Abstract
The application of modern high-throughput genomics to the study of cancer genomes has exploded in the past few years, yielding unanticipated insights into the myriad and complex combinations of genomic alterations that lead to the development of cancers. Coincident with these genomic approaches have been computational analyses that are capable of multiplex evaluations of genomic data toward specific therapeutic end points. One such approach is called “immunogenomics” and is now being developed to interpret protein-altering changes in cancer cells in the context of predicted preferential binding of these altered peptides by the patient’s immune molecules, specifically human leukocyte antigen (HLA) class I and II proteins. One goal of immunogenomics is to identify those cancer-specific alterations that are likely to elicit an immune response that is highly specific to the patient’s cancer cells following stimulation by a personalized vaccine. The elements of such an approach are outlined herein and constitute an emerging therapeutic option for cancer patients.
Cancer immunologists developed experimentally testable hypotheses around the idea that mutated proteins in cancer cells provided favored targets for immune response in the mid-1980s (De Plaen et al. 1988; Monach et al. 1995). Their hypotheses were inspired by earlier observations of immune-capable mice that developed spontaneous cancers and, after removal and rechallenge, exhibited resistance to their original cancers (Foley 1953; Prehn and Main 1957; Old 1982). This implied that cancer-specific immunity was a plausible resistance mechanism, but the mechanism for immunity was unclear. Experimental approaches to test for the presence of tumor-specific mutant antigens or “neoantigens” were aided by discoveries in immunology that elucidated the role and functions of the human leukocyte antigen (HLA) proteins (Bjorkman et al. 1987; Babbitt et al. 2005), and by the advent of molecular cloning techniques. This combination led to the cloning and identification of the first tumor neoantigen by Boon and colleagues, a peptide called MAGE-A1 that was derived from a melanoma patient sample (van der Bruggen et al. 1991). As exciting as this proof of hypothesis was at the time, the laborious and lengthy procedures required to identify a single tumor-specific neoantigen that was highly unique to the patient’s disease meant there was no clear trajectory for this approach in a clinical setting.
IMMUNOGENOMICS AND NEOANTIGEN PREDICTION
Several recent developments have made it more plausible to revisit the notion of identifying tumor-unique neoantigenic peptides, however. In particular, the development and widespread use of next-generation sequencing (NGS) platforms has figured prominently in this trend (Mardis 2017). Introduced in the mid-2000s, these instruments and their accompanying techniques led to the first cancer whole-genome sequencing study that compared a tumor with normal genome for a single patient, establishing that somatic mutations could be discovered in the comparison (Ley et al. 2008). Selective hybridization-based techniques known as “hybrid capture” emerged in 2009 and permitted isolation and sequencing of only the known protein-coding exons from a whole-genome library (the exome). This “whole-exome sequencing” approach significantly simplified data analysis for the identification of DNA-level changes that were somatic (i.e., specific to the tumor cells) and led to changes in the amino acid sequences of the resulting proteins. The foundational underpinning for mutation discovery lies in the reference human genome sequence, which serves as a template for NGS read alignment. Over time, the increasing length of NGS reads, the advent of read-pair data (both ends of each library fragment are sequenced), and the improved mapping algorithms have led to overall improvements in the quality of NGS read alignment on the highly repetitive and complex human genome reference.
Beyond these advances, the most important step in this process—namely, the proper identification of variants in aligned NGS read data—has also improved over time. There are various algorithmic approaches to somatic variant identification from NGS data that typically are highly tuned to the type of variant being detected. The basic workflow involves detecting variants in the aligned read data from the tumor sample, and separately in the normal sample, and then comparing the variants at each locus to eliminate those shared between tumor and normal from further consideration because they are constitutional or “germline” in origin (Ding et al. 2010). In practice, each variant identification algorithm has strengths and weaknesses (Griffith et al. 2015), and therefore most data analysis approaches to somatic variant identification use more than one variant caller and consider most strongly the shared or consensus variant lists called by each algorithm as likely true positives. Furthermore, variant detection accuracy is a function of data coverage, so most variant detection approaches utilize a pre- or post-variant call filtering scheme to remove all variants identified at loci with suboptimal coverage depth. Generally speaking, detection of somatic single-nucleotide variants (SNVs) or point mutations is the most straightforward variant type to identify. Detecting the insertion or deletion (indel) of one or more nucleotides is more difficult, as gapped alignment or assembly of the sequence read data that define the indel is required. Here, some proportion of the indel-containing reads will not map without special alignment algorithms, because of the need to produce a gapped alignment with sufficient length of sequence similarity to anchor the read accurately to the surrounding sequence. As a result, coverage is almost always lower at indel variant sites because of read mapping difficulties. Finally, structural variants are the most difficult to detect with high accuracy. These variants include chromosomal-scale events such as inversions, translocations, and deletions and are detected based on mismapped read-pair data following NGS read alignment. Both indels and structural variants, when they occur in protein-coding genes, can provide important and potentially immunogenic changes to the amino acid sequences of expressed proteins in a cancer cell.
The next step is annotating the variants that have been identified, by interpreting their altered nucleotide sequences in the context of their protein-coding impact. This is straightforward in SNVs, but indels that result in a shift in the open reading frame (frameshift) and gene-fusing structural variants are more difficult to interpret properly. Automated annotation software such as Variant Effect Predictor (VEP) (McLaren et al. 2016) or ANNO-VAR (Wang et al. 2010) typically is used for variant interpretation. For many genes in the human genome, multiple transcripts have been identified, which complicates the annotation process because one must specify which transcript is to be annotated. In the absence of adequate RNA-seq data to infer the correct transcript, one typically annotates from the transcript with the highest evidence for expression. Tissue-specific expression data sets, such as GTEx (www.gtexportal.org/), also can be consulted for transcript information, if desired. Once the variants and their proper notation are determined along with their flanking sequences, the results are parsed for use in the neoantigen detection schema described below. Typically for major histocompatibility complex (MHC) class I restricted neoantigen discovery, we tile through the variant-containing region of each protein with peptides of eight to 11 amino acids to generate the test sequences for neoantigen evaluation. For MHC class II restricted neoantigen discovery, the tiled peptides are 15 amino acids in length.
Stringent alignment of NGS reads from the normal tissue comparator onto the HLA coding genes permits the haplotype of each patient to be determined from NGS exome data. There are several approaches to HLA typing from NGS data, including HLAminer (Warren et al. 2012), OptiType (Szolek et al. 2014), and ATHLATES (Liu et al. 2013). HLAminer and OptiType also can call HLA haplotypes from RNA-seq data. In high-mutation-load tumor types, it also is advised to evaluate and annotate the HLA gene sequences from the tumor tissue NGS data, because there may be inactivating (nonsense or frameshift) mutations in one or more of these genes that would preclude further consideration of these mutation-inactivated HLA proteins in neoantigen prediction.
The final step in neoantigen discovery is the prediction of binding affinities for each novel somatic mutation-translated set of peptides to the patient’s HLA proteins. The development of modeling algorithms that calculate HLA binding affinities has been an active area of research that has resulted in several approaches such as analysis by neural network-based algorithms that are trained on measured binding affinities (e.g., NetMHC), scattering-matrix method (SMM)-based approaches, and others (Peters and Sette 2005; Lundegaard et al. 2008; Srivastava et al. 2013). The large number of algorithms indicates the attendant difficulty of identifying neoantigens, although precision has improved over time, especially for class I HLA proteins, for which more experimental data exist, even for the more rare haplotypes. Class II neoantigen binding is more difficult to predict because of the nature of the peptide binding site, which is open at both ends, permitting the flanking amino acids around the nine-amino-acid-core binding motif to influence binding affinity in a context-dependent manner (Paul et al. 2015).
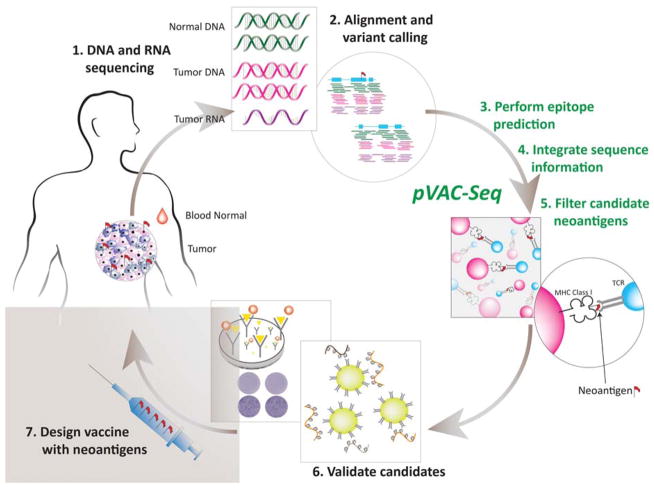
Because of the complexities of the aforementioned set of steps required to generate predicted neoantigens, our group published a computational pipeline called pVAC-Seq (personalized Variant Antigens by Cancer Sequencing) that is designed to perform these steps in sequence (Hundal et al. 2016). pVAC-Seq requires several input data, including an annotated list of somatic variants, comparison sequences from the native peptide correlates, predicted HLA class I haplotypes for the patient, and RNA-seq-derived data regarding mutant expression levels for each of the input peptides. The overall workflow for neoantigen prediction is represented in Figure 1.
Figure 1.
Generalized workflow diagram for identification of tumor-specific mutant antigens (“neoantigens”) from next-generation sequencing (NGS) data. MHC, major histocompatibility complex; TCR, T-cell receptor.
Neoantigen prediction can be used clinically to achieve several therapeutic goals, depending on the nature of the study. Two case reports are presented to illustrate the clinical utility of neoantigen prediction to (1) predict neo-antigen load as a consequence of an identified mismatch repair gene defect and as an indicator of likely response to checkpoint blockade therapy, (2) identify changes in a temporal series of tumors from the same individual patient, and (3) design a personalized vaccine for a patient with recurrent cancer.
CASE REPORTS
We recently reported (Johanns et al. 2016) on an individual case of a male patient (early 30s) with a history of colon polyps who presented with a brain tumor that was removed by craniotomy and diagnosed as a glioblastoma multiforme (GBM), World Health Organization (WHO) grade IV with primitive neuroectodermal tumor (PNET) features. Following resection, the patient received radiation and temozolomide (TMZ) treatment per standard of care, but new symptoms subsequent to this treatment during high-dose maintenance TMZ led to the laminectomy and diagnosis by histopathology of a drop spinal metastasis. Intermediate to the primary and secondary diagnoses, a panel gene test (Foundation One) identified an abnormally elevated mutation rate, and a reflex assay of blood DNA for germline susceptibility identified a known pathogenic mutation in polymerase epsilon (POLE L424 V). Based on this result, the patient received a PD-1 checkpoint blockade therapy (pembrolizumab) for about 3 wk following the removal of the drop metastasis. Upon experiencing further complications after 3 wk of pembrolizumab therapy, the patient was subsequently diagnosed with a second drop metastasis lower on the spinal cord. This tumor also was resected by laminectomy and diagnosed as a GBM with PNET features by histopathology. Since all three occurrences of this patient’s cancer were banked, we isolated DNA and RNA from each sample. An exome capture-based sequencing comparison of all three tumors to a matched blood normal from the patient coupled with RNA-seq from each of the three tumors were used to characterize the patient’s changes in tumor heterogeneity and neoantigen load over time. We also performed immunohistochemistry (IHC) on slides from each tumor resection sample to identify whether the use of checkpoint blockade therapy had influenced the tumor infiltration of different immune molecules behind the blood–brain barrier.
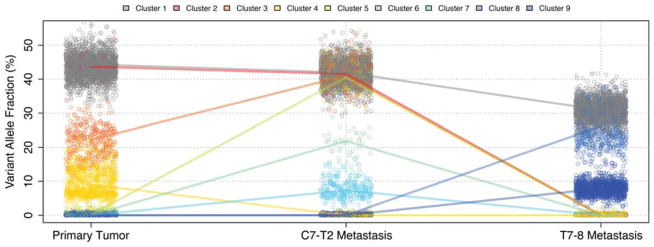
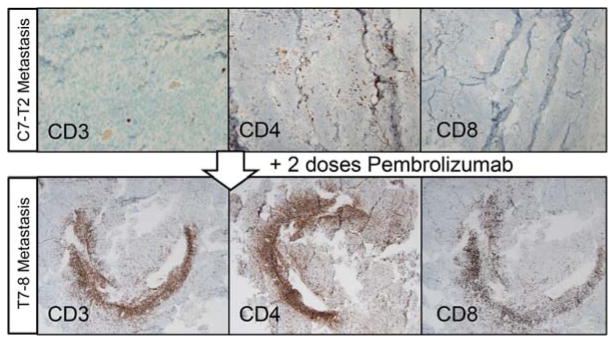
As demonstrated in our retrospective sequencing and analysis, genomic characterization of a tumor series can verify an anticipated “hyper” mutation rate for patients with a POLE pathogenic mutation and can illustrate response to treatment in the form of dramatic changes in tumor heterogeneity and subclonal tumor cell populations. This was especially evident in the comparison of the first drop metastasis to the second, with an anti PD-1 therapy intermediate (Fig. 2). Similarly, we utilized our genomic data to identify significant flux in the neoantigens identified in all three tumors. Our comparative analysis of RNA expression data obtained from each tumor illustrated the significant changes in immune cell infiltrates following anti-PD1 checkpoint blockade therapy, and these were supported by IHC studies that further illustrated the significant influx of immune cells following anti-PD1 therapy (Fig. 3). Importantly, this study provided the first combined genomic and molecular evidence that checkpoint blockade could influence immune activity behind the blood–brain barrier in a central nervous system (CNS) tumor.
Figure 2.
Comparison of tumor heterogeneity and subclonal population variation in sequential samples from a single patient. (Reprinted, with permission, from Johanns et al. 2016, © AACR.)
Figure 3.
Immunohistochemistry-based evaluation of sequential glioblastoma multiforme (GBM) samples for immune infiltration before and after anti PD-1 therapy. (Reprinted, with permission, from Johanns et al. 2016, © AACR.)
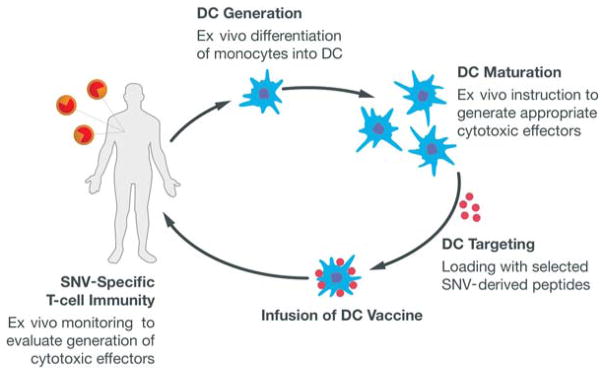
We also have used immunogenomic methods to identify neoantigens in individual patient cancers for the purposes of designing a personalized vaccine-based therapeutic (Carreno et al. 2015). This work built upon proof-of-concept studies completed with Robert Schreiber’s laboratory using carcinogen-induced sarcoma mouse models with a high mutational load. These studies investigated whether the combination of exome sequencing and RNA sequencing could identify neoantigens in this tumor model (Matsushita et al. 2012) and further evaluated whether a peptide vaccine based on the identified neoantigens could be used to vaccinate the mouse and lead to the eradication of the tumor (Gubin et al. 2014). In the human vaccine trial, the pVAC-Seq pipeline (Hundal et al. 2016) identified neoantigens from melanoma samples in three patients with BRAF-mutated melanomas that had received prior treatment with a BRAF small-molecule-inhibitor therapy and with an anti-CTLA4 checkpoint blockade therapy (ipilimumab). After identifying neoantigens, the vaccines were produced by isolating dendritic cells from each patient and culturing them to maturity based on established in vitro methods, and then coupling the mature dendritic cells with synthetic peptides for each unique neoantigen plus two control peptides (based on gp-100 shared antigens). These dendritic cell cultures were infused into the patients in a series of three vaccine infusions at 2-wk intervals. The overall workflow is shown in Figure 4.
Figure 4.
Overall workflow for creating a dendritic cell (DC) vaccine based on mis-sense somatic point mutations (single-nucleotide variants [SNVs]) that generate putative neoantigens.
Postvaccination, we then monitored each patient’s response using flow cytometry–based dextramer assays designed to specifically identify which of the neoantigens were eliciting a specific CD8+ T-cell response. In each case, we saw strong responses from three of the seven neoantigens utilized per patient (each set of neoantigens is completely unique to each patient), and we saw strong responses from our control gp-100 peptides. To further investigate the T-cell response, we evaluated the complexity of the T-cell receptor (TCR) repertoire using the Adaptive Biosciences immunoSEQ assay (http://www.adaptivebiotech.com/immunoseq), which also uses next-generation sequencing to provide the TCR data for analysis. Our results demonstrated that, for neoantigens with a discernable response by dextramer-flow cytometry assay, the corresponding T cells showed increased diversity in the TCR repertoire post-vaccine compared with pre-vaccine.
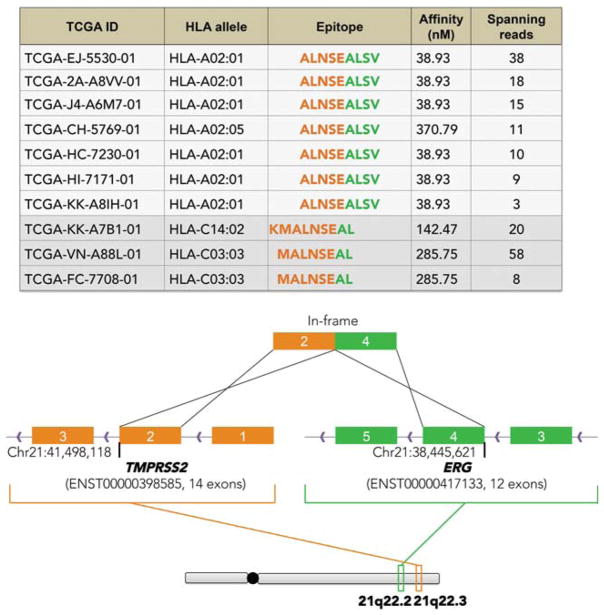
These initial studies have led to new clinical trials of personalized vaccines in tumor types with a lower average mutational load than UV-associated melanomas, such as triple-negative breast cancer. Here, the challenge is to identify a sufficient number of neoantigens using the pVAC-Seq pipeline, for the purposes of designing the vaccine. To address this, we have been expanding the types of variants considered in our analysis beyond SNVs (point mutations). For example, although they are less frequent and more difficult to identify precisely, insertion or deletion (indel) variants can shift the open reading frame of a protein, thereby creating a novel tumor-unique peptide that may be a neoantigen. Similarly, the identification of structural variants that generate fusion proteins can also potentially encode a neoantigenic peptide at the junction sequence of the two proteins. We recently used an approach to mine fusion transcripts from cancer RNA-seq data for purposes of evaluating neoantigenicity of predicted fusions. The implementation of this approach, called INTEGRATE-Neo, was tested in the context of TCGA prostate cancer data (exome and RNA-seq) for a common fusion peptide created by the translocation that fuses TMPRSS2 to ERG in ~50% of prostate cancers. As shown in Figure 5, the HLA binding energy calculated for this novel fusion peptide varies across patients according to HLA haplotype. Like class I neoantigens, we also wish to evaluate class II restricted neoantigens and add these to our vaccines. We are actively pursuing these and other changes to our neoantigen discovery pipeline (https://github.com/griffithlab/pVAC-Seq).
Figure 5.
Evaluation of human leukocyte antigen (HLA) binding affinity for a common fusion peptide in prostate cancer (TMPRSS2-ERG) as evaluated by INTEGRATE-Neo. (Reprinted from Zhang et al. 2016, by permission of Oxford University Press.)
Finally, we would like to expand our personalized neo-antigen vaccine approaches to tumor types with significantly lower mutational burden, such as pediatric cancers. In particular, the opportunity to pursue these studies lies in patients emerging with recurrent disease, post-standard-of-care therapies such as radiation and alkylating chemotherapies like TMZ. In a study published by Costello’s laboratory, ~30% of pediatric glioma patients with recurrent tumors post-radiation and TMZ demonstrated a significant elevation in their tumor mutational burden posttherapy (van Thuijl et al. 2015). These children might represent reasonable subjects for a clinical trial to evaluate a personalized neoantigen vaccine in the recurrent setting. In this regard, since the TMZ-induced mutations are subclonal (i.e., not in the founder clone), we would need to include a combination of founder clone variants from the primary tumor and newer, TMZ-induced subclonal variants to increase the likelihood of clinical benefit.
REMAINING QUESTIONS IN IMMUNOGENOMICS
Although these new applications of genomics to clinical cancer diagnostics and therapeutics are exciting and potentially transformational, we have many remaining questions to answer. Examples include questions about the optimal number and types of neoantigens that are ideal for vaccine efficacy, the type of vaccine platform (dendritic cell–based, peptide-based, DNA-based, and so on) that is scalable, rapid to produce, and affordable so that many patients can benefit (if indeed, benefit is demonstrated). Better information about how patients respond to a vaccine across the spectrum of tumor burden is needed, so we can more effectively design our clinical trials. For example, are patients with advanced tumor burden less likely to have a clinical response or benefit because the immune system, although activated by the vaccine, cannot respond fast enough to eradicate their cancers? Conversely, do patients require some minimal level of tumor burden to achieve a benefit by giving the T cells elicited by the vaccine a target to attack? And finally, with a tumor burden eliminated by surgery, will the immune surveillance remain? Or will surveillance potentially be eluded by the emergence of new tumors that have suppressed the expression of the neoantigens targeted by the vaccine? Since this phenomenon of neoepitope editing already has been demonstrated in acquired resistance to checkpoint blockade therapies (Anagnostou et al. 2016), it is likely also in the therapeutic vaccine setting.
CONCLUSION
The evolving applications of modern genomics and computational analysis to the immunology-based study of cancer have transformed our studies since these central concepts of cancer immunology were hypothesized in the 1980s. Certainly, the facility with which somatic mutations can be identified in individual tumor samples has encouraged progress, but vital contributions were required toward the algorithmic evaluation of mutant peptides in the context of each patient’s HLA haplotypes and in devising novel immunotherapeutics. Immunogenomics has been demonstrated to be clinically informative in several early studies, outlined here, and in research that builds the case for clinical translation. As our level of sophistication increases in identifying neoantigens across the spectrum of mutation types and HLA classes, foundational knowledge about cancer immunology will result as will our understanding of how best to apply this information to clinical care.
Acknowledgments
We acknowledge our clinical collaborators on the case studies reported, including Beatriz Carreno, Gerald Linette, Gavin Dunn, and Tanner Johanns, and we thank Robert Schreiber for his incredible guidance and encouragement to pursue these studies. We acknowledge the patients who were studied herein and thank them for their courage and willingness to contribute to clinical research.
References
- Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2016;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. 1985. J Immunol. 2005;175:4163–4165. [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH, Mardis ER, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Plaen E, Lurquin C, Van Pel A, Mariamé B, Szikora JP, Wölfel T, Sibille C, Chomez P, Boon T. Immunogenic (tum−) variants of mouse tumor P815: Cloning of the gene of tum− antigen P91A and identification of the tum− mutation. Proc Natl Acad Sci. 1988;85:2274–2278. doi: 10.1073/pnas.85.7.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: Accomplishments and challenges. Hum Mol Genet. 2010;19:R188–R196. doi: 10.1093/hmg/ddq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EJ. Antigenic properties of methylcholanthrene-induced tumors in mice of the strain of origin. Cancer Res. 1953;13:835–837. [PubMed] [Google Scholar]
- Griffith M, Miller CA, Griffith OL, Krysiak K, Skidmore ZL, Ramu A, Walker JR, Dang HX, Trani L, Larson DE, et al. Optimizing cancer genome sequencing and analysis. Cell Syst. 2015;1:210–223. doi: 10.1016/j.cels.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanns Tanner M, Miller CA, Dorward IG, Tsien C, Chang E, Perry A, Uppaluri R, Ferguon C, Schmidt RE, Dahiya S, et al. Immunogenomics of hypermutated glioblastoma: A patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6:1230–1236. doi: 10.1158/2159-8290.CD-16-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, Pfeifer JD. ATHLATES: Accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: Accurate web accessible predictions of human, mouse and monkey MHC Class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36(Web Server):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. DNA sequencing technologies: 2006–2016. Nat Protoc. 2017;12:213–218. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- Old LJ. Cancer immunology: The search for specificity. Natl Cancer Inst Monogr. 1982;60:193–209. [PubMed] [Google Scholar]
- Paul S, Lindestam Arlehamn CS, Scriba TJ, Dillon MB, Oseroff C, Hinz D, McKinney DM, Carrasco Pro S, Sidney J, Peters B, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Methods Immunol. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–778. [PubMed] [Google Scholar]
- Srivastava A, Ghosh S, Anantharaman N, Jayaraman VK. Hybrid biogeography based simultaneous feature selection and MHC class I peptide binding prediction using support vector machines and random forests. J Immunol Methods. 2013;387:284–292. doi: 10.1016/j.jim.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: Precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, Malmström A, Hallbeck M, Heimans JJ, Kloezeman JJ, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL, Choe G, Freeman DJ, Castellarin M, Munro S, Moore R, Holt RA. Derivation of HLA types from Shotgun sequence datasets. Genome Med. 2012;4:95. doi: 10.1186/gm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mardis ER, Maher CA. INTEGRATE-neo: A pipeline for personalized gene fusion neoantigen discovery. Bioinformatics. 2016;2016 doi: 10.1093/bioinformatics/btw674. pii: btw674. [DOI] [PMC free article] [PubMed] [Google Scholar]