMed] [Google Scholar]
- 14.Preston AM, Gurisik E, Bartley C, Laybutt DR, Biden TJ. Diabetologia. 2009;52:2369–2373. doi: 10.1007/s00125-009-1506-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang XZ, et al. Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oslowski CM, Urano F. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adolph TE, et al. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyce M, et al. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 19.Thaiss CA, Zmora N, Levy M, Elinav E. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 20.Adachi O, et al. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 21.Vaishnava S, et al. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou B, Reizis B, DeFranco AL. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mombaerts P, et al. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 24.Sun Z, et al. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 25.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J, et al. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinchuk V, Wu Y, Grossenbacher-Zinchuk O. Sci Rep. 2013;3:1365. doi: 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


