Summary
Utilization of negative checkpoint regulators (NCRs) for cancer immunotherapy has garnered significant interest with the completion of clinical trials demonstrating efficacy. While the results of monotherapy treatments are compelling, there is increasing emphasis on combination treatments in an effort to increase response rates to treatment. One of the most recently discovered NCRs is VISTA (V-domain Ig-containing Suppressor of T cell Activation). In this review, we describe the functions of this molecule in the context of cancer immunotherapy as a target for the management of inflammatory diseases. We also discuss factors that may influence the use of anti-VISTA antibody in combination therapy and how genomic analysis may assist in providing indications for treatment.
Keywords: VISTA, PD-1H, cancer, immunotherapies
Introduction
The approval of antibodies targeting cytotoxic T lymphocyte antigen 4 (CTLA-4) and the programmed death receptor 1 (PD-1) – programmed death ligand 1 (PD-L1) pathway revitalized interest in utilizing antibodies against negative checkpoint regulators to counter the normally suppressive effects of these molecules. VISTA (1) (also known as PD-1H (2), DD1α (3), c10orf54, Gi24 (4), Dies1 (5), and SISP1(6)), is one such NCR that is currently in phase I clinical trials (NCT02671955). Here we summarize the literature and the potential use of antagonist anti-VISTA antibodies in immuno-oncology and agonists for the management of inflammation. In addition, we discuss how antibodies targeting NCRs may be used in combination with other agents and how genomic analysis can assist in providing indications for usage.
VISTA structure and expression
VISTA is a type I transmembrane protein consisting of a single N-terminal immunoglobulin (Ig) V domain, an approximately 30 amino acid (aa) stalk, a transmembrane domain, and a 95 aa cytoplasmic tail (1, 2). Phylogenetic analysis of the full VISTA molecule shares similarities with PD-1, CD28, and CTLA-4, with the highest identity with PD-1 (2). However, a close comparison of VISTA with the other members of the CD28 family reveals some differences. Unlike the genomically clustered group CD28/CTLA4/ICOSVISTA is isolated on chromosome 10 (10q22.1) with no neighboring Ig superfamily members. Interestingly, it is located within a large intron of the CDH23 gene in all genomes starting with the most primitive predicted ortholog in ray-finned fish. VISTA is the most conserved among the B7 members and shows 76% identity between mouse and human and unparallelled 31% sequence identity (59.4% identity in the cytoplasmic tail) between mouse and zebra fish counterparts. The cytoplasmic tail shares 90.6% identity between mouse and human suggesting a tightly conserved functional role (1, 2). By comparison the human and mouse PD-1 tails only share 59% identity.
In contrast to results using the whole protein, analysis of the IgV domain of VISTA shows that this has the greatest homology with programmed death ligand 1 (PD-L1). Subsequent sequence prediction and modeling after PD-L1 shows that the IgV domain of VISTA possesses the canonical disulfide bond between the putative B and F strands. However, it also uniquely has four additional invariant cysteines (three predicted to be within the IgV domain and an additional one in the stalk region) (1). Indeed, the VISTA IgV domain is the most divergent among both B7 member and IgV domains in general (7). While it is possible that the conserved cysteine residues contribute to dimerization, efforts to identify multimeric complexes have been unsuccessful (data not shown).
Within the conserved cytoplasmic tail, VISTA resembles CD28 and CTLA-4. While it does not possess a classic ITIM/ITAM motif, setting it apart from other B7 co-receptor molecules, VISTA has a conserved Src homology 2 (SH2)-binding (YxxQ, potentially capable of binding STAT proteins) motif in the middle of the cytoplasmic tail and three C-terminal SH3-binding domains (PxxP, two in CD28 and one in CTLA-4). It remains to be tested whether the motifs within the VISTA tail actually recruit SH2/SH3 domain adapter proteins as was demonstrated for CD28 and CTLA-4. Taken together, these data suggest that VISTA may act as both a ligand and receptor in regulating immune responses (1–3, 8–12). Emerging studies from a number of labs support this concept.
In mice, VISTA mRNA is expressed in embryonic stem cells at the blastocyte stage of development. Studies suggest it regulates signaling of bone morphogenetic protein 4, which subsequently impact in vitro stem cell differentiation (5, 13, 14). In adult mice at steady state, mRNA for VISTA is primarily confined to hematopoietic tissues including bone marrow, thymus, spleen, and lymph node. The lung and small intestine also have high levels of expression, which is probably due to the presence of leukocyte infiltrate in these tissues. Low but detectable mRNA levels of VISTA are also observed in the heart, brain, muscle, kidney, testis, and placenta (1, 2). However, extensive immunohistological analysis in mice support the conclusion that VISTA protein is exclusively expressed within the hematopoietic compartment (data not shown).
Within the hematopoietic compartment, overall the highest levels of protein expression of VISTA are found in myeloid cells. This includes expression on macrophages, conventional dendritic cells, monocytes, and circulating neutrophils. Within the CD4 T cell compartment, VISTA expression is highest in naïve cells and FoxP3+ regulatory T cells (Treg). Memory CD4 T cells also express VISTA, albeit at a slightly decreased intensity. In addition, CD8 T cells and natural killer (NK) cells also have low, but detectable, surface expression of VISTA, while B cells do not express this molecule (1, 2). Interestingly, another group identified VISTA as a downstream target of p53 activity in response to stress. This observation suggests that surface VISTA is induced in apoptotic cells that sustained DNA damage (3).
Consistent with the mouse data, in humans VISTA is primarily, if not exclusively, found in hematopoietic tissues. Myeloid cells, including patrolling (CD14dimCD16+) and inflammatory (CD14+CD16+/−) monocytes, and lymphoid and myeloid dendritic cell populations have the highest expression, with intermediate levels on neutrophils (11). Monocytes from HIV infected individuals have elevated levels in comparison to healthy controls (8). In contrast to T cells in mice, CD4 and CD8 T cells express VISTA to a similar extent. Dim expression of VISTA is found on CD56lo NK cells (11).
Phenotype of VISTA deficient mice
Lexicon Pharmaceuticals generated VISTA deficient mice on a mixed genetic background by targeting exon 1 for deletion. Preliminary studies showed a slight increase in CD4 T cell frequency in blood (15). Two groups independently crossed these mice to a C57BL/6 background and performed a more extensive analysis of them (9, 12). Both groups observed that VISTA deficient mice had similar numbers of T cells in the spleen; however, the frequency of activated cells within this population becomes elevated over time in comparison to age-matched controls (9, 12). Consistent with this phenotype, after re-activation in vitro, T cells produced more IFNγ, TNFα, and IL-17A (9, 12). In addition, deficient mice had elevated frequencies of myeloid cells in the spleen and higher plasma levels of the chemokines eotaxin, IP-10, MCP-1, and MIG (12). Histology of 12-month old mice showed increased immune cell infiltration in the liver, lung, and pancreas in comparison to controls but lacked overt autoimmunity. However, when VISTA deficient mice were crossed to 2D2 transgenic mice, expressing a TCR that recognizes myelin oligodendrocyte protein, they had a high incidence of spontaneous autoimmune encephalomyelitis (12).
Lee and co-workers also created VISTA deficient mice using a different approach. They floxed exons 2 and 3 with loxP sites and intercrossed them with EIIa-cre mice, which can lead to germline deletion of VISTA. Subsequent VISTA heterozygous mice were backcrossed onto a C57BL/6 background and then intercrossed with each other to obtain VISTA deficient mice. In contrast to the Lexicon VISTA deficient mouse, these mice do develop overt autoimmunity with age, manifesting as increased skin inflammation, anti-nuclear antibody titers, and immune complex deposition in the kidneys (3). The basis for this difference is currently unknown; however, one possibility is that the Lee mice may contain residual 129 genes as they were only backcrossed 7 generations (3). Another is that the normal flora in these mice may be different and subsequently influencing the immune microenvironment to be more inflammatory.
VISTA functions as both a ligand and a receptor
The notion that VISTA functions as a ligand initially developed from a series of in vitro assays measuring T cell responses to stimulation in the presence of VISTA. Mouse and human CD4 and CD8 T cells stimulated with anti-CD3 in the presence of VISTA-Ig fusion protein proliferated less and produced reduced amounts of IFNγ and IL-2 (1, 3, 11). Additional experiments using A20 over-expressing VISTA-RFP or VISTA-transduced bone marrow dendritic cells as antigen presenting cells (APC) also demonstrated reduced proliferation of antigen-specific CD4 T cells as compared to priming elicited by cells expressing control RFP protein (1). Furthermore, myeloid-derived suppressor cells (MDSC) isolated from mice infected with LP-BM5 retrovirus can inhibit B cell proliferation in vitro in a VISTA dependent manner as demonstrated through utilization of VISTA deficient mice and treatment with an anti-VISTA blocking antibody (16). Taken together these studies indicate that VISTA engagement with an unknown VISTA counter-receptor negatively regulates their activation.
A series of studies from the Chen lab have convincingly demonstrated that the engagement of VISTA on the T cell surface by antibody can directly induce T cell suppression. In multiple murine models of GVHD, treatment with the anti-VISTA antibody (clone MH5A) on day -1 and day 0 relative to lethal irradiation of recipient mice significantly increased survival of mice and decreased expansion of donor CD4 and CD8 T cells in the spleen and liver (2, 10). As the numbers of endogenous T cells was similar at early time points after treatment, it is thought that this antibody does not deplete VISTA positive cells and instead functions as a VISTA agonist. In addition, studies utilizing VISTA deficient mice show that expression of VISTA on the donor T cells, but not the host, is necessary for antibody-induced suppression of disease (10). These findings suggest that VISTA can function as a negative checkpoint regulator on T cells. We have now reproduced these findings with other antibodies to VISTA in our anti-VISTA library. As such, there are antibodies to VISTA that enhance immunity (clone 13F3) and anti-VISTA antibodies that induce profound suppression (MH5A). Isotype and fine specificity of these antibodies are now being closely evaluated for the determinants that are critical for VISTA antagoinism vs agonism.
Since both mouse and human myeloid populations express high densities of VISTA (1, 2, 8, 11), it is conceivable that in addition to VISTA potentially serving as a ligand for T cells, it may also be involved in reverse signaling within the myeloid compartment. Transient over-expression of human VISTA in CD14+ monocytes resulted in increased production of IL-6, IL-8, IL-1β, and TNFα. This activity requires the presence of the cytoplasmic domain, suggesting that VISTA engagement on monoctyes results in signaling. In addition, activated CD14+ CD16+ monocytes isolated from HIV positive individuals express elevated levels mRNA for many of these same cytokines (8). Although VISTA lacks recognized ITIM or ITSM in the cytoplasmic domain, the protein sequence contains potential protein kinase C binding sites and proline rich motif, which may function as a platform to interact with other complexes (2). Furthermore, studies of PD-L1 and PD-L2 on dendritic cells suggest that engagement of these ligands can have cell intrinsic effects (17–22). Additional work is needed to further examine downstream signaling effects of VISTA on myeloid cells.
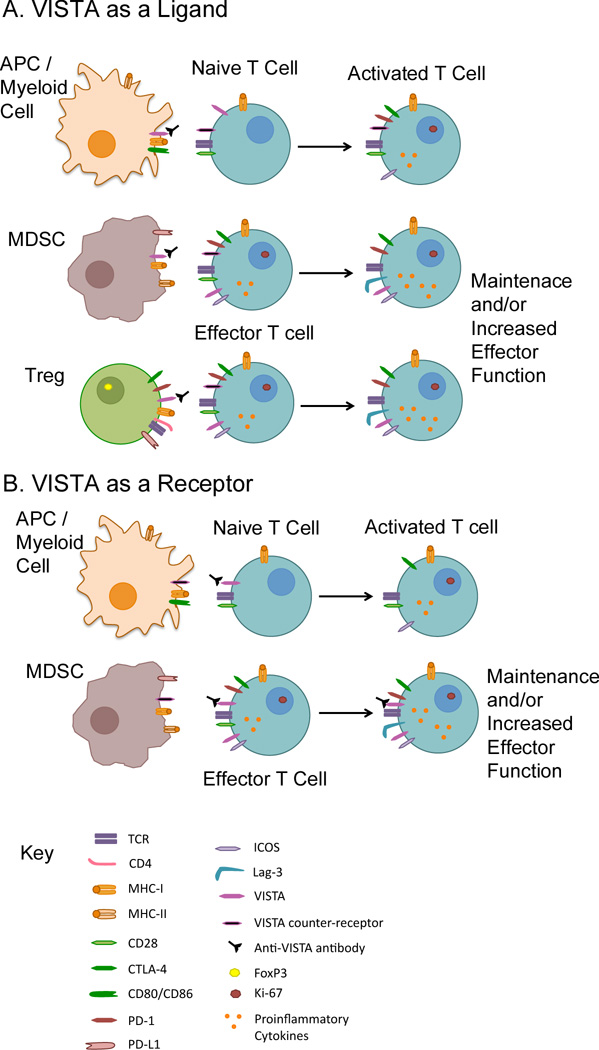
While we present the potential roles of VISTA as a ligand and receptor separately, these functions are not mutually exclusive. Indeed, both in vitro and in vivo mixing experiments that utilize VISTA deficiency on T cells and APCs show that optimal T cell activation occurs in its absence on both cellular compartments (9, 12). Therefore in the context of cancer immunotherapy, blocking antibodies can function to prevent its engagement of an unknown counter-receptor on T cells or by directly targeting VISTA expressed on naïve and effector T cells to prevent it from providing negative signaling upon counter-receptor engagement (Figure 1). Likewise, agonistic antibodies to VISTA that trigger immune suppression may elicit suppressive activities from both T cells and myeloid cells.
Figure 1. VISTA function as a ligand and a receptor.
Potential interactions that may occur are shown and how targeting with a blocking anti-VISTA antibody can affect these results.
The tumor microenvironment and VISTA
The relationship between the development of cancer and host immunity has long been debated and continuously remodeled. Originating from the concept of immunosurveillance, that the immune system can identify and eliminate cancers by way of recognizing non-self antigens, we now understand the relationship to be far more nuanced and complex. Tumor immuno-editing is a dynamic process in which host immunity can either eliminate or at least contain the tumor growth (equilibrium) but also, it can potentiate tumor growth leading to the escape of a clinically overt cancer (23). The understanding that host immunity can exert deleterious effects offered profound insight on vastly unsuccessful immunotherapy approaches that relied solely on generating anti-tumor responses. Despite eliciting potent anti-tumor responses, activated T cells are thwarted upon trafficking into the immunosuppressive tumor microenvironment (TME) (24).
Immune-mediated tumor evasion represents a wide network of active mechanisms that shape anti-tumor mechanisms into those that potentiate tumor growth and allow the tumor to escape the immune system. The emergence of an immunosuppressive tumor microenvironment stems from multiple factors. Here, we will focus predominantly on the development of inhibitory mechanisms and the potential involvement of VISTA in these mechanisms.
NCRs deliver co-inhibitory signals to dampen T cell response. The B7 family represents a class of structurally related co-inhibitory molecules belonging to the Ig superfamily. Active research has shed light on the expanding B7 family. Their expression is found to be upregulated in multiple different human cancers and murine models. In particular, NCRs such as CTLA-4 and the PD-1 - PD-L1 axis have emerged as a fundamental target for reversing immunosuppression largely due to the profound clinical responses in a spectrum of cancers (25–29). VISTA must be included in this cast of NCRs as its expression is also heightened in the TME.
NCR expression is regulated at many stages in the development of the anti-tumor host response driven by a mix of constitutive oncogenic signaling as well as by adaptive immune resistance. CD80 and CD86 are the binding partners for CTLA-4. They are expressed on APCs and as a result, CTLA-4 is viewed to function primarily within secondary lymphoid organs where T cell activation occurs (25). CTLA-4 is not expressed on naïve or memory cells, which provides full opportunity to CD28 to engage CD80/86 and allow co-stimulation to dominate. However, upon TCR signaling, CTLA-4 mRNA is stabilized, protein is mobilized and subsequently expressed on the surface. Upon outcompeting CD28 and ligating with CD80/CD86, CTLA-4 can inhibit T cell responses in both cell-autonomous and cell-extrinsic ways (26).
In contrast to CTLA-4, it is viewed that PD-1 pathway acts to restrain T cell responses in the peripheral tissues, such as at the tumor bed where ligand and receptor are both in abundance (27, 28). Within the TME, PD-1 – PD-L1 engagement leads to many downstream mechanisms that in general support tumor growth. There are two proposed broad mechanisms of PD-L1 regulation that govern its expression within the TME: innate immune resistance and adaptive immune resistance. The regulation of PD-L1 by oncogenes is known as innate immune resistance and is constitutively driven and is expressed independent of inflammatory signals within the TME. Consistent with its role as a negative feedback regulator of effector responses, during adaptive immune resistance PD-L1 is upregulated by inflammatory mediators within the TME and heavily dependent on IFNγ or TLR-mediated signaling pathways (34–37).
VISTA is a member of the B7 family of NCR and represents a new target for immunotherapy. Unlike PD-L1, which is detected in both tumor and hematopoietic lineages, VISTA expression is constitutive in vivo and is primarily, if not exclusively, expressed within the hematopoietic lineage (1, 29). In multiple mouse models, VISTA expression is upregulated in the TME and plays a critical role in shaping anti-tumor immunity (29). In particular, VISTA expression is higher on tumor infiltrating myeloid cells such as myeloid DCs and MDSCs, and on tumor infiltrating Tregs compared to those in the periphery (29). On MDSCs, VISTA increased almost 10-fold on tumor infiltrating leukocytes compared to those in the peripheral lymph node (29). This heightened expression within the TME suggests that factors present, such as hypoxia, may upregulate VISTA expression. Importantly, this indicates that tumors with infiltrating immune cells may harbor abundant levels of VISTA available for therapeutic targeting.
Multiple pre-clinical studies have been performed to examine potential role of VISTA in mediating an immunosuppressive TME. Wang et al. initially demonstrated this in a murine model of fibrosarcoma. They showed that over-expression of VISTA in tumor cells significantly increased tumor growth due to a direct or indirect impact of the ligand activity of VISTA on suppressing T cell immunity (1). In a glioma tumor model with radiotherapy, VISTA−/− mice were more resistant to tumor growth than wild-type mice. This was mediated by a CD4 dependent mechanism, as CD4 depletion prevented resistance (9). Work by Le Mercier et al. showed that VISTA monotherapy with a blocking monoclonal antibody significantly reduced growth in many different solid tumor models (B16/OVA melanoma, B16/BL6 melanoma, MB49 bladder carcinoma, and PTEN/BRAF inducible melanoma) regardless of their immunogenic status or origin (transplantable or inducible). It should be noted that agonistic antibodies to VISTA (those that induce immune suppression) do not enhance immunity in syngeneic tumor models (data not shown). Overall anti-Vista monotherapy (clone 13F3) reshapes the suppressive nature of the TME by reducing the number of MDSCs and tumor specific Tregs. Moreover, anti-VISTA treatment increases the proliferation of TIL and promotes T cell effector function (29). More recently, Kondo et al. investigated a model of squamous carcinoma. Although they did not see striking tumor regression (this could be explained by the fact that they used a different anti-VISTA antibody), treatment did significantly increase CD8 T cell activation and effector function (30). Furthermore, a study comparing VISTA−/−, PD-1−/−, and VISTA−/−PD1−/− mice demonstrated that the PD1 and VISTA checkpoints are non-redundant in antigen-specific responses and in chronic inflammation. Consistent with this finding, autoimmunity was exacerbated in the double deficient mice both in spontaneous disease on a wild-type background, or on a CNS-disease susceptible 2D2 background (31). Consequently, VISTA represents a non-redundant target in the context of PD-1 pathway for blockade in cancer which may impact its clinical potential as a therpeutic target. These promising results of tumor studies in mice led to a phase I anti-VISTA monotherapy clinical trial (NCT02671955) which is currently ongoing. However, the fact that VISTA and PD-1 appear to regulate non-redundant pathways of immune suppression in the TME offers significant excitement for combination therapy.
Targeting VISTA in the context of contemporary antibody combination therapy in immuno-oncology
It is now becoming apparent that multiple pathways exist to maintain the immunosuppressed nature of the tumor microenvironment. In order to reach higher response rates treatment strategies must impact multiple suppressive arms concurrently (depicted in Figure 2) (32). The advent of combination immunotherapy has introduced new challenges. First, the mechanisms of each individual component therapy should be evaluated in order to allow the best selection of synergistic combinations. Second, to improve the frequency of clinical responses to therapy predictive biomarkers that will allow the tailored selection of therapies towards a patient’s individual needs are extremely important. Finally, as the composition of the TME and the anti-tumor immune response evolves during immunotherapy, response biomarkers are warranted. This is especially true given that immunotherapy may induce delayed responses or ‘mixed tumor regression’ in which within a single patient some metastases may respond while others do not. Below is a discussion of the current understanding of the mechanism of action and responses of monotherapy and combination therapy, and the potential value that anti-VISTA could add to such approaches.
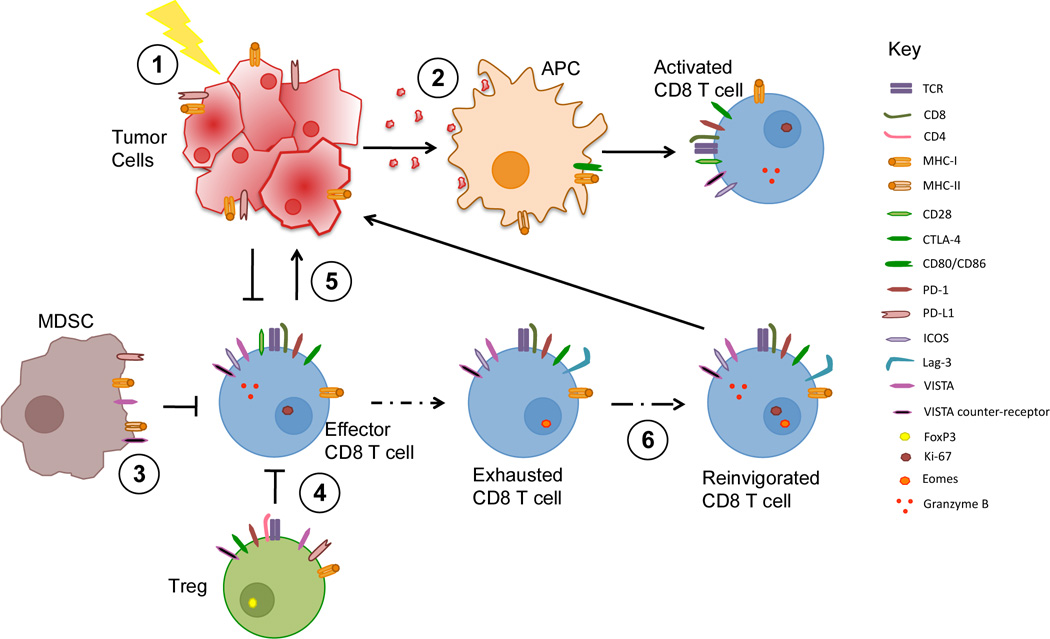
Figure 2. Mechanisms of immunotherapy.
Anti-tumor T cell responses can be modulated by targeting different cell types and arms of immunity. 1 | Immunological Cell Death: tumor cell death induced by radiotherapy or some chemotherapies provide antigen and/or released adjuvant factors that elicit tumor-specific immunity. These treatments may also increase the diversity of the TCR repertoire. 2 | Vaccines: peptides, proteins and allogeneic or autologous tumor cell vaccines may also be used to elicit tumor-specific responses. Some vaccines use cells modified to provide additional adjuvant activity. For example, Gvax, consists of tumor cells modified to release GM-CSF to enhance antigen presentation. 3| Enhanced Innate Immunity: Myeloid cells within tumors may contribute to creating an immunosuppressed environment. Treatments may modulate myeloid activity toward a more inflammatory type sustaining adaptive immune responses 4 | Treg Modulation. Some treatments can either interfere with Treg activity or alter the T effector to Treg balance. 5 | Effector T cell mobilization. Immunotherapies may increase the infiltration of tumors with T cells 6 | Release from Exhaustion. Blockade of negative checkpoints may allow reinvigoration of exhausted cells and restore proliferation and effector function. PD-1 - PD-L1 is the principal pathway, but other negative checkpoints such as LAG-3, TIGIT and TIM-3 contribute to dysfunction of exhausted cells.
CTLA-4
CTLA-4 is the prototypical NCR functioning by interfering with costimulation, providing a brake to the generation of T cell immune responses early in T cell activation. As stated above, CTLA-4 is upregulated upon T cell activation. Targeting of CTLA-4 with the monoclonal antibody ipililumab was the first to reach FDA approval (Yervoy). Following treatment of metastatic melanoma, ipililumab was found to increase the frequency of NY-ESO-1-specific T cells and the frequency of polyfunctional T cells producing IFNγ, MIP-1β and TNFα (33). This increase in T cell frequency may be a biomarker for response to anti-CTLA-4. In particular, responding patients have increased T cells expressing inducible costimulator (ICOS), a costimulator of effector T cells that can also enhance Treg functions (43–45). Consistent with this observation, responses mediated by CTLA-4 blockade in a model of B16/BL6 melanoma were greatly impaired in mice deficient in either ICOS or the ICOS ligand (ICOSL) (34). Futhermore, in ipilimumab treated patients, ICOShi T cells had elevated T-bet expression that was phosphoinositide-3-kinase (PI3K) dependent. ICOS knock down led to reduction in PI3K-signaling and T-bet expression (35). These data support ICOS’s status as a biomarker for response, and may suggest combination with agonist therapies towards the ICOS pathway.
As Tregs constitutively express CTLA-4, the question arises as to whether the mechanism for anti-CTLA-4 therapy might be through depletion or blockade of Treg. To address this question, Peggs et al. created mice in which the extracellular domain of the CTLA-4 protein was replaced with human version, so that antibody could be targeted to either the Treg or T effector compartment, either in vitro, or in reconstitution of RAG−/− mice. These data showed that CTLA-4 blockade targets both the effector and Treg populations. Further, in a B16/BL6 tumor model with vaccine, full therapeutic effect was only seen if the antibody could recognize CTLA-4 on the both the Treg and T effector populations (36). In murine tumor models, the efficacy of anti-CTLA-4 was shown to be increased if the antibody was on an IgG2a backbone, or to a lesser extent IgG2b. This was because this resulted in a loss of intratumoral Tregs with an increase in effector T cell populations and an improved T effector to Treg ratio (37).
However, in humans, while there seems to be some agreement towards a role for ipilimumab on Tregs, whether this activity involves depletion is more controversial. A recent study collected PBMCs and tumor biopsies from patients undergoing ipilimumab therapy at baseline and 4 weeks post treatment and looked at antibody-dependent cell-mediated cytotoxicity (ADCC) ex vivo. They found that ipilimumab caused CD16-dependent, ADCC-dependent killing of Tregs through CD16+ non-classical monocytes (38). Additionally, patients responding to ipilimumab therapy had higher baseline levels of circulating and tumor CD16+ monocytes than non-responsive patients, suggesting that this may be useful as part of a panel of biomarkers. However, a study that looked at CD4+CD25+ T cells in peripheral blood from patients with stage IV melanoma or renal cell cancer undergoing CTLA-4 blockade found no difference in suppressive ability of these cells in vitro. They also saw no decrease in FoxP3 gene expression within CD4 T cells, and concluded that anti-CTLA-4 was not depleting Tregs (39). Kavanagh et al. reported that ipilimumab increased the frequency and proliferation (Ki-67+) of both activated effector CD4 and FoxP3+ Tregs in the blood of patients, and concluded that ipilimumab does not deplete Tregs (40). However, this study did not report on intratumoral Tregs.
There are two important aspects of VISTA biology as they relate to CTLA-4 and the regulation of T cell activation. First, as stated above, multiple labs have shown that VISTA-Ig can suppress the activation of resting T cells (1, 3, 11, 31). This effect of VISTA on T cells is through a putative VISTA counter-receptor expressed on resting T cells. In addition, studies from the Chen lab have repeatedly documented that VISTA engagement, as a receptor expressed on resting T cells, can suppress T cell activation (2, 9, 10). This is possible because VISTA expression peaks on activated CD4 T cells within one day before dropping rapidly (data not shown). This finding is consistent with the observation that treatment with an agonist anti-VISTA antibody given on day 3 (versus day -1 and 0) is ineffective in preventing GVHD lethality (10). In contrast regulation of CTLA-4 is exerted shortly after T cell activation. As such, an antagonistic antibody to VISTA, in both cases would enhance the activation and perhaps clonal diversification of the tumor T cell repertoire. Studies to examine the breadth of the tumor specific T cell repertoire in anti-VISTA treated mice and humans is of significant interest.
PD-1 – PD-L1
The PD-1 pathway followed ipilimumab into the clinic with FDA approval for anti-PD-1 antibodies nivolumab (Opdivo, Bristol-Myers Squibb) and pembrolizumab (Keytruda, Merck) in 2015 and 2016 respectively. PD-1 is a NCR, but in contrast to CTLA-4, PD-1 blocks PI3K activation while CTLA-4 binds to phosphatase PP2A leading to inhibition of Akt phosphorylation (41). The expression of the ligands PD-L1 and PD-L2 is broader and can be induced on many cell types, including tumor cells, after exposure to IFNγ (42). The mechanism of action of PD-1 blockade appears to be localized to the periphery and the tumor site itself, in releasing exhausted cells allowing restoration of effector function (43).
Initial phase I trials reported that PD-L1 may serve as a predictive biomarker for targeting the PD-1 pathway (44). However, it is now becoming clear that some patients will respond even if their tumors are characterized as PD-L1 negative or low (45). Furthermore, the observed increased response rate doesn’t translate to a survival benefit (46). In metastatic melanoma patients undergoing anti-PD-1 therapy with pembrolizumab, it was found that while PD-L1 expression correlates with outcomes, a better predictor of response is a high density of CD8 cells in the margin of the tumor (43).
Although there is ample evidence supporting a role for VISTA in suppressing the activities of resting T cells, its role in suppressing effector cell function within the TME is yet to be resolved. Synergistic effects of anti-VISTA and anti-PD-L1 occurred in CT26 colon cancer in mice which led to reduced tumor growth and increased long term survival in comparison to monotherapy treatment of each. This also correlated with increased production of IFNγ, TNFα, and granzyme B by CD8 T cell from tumor draining lymph nodes after in vitro re-stimulation (31). In a murine model of squamous cell carcinoma using a different anti-VISTA antibody clone, no benefit on tumor growth was observed in combining anti-VISTA and anti-PD-1 treatment. However, there was a striking increase in CD8 T cell effector function when anti-VISTA was used alone or in combination with anti-PD-1 in comparison to controls (30). Results with both of these studies suggest that the effects of targeting VISTA and the PD-1 – PD-L1 axis are non-redundant. However, the extent to which the effects of targeting VISTA are due to effects on CD8 T cells, Tregs, and tumor infiltrating myeloid cells is yet to be determined.
PD-1 and CTLA-4 in combination
Several pre-clinical and clinical studies have been performed to examine dual blockade of both of these NCRs. In a B16 model of melanoma, vaccination with irradiated cells expressing GM-CSF (Gvax) or Flt3-ligand (Fvax) along with a combination of anti-PD-1 and anti-CTLA-4 was much more effective than when the vaccine was used with either antibody alone. Following treatment, the fraction of TILs expressing CTLA-4 and PD-1 increased, but these cells showed characteristics of reinvigoration including the generation of IFNγ/TNFα double producers. The authors surmised that blockade allows the tumor-specific T cells that would otherwise be anergic to maintain effector functions (47).
Recently, an in depth study of mechanisms of action and the mechanisms of resistance of anti-CTLA-4, anti-PD-1, and irradiation was described by Twyman-Saint Victor et al. using both mouse models and human patient data. They found that anti-CTLA-4 primarily decreased Tregs, PD-L1 blockade reinvigorated exhausted T cells, and radiation diversified the TCR repertoire of TILs. In addition, dual checkpoint blockade with anti-CTLA-4 and anti-PD-L1 was inferior to dual checkpoint blockade plus radiation. When looking at biomarkers, the best responses occurred when reinvigoration (Ki-67hi and GzmBhi) within PD1+ CD8 T cells was high but the proportion of exhausted (PD-1+ and Eomes+) CD8 T cells were low. In addition, patients with PD-L1hi melanoma had high levels of persistent exhaustion and did not respond to treatment (48).
Another study in patients treated with anti-PD-1 therapy (either pembrolizumab or nivolumab) found that the proportion of CD8 T cells within the tumor that are PD-1hi CTLA-4hi was strongly correlated with responsiveness (49). As CTLA-4 blockade appears to increase the frequency of responding antigen-specific T cells (33, 50), and this is a predictive biomarker for PD-1 therapy, the mechanisms of these two treatments is highly complementary.
Response rates for CTLA-4 and PD-1 from clinical trial are promising. A phase II trial of patients with advanced melanoma showed an overall response rate (ORR) of 61% with the combination of nivolumab and ipilimumab as compared to 11% for ipilimumab alone (51). Based on these results, the combination was granted accelerated FDA approval for patients with BRAF V600 wild-type unresectable or metastatic melanoma. Subsequently, a phase III study tested either nivolumab alone, nivolumab plus ipilimumab, or ipilimumab alone in a 1:1:1 ratio of 945 untreated advanced melanoma patients (52). Nivolumab alone or the combination of nivolumab and ipilimumab resulted in significantly longer progression-free survival and higher ORR than did treatment with ipilimumab alone. In patients with PD-L1 negative tumors, the combination was better than either treatment alone. In PD-L1 positive tumors, both the nivolumab alone and combination groups were effective in increasing survival, although there was some difference in ORR (52).
As discussed above, PD-L1 expression may be an indication of the presence of a responding T cell pool, which may be effective in the presence of PD-1 blockade that releases the cells from exhaustion. In PD-L1 negative tumors, CTLA-4 blockade may first be required to increase the responding T cell pool. As PD-L1 positive patients may benefit less from ipilimumab, but may still suffer from the increased adverse events of combination therapy, PD-L1 expression may act as a biomarker against the use of ipilimumab (52). A recent study by Kim et al. have underscored the role of MDSCs in murine models in which tumors were resistant to anti-CTLA-4 and anti-PD-1 blockade (53). It is under these conditions, that anti-VISTA, with its putative impact on the myeloid compartment, may exert therapeutic benefit.
A role for VISTA in myeloid targeting therapies in combination blockade
While there are multiple cells and molecules that subvert the full potential of immunotherapy, targeting of MDSCs may offer its distinct advantage in combination therapy to lead to an overall enhancement of anti-tumor T cell function. As early as the 1980s, dysregulated myelopoiesis and expansion of immature precursors was first observed in the setting of cancer (54). Now, MDSCs have been documented to be greatly expanded in many mouse models (55, 56) and in human cancers such as colon, breast, melanoma, and pancreatic, among others (64, 65). They inhibit T cell activity through multiple different mechanisms ultimately leading to increased tumor burden (54) including nutrient depletion (57, 58), disrupting TCR signal transduction (59), abrogating T cell migration (60, 61), and utilizing the PD-L1 – PD-1 pathway as a mechanism to suppress T cells (62).
In late intervention of CT26 colon cancer and 4T1 breast cancer in mice, at tumor volumes at which CTLA-4 and PD-1 combination blockade fails, it appears to be a result of granulocytic MDSC function. Treatments that reduce tumor granulocytic MDSCs (anti-Ly6G or epigenetic-modulating drugs) used in combination with dual checkpoint blockade allowed rejection of even very large tumors (53). Similarly, a treatment that blocked CSF-1 causing CD206-hi tumor associated macrophage (TAM) death, was highly synergistic with dual blockade in large tumors (63). On the receptor side, CSFR blockade decreased M2 polarization of TAMs. This improved survival in a PDGF-B-driven glioma model, and reduced growth of proneural glioma xenografts in NOD-SCID mice (64). Due to the high density of VISTA expression on MDSC, future studies examining the impact of anti-VISTA therapy used in similar combination blockade would be useful in determining if the treatment is modulating the myeloid compartment. Transfection of VISTA into monocytes altered cytokine expression to be more pro-inflammatory and this required the presence of its cytoplasmic domain (8). This would be consistent with VISTA functioning as a receptor on myeloid cells after engagement with its counter-receptor. We envision that VISTA is a criticial immunoregulatory receptor on MDSCs that can control their suppression activity within the TME and be a valuable complementary therapy to combination blockade of PD-1 and CTLA-4.
Combination strategies with other negative checkpoint molecules
T cells that express PD-1 along with other inhibitory molecules including CTLA-4, T cell immunoglublin and mucin-domatin containing-3 (TIM-3) (65, 66), lymphocyte-activation gene 3 (LAG-3) (67, 68), B and T lymphocyte associated (BTLA) (79, 80), T cell Ig and ITIM domain (TIGIT) (81, 82), and CD39 (69) are thought to be the most dysfunctionally exhausted cells (70). While PD-1 is the most dominant regulator of exhaustion, other negative signaling molecules likely control the severity of exhaustion, and combination blockade of these with PD-1 may increase the reinvigoration of exhausted cells in the tumor setting. TIM-3 is expressed by the most severely exhausted PD-1+ CD8+ T cells in tumors (65). In the CT26 mouse model of colon cancer, antibody blockade of PD-L1 and TIM-3 was much more effective in reducing tumor burden than either treatment alone (65). Similarly, in vitro blockade of both TIGIT and PD-1 increased T cell function additively (71). Furthermore, blockade of TIGIT was effective in mouse preclinical tumor models, and was also synergistic with blockade of TIM-3 (72) or blockade of PD-L1 (73). Interestingly, similarly to CTLA-4 and CD28, TIGIT competes with a costimulatory molecule CD226 for its ligand, poliovirus receptor, and actually impairs function of CD226 through interfering with its homodimerization (73). Given that many checkpoint molecules function in multiple directions, or have different binding partners, combination blockade should take into consideration how interacting pathways are affected, including those of positive costimulatory regulators.
Combination therapies with agonist antibodies
In addition to antibody blockade of negative checkpoints, some treatment strategies employ agonist antibodies that target costimulatory molecules to convey activating signals to T cells. These agents appear to work best when combined with blockade of negative checkpoints – “removing the brakes” (74, 75). Costimulatory molecules include those in the tumor necrosis factor receptor (TNFR) superfamily and the Ig superfamily. Agonists for CD137, OX40, GITR, CD40, CD27 and ICOS are currently in clinical trials.
As described above, checkpoint blockade with ipilimumab induces ICOS+ cells response (43–45), which makes this an obvious candidate for combination therapy. Preclinical cancer models have shown improved anti-tumor immune responses with ICOS costimulatory antibody (76), tumor cell vaccines engineered to express ICOS ligand (77). Clinical trials are underway for ICOS agonist with nivolumab (NCT02904226).
Additional work has focused on effects of TNFR superfamily members. One study tested an allogeneic cell-based vaccine that secreted Fc-OX40L, Fc-ICOSL, or Fc-4-1BBL versus systemic agonist antibody and found that OX40 was the most effectively targeted pathway (78). In murine tumor models, OX40 agonist antibody has enhanced survival as monotherapy (79) or combined with CTLA-4 blockade (80). Treatment expanded Tbethi Eomeshi Granzyme B+ CD8 T cells and both Th1 and Th2 CD4 T cells (80). OX40 agonist has shown promise in the clinic, and has entered multiple clinical trials both as a monotherapy or in combination with other immunotherapies including anti-CTLA4 and PD-L1 blockade (NCT02705482, NCT02221960, NCT02205333, completed) and anti-CD137 (anti-4-1BB) agonist (NCT02315066). Anti CD137 (4 1BB) antibody has been effective in monotherapy in preclinical models including melanoma, breast and colon carcinomas (81). However, early phase I trials using this antibody were terminated because of cases of severe hepatotoxicity (82). Newer trials are currently underway using lower doses of anti CD137 antibodies to treat patients with melanoma, renal cell carcinoma, and ovarian cancer.
Perspectives on combination therapy with anti-VISTA
As depicted in Figure 2, there are multiple mechanisms that immunotherapies can target. In theory, the best results would occur when mechanisms are non-overlapping. As discussed, effects of an antagonist anti-VISTA antibody appear to be non-overlapping with CTLA-4 and PD-1 – PD-L1 pathways (30, 31). Work studying mechanisms of action of antibody targeting of CTLA-4 and PD-L1 suggest they function through decreasing Treg and increasing reinvigoration, respectively (48), therefore anti-VISTA is likely to have independent functions. Current data shows a marked increase in effector function (29–31); however, this work does not distinguish if this is a result of increasing the T cell repertoire, decreasing exhaustion, and/or increasing reinvigorization. All of these effects may also be indirect consequences of targeting VISTA on myeloid cells, such as MDSCs, which have the most robust expression of this molecule (1, 2, 11, 29), and future studies are needed to determine if it impacts their expansion and/or suppressor function. If this is the major effect, cancers characterized by high infiltration of MDSCs may be particularly responsive to treatment.
Genomic approaches to immunotherapy response
The widespread availability of patient genomic data has made computational approaches to studying tumor immunology increasingly accessible. Multi-dimensional datasets such as the Cancer Genome Atlas (TCGA) integrate expression, sequencing, and methylation data from large numbers of patients with multiple tumor types. These datasets enable simple pan-cancer comparisons to be made, such as the landscape of VISTA expression across multiple tumor types (Figure 3A). In addition to these superficial comparisons, several methods have been developed that use these data to more deeply characterize the immune response profile of each patient, pushing the field of tumor immunology into the realm of personalized medicine. These methods have utilized patient gene expression and DNA sequencing data to identify the immune cells present in a patient’s tumor, as well as the tumor antigens that are likely driving their infiltration. While these approaches have not yet been used to predict anti-VISTA response, they have been applied in several studies to identify biomarkers indicative of response to anti-CTLA-4 and anti-PD1 therapy. We outline these methods and their applications to immunotherapy below with the goal of providing a roadmap for future analyses of anti-VISTA response.
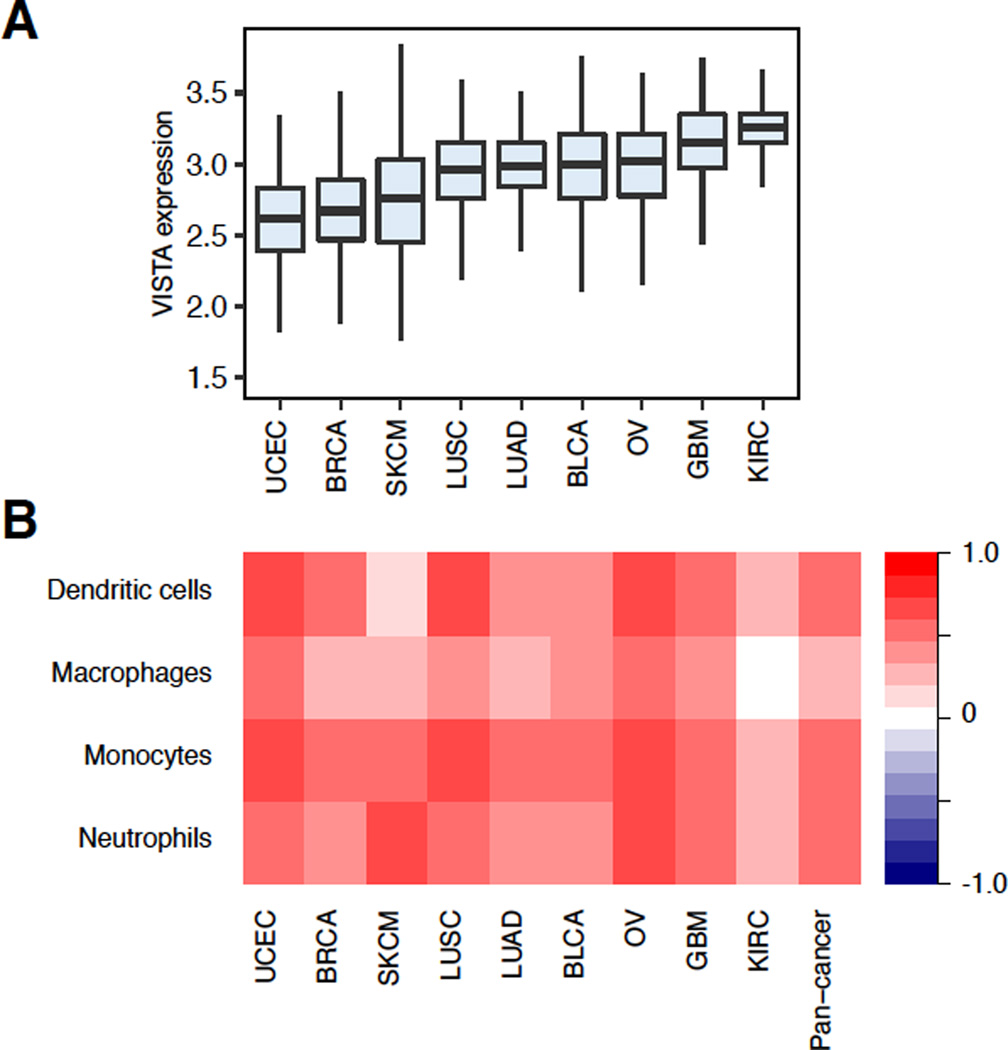
Figure 3. Association between VISTA expression and myeloid infiltration across tumor types.
A. Boxplots comparing the distribution of VISTA expression across 9 different TCGA tumor types. Each box spans quartiles with bold lines representing the median VISTA expression level in each tumor type. All outliers were excluded in the plot. B. Spearman correlations between VISTA expression and infiltration levels of different myeloid cell types. Myeloid cell infiltration was inferred using patient gene expression data (87).
Inferring infiltration of immune cell subsets from genomic data
Tumors are infiltrated by a variety of immune cell types that each play distinct roles in the tumor’s growth and survival. By quantifying the infiltration levels of distinct immune cell subsets, a wealth of information can be obtained regarding patient prognosis and potential immunotherapy response. Traditionally, histopathological techniques have been used to assess the types of immune cells present in the patient’s tumor. However, these techniques are time consuming and require a large number of antibodies in order to comprehensively define a patient’s tumor microenvironment. To address this issue, several methods now exist that use genomic data to delineate the tumor infiltrating lymphocytes of a patient’s tumor. These methods primarily function by examining the expression levels of lymphocyte-specific genes in a patient’s gene expression profile.
One approach to expression-based inference of immune infiltration is to examine the distribution of lymphocyte-specific genes in a patient’s ranked gene expression profile. There are currently two methods that enable these kinds of analyses. The first method, single sample gene set enrichment analysis (ssGSEA), requires a binary list of lymphocyte-specific genes, known as a gene signature, and examines how these genes are distributed in a ranked patient gene expression profile (83). A higher enrichment score indicates higher lymphocyte-specific gene expression in a sample, and thus higher infiltration for that lymphocyte. An early method, known as ESTIMATE, used this approach with a general immune infiltration signature to infer non-specific immune infiltration levels in 12 TCGA tumor types (84). In another study, ssGSEA was applied in colorectal cancer to examine the features associated with the infiltration of different immune cell types and characeterized the factors driving immunogenicity (85). In this study, differentially expressed genes identified across 28 different immune cell subsets were used as gene signatures to infer the infiltration levels of several different cell types. This study demonstrated the potential of ssGSEA approaches and allowed for a thorough characterization of the factors driving immunogenicity in colorectal cancer. The second enrichment-based method, known as binding association with sorted expression (BASE) foregoes gene signatures and instead utilizes the entire transcriptome of each reference lymphocyte to calculate a lymphocytic infiltration score (86). This method, which was originally developed to infer transcription factor activity from ChIPseq data, converts each lymphocyte’s gene expression profile into a series of weights that inform how differentially expressed each gene is in that cell type. By accounting for the entire transcriptome of a reference lymphocyte, this method avoids the biases involved in selecting a gene signature and uses a greater dynamic range to determine cell-specific gene expression. This method was utilized to examine the landscape of immune infiltration in breast cancer, finding several cell types associated with patient survival, mutation burden, and molecular subtype (87). Recently, this method was used to demonstrate a positive relationship between VISTA expression and infiltration from different myeloid cell lineages across cancer types (Figure 4B).
While enrichment-based methods of inferring immune infiltration have been successful at performing sample-sample comparisons, they cannot accurately quantify the cellular fractions in a given sample. For this task, several deconvolution methods exist that use gene expression data to infer the proportions of each cell type present in bulk cell mixtures (88–91). One method in particular, CIBERSORT, has proven to be effective at enumerating the different infiltrating immune cell types from bulk tumor expression data collected from microarrays (92) and was utilized to identify cellular proportions that were predictive of patient survival in an assembly of 166 independent datasets encompassing 39 different cancer types (93).
In addition to expression-based methods, several studies have also shown the capability of using DNA methylation data to infer immune infiltration from patient Illumina Infinium 25k or 450k methylation arrays (94–96). These methods are dependent on identifying the differentially methylated CpGs that are unique to distinct immune cell subsets. While promising, these approaches are not as developed as expression-based methods due to the lack of immune cell-specific methylation data. As more epigenetic data is compiled on both patients and immune cells, these methods may serve as good complementary analyses to be performed alongside expression-based analyses.
Together, these methods can be applied to quickly assess the levels of patient tumor immune infiltration for a large number of cell types. When used in conjunction with immunotherapy response data, these methods can be used to identify the cell types likely involved in response or resistance to any given cancer treatment. As clinical trials on various therapies, including anti-VISTA, continue to generate data, these methods will greatly enhance our ability to provide individualized care to increase clinical responses.
Computational approaches to identifying neoantigens
While understanding the lymphocytes present in a patient’s tumor can greatly benefit our understanding of immunotherapy response, features intrinsic to each patient’s disease provide useful information regarding the tumor’s immunogenicity. Mutations specific to each patient alter the structure of some proteins into non-self forms the immune system does not recognize. Short fragments of these mutated proteins, known as neoantigens, are capable of binding to the major histocompatibility complex (MHC) class I proteins used by antigen presenting cells to educate CD8 T cells. Neoantigens have been shown to associate with increased immune infiltration in a variety of tumor types, making them good candidates for predicting immunotherapy response (97, 98).
The variable nature of both somatic mutations and the MHC loci, known as human leukocyte antigen (HLA) in humans, makes the identification of neoantigens highly specific for a given patient. As such, there have now been several tools developed to aid in the identification of neoantigens from individual tumor DNA sequencing data. These tools typically perform one of three steps necessary in neoantigen identification: somatic mutation identification (99–105), HLA typing (106–111), and neoantigen-MHC binding affinity prediction (112). In addition, some pipelines have been created such as, pVAC-Seq, to integrate several of these steps together while also accounting for the coverage and expression information that may confound sequencing based studies (113). As these approaches continue to evolve and neoantigen prediction becomes more routine, our ability to identify biomarkers indicative of immune infiltration and response to immunotherapeutic approaches should be greatly improved.
Integrating genomic data to predict immunotherapy response
While there are currently no studies that use patient genomic data to predict anti-VISTA immunotherapy response, there have been several studies that use gene expression or DNA sequencing information collected from pre-treatment patient cohorts to identify the genomic features associated with response to anti-CTLA-4 and anti-PD-1 therapy. In a study on CTLA-4 blockade response, whole exome sequencing was performed on 110 pre-treatment melanoma tumor biopsies and their matching germline tissue samples. In addition to this, RNAseq data was obtained from 40 of these biopsies, enabling comprehensive genomic analyses to be performed on a small patient cohort. This study found using several of the methods described above that higher mutation and neoantigen counts were associated with clinical benefit to treatment with ipilimumab. Additionally, they found through gene expression analyses that the expression of cytolytic genes, which are indicative of cytotoxic cell infiltration, was also linked to improved ipilimumab response (114).
Similar studies have been performed to examine the genomic correlates of response to anti-PD-1 therapy. One analysis examined the relationship between mutation burden and response to pembrolizumab in non-small cell lung cancer (NSCLC). This group found that higher mutation load was associated with improved progression-free survival, and sensitivity to treatment (115). In a later study focusing on melanoma, whole exome sequencing was performed on a collection of 38 pre-treatment or early on-treatment tumors and of these 38, RNA sequencing information was collected from 27 of them. Interestingly, this study did not find a relationship between mutation load and improved anti-PD-1 response. However, clinical benefit was associated with mutations in the DNA repair gene BRCA2. Additionally, this group showed that genes involved in mesenchymal transition, angiogenesis, and T cell suppression were more highly expressed in the non-responsive group relative to the responsive group (116).
In addition to these studies, a follow-up study was performed to examine the effect of neoantigen intratumor heterogeneity on tumor immune response and found, using the anti-PD-1 NSCLC and anti-CTLA-4 melanoma datasets mentioned above, that clonal neoantigen burden was associated with overall survival and response to CTLA-4 and PD-1 blockade in melanoma and non-small cell lung cancer, respectively (117). This analysis demonstrates the high level of complexity behind the tumor immune response and its role on immunotherapy sensitivity. However, the genomic data obtained from the small patient cohorts included in the study led to significant insights that could not have been obtained through other methods.
Together, these analyses show the potential of using patient genomic data to identify biomarkers for immunotherapy response. Interestingly, there was a high amount of discordance between each of the studies, demonstrating the need for additional immunotherapy datasets encompassing larger numbers of patients. The differences between genomic correlates associated with anti-PD-1 and anti-CTLA4 blockade additionally suggests that response to each type of immunotherapy may be dependent on unique molecular mechanisms. Going forward, similar studies on anti-VISTA could provide a large amount of information regarding the molecular mechanisms involved in anti-VISTA response, as well as predictive biomarkers that could be used to identify patients most likely to be responsive to the drug.
Conclusions
Due to the success of targeting the negative checkpoint regulators CTLA-4, PD-1, and PD-L1 to treat cancer, a spotlight has been cast on this entire family of molecules. VISTA has several unique features that suggest that it functions non-redundantly to PD-1. Consequently, therapeutic treatment with antibody targeting it may be beneficial not only as a monotherapy, but also in combination with other treatments. Further studies are also needed to help identify genetic signatures that can provide further insight into the mechanism of action of therapy and identification of patients most likely to respond to treatment.
Acknowledgments
This work is supported by the NIH Centers of Biomedical Research Excellence (COBRE) grant GM103534 (to CC).
Footnotes
Disclosures. I. Le Mercier is an employee of ImmuNext. R.J. Noelle is a CSO, has commerical research support, and has ownership interests (including patents) in ImmuNext.
References
- 1.Wang L, Rubinstein R, Lines JL, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208:577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flies DB, Wang S, Xu H, Chen L. Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J Immunol. 2011;187:1537–1541. doi: 10.4049/jimmunol.1100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon KW, Byun S, Kwon E, et al. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science. 2015;349:1261669. doi: 10.1126/science.1261669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakr MA, Takino T, Domoto T, et al. GI24 enhances tumor invasiveness by regulating cell surface membrane-type 1 matrix metalloproteinase. Cancer Science. 2010;101:2368–2374. doi: 10.1111/j.1349-7006.2010.01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aloia L, Parisi S, Fusco L, Pastore L, Russo T. Differentiation of embryonic stem cells 1 (Dies1) is a component of bone morphogenetic protein 4 (BMP4) signaling pathway required for proper differentiation of mouse embryonic stem cells. J Biol Chem. 2010;285:7776–7783. doi: 10.1074/jbc.M109.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang YS. SISP-1, a novel p53 target gene and use thereof. Google Patents. 2010 [Google Scholar]
- 7.Flajnik MF, Tlapakova T, Criscitiello MF, Krylov V, Ohta Y. Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7's historical relationship with the MHC. Immunogenetics. 2012;64:571–590. doi: 10.1007/s00251-012-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharaj P, Chahar HS, Alozie OK, et al. Characterization of Programmed Death-1 Homologue-1 (PD-1H) Expression and Function in Normal and HIV Infected Individuals. PLoS ONE. 2014;9:e109103. doi: 10.1371/journal.pone.0109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flies DB, Han X, Higuchi T, Zheng L, Sun J, Ye JJ, Chen L. Coinhibitory receptor PD-1H preferentially suppresses CD4(+) T cell-mediated immunity. J Clin Invest. 2014;124:1966–1975. doi: 10.1172/JCI74589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flies DB, Higuchi T, Chen L. Mechanistic Assessment of PD-1H Coinhibitory Receptor-Induced T Cell Tolerance to Allogeneic Antigens. J Immunol. 2015;194:5294–52304. doi: 10.4049/jimmunol.1402648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lines JL, Pantazi E, Mak J, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74:1924–1932. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Le Mercier I, Putra J, et al. Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc Natl Acad Sci U S A. 2014;111:14846–14851. doi: 10.1073/pnas.1407447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battista M, Musto A, Navarra A, Minopoli G, Russo T, Parisi S. miR-125b regulates the early steps of ESC differentiation through dies1 in a TGF-independent manner. Int J Mol Sci. 2013;14:13482–13496. doi: 10.3390/ijms140713482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parisi S, Battista M, Musto A, Navarra A, Tarantino C, Russo T. A regulatory loop involving Dies1 and miR-125a controls BMP4 signaling in mouse embryonic stem cells. FASEB J. 2012;26:3957–3968. doi: 10.1096/fj.12-211607. [DOI] [PubMed] [Google Scholar]
- 15.Tang T, Li L, Tang J, et al. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 16.Green KA, Wang L, Noelle RJ, Green WR. Selective Involvement of the Checkpoint Regulator VISTA in Suppression of B-Cell, but Not T-Cell, Responsiveness by Monocytic Myeloid-Derived Suppressor Cells from Mice Infected with an Immunodeficiency-Causing Retrovirus. J Virol. 2015;89:9693–9698. doi: 10.1128/JVI.00888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blocki FA, Radhakrishnan S, Van Keulen VP, et al. Induction of a gene expression program in dendritic cells with a cross-linking IgM antibody to the co-stimulatory molecule B7-DC. FASEB J. 2006;20:2408–2410. doi: 10.1096/fj.06-6171fje. [DOI] [PubMed] [Google Scholar]
- 18.Van Keulen VP, Ciric B, Radhakrishnan S, et al. Immunomodulation using the recombinant monoclonal human B7-DC cross-linking antibody rHIgM12. Clin Exp Immunol. 2006;143:314–321. doi: 10.1111/j.1365-2249.2005.02992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuipers H, Muskens F, Willart M, et al. Contribution of the PD-1 ligands/PD-1 signaling pathway to dendritic cell-mediated CD4+ T cell activation. Eur J Immunol. 2006;36:2472–2482. doi: 10.1002/eji.200635978. [DOI] [PubMed] [Google Scholar]
- 20.Radhakrishnan S, Nguyen LT, Ciric B, et al. Immunotherapeutic potential of B7-DC (PD-L2) cross-linking antibody in conferring antitumor immunity. Cancer Res. 2004;64:4965–4972. doi: 10.1158/0008-5472.CAN-03-3025. [DOI] [PubMed] [Google Scholar]
- 21.Radhakrishnan S, Nguyen LT, Ciric B, Van Keulen VP, Pease LR. B7-DC/PD-L2 cross-linking induces NF-kappaB-dependent protection of dendritic cells from cell death. J Immunol. 2007;178:1426–1432. doi: 10.4049/jimmunol.178.3.1426. [DOI] [PubMed] [Google Scholar]
- 22.Hobo W, Maas F, Adisty N, de Witte T, Schaap N, van der Voort R, Dolstra H. siRNA silencing of PD-L1 and PD-L2 on dendritic cells augments expansion and function of minor histocompatibility antigen-specific CD8+ T cells. Blood. 2010;116:4501–4511. doi: 10.1182/blood-2010-04-278739. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 24.Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015;4:e1016700. doi: 10.1080/2162402X.2015.1016700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topalian Suzanne L, Drake Charles G, Pardoll Drew M. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolchok JD, Saenger Y. The mechanism of anti-CTLA-4 activity and the negative regulation of T-cell activation. Oncologist. 2008;13 Suppl 4:2–9. doi: 10.1634/theoncologist.13-S4-2. [DOI] [PubMed] [Google Scholar]
- 27.Topalian Suzanne L, Drake Charles G, Pardoll Drew M. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Han X. Anti–PD-1/PD-L1 therapy of human cancer: past, present, and future. The Journal of Clinical Investigation. 125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Mercier I, Chen W, Lines JL, et al. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer Res. 2014;74:1933–1944. doi: 10.1158/0008-5472.CAN-13-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo Y, Ohno T, Nishii N, Harada K, Yagita H, Azuma M. Differential contribution of three immune checkpoint (VISTA, CTLA-4, PD-1) pathways to antitumor responses against squamous cell carcinoma. Oral Oncol. 2016;57:54–60. doi: 10.1016/j.oraloncology.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Yuan Y, Chen W, et al. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc Natl Acad Sci U S A. 2015;112:6682–6687. doi: 10.1073/pnas.1420370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth MJ, Ngiow SF, Ribas A, Teng MWL. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 33.Yuan J, Gnjatic S, Li H, et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proceedings of the National Academy of Sciences. 2008;105:20410–20415. doi: 10.1073/pnas.0810114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu T, He Q, Sharma P. The ICOS/ICOSL Pathway Is Required for Optimal Antitumor Responses Mediated by Anti–CTLA-4 Therapy. Cancer Research. 2011;71:5445–5454. doi: 10.1158/0008-5472.CAN-11-1138. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Fu T, Suh W-K, et al. CD4 T cells require ICOS-mediated PI3K-signaling to increase T-bet expression in the setting of anti-CTLA-4 therapy. Cancer immunology research. 2014;2:167–176. doi: 10.1158/2326-6066.CIR-13-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti–CTLA-4 antibodies. The Journal of Experimental Medicine. 2009;206:1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, Korman AJ. Anti-CTLA-4 Antibodies of IgG2a Isotype Enhance Antitumor Activity through Reduction of Intratumoral Regulatory T Cells. Cancer Immunology Research. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 38.Romano E, Kusio-Kobialka M, Foukas PG, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proceedings of the National Academy of Sciences. 2015;112:6140–6145. doi: 10.1073/pnas.1417320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maker AV, Attia P, Rosenberg SA. Analysis of the Cellular Mechanism of Antitumor Responses and Autoimmunity in Patients Treated with CTLA-4 Blockade. The Journal of Immunology. 2005;175:7746–7754. doi: 10.4049/jimmunol.175.11.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kavanagh B, O'Brien S, Lee D, et al. CTLA4 blockade expands FoxP3+ regulatory and activated effector CD4+ T cells in a dose-dependent fashion. Blood. 2008;112:1175–1183. doi: 10.1182/blood-2007-11-125435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA-4 and PD-1 Receptors Inhibit T-Cell Activation by Distinct Mechanisms. Molecular and Cellular Biology. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenwald RJ, Freeman GJ, Sharpe AH. THE B7 FAMILY REVISITED. Annual Review of Immunology. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 43.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New England Journal of Medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbst RS, Soria J-C, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert C, Long GV, Brady B, et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. New England Journal of Medicine. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 47.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proceedings of the National Academy of Sciences. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest. 2016;126:3447–3452. doi: 10.1172/JCI87324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liakou CI, Kamat A, Tang DN, et al. [Google Scholar]
- 51.Postow MA, Chesney J, Pavlick AC, et al. Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma. New England Journal of Medicine. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. New England Journal of Medicine. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K, Skora AD, Li Z, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Youn J-I, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. The Journal of Immunology. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Youn J-I, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Journal of Leukocyte Biology. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells Inhibit T-Cell Activation by Depleting Cystine and Cysteine. Cancer Research. 2010;70:68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagaraj S, Gupta K, Pisarev V, et al. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanson EM, Clements VK, Sinha P, Ilkovitch D, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells Down-Regulate L-Selectin Expression on CD4+ and CD8+ T Cells. The Journal of Immunology. 2009;183:937–944. doi: 10.4049/jimmunol.0804253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molon B, Ugel S, Del Pozzo F, et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. The Journal of Experimental Medicine. 2011;208:1949–1962. doi: 10.1084/jem.20101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noman MZ, Desantis G, Janji B, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. The Journal of Experimental Medicine. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Y, Knolhoff BL, Meyer MA, et al. CSF1/CSF1R Blockade Reprograms Tumor-Infiltrating Macrophages and Improves Response to T Cell Checkpoint Immunotherapy in Pancreatic Cancer Models. Cancer research. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. The Journal of Experimental Medicine. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin H-T, Anderson AC, Tan WG, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proceedings of the National Academy of Sciences. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyazaki T, Dierich A, Benoist C, Mathis D. Independent modes of natural killing distinguished in mice lacking Lag3. Science. 1996;272:405–408. doi: 10.1126/science.272.5260.405. [DOI] [PubMed] [Google Scholar]
- 68.Okazaki T, Okazaki I-m, Wang J, et al. PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. The Journal of Experimental Medicine. 2011;208:395–407. doi: 10.1084/jem.20100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta PK, Godec J, Wolski D, et al. CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog. 2015;11:e1005177. doi: 10.1371/journal.ppat.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chauvin J-M, Pagliano O, Fourcade J, et al. TIGIT and PD-1 impair tumor antigen–specific CD8+ T cells in melanoma patients. The Journal of Clinical Investigation. 125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurtulus S, Sakuishi K, Zhang H, et al. Mechanisms of TIGIT-driven immune suppression in cancer. Journal for Immunotherapy of Cancer. 2014;2:O13-O. [Google Scholar]
- 73.Johnston Robert J, Comps-Agrar L, Hackney J, et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8+ T Cell Effector Function. Cancer Cell. 26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 74.Tirapu I, Mazzolini G, Rodriguez-Calvillo M, Arina A, Palencia B, Gabari I, Melero I. Effective tumor immunotherapy: start the engine, release the brakes, step on the gas pedal,...and get ready to face autoimmunity. Arch Immunol Ther Exp (Warsz) 2002;50:13–18. [PubMed] [Google Scholar]
- 75.Sanmamed MF, Pastor F, Rodriguez A, Perez-Gracia JL, Rodriguez-Ruiz ME, Jure-Kunkel M, Melero I. Agonists of Co-stimulation in Cancer Immunotherapy Directed Against CD137, OX40, GITR, CD27, CD28, and ICOS. Seminars in Oncology. 2015;42:640–655. doi: 10.1053/j.seminoncol.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 76.Nelson MH, Kundimi S, Bowers JS, et al. The inducible costimulator augments Tc17 cell responses to self and tumor tissue. Journal of immunology (Baltimore, Md : 1950) . 2015;194:1737–1747. doi: 10.4049/jimmunol.1401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan X, Quezada SA, Sepulveda MA, Sharma P, Allison JP. Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. The Journal of Experimental Medicine. 2014;211:715–725. doi: 10.1084/jem.20130590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fromm G, de Silva S, Giffin L, Xu X, Rose J, Schreiber TH. Gp96-Ig/Costimulator (OX40L, ICOSL, or 4-1BBL) Combination Vaccine Improves T-cell Priming and Enhances Immunity, Memory, and Tumor Elimination. Cancer Immunology Research. 2016;4:766–778. doi: 10.1158/2326-6066.CIR-15-0228. [DOI] [PubMed] [Google Scholar]
- 79.Shibahara I, Saito R, Zhang R, et al. OX40 ligand expressed in glioblastoma modulates adaptive immunity depending on the microenvironment: a clue for successful immunotherapy. Molecular Cancer. 2015;14:41. doi: 10.1186/s12943-015-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Redmond WL, Linch SN, Kasiewicz MJ. Combined Targeting of Costimulatory (OX40) and Coinhibitory (CTLA-4) Pathways Elicits Potent Effector T Cells Capable of Driving Robust Antitumor Immunity. Cancer Immunology Research. 2014;2:142–153. doi: 10.1158/2326-6066.CIR-13-0031-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melero I, Shuford WW, Newby SA, et al. Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat Med. 1997;3:682–685. doi: 10.1038/nm0697-682. [DOI] [PubMed] [Google Scholar]
- 82.Li S-Y, Liu Y. Immunotherapy of melanoma with the immune costimulatory monoclonal antibodies targeting CD137. Clinical Pharmacology : Advances and Applications. 2013;5:47–53. doi: 10.2147/CPAA.S46199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics. 2007;8:452. doi: 10.1186/1471-2105-8-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varn FS, Andrews EH, Mullins DW, Cheng C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat Commun. 2016;7:10248. doi: 10.1038/ncomms10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qiao W, Quon G, Csaszar E, Yu M, Morris Q, Zandstra PW. PERT: a method for expression deconvolution of human blood samples from varied microenvironmental and developmental conditions. PLoS Comput Biol. 2012;8:e1002838. doi: 10.1371/journal.pcbi.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
- 90.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29:2211–2212. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 91.Liebner DA, Huang K, Parvin JD. MMAD: microarray microdissection with analysis of differences is a computational tool for deconvoluting cell type-specific contributions from tissue samples. Bioinformatics. 2014;30:682–689. doi: 10.1093/bioinformatics/btt566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koestler DC, Christensen B, Karagas MR, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Accomando WP, Wiencke JK, Houseman EA, Nelson HH, Kelsey KT. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome Biol. 2014;15:R50. doi: 10.1186/gb-2014-15-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 103.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Warren RL, Choe G, Freeman DJ, Castellarin M, Munro S, Moore R, Holt RA. Derivation of HLA types from shotgun sequence datasets. Genome Med. 2012;4:95. doi: 10.1186/gm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boegel S, Lower M, Schafer M, et al. HLA typing from RNA-Seq sequence reads. Genome Med. 2012;4:102. doi: 10.1186/gm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim HJ, Pourmand N. HLA typing from RNA-seq data using hierarchical read weighting [corrected] PLoS One. 2013;8:e67885. doi: 10.1371/journal.pone.0067885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai Y, Ni M, Cooper B, Wei Y, Fury W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics. 2014;15:325. doi: 10.1186/1471-2164-15-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nielsen M, Lundegaard C, Blicher T, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]