Abstract
It has long been known that pseudouridine (Ψ) is the most abundant modified nucleotide in stable RNAs, including tRNA, rRNA, and snRNA. Recent studies using massive parallel sequencing have uncovered the presence of hundreds of Ψs in mRNAs as well. In eukaryotes and archaea, RNA pseudouridylation is introduced predominantly by box H/ACA RNPs, RNA–protein complexes each consisting of a single RNA moiety and four core proteins. It has been well established that Ψ plays an essential role in regulating the structure and function of stable RNAs in several model organisms, including yeast, Xenopus laevis, and humans. However, the functional role of Ψ in mRNA remains to be elucidated. One possibility (and true for stop/termination codons) is that Ψ influences decoding during translation. It is imperative, therefore, to establish a system, in which one can site-specifically introduce pseudouridylation into target mRNA and biochemically test the impact of mRNA pseudouridylation on protein translation. Here, we present a method for (1) site-specific conversion of uridine into Ψ in mRNA by designer box H/ACA RNP, (2) detection of Ψ in target mRNA using site-specific labeling followed by nuclease digestion and thin layer chromatography, and (3) analysis of recoding of pseudouridylated premature termination codon in mRNA during translation.
1. INTRODUCTION
Once dubbed the fifth nucleotide, pseudouridine (Ψ) is the most abundant posttranscriptionally modified nucleotide, constituting ~5% of total RNA ribonucleotides (Davis & Allen, 1957). The conversion of uridine to Ψ (pseudouridylation) requires two distinct chemical reactions: the breaking of the C1′—N1 glycosidic bond and the making of a new carbon–carbon (C1′—C5) bond that relinks the base to the sugar (Wu, Yu, Kantartzis, & Yu, 2011). Pseudouridylation is a true isomerization reaction, which creates an extra hydrogen bond donor.
Earlier biophysics studies using NMR revealed that Ψ, when present in RNA, favors the formation of 3′-endo conformation and promotes base stacking, thus stabilizing RNA structures (Davis, 1995; Lee & Tinoco, 1980). In line with this, computational simulation studies have independently established the role for Ψ in stabilizing the structure of the anticodon stem loop of tRNA (Bilbille et al., 2009; Durant & Davis, 1999). In addition, it has been reported that loss of a single Ψ in the tRNA anticodon stem loop results in a decrease in the efficiency of recoding (Lecointe et al., 2002). Furthermore, experimental results indicate that Ψs in U2 snRNA contribute to the formation of active U2 snRNP and the spliceosome, and are thus important for splicing in Xenopus oocytes (Yu, Shu, & Steitz, 1998). Likewise, Ψs in rRNAs are concentrated in functional important regions, such as peptidyl transferase center and decoding center, and play important roles in protein synthesis. Specifically, abolishing these Ψs severely affects ribosome functions, resulting in reduced rate of translation and increased stop-codon readthrough and frame shifting (King, Liu, McCully, & Fournier, 2003; Liang, Liu, & Fournier, 2007).
In eukaryotes and archaea, pseudouridylation is introduced predominantly by box H/ACA RNPs, each of which contains a unique small RNA (box H/ACA RNA) and four core proteins (NAP57/dyskerin/Cbf5, Nhp2, Nop10, and Gar1) (Karijolich & Yu, 2008). The RNA component serves as a guide that specifies, through base-paring interaction with its substrate RNA, the target uridine for pseudouridylation (Ge & Yu, 2013). One of the four core proteins, NAP57/dyskerin/Cbf5, catalyzes the chemical reactions, converting the target uridine to Ψ (Huang, Karijolich, & Yu, 2011).
Based on this guide-substrate base-pairing scheme, Karijolich and Yu designed an artificial box H/ACA RNA to introduce Ψ into mRNA at a premature termination codon (PTC). They demonstrated that Ψ was indeed incorporated into TRM4 mRNA at the PTC, and remarkably, pseudouridylated PTC promotes nonsense suppression by altering ribosome decoding (Fernandez et al., 2013; Karijolich & Yu, 2011). Recently, three groups developed Ψ-seq and identified hundreds of naturally occurring Ψs in both yeast and human mRNAs (Carlile et al., 2014; Lovejoy, Riordan, & Brown, 2014; Schwartz et al., 2014). Many of these novel Ψs reside in the coding regions, and the majority of them respond to environmental stress, indicating functional significance (Carlile et al., 2014).
Here, we present a protocol that details how to introduce and detect Ψs in mRNA, and how to analyze the protein products generated by Ψ-mediated recoding (nonsense suppression). Starting with the plasmid bearing the C-terminally tagged TRM4 gene, we create a PTC in this gene using site-directed mutagenesis. We then construct a second plasmid containing an artificial box H/ACA RNA gene (based on snR81) (Ma et al., 2005) designed to target the uridine of the PTC in TRM4 mRNA for pseudouridylation. Transformation of yeast cells with these two plasmids allows site-specific pseudouridylation at the PTC, leading to the production of a full-length PTC-readthrough protein, which can readily be detected by Western blotting analysis.
2. INCORPORATION OF PSEUDOURIDINE IN TRM4 mRNA
2.1 Overview
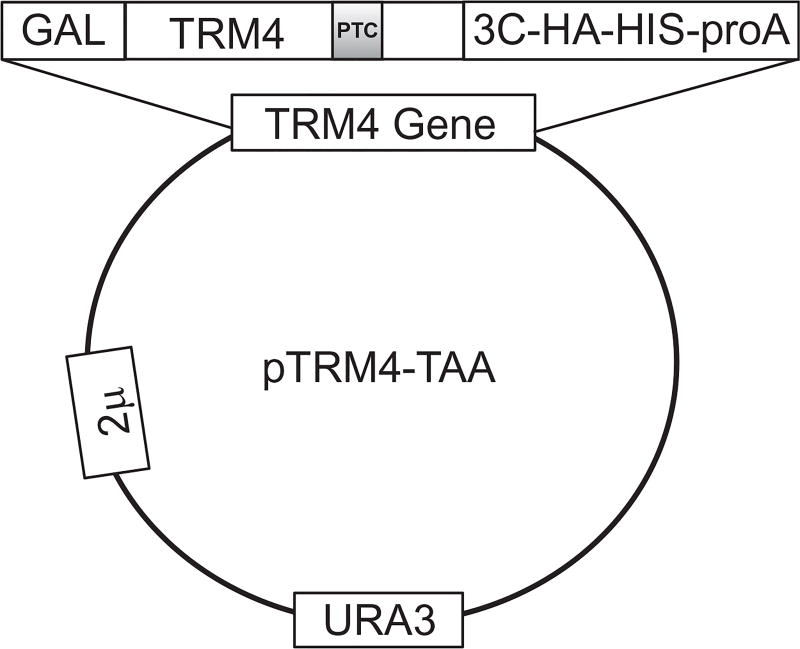
All naturally occurring ORF stop codons, UAA, UGA, and UAG, contain a uridine in the first position. To study Ψ-mediated nonsense suppression, we choose a plasmid containing a C-terminally tagged (3C-HA-HIS6X-Protein A) reporter gene, TRM4, which, when transformed into yeast cells, is highly expressed (Chernyakov, Whipple, Kotelawala, Grayhack, & Phizicky, 2008). Using PCR-based site-directed mutagenesis, we then introduce a PTC into the ORF of TRM4 at codon 602 (TTT-to-TAA change), as shown in Fig. 1.
Figure 1.
Schematic representation of pTRM4-TAA expression plasmid. The wild-type TRM4 gene is mutated at F602 position (TTT converted to TAA) to generate a premature termination codon (PTC) (indicated). The plasmid-borne TRM4 gene is fused with a multicomponent C-terminal tag to facilitate the detection and purification of full-length Trm4 product. The tag contains a 3C protease cleavage site that can be used to release the purified protein from the tag. The expression of TRM4 gene is under the control of Gal promoter, which is galactose-inducible. URA3 is an auxotroph selective marker in yeast. This is a high copy 2 µ plasmid.
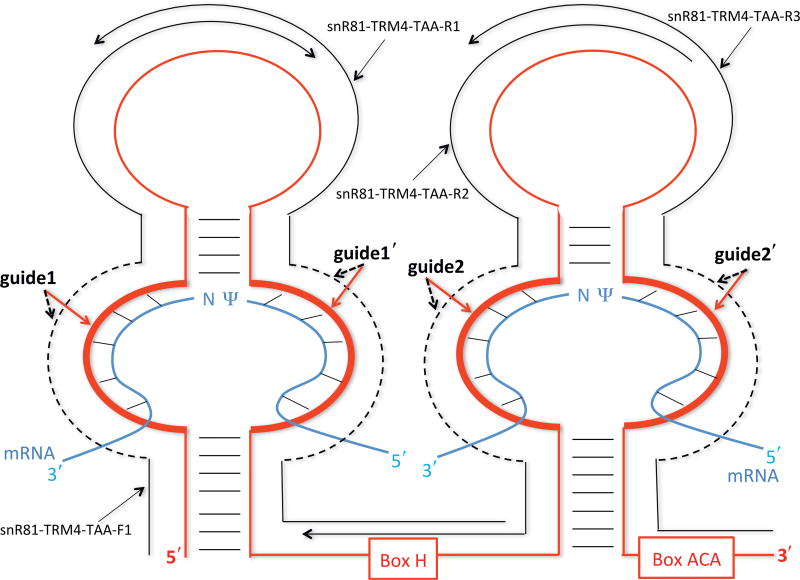
To introduce Ψs into PTC, we generate an artificial box H/ACA guide RNA (based on yeast snR81 box H/ACA RNA) (Ma et al., 2005), in which the guide sequences are designed to target the uridine of the PTC codon for pseudouridylation. The artificial box H/ACA RNA is amplified by PCR with four overlapping primers. The first primer (snR81-TRM4-TAA-F1) is a sense forward primer and the other three (snR81-TRM4-TAA-R1, snR81-TRM4-TAA-R2, and snR81-TRM4-TAA-R3) are antisense reverse primers (Figs. 2 and 3). Within each primer, there is a tract of nucleotides that represents the guide sequence (indicated as guide1, guide1′, guide2, and guide2′ in Figs. 2 and 3). Guide1 and guide1′ form the first (5′) pseudouridylation pocket, and guide2 and guide2′ form the second (3′) pseudouridylation pocket in the guide RNA (Fig. 2). The guide sequences in both pseudouridylation pockets of the artificial guide RNA (snR81-TRM4-TAA) are designed to target the PTC site in TRM4-TAA. Similar to snR81-TRM4-TAA construct, a control snR81 guide RNA construct is generated by PCR with four overlapping primers, snR81-Control-F1, snR81-Control-R1, snR81-Control-R2, and snR81-Control-R3. The guide sequences in snR81-Control do not target the PTC of TRM4-TAA and have no targets in yeast genome.
Figure 2.
Schematic representation of box H/ACA guide RNA and its interaction with its substrate RNAs. The guide RNA (red) folds into a hairpin-hinge-hairpin-tail structure. The pseudouridylation pockets responsible for target site specification are the internal loops (thick red lines) within the hairpin structures. The H and ACA boxes and the guide sequences (guide1, guide1′, guide2, and guide2′) are indicated. The mRNA substrates (blue) are paired with the guide sequences of the guide RNA, as indicated by short lines between strands. The pseudouridylated uridine (Ψ) and its downstream residue (N) in the mRNA substrates are left unpaired. The four overlapping primers, used for guide RNA construction (see text), are also shown (black lines). The dotted black lines denote specific guide sequences.
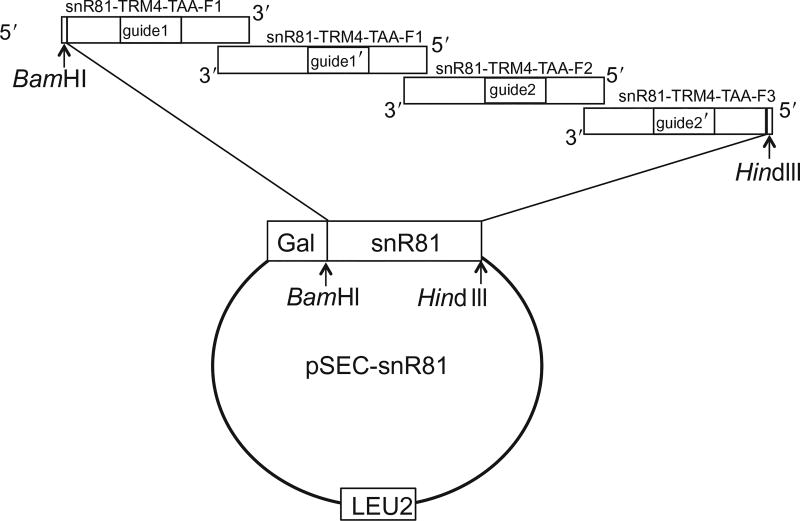
Figure 3.
The strategy for cloning artificial snR81 using four primers overlapping PCR (also see Fig. 2). The construction is based on naturally occurring yeast snR81 box H/ACA RNA. The 5′-most primer is sense-strand sequence (forward), while the other primers have antisense sequences (reverse). The guide sequences (guide1, guide1′, guide2, and guide2′) are changed to base pair with substrate mRNAs, while the rest of the snR81 sequence remain unchanged. The amplified artificial snR81 is digested with BamHI and HindIII and subsequently cloned into pSEC plasmid under the control of the Gal promoter. LEU2 is an auxotroph selective marker in yeast.
Finally, yeast BY4741 strain is cotransformed with the TRM4-TAA reporter and one of the guide RNA constructs (control or PTC-specific). It is expected that only the PTC-specific guide RNA (snR81-TRM4-TAA) will direct TRM4-TAA mRNA pseudouridylation at the PTC codon.
2.2 Construction of pTRM4-TAA Expression Plasmid (PCR-Based Mutagenesis)
2.2.1 Buffers, Reagent, and Solutions
pTRM4-WT: a plasmid bearing the wild-type TRM4 sequence with C-terminally tag (gift from Eric Phizicky at University of Rochester Medical Center) (Huang, Wu, & Yu, 2012)
TRM4-TAA-F1 primer: 5′-AAA TTA AGC TCT GGT TGC GCC TAA ATT GAT GTG TCA AGA-3′
TRM4-TAA-R1 primer: 5′-TCT TGA CAC ATC AAT TTA GGC GCA ACC AGA GCT TAA TTT-3′
10 mM dNTPs (Fermentas, cat. no. R0181)
6× DNA loading dye (Fermentas, cat. no. R0611)
Pfu DNA polymerase (5 U/µL) and 10× buffer (Stratagene, cat. no. 600250)
DpnI restriction endonuclease (Fermentas, cat. no. ER1705)
XL1-blue competent cells (Stratagene, cat. no. 200521)
100 mg/mL Ampicillin (IBI Scientific, cat. no. IB02040)
5× TBE buffer: 445 mM Tris, 445 mM boric acid, 16 mM EDTA
0.5× TBE buffer: mix 100 mL of 5× TBE buffer with 900 mL of ddH2O
LB liquid medium: 10 g of NaCl, 10 g of peptone, 5 g of yeast extract, fill to 1 L with ddH2O and autoclave
LB-ampicillin solid medium: 20 g of agar, 10 g of NaCl, 10 g of peptone, 5 g of yeast extract, fill to 1 L with ddH2O and autoclave. Allow to cool to ~55 °C, and add 1 mL of 100 mg/mL ampicillin. Gently mix and pour about 25 mL of medium in Petri dishes to achieve a bed height of ~0.5 cm 70% Ethanol: mix 700 mL of ethanol with 300 mL of ddH2O
2.2.2 Protocol
Purify pTRM4-WT plasmid using miniprep kit from Qiagen. Prepare the plasmid stock solution at 1 µg/µL, and dilute with ddH2O to 10 ng/µL working concentration.
Prepare DNA oligonucleotide primers with ddH2O to 10 µMworking concentration.
- Mix the following components in a 0.2-mL PCR tube:
5 µL 10× Pfu DNA polymerase buffer 2 µL 10 mM dNTPs 2 µL 10 ng/µL pTRM4-WT plasmid 2 µL 10 µM TRM4-TAA-F1 primer 2 µL 10 µM TRM4-TAA-R1 primer 1 µL 5U/µL Pfu DNA polymerase Add ddH2O to a final reaction volume of 50 µL. In a separate 0.2-mL PCR tube, prepare a control PCR with ddH2O substituting the primers.
- Perform the PCR cycles as follows:
- A: 95 °C for 2 min (1 cycle)
-
B: 95 °C for 30 s55 °C for 1 min68 °C for 9 min(Repeat B for 28 cycles)
-
C: 72 °C for 2 min (1 cycle)4 °C for indefinite time
Take 10 µL of the PCR product and add 2 µL of 6× DNA loading dye before loading the samples onto 1% agarose gel containing 0.05% (v/v) ethidium bromide.
Carry out electrophoresis in 0.5× TBE buffer for 30 min (at 10 V/cm gel length). Visualize the PCR products under UV light. The control PCR should have no visible bands.
Once the successful amplification is confirmed, add 2 µL (10 U/µL) of DpnI (specific for methylated sites) to the PCR to linearize (remove) the wild-type plasmid template (pTRM4-WT) (the wild-type plasmid template is methylated, whereas the PCR product is not) and incubate at 37 °C for 2 h.
Precipitate the PCR product by adding 200 µL of 100% ethanol and 5 µL of 3 MNaOAc, followed by centrifugation at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the PCR product in 10 µL of ddH2O.
Under sterile conditions, add 2 µL of PCR products to 100 µL of XL-blue competent cells (prealiquoted in 1.5-mL tube). Incubate on ice for 20 min.
Heat shock the sample for 90 s at 42 °C and immediately return to ice for 2 min.
Add 900 µL of autoclaved LB liquid medium to the sample and shake at 200 rpm for 1 h at 37 °C.
Plate the sample onto a LB-ampicillin solid medium and incubate at 37 °C for 16 h.
When colonies appear, pick 5–10 individual colonies and prepare plasmid DNA from each colony. Sequence the candidate plasmids using a proper primer. Choose one plasmid with correct sequence (codon TTT converted to TAA at F602 position) and label it as pTRM4-TAA. Adjust the concentration of the plasmid to 1 µg/µL.
2.3 Construction of TRM4-TAA-Targeting snR81 Expression Plasmid
2.3.1 Buffers, Reagent, and Solutions
pSEC plasmid: plasmid with SnoRNA expression cassette. A high copy E. coli–yeast shuttle plasmid with LEU2 as the auxotroph selective marker in yeast.
snR81-TRM4-TAA-F1: 5′-GCG GGA TCC GGG ACT GCA TCA ATT GCG GCG AGG CAG CCC ACA TCA AGT GGA ACT ACA C-3′
snR81-TRM4-TAA-R1: 5′-TGT TAG GAT TGC TCT TGG GAC CGT TGC GCC GCG ACA AGG AAG TCT GTG TAG TTC CAC TTG-3′
snR81-TRM4-TAA-R2: 5′-CCA TCC GTG GAC TGT ACA GGT TCA GCG GGG GAA TTG ATG TTT GCT TGT TAG GAT TGC TCT-3′
snR81-TRM4-TAA-R3: 5′-GCG AAG CTT AGA TGT GAA AAA GCG CGC CCC CCC GAA TCA TAT AAC TTC TGC ACC ATC CGT GGA CTG T-3′
snR81-Control-F1: 5′-GCG GGA TCC GGG ACT GCT CGA TTA GCG GCG AGG CAG CCC ACA TCA AGT GGA ACT ACA C-3′
snR81-Control-R1: 5′-TGT TAG GAT TGC TCT TGG GAC CGA TTA CTC GCG ACA AGG AAG TCT GTG TAG TTC CAC TTG-3′
snR81-Control-R2: 5′-CCA TCC GTG GAC TGT ACA GGT TCA GCG GGG GTA ATC GAT TTT GCT TGT TAG GAT TGC TCT-3′
snR81-Control-R3: 5′-GCG AAG CTT AGA TGT GAA AAA GCG ACT CCC CCC GAA TCA TAT AAC TTC TGC ACC ATC CGT GGA CTG T-3′
Taq DNA polymerase (5 U/µL) (Fermentas, cat. no. EP0401)
10× Taq DNA polymerase buffer (Fermentas, cat. no. B34)
10 mM dNTPs (Fermentas, cat. no. R0181)
6× DNA loading dye (Fermentas, cat. no. R0611)
BamHI restriction endonuclease (Fermentas, cat. no. ER0051)
HindIII restriction endonuclease (Fermentas, cat. no. ER0501)
T4 DNA ligase (1 U/µL) (Fermentas, cat. no. EL0011) and 5× buffer (cat. no. B69)
DH5α competent cells (Invitrogen, cat. no. 18265-017)
Phenol (Alfa Aesar, cat. no. A15760-0E)
Chloroform (J.T. Baker, cat. no. 9182-01)
Isoamyl alcohol (Sigma-Aldrich, cat. no. I9392)
Ampicillin (IBI Scientific, cat. no. IB02040)
Glycogen (Sigma-Aldrich, cat. no. G0885)
5× TBE buffer: 445 mM Tris, 445 mM boric acid, 16 mM EDTA
0.5× TBE buffer: mix 100 mL of 5× TBE buffer with 900 mL of ddH2O
Qiagen Gel Extraction kit (Qiagen, cat. no. 28704)
LB liquid medium: 10 g of NaCl, 10 g of peptone, 5 g of yeast extract, fill to 1 L with ddH2O and autoclave
LB-ampicillin solid medium: 20 g of agar, 10 g of NaCl, 10 g of peptone, 5 g of yeast extract, fill to 1 L with ddH2O and autoclave.
Allow to cool, add 1 mL of 100 mg/mL of ampicillin, and mix well.
Pour about 20–25 mL of medium in Petri dishes to achieve a bed height of ~0.5 cm
PCA (phenol/chloroform/isoamyl alcohol=25/24/1 [v/v/v]): saturated (1:1 [v/v]) with 20 mM Tris–HCl, pH 8.0
G50 buffer: 20 mMTris–HCl at pH 7.5, 300 mMsodium acetate, 2 mM EDTA, 0.2% SDS
70% Ethanol: mix 700 mL of ethanol with 300 mL of ddH2O
2.3.2 Protocol
Prepare following DNA oligonucleotide primers with ddH2O to 10 µM working concentration: snR81-TRM4-TAA-F1, snR81-TRM4-TAA-R3, snR81-Control-F1, and snR81-Control-R3.
Prepare following DNA oligonucleotide primers with ddH2O to 1 µM working concentration: snR81-TRM4-TAA-R1, snR81-TRM4-TAA-R2, snR81-Control-R1, and snR81-Control-R2.
- Mix the following buffers and reagents in a 0.2-mL PCR tube (to generate snR81-TRM4-TAA). The ratio of four primers is optimized to yield the PCR product at expected size.
5 µL 10× Taq DNA polymerase buffer 2 µL 10 mM dNTPs 2 µL 10 µM snR81-TRM4-TAA-F1 primer 1 µL 1 µM snR81-TRM4-TAA-R1 primer 1 µL 1 µM snR81-TRM4-TAA-R2 primer 2 µL 10 µM snR81-TRM4-TAA-R3 primer 1 µL 5 U/µL Taq DNA polymerase Add ddH2O to a final reaction volume of 50 µL. - In a separate 0.2-mL PCR tube, prepare the following buffers and reagents (to generate snR81-Control):
5 µL 10× Taq DNA polymerase buffer 2 µL 10 mM dNTPs 2 µL 10 µM snR81-Control-F1 primer 1 µL 1 µM snR81-Control-R1 primer 1 µL 1 µM snR81-Control-R2 primer 2 µL 10 µM snR81-Control-R3 primer 1 µL 5 U/µL Taq DNA polymerase Add ddH2O to a final reaction volume 50 µL. - Perform the PCR as follows:
- A: 95 °C for 2 min (1 cycle)
-
B: 95 °C for 30 s42 °C for 30 s72 °C for 30 s(Repeat B for 35 cycles)
-
C: 72 °C for 2 min (1 cycle)4 °C for indefinite time
Take 10 µL of the PCR product and add 2 µL of 6× DNA loading dye before loading the samples on 1% agarose gel containing 0.05% (v/v) of ethidium bromide (to check the PCR products).
Conduct electrophoresis in 0.5× TBE buffer for 30 min (at 10 V/cm gel length). Visualize the PCR products under UV light.
Once the success of PCR amplification is confirmed, precipitate the remaining PCR products by adding 200 µL of 100% ethanol and 4 µL of 3 M NaOAc to each tube, and spinning at maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve each PCR product in 10 µL of ddH2O.
The dissolved PCR products are then mixed with 2 µLof 6× DNA loading dye and loaded on a 2% agarose gel (with 0.05% (v/v) ethidium bromide). The bands at expected position (size) are excised and purified using Qiagen Gel Extraction kit (refer to kit manual for detailed procedure).
The PCR products (snR81-TRM4-TAA and snR81-Control) are each eluted with 40 µL of ddH2O. The final concentration of the DNA is adjusted to ~1 µg/µL using ddH2O.
- Digest the PCR products with BamHI by mixing the following buffer and reagents in a 1.5-mL tube:
5 µL 1 µg/µL PCR product (snR81-TRM4-TAA or snR81-Control) 5 µL 10× BamHI digestion buffer 2.5 µL BamHI (10 U/µL) Add ddH2O to a final reaction volume of 50 µL. - In a separate 1.5-mL tube, digest, in parallel, the pSEC plasmid with BamHI:
5 µL 1 µg/µL pSEC plasmid 5 µL 10× BamHI digestion buffer 2.5 µL BamHI (10 U/µL) Add ddH2O to a final reaction volume of 50 µL. Seal the caps of both tubes with parafilm and incubate for 2 h in a 37 °C water bath.
Add 450 µL of G50 buffer and 500 µL of PCA to each tube. Vigorously vortex for 30 s.
Spin the tubes at the maximum speed (~14,000 × g) in a bench-top centrifuge for 1 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin the tubes at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 10 µL of ddH2O.
- Digest the BamHI-digested PCR product with HindIII by mixing the following buffer and reagents in a new 1.5-mL tube:
10 µL 1 µg/µL BamHI-digested PCR product (snR81-TRM4-TAA or snR81-Control) 5 µL 10× HindIII digestion buffer 2.5 µL HindIII (10 U/µL) Add ddH2O to a final reaction volume of 50 µL. - In parallel, digest the BamHI-digested pSEC with HindIII in a separate 1.5-mL tube:
10 µL 1µg/µL BamHI-digested pSEC plasmid 5 µL 10× HindIII digestion buffer 2.5 µL HindIII (10 U/µL) Add ddH2O to a final reaction volume of 50 µL. Seal the caps of both tubes with parafilm and incubate for 2 h in 37 °C water bath.
Add 450 µL of G50 buffer and 500 µL of PCA to each tube. Vigorously vortex for 30 s.
Spin the tubes at the maximum speed (~14,000 × g) in a bench-top centrifuge for 5 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin the tubes at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 10 µL of ddH2O.
- Prepare the ligation reaction in a new 1.5-mL tube:
5 µL Digested PCR product (snR81-TRM4-TAA or snR81-Control) 1 µL Digested pSEC plasmid 2 µL 5× T4 DNA ligase buffer 2 µL T4 DNA ligase (1 U/µL) Seal the tube cap with parafilm and incubate for overnight in a 16 °C water bath.
Under sterile conditions, add 5 µL of the ligation reaction to 100 µL of DH5α competent cells (prealiquoted in 1.5-mL tube), mix, and lay on ice for 10 min.
Heat shock the cells at 42 °C for 45 s and immediately return to ice for 2 min.
Add 900 µL of autoclaved ampicillin-free LB liquid medium to the tube and shake at 200 rpm for 1 h at 37 °C.
Spin the tube at ~4000 × g in a bench-top centrifuge for 5 min to pellet the cells. Remove supernatant.
Resuspend the pellet in 100 µL of ampicillin-containing LB liquid medium and plate the cells onto a LB-ampicillin solid medium. Incubate for overnight at 37 °C.
Pick 5–10 individual colonies for each sample (snR81-TRM4-TAA and snR81-Control) and prepare plasmid DNA from each colony. Sequence the candidate plasmids using a proper primer. Choose one correct plasmid for snR81-TRM4-TAA and one for snR81-Control, and label them as psnR81-TRM4-TAA and psnR81-Control, respectively. The final concentration of the plasmid is adjusted with ddH2O to 1 µg/µL (working concentration).
2.4 Cotransformation of Yeast with pTRM4-TAA and psnR81-TRM4-TAA
2.4.1 Buffers, Reagent, and Solutions
S. cerevisiae strain BY4741 (Open Biosystems, cat. no. YSC1048)
Yeast extract (BD Diagnostics, cat. no. 90000-444)
Peptone (BD Diagnostics, cat. no. 90000-264)
Yeast nitrogen base (AMRESCO, cat. no. 97064-322)
Ammonium sulfate (EMD Millipore, cat. no. EM-AX1385-1)
l-Isoleucine (Sigma-Aldrich, cat. no. I2752)
l-Valine (Sigma-Aldrich, cat. no. V0500)
Adenine hemisulfate salt (Sigma-Aldrich, cat. no. A9126)
l-Arginine monohydrochloride (Sigma-Aldrich, cat. no. A5131)
l-Histidine monohydrochloride (Sigma-Aldrich, cat. no. H8125)
l-Lysine monohydrochloride (Sigma-Aldrich, cat. no. L5626)
l-Methionine (Sigma-Aldrich, cat. no. M9625)
l-Phenylalanine (Sigma-Aldrich, cat. no. P2126)
l-Tryptophan (Sigma-Aldrich, cat. no. T0254)
l-Tyrosine (Sigma-Aldrich, cat. no. T3754)
Galactose (J.T. Baker, cat. no. M672-07)
Lithium acetate (Alfa Aesar, cat. no. AA13417-30)
Polyethylene glycol PEG-3350 (J.T. Baker, cat. no. JTU221-8)
- Synthetic leucine/uracil double drop-out powder:
Yeast nitrogen base 25.1 g/15 L Ammonium sulfate 75.4 g/15 L Isoleucine 450 mg/15 L Valine 2.25 g/15 L Adenine 300 mg/15 L Arginine 300 mg/15 L Histidine 300 mg/15 L Leucine 0 mg/15 L Lysine 450 mg/15 L Methionine 300 mg/15 L Phenylalanine 750 mg/15 L Tryptophan 300 mg/15 L Tyrosine 450 mg/15 L Uracil 0 mg/15 L YPD liquid medium: 10 g of yeast extract, 20 g of peptone, and 20 g of dextrose, fill with ddH2O to 1 L and autoclave
One-step-transformation buffer: 100 mM lithium acetate, 50% (w/v) PEG-3350 solution
SGal-LEU-URA double drop-out liquid media: 7.5 g of synthetic leucine/uracil double drop-out powder, 20 g of galactose, fill to 1 L with ddH2O and autoclave
SGal-LEU-URA double drop-out solid media: 7.5 g of synthetic leucine/uracil double drop-out powder, 20 g of galactose, fill to 1 L with ddH2O and autoclave. Pour about 20–25 mL of medium in Petri dishes to achieve a bed height of ~0.5 cm
2.4.2 Protocol
Under sterile conditions, pick a single yeast colony to grow in 5 mL of YPD liquid medium. Shake at 200 rpm for overnight at 30 °C.
The cells are diluted to ~0.5 OD (600 nm) with 5 mL of fresh YPD liquid medium and continue propagation.
When the OD600nm reaches 2.0, precipitate the cells by centrifugation at 2500 × g at 4 °C for 5 min in an SH3000 rotor (Sorvall RC-5C Plus centrifuge).
Carefully remove all supernatant YPD medium and resuspend the cell pellets in 200 µL of one-step-transformation buffer.
- In a new 1.5-mL tube, prepare the transformation mixture:
50 µL Resuspended cells 1 µL 1 µg/µL pTRM4-TAA plasmid 1 µL 1 µg/µL psnR81-TRM4-TAA plasmid Seal the tube cap with parafilm and incubate in a 42 °C water bath for 30 min.
Supply 300 µL of fresh YPD media to the mixture and shake at 200 rpm in a 30 °C-shaker for 1 h.
Pellet the cells by centrifugation at 2500 × g for 5 min in a bench-top centrifuge.
Carefully remove all supernatant YPD medium, and resuspend the cells in 100 µL of ddH2O.
Spread the cells on SGal-LEU-URA solid media and incubate at 30 °C for 2–3 days in order to see colonies.
3. DETECTION OF PSEUDOURIDINE IN THE TRM4 mRNA
3.1 Overview
After the TRM4-PTC mRNA and corresponding guide RNA are coexpressed in yeast, TRM4 mRNA pseudouridylation at the PTC site is assessed. Here we present a protocol for purification, site-specific cleavage, isotope-labeling, and nuclease digestion of TRM4 mRNA, and analysis of U-to-Ψ conversion at the PTC by thin layer chromatography (TLC). [Note, the conventional approach—CMCT modification coupled with primer extension—is not sensitive enough to allow clear detection and accurate quantification of Ψ in low abundance mRNA.]
The first step in this protocol is to extract a large amount of total RNA from yeast cells. From this pool of RNA, TRM4 mRNA is further enriched by oligonucleotide affinity chromatography, where TRM4 mRNA, when hybridized to a biotinylated antisense TRM4 oligonucleotide, is pulled-down by streptavidin-coupled agarose beads.
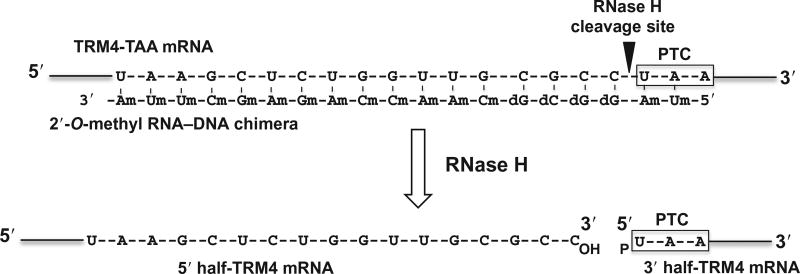
The second step is to cleave the enriched TRM4 mRNA specifically at the site 5′ of the PTC uridine. This is achieved by site-specific RNase H cleavage directed by 2′-O-methyl RNA–DNA chimera. Briefly, an antisense TRM4 2′-O-methyl RNA–DNA chimera is designed, which contains, from 5′ to 3′, two 2′-O-methyl RNA nucleotides (complementary to the first two nucleotides of the PTC), four deoxynucleotides (complementary to the four nucleotides immediately upstream of the PTC), and thirteen 2′-O-methyl RNA nucleotides (complementary to the 13 nucleotide sequence that is 5 nucleotides upstream of the PTC) (Fig. 4; Yu, 1999). This chimeric oligo, when annealed to TRM4 mRNA, is able to direct RNase H to cleave the phosphodiester bond 3′ of the substrate nucleotide (immediately preceding the uridine of PTC) that base pairs with the 5′-most deoxynucleotide of the chimera. The resulting 3′-half fragment of the mRNA begins with the PTC codon (UAA) and carries a 5′ phosphate (pU) (Fig. 4).
Figure 4.
Schematic representation of site-specific RNase H cleavage of TRM4 mRNA directed by 2′-O-methyl RNA–DNA chimera. The target sequence of TRM4 mRNA is shown and the PTC (UAA) is indicated. The antisense TRM4 2′-O-methyl RNA–DNA chimera is also shown. d represents deoxy, and m stands for 2′-O-methyl. The arrow indicates the RNase H cleavage site.
The third step is to radiolabel the U residue of the PTC codon of the 3′-half fragment. To this end, the 3′-half fragment is dephosphorylated by calf intestine phosphatase (CIP) and rephosphorylated by T4 polynucleotide kinase (PNK) in the presence of [γ32P]ATP (Huang & Yu, 2013).
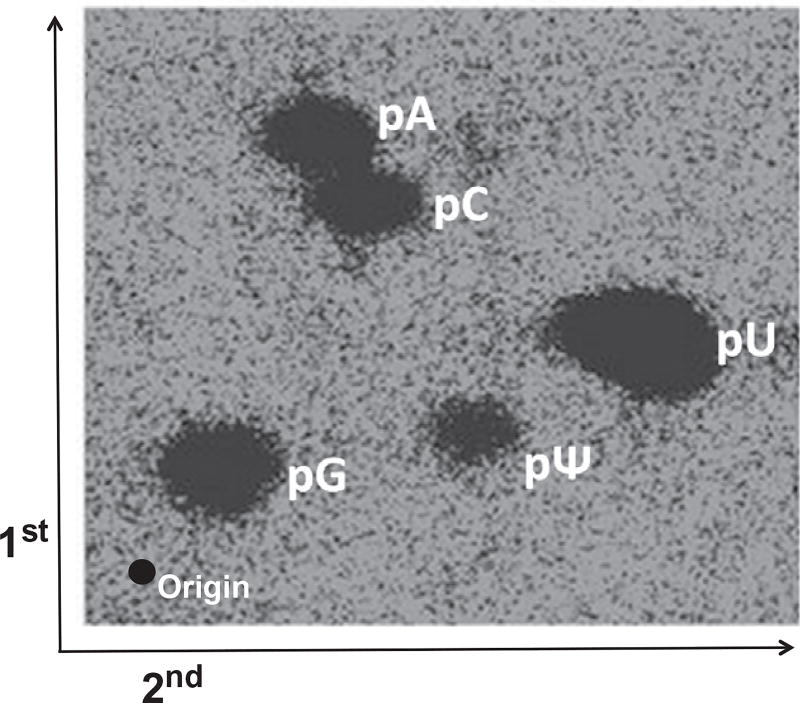
The last step is to purify the radiolabeled 3′-half fragment of TRM4 mRNA by gel electrophoresis, digest the purified RNA with nuclease P1, and analyze Ψ/U ratio using TLC (Fig. 5). In order to precisely locate the Ψ on the TLC plate, the nuclease P1 digested sample is spiked with 5′-32P-adenosine-5′-monophosphate, 5′-32P-cytidine-5′-monophosphate, and 5′-32P-guanosine-5′-monophosphate before being spotted on the TLC plate. The percentage of pseudouridylation at the PTC is calculated using the formula: Ψ/(Ψ+U) (Fig. 5).
Figure 5.
TLC analysis of uridine-to-Ψ conversion at the PTC within the TRM4 mRNA transcript. The PTC-containing TRM4 mRNA, coexpressed with a PTC-specific guide RNA, is purified by oligonucleotide affinity chromatography. The RNA is cleaved by RNase H (directed by a specific 2′-O-methyl RNA–DNA chimera) at the site 5′ of the PTC (UAA). The resulting 3′-half fragment is 5′ radiolabeled with 32P through dephosphorylation and rephosphorylation (see text). The labeled RNA is digested with nuclease P1 to completion. The digested sample is mixed with an equal amount of 5′-32P-adenosine-monophosphate, 5′–32P-cytidine-monophosphate, and 5′–32P-guanosine-monophosphate, and analyzed by two-dimensional TLC. The first and second dimensions are indicated. The origin and the positions of each 5′-phosphorylated nucleotide are also indicated.
3.2 Isolation of Total RNA from S. cerevisiae
3.2.1 Buffers, Reagent, and Solutions
1× RIB buffer: 0.2 M Tris–HCl pH 7.5, 0.5 M NaCl, 0.01 M EDTA, 1% SDS
PCI-RIB (phenol/chloroform/isoamyl alcohol=50/49/1 [v/v/v]): saturated (1:1 [v/v]) with 1× RIB buffer
Sterile acid-washed glass beads
3.2.2 Protocol
Pick and grow a cotransformed yeast colony in 100 mL of SGal-LEU-URA liquid medium. Shake at 200 rpm at 30 °C and monitor OD600nm.
When the OD600nm reaches 2.0, precipitate cells by centrifugation at 2500 × g at 4 °C for 5 min in an SH3000 rotor (Sorvall RC-5C Plus centrifuge).
Resuspend cell pellet (~200 µL) in 400 µL of 1× RIB buffer and transfer to a new 2-mL screw-cap tube.
Add 400 µL of sterile acid-washed glass beads and 400 µL of PCI-RIB and place on ice. Seal the tube cap with parafilm to avoid leakage during vortexing.
Vortex vigorously for 40 s and then immediately place the tube on ice for 40 s.
Repeat step 5 five times.
Spin the 2-mL screw-cap tube at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min and transfer the supernatant to a new 1.5-mL tube.
Add 400 µL of PCI-RIB and vortex for 30 s.
Spin the 1.5-mL tube at maximum speed (~14,000 × g) in a benchtop centrifuge for 5 min and transfer the supernatant to a new 1.5-mL tube.
Repeat steps 8 and 9 one more time.
Carefully remove the upper aqueous phase to a new 1.5-mL tube and add 2× volume of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Precipitate the total RNA by centrifugation at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the RNA pellet with 70% ethanol and air-dry. Dissolve the RNA pellet in ddH2O to a final concentration of 1 µg/µL.
3.3 Purification of TRM4 mRNA Using Biotinylated-TRM4 Antisense Oligo and Streptavidin Agarose Beads
3.3.1 Buffers, Reagent, and Solutions
Biotin-streptavidin binding buffer: 0.1 M phosphate, 0.15 M NaCl, 0.1% SDS, 1% NP-40, pH 7.2
Biotinylated-TRM4 antisense oligo (IDT): biotin-5′-CCACTCTTGTTGGTTCACCAGTGGC-3′
Streptavidin agarose bead (Pierce)
Elution buffer: 0.1 M glycine–HCl pH 7.0
3.3.2 Protocol
Mix 10 µL of 10 µM biotinylated-TRM4 antisense oligo with 100 µL of 1 µg/µL purified total RNA in a 1.5-mL tube and place in a 65 °C heating block for 10 min.
Transfer the tube to room temperature (place it on bench top) for 30 min to let the mRNA anneal to biotinylated-TRM4 antisense oligo.
In the mean time, prepare 50 µL bed volume of streptavidin agarose beads in a new 1.5-mL tube.
Spin the tube at medium speed (~3000 × g) in a bench-top centrifuge for 1 min and carefully discard supernatant.
Add 200 µL of biotin-streptavidin binding buffer to the beads and mix by inverting the tube.
Spin the tube at the medium speed (~3000 × g) in a bench-top centrifuge for 1 min and carefully discard supernatant.
Repeat steps 5 and 6 one more time.
Add the annealed mRNA/biotinylated-TRM4 antisense oligo mixture and 100 µL of biotin-streptavidin binding buffer to the streptavidin agarose beads and nutate at room temperature for 2 h.
Spin the tube at medium speed (~3000 × g) in a bench-top centrifuge for 1 min and carefully discard supernatant.
Add 200 µL of biotin-streptavidin binding buffer to the beads and mix by inverting the tube.
Repeat steps 9 and 10 three more times.
Add 200 µL of elution buffer to the beads and heat in a 65 °C heating block for 10 min. Mix by inverting the tube several times during the 10 min heating period.
Immediately spin the tube at the medium speed (~3000 × g) in a bench-top centrifuge for 1 min and transfer the supernatant to a new 1.5-mL tube.
Wash and elute the beads again by repeating steps 12 and 13 one more time.
Combine the supernatant from steps 13 and 14 and add 800 µL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Precipitate the TRM4mRNA by centrifugation at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Dissolve the pellet in a small volume of ddH2O.
3.4 Site-Specific RNase H Cleavage of TRM4 mRNA Directed by 2′-O-Methyl RNA–DNA Chimeras
3.4.1 Buffers, Reagent, and Solutions
RNase H (Amersham)
10× Buffer for RNase H: 200 mMTris–HCl (pH 7.5), 100 mMMgCl2, 1 M KCl, 250 mM DTT, 50% sucrose
2′-O-Methyl RNA–DNA chimera oligo (IDT): 5′-Um-Am-dG-dG-dC-dG-Cm-Am-Am-Cm-Cm-Am-Gm-Am-Gm-Cm-Um-Um-Am-3′
RNase inhibitor (Fermentas, cat. no. EO0381)
Urea (Sigma, cat. no. U5378)
40% Acrylamide (acrylamide:bis acrylamide=19:1) (Fisher Scientific, cat. no. BP1406-1)
10% Ammonium persulfate (APS) (Fisher Scientific, cat. no. M2300)
N,N,N′,N′-Tetramethylethylenediamine (TEMED) (Sigma, cat. no. T9281)
GeneRuler Low Range (size markers) (Thermo, cat. no. SM1193)
5× TBE buffer: 445 mM Tris, 445 mM boric acid, 16 mM EDTA
0.5× TBE buffer: 44.5 mM Tris, 44.5 mM boric acid, 1.6 mM EDTA (prepare it by making a 1/10 dilution of 5× TBE buffer with ddH2O)
2× RNA sample buffer: 95% formamide, 0.025% SDS, 0.025% bromophenol blue, 0.025% xylene cyanol, 0.5 mM EDTA
G50 buffer: 20 mMTris–HCl at pH 7.5, 300 mMsodium acetate, 2 mM EDTA, 0.2% SDS
PCA (phenol/chloroform/isoamyl alcohol=25/24/1 [v/v/v]): saturated (1:1 [v/v]) with 20 mM Tris–HCl, pH 8.0
2× RNA loading dye: 95% formamide, 0.5 mM EDTA, 0.01% bromophenol blue, 0.005% xylene cyanol
3.4.2 Protocol
Mix 6 µL of TRM4 mRNA (~100 pmol) with 1 µL of complementary 2′-O-methyl RNA–DNA chimera (~200 pmol) in a 1.5-mL tube and place in a 95 °C heating block for 3 min.
Place the tube on bench top for 10 min to let the TRM4 mRNA anneal to the 2′-O-methyl RNA–DNA chimera.
Add 1 µL of 10× RNase H buffer, 1 µL of RNase inhibitor (40 U), and 1 µL of RNase H (2 U) to the annealed TRM4 mRNA/2′-O-methyl RNA–DNA chimera mixture.
Seal the tube cap with parafilm and incubate at 37 °C for 1 h.
Add 490 µL of G50 buffer and 500 µL of PCA and mix by vortexing for 30 s.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 3 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 5 µL of ddH2O.
- To facilitate the next dephosphorylation step, the 3′ mRNA fragments can be separated through gel electrophoresis (Adachi & Yu, 2014). Prepare a 6% acrylamide gel solution following the recipe below:
14.41 g Urea 3 mL 5× TBE 4.5 mL 40% Acrylamide (acrylamide:bis acrylamide=19:1) Bring up to 30 mL with ddH2O. Add 300 µL of freshly made 10% APS and 20 µL of TEMED to the solution and mix gently.
Immediately pour the gel solution in between two 20 × 30 cm glass plates (tape-sealed) with 0.42 mm thick spacers. And insert a comb into the gel (from the top).
Allow the gel to polymerize at room temperature for at least 20 min.
Remove the comb and clean up the wells with 0.5× TBE. Remove the tape on the bottom of the glass plates and prerun the gel in 0.5× TBE buffer at 20 W (constant power) for 30 min.
Add 5 µL of RNA loading dye to the above RNase H-treated RNA sample (5 µL), and heat it at 95 °C for 3 min and immediately chill on ice.
Load the RNA sample into a well of the gel and load a size marker into an adjacent well.
Carry out electrophoresis at 20W for about 60 min.
Turn off the power supply when the bromophenol blue dye reaches the bottom of the gel.
Expose the gel to a phosphor imaging screen and scan the PhosphorImager.
Locate the 3′ half RNA on the gel according to the size marker and cut out the RNA band from the gel with a clean scalpel.
Place the gel slice in a 1.5-mL tube and add 400 µL of G50 buffer, and then place the tube in dry ice for 5 min.
Place the tube on a rotator and rotate at room temperature overnight.
Add 500 µL of PCA to the tube and mix by vortexing for 30 s.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 5 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 10 µL of ddH2O.
3.5 Dephosphorylation of the 3′ mRNA Fragment of TRM4 to Generate a 5′-Hydroxyl End
3.5.1 Buffers, Reagent, and Solutions
CIP (Thermo Scientific)
10× Buffer for CIP
3.5.2 Protocol
- Prepare the dephosphorylation reaction with the following buffer and reagents in a 1.5-mL tube:
2 µL 10× Buffer for CIP 10 µL Cleaved 3′ RNA fragment 1 µL RNase inhibitor (40 U) 2 µL CIP (2 U) Add ddH2O to a final reaction volume of 20 µL. Seal the tube cap with parafilm and incubate at 37 °C for 30 min.
Add 480 µL of G50 buffer and 500 µL of PCA and mix by vortexing for 30 s.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 5 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 10 µL of ddH2O.
3.6 Rephosphorylation of the 3′ TRM4 mRNA Fragment with [γ-32P]ATP
3.6.1 Buffers, Reagent, and Solutions
T4 PNK (Thermo Scientific)
10× Buffer for T4 PNK forward reaction
[γ32P]-ATP (3000 Ci/mmol, 10 µCi/µL) (Perkin Elmer)
3.6.2 Protocol
- Set up a rephosphorylation reaction in a 1.5-mL tube using the following buffer and reagents:
2 µL 10× Buffer for T4 PNK forward reaction 10 µL Dephosphorylated 3′ RNA fragment 2 µL [γ32P]-ATP (3000 Ci/mmol, 10 µCi/µL) 1 µL RNase inhibitor (40 U) 2 µL T4 PNK (20 U) Add ddH2O to a final reaction volume of 20 µL. Seal the tube cap with parafilm and incubate for 30 min in a 37 °C water bath.
Add 480 µL of G50 buffer and 500 µL of PCA and mix by vortexing for 30 s.
Spin the tube at maximum speed (~14,000 × g) in a bench-top centrifuge for 3 min.
Carefully transfer the upper aqueous phase to a new 1.5-mL tube. Add 1 mL of 100% ethanol and 1 µL of 10 mg/mL glycogen (as carrier).
Place the tube in dry ice for 5 min.
Spin at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min.
Wash the pellet with 70% ethanol and air-dry. Carefully dissolve the pellet in 10 µL of ddH2O.
For higher purity, the 3′ mRNA fragment is separated on a 6% urea-PAGE gel as described in Section 3.4.2, steps 11–29.
3.7 Nuclease P1 Digestion and TLC Analysis of Pseudouridine in TRM4 mRNA
3.7.1 Buffers, Reagent, and Solutions
10× Nuclease P1 digestion buffer: 200 mM acetic acid, 10 mM CaCl2, pH 6.0
Nuclease P1 (0.2 µg/µL): 0.1 µg of nuclease P1 is sufficient to digest 10 pmol of RNA substrate (at 37 °C for 1 h)
TLC PEI cellulose F plate (EMD)
First dimension developing buffer: isobutyric acid/ddH2O/NH4OH (66:33:1) [v/v/v]
Second dimension developing buffer: isopropanol/HCl/ddH2O (70:15:15) [v/v/v]
3.7.2 Protocol
- Set up the following nuclease P1 digestion reaction in a 1.5-mL tube:
1 µL 10× Nuclease P1 digestion buffer 1 µL Rephosphorylated 3′ RNA fragment 1 µL Nuclease P1 (0.2 µg/µL) Add ddH2O to a final reaction volume of 10 µL. Seal the tube cap with parafilm and incubate at 37 °C for 2 h.
The nuclease P1 digestion reaction is then spiked with 0.1 µL of each of the following controls: 5′-32P-adenosine-monophosphate, 5′-32P-cytidine-monophosphate, and 5′-32P-guanosine-monophosphate.
Spot 2 µL of spiked P1 reaction at the origin (one of the corners) of the TLC PEI cellulose F plate (20 × 20 cm2) and place the plate in a chemical hood until it is completely dry.
Place the plate (with the origin at the bottom left) in a chromatography tank containing 75 mL of first dimension developing buffer and allow the solvent to ascend.
Remove the plate from the tank, when the solvent front is about 1 cm from the top of the plate (it takes ~10 h).
Leave the plate in a chemical hood and let it air-dry for overnight.
Rotate the plate by 90° (counterclockwise) and place it (with the origin at the bottom right) in the chromatography tank with 75 mL of second dimension developing buffer. Allow the solvent to ascend.
Remove the plate from the tank, when the solvent front is about 1 cm from the top of the plate (it takes ~10 h).
Let the plate air-dry in a chemical hood for overnight.
Cover the plate with Saran Wrap and expose it to a PhosphorImager (Molecular Dynamics) for overnight. Visualize the 32PΨ and 32PU spots, and quantify the U-to-Ψ conversion efficiency.
4. RECODING OF PSEUDOURIDYLATED PTC CODON
4.1 Overview
To study Ψ-mediated stop-codon recoding, we take advantage of multiple tags at the C-terminus of Trm4. If the Ψ-containing PTC behaves similarly to an unmodified stop codon—terminating ribosome translation, no fulllength Trm4 protein (including the C-terminal tags) will be generated. Consequently, no signal will appear when probed with anti-C-terminal tag antibodies (Western blot). If, however, pseudouridylation of the PTC codon leads to nonsense suppression, a full-length readthrough product will be generated and detected by Western blot (anti-C-terminal tag antibodies). The C-terminal tags can also be used to purify (immunoprecipitate) the readthrough product, which can then be analyzed by mass spectrometry to determine the amino acid(s) incorporated at the pseudouridylated stop codon (This is not discussed here).
The first step is to prepare the cell extract for SDS-PAGE gel electrophoresis. Since yeast has resilient cell wall, we need to lyse cells by mixing glass beads with cells and vortex at high frequency. The glass bead-beating technique is ideal for small-scale preparation (discussed below). For medium and large scale, homogenizer or French press should be used (not discussed here).
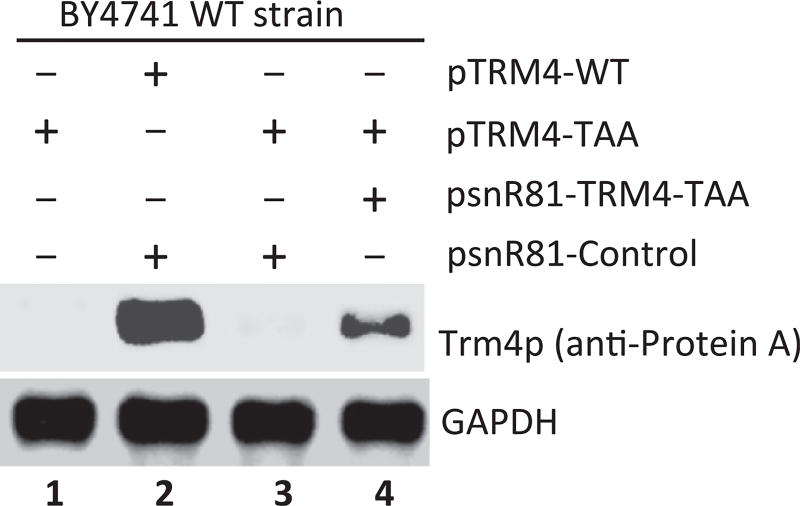
The second step is to run SDS-PAGE and Western blotting. Since the C-terminal tag contains Protein A, monoclonal anti-Protein A antibody is used to probe the full-length readthrough Trm4 protein (Fig. 6). Any housekeeping gene products in S. cerevisiae could be used as loading controls. GAPDH is used in this study (Fig. 6).
Figure 6.
Ψ-Mediated nonsense suppression detected by Western blotting. Equal amounts of total protein are loaded (lanes 1–4), and anti-Protein A and anti-GAPDH (loading control) are used for blotting. Readthrough of PTC is visualized (lane 4), where a cognate (PTC-specific) guide RNA designed to target the PTC for pseudouridylation is coexpressed. In contrast, no significant suppression is observed when no guide RNA (lane 1) or a guide RNA-containing random guide sequences is coexpressed (lane 3). Lane 2 is a positive control where the wild-type TRM4 mRNA (with no PTC) is expressed.
4.2 Preparation of Cell Extract from S. cerevisiae
4.2.1 Buffers, Reagent, and Solutions
Acid-washed glass beads, 0.5 mm (BioSpec Products Inc., cat. no. 11079105)
2× Cell extract preparation buffer: 100 mM Tris–HCl at pH 7.5, 300 mM KCl, 3 mM MgCl2, 1mM EDTA, 2 mM DTT (add before use), 1 mM PMSF (add before use), and 20% glycerol
4.2.2 Protocol
Pick a cotransformed yeast colony and grow in 100 mL of SGal-LEU-URA liquid medium. Shake at 200 rpm at 30 °C and monitorOD600nm.
When OD600nm reaches 2.0, pellet the cells by centrifugation at 2500 × g at 4 °C for 5 min in an SH3000 rotor (Sorvall RC-5C Plus centrifuge).
Transfer the cell pellet (~200 µL) to a new 2-mL screw-cap tube and add 200 µL of 2× cell extract preparation buffer and 200 µL of glass beads. Seal the tube cap with parafilm to avoid leakage.
Vortex vigorously for 1 min and then immediately place the tube on ice for 30 s.
Repeat step 4 for seven more times.
Spin the tube at the maximum speed (~14,000 × g) in a bench-top centrifuge for 10 min and transfer the supernatant to a new 1.5-mL tube.
Repeat step 6 for four more times until there is no cell pellet accumulated at the bottom of the new tube after spin.
Aliquot the supernatant into 1.5-mL tubes (10 µL each). Use one tube at a time for Western blotting.
4.3 Detection of Full-Length Readthrough Trm4 Protein by Western Blotting
4.3.1 Buffers, Reagent, and Solutions
2× Loading dye: 0.5 MTris–HCl (pH 6.8), 4.4% (w/v) SDS, 20% (v/v) glycerol, 2% (v/v) 2-mercaptoethanol, and 2% (v/v) bromophenol blue
4–15% Tris–HCl Ready Gels (Bio-Rad, cat. no. 161-1104)
10× Tris–glycine–SDS buffer (Bio-Rad, cat. no. 161-0732)
10× Tris–glycine buffer (Bio-Rad, cat. no. 161-0734)
Monoclonal anti-Protein A, mouse (Sigma-Aldrich, cat. no. P2921)
Antibody against GAPDH (Pierce, cat. no. MA5-15738)
oat anti-mouse IgG (H+L)-AP conjugate (Bio-Rad, cat. no. 170-6520)
1-Step NBT/BCIP (Pierce, cat. no. 34042)
Western blot running buffer: mix 100 mL of 10× Tris–glycine–SDS buffer with 900 mL of ddH2O
Western blot transferring buffer: mix 100 mL of 10× Tris–glycine buffer with 200 mL of methanol and 700 mL of ddH2O
Western blot washing buffer: mix 100 mL of 10× Tris–glycine buffer with 100 µL of Tween-20 and 899.9 mL of ddH2O
Western blot blocking buffer: dissolve 5 g of BSA in 100 mL of ddH2O, pass through 0.2-µm filter
4.3.2 Protocol
Leave the following buffers and equipment at 4 °C for overnight: 1 L of Western blot running buffer, 1 L of Western blot transferring buffer, gel electrophoresis apparatus, and gel transfer box.
- Prepare the following loading mixture in a 1.5-mL tube:
10 µL 2× loading dye 1 µL Cell extract (~10 µg) 9 µL ddH2O Seal the tube cap with parafilm and place in a 95 °C heating block for 5 min.
Briefly centrifuge the tube in a bench-top centrifuge to collect condensation.
Load 8 µL of sample on the 4–15% Tris–HCl Ready Gel and carry out electrophoresis at 110 V at 4 °C until the bromophenol blue dye reaches the bottom of the gel.
In a container with Western blot transferring buffer, place the gel onto a piece of Whatman nitrocellulose membrane (pore size: 0.1 µm) and sandwich the gel/membrane with two pieces of Whatman filter paper.
Place the gel/membrane sandwich into a transfer cassette, with gel facing the anode and membrane facing the cathode.
Transfer the protein from gel to membrane at constant 100 V at 4 °C for 2 h.
Separate the membrane from the gel, place the membrane (with the protein side up) in a small plastic box containing 20 mL of 5% BSA solution, and shake it (low rpm) at room temperature for 1 h.
Transfer and soak the membrane in 20 mL of Western blot washing buffer and shake at room temperature for 10 min.
Take 5 µL of monoclonal anti-Protein A and mix with 10 mL of primary hybridization buffer. Soak the membrane in this primary hybridization buffer and shake at 4 °C for 16 h.
Transfer and soak the membrane in 20 mL of Western blot washing buffer (fresh) and shake at room temperature for 10 min.
Repeat step 12 for three more times.
Mix 1 µL of goat anti-mouse (H+L)-AP conjugated IgG with 10 mL of freshly made secondary hybridization buffer. Soak the membrane in this secondary hybridization buffer and shake at room temperature for 1 h.
Transfer and soak the membrane in 20 mL of freshly made Western blot washing buffer and shake at room temperature for 10 min.
Repeat step 15 for three more times.
Transfer and soak the membrane in 10 mL of 1-Step NBT/BCIP solution and shake while shielded from light at room temperature for 5–15 min to visualize protein bands.
Terminate the developing reaction by transferring and soaking the membrane in 20 mL of ddH2O.
Air-dry the membrane in darkness before scanning.
Acknowledgments
We thank the members of the Yu laboratory for inspiring discussions. This work was supported by grants GM104077 and AG039559 (to Y.-T.Y.) from the National Institutes of Health.
References
- Adachi H, Yu YT. Purification of radiolabeled RNA products using denaturing gel electrophoresis. Current Protocols in Molecular Biology. 2014;105 doi: 10.1002/0471142727.mb0420s105. http://dx.doi.org/10.1002/0471142727.mb0420s105, 4.20.1–4.20.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbille Y, Vendeix FA, Guenther R, Malkiewicz A, Ariza X, Vilarrasa J, et al. The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Research. 2009;37(10):3342–3353. doi: 10.1093/nar/gkp187. http://dx.doi.org/10.1093/nar/gkp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. http://dx.doi.org/10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes & Development. 2008;22(10):1369–1380. doi: 10.1101/gad.1654308. http://dx.doi.org/10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Research. 1995;23(24):5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. The Journal of Biological Chemistry. 1957;227(2):907–915. [PubMed] [Google Scholar]
- Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+–C base-pair and by pseudouridine. Journal of Molecular Biology. 1999;285(1):115–131. doi: 10.1006/jmbi.1998.2297. http://dx.doi.org/10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500(7460):107–110. doi: 10.1038/nature12302. http://dx.doi.org/10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Yu YT. RNA pseudouridylation: New insights into an old modification. Trends in Biochemical Sciences. 2013;38(4):210–218. doi: 10.1016/j.tibs.2013.01.002. http://dx.doi.org/10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Karijolich J, Yu YT. Post-transcriptional modification of RNAs by artificial box H/ACA and box C/D RNPs. Methods in Molecular Biology. 2011;718:227–244. doi: 10.1007/978-1-61779-018-8_14. http://dx.doi.org/10.1007/978-1-61779-018-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wu G, Yu YT. Inducing nonsense suppression by targeted pseudouridylation. Nature Protocols. 2012;7(4):789–800. doi: 10.1038/nprot.2012.029. http://dx.doi.org/10.1038/nprot.2012.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Yu YT. Synthesis and labeling of RNA in vitro. Current Protocols in Molecular Biology. 2013;4(Unit4.15) doi: 10.1002/0471142727.mb0415s102. http://dx.doi.org/10.1002/0471142727.mb0415s102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. Insight into the protein components of the box H/ACA RNP. Current Proteomics. 2008;5(2):129–137. doi: 10.2174/157016408784911936. http://dx.doi.org/10.2174/157016408784911936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–398. doi: 10.1038/nature10165. http://dx.doi.org/10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Molecular Cell. 2003;11(2):425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. The Journal of Biological Chemistry. 2002;277(34):30445–30453. doi: 10.1074/jbc.M203456200. http://dx.doi.org/10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- Lee CH, Tinoco I, Jr Conformation studies of 13 trinucleoside diphosphates by 360 MHz PMR spectroscopy. A bulged base conformation. I. Base protons and H1′ protons. Biophysical Chemistry. 1980;11(2):283–294. doi: 10.1016/0301-4622(80)80031-7. http://dx.doi.org/0301-4622(80)80031-7 [pii] [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Molecular Cell. 2007;28(6):965–977. doi: 10.1016/j.molcel.2007.10.012. http://dx.doi.org/10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9(10):e110799. doi: 10.1371/journal.pone.0110799. http://dx.doi.org/10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. The EMBO Journal. 2005;24(13):2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. http://dx.doi.org/10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yu AT, Kantartzis A, Yu YT. Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation. Wiley Interdisciplinary Reviews RNA. 2011;2(4):571–581. doi: 10.1002/wrna.77. http://dx.doi.org/10.1002/wrna.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT. Construction of 4-thiouridine site-specifically substituted RNAs for cross-linking studies. Methods. 1999;18(1):13–21. doi: 10.1006/meth.1999.0752. http://dx.doi.org/10.1006/meth.1999.0752. [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. The EMBO Journal. 1998;17(19):5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]