Abstract
Marine environments are largely unexplored and can be a source of new molecules for the treatment of many diseases such as malaria, cancer, tuberculosis, HIV etc. The Marine environment is one of the untapped bioresource of getting pharmacologically active nonribosomal peptides (NRPs). Bioprospecting of marine microbes have achieved many remarkable milestones in pharmaceutics. Till date, more than 50% of drugs which are in clinical use belong to the nonribosomal peptide or mixed polyketide-nonribosomal peptide families of natural products isolated from marine bacteria, cyanobacteria and fungi. In recent years large numbers of nonribosomal have been discovered from marine microbes using multi-disciplinary approaches. The present review covers the NRPs discovered from marine microbes and their pharmacological potential along with role of genomics, proteomics and bioinformatics in discovery and development of nonribosomal peptides drugs.
Keywords: microbe derived-compounds, marine natural products, nonribosomal peptides, antimicrobial, anticancer
Marine ecosystem resources for new drug discovery
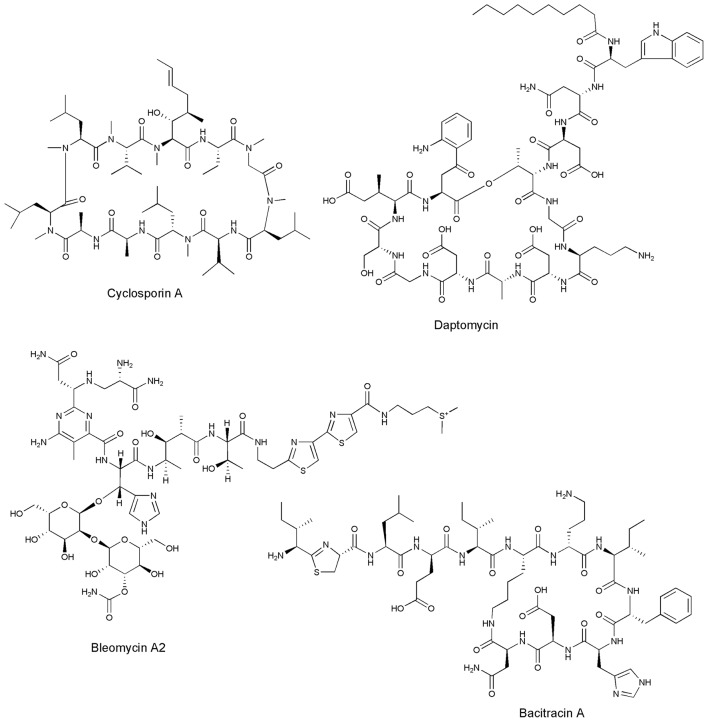
The marine ecosystem is most complex and largest aquatic systems on earth. It includes oceans, intertidal ecology, salt marsh, lagoons, estuaries, coral reefs, mangroves, deep sea, sea floor etc. Marine ecosystem has a enormous variety of organisms that are different in their physiology and adaptations and most of the marine life is found in coastal habitats (Hedgepeth, 1957). According to the Global Biodiversity Assessment by the United Nations Environment Program, oceans consist of 178,000 marine species in 34 phyla. It is estimated that 102 fungi, 103 bacteria and 107 viruses are likely to exist in one milliliter of seawater (Kubanek et al., 2003). Marine organisms comprise around 50% of the total biodiversity on earth. These organisms have shown remarkable contribution in the discovery and production of novel biomolecules (Jimeno et al., 2004; Vignesh et al., 2011). During 1981–2002 50% of US- FDA approved drugs are reported from either marine bioactive compounds or their synthetics analogs (Vinothkumar and Parameswaran, 2013). Cytosine arabinoside, Ara-C (anticancer) and adenine arabinoside, Ara-A (antiviral) were first discovered in the early 1950s and approved by Food and Drug Administration (US-FDA). These drugs were isolated from Caribbean sponge (Cryptotheca crypta), as spongouridine and spongothymidine. Blunt et al. (2015) reported more than 20,000 natural bioactive compounds have been obtained from marine environment in last 50 years (Blunt et al., 2015). Out of these 9 were approved as drugs and many of them are still in clinical trials. It is well documented that more than 50% of drugs that are in clinical use today belong to the nonribosomal peptides or mixed polyketide-NRP families (Hranueli et al., 2010; Agrawal et al., 2016; Table 1). Marine microbes contributes 70% of discovery of NRPs with antimicrobial, antiviral, cytostatic, immunosuppressant, antimalarial, antiparasitic, animal growth promoters and natural insecticides activities etc. (Vinothkumar and Parameswaran, 2013). Which makes marine microbial an important bioresource for getting NRPs with numerous pharmaceutical applications. The examples of some NPR based drugs which are now in the market are Daptomycin (antibiotics), Bleomycin (antitumor), Bacitracin (antibiotics for skin infections), Cyclosporin (antifungal and immunosuppressant drugs) (Figure 1) (Strieker et al., 2010). Norine is the first database entirely dedicated to NRPs and contains more than 1186 entries (Caboche et al., 2008, 2009). In this review we focus on antimicrobial and anticancer NRPs reported from marine microbes with their biological targets.
Table 1.
List of some marine derived NRPs and their present status (Newman and Cragg, 2004; Fenical, 2006; Jimenez et al., 2009; Petit and Biard, 2013).
| Metabolite | Source | Pharmacological activity | R&D stage |
|---|---|---|---|
| Ecteinascidin 743 (YondelisTM) | Ecteinascidia turbinate (Sea squirt) | Anticancer | Market |
| Cephalosporine | Cephalosporium acremonium (Fungi) | Antibiotic | Market |
| Bengamide derivative (LAF389) | Jaspis sp. (Sponge) | Anticancer | Phase I |
| Hemiasterlin derivative (HTI-286) | Cymbastella sp. (Sponge) | Anticancer | Phase I |
| Dehydrodidemnine B (AplidineTM) | Aplidium albicans (Tunicate) | Anticancer | Phase II |
| Dolastatin 10 | Dolabella auricularia (Mollusc and Cyanobacteria) | Anticancer | Phase II |
| Kahalalide F | Elysia rufescens (Sea slug) | Antitumor | Phase II |
| Bryostatin 1 | Bugula neritina (Bryozoan) | Anticancer | Phase III |
| Diazonamide | Diazona angulata (Tunicate) | Anticancer | Preclinical |
| Thiocoraline | Mi Cromonospora marina (Bacteria) | Anticancer | Preclinical |
| Vitilevuamide | Didemnum cucliferum and Polysyncraton lithrostrotum (Tunicates) | Anticancer | Preclinical |
Figure 1.
Structures of marketed NRPs.
Nonribosomal peptide and their bio combinatorial synthesis
An extensive literature on biosynthesis of non-ribosomal peptides is available in previous reviews (Sieber and Marahiel, 2003; Finking and Marahiel, 2004; Caboche et al., 2009; Strieker et al., 2010; Pfennig and Stubbs, 2012). Here we just summarized how NPRs are synthesized biologically, biomolecular structural architecture and enzymatic machinery of non-ribosomal peptide synthetases (NRPSs). NRPs are peptide secondary bioactive metabolites synthesized by a multi-modular enzyme complex called nonribosomal peptide synthetases (NRPSs) found only in bacteria, cyanobacteria and fungi (Matsunaga and Fusetani, 2003; Nikolouli and Mossialos, 2012). NRPs are formed from a series of enzymatic transformations employing a much more diverse set of precursors and biosynthetic reactions. NRPSs utilize both proteinogenic and nonproteinogenic amino acids (not encoded by DNA) as building blocks for the growing peptide chain (Finking and Marahiel, 2004; Felnagle et al., 2008). Moreover, these secondary bioactive metabolite peptides contain unique structural features, such as D-amino acids, N-terminally attached fatty acid chains, N- and C-methylated residues, N- formylated residues, heterocyclic elements, and glycosylated amino acids, as well as phosphorylated residues etc.; (Sieber and Marahiel, 2003). As a result, NRPs exhibit a broad spectrum of biological activities, ranging from antimicrobial to anticancer (Hur et al., 2012). The macrocyclic structure is a common feature of nonribosomally synthesized bioactive peptides, which is responsible for reduction in structural flexibility and may, therefore, constrain them into the biologically active conformation (Sieber and Marahiel, 2003; Grünewald and Marahiel, 2006).
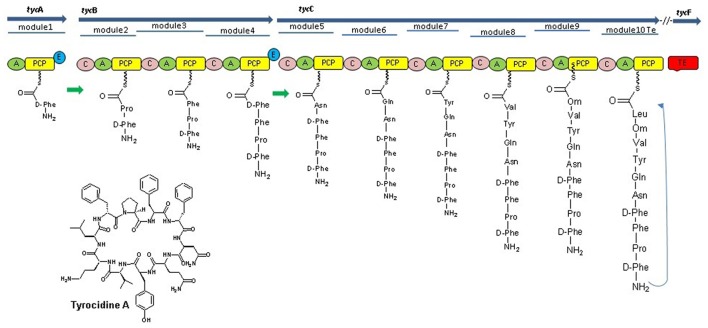
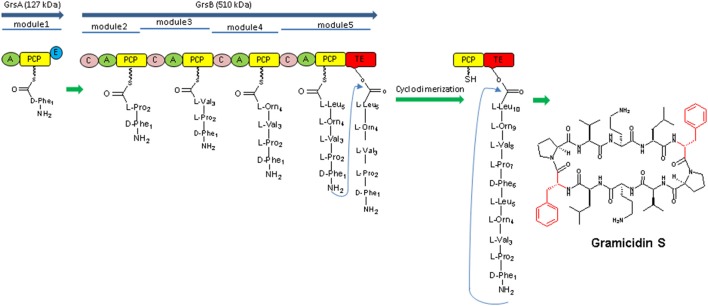
The discovery of NRPs began when Tatum and colleagues (Mach et al., 1963) provided first evidence that tyrocidine, a cyclic decapeptide produced by Bacillus brevis, was biosynthesized by a mechanism independent of the ribosome (Mankelow and Neilan, 2000). They found that protein synthesis in B. brevis was inhibited by using ribosome targeting antibiotics like chloramphenicol and chlortetracycline, however, the biosynthesis of tyrocidine was not obstructed by the same. Additional biochemical analyses demonstrated that gramicidin S, a cyclic decapeptide produced by B.brevis, did not include tRNA molecules or aminoacyl-tRNA-synthetases (Nikolouli and Mossialos, 2012; Figure 2). Further work by Lipmann established that the production of cyclic decapeptide, gramicidin is an ATP-dependent reaction, catalyzed by these enzymes incorporating amino acids in a two-step process by their modules and their respective domains. The first step involves release of pyrophosphate (PPi) and the second step releases adenosine monophosphate (AMP), with the end result being an amino acid covalently linked to the enzyme (Wu et al., 2003). These finding suggested that tyrocidine and gramicidin S peptide synthesis did not involve ribosomal machinery for their synthesis, which leads to discovery of the NRPs and NRPSs. These data also gave the first indication of an amino acid as a “carrier” being involved in NRPS enzymology (Felnagle et al., 2008; Condurso and Bruner, 2012; Figure 3).
Figure 2.
Tyrocidine biosynthesis in bacteria B. brevis nonribosomal peptide synthetases of tyrocidine synthesis mainly consist, three NRPSs TycA, TycB, and TycC, which contain 10 modules (TycA comprises one module, TycB three, and TycC six modules) each of those responsible for the incorporation of a cognate amino acid into the growing chain with the help of their domains. The Te domain at the last module of TycC catalyzes peptide cyclization and thereby release of the final product (Mootz et al., 2000).
Figure 3.
The Gramicidin S biosynthetic machinery the enzymatic assembly consists of two NRPSs (GrsA and GrsB) and their modules, respectively. Each module is responsible for the incorporation of one monomeric amino acid. The thioesterase domain (TE domain) catalyzes the dimerization of two assembled pentapeptides and subsequent cyclization, resulting in gramicidin S (Hoyer et al., 2007).
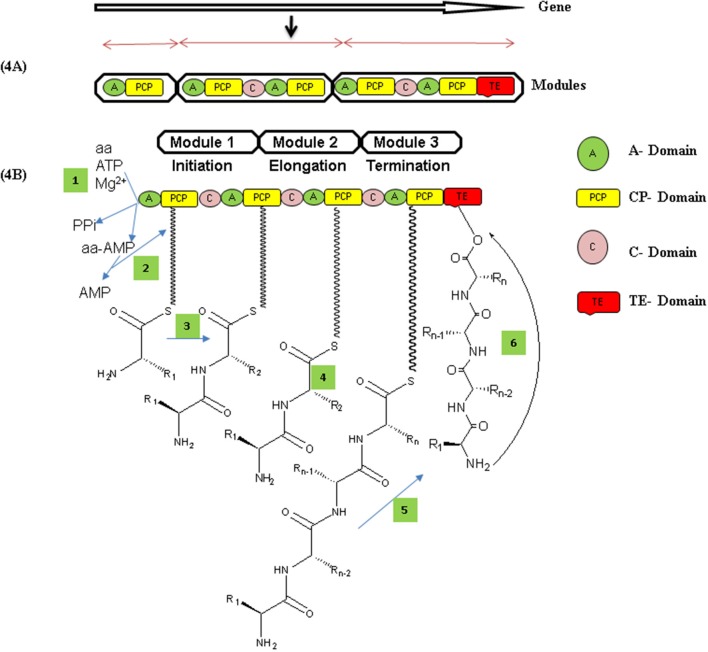
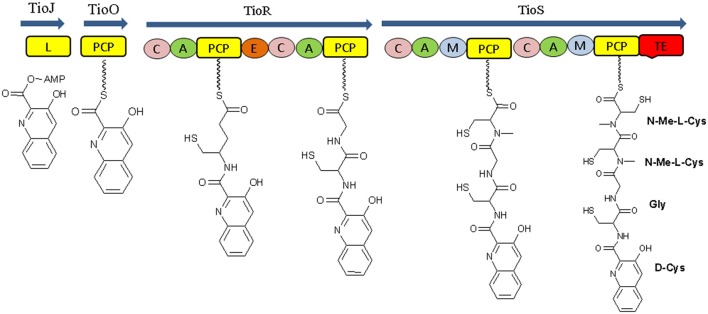
The biosynthetic study of NRP compounds is challenging if we consider their complexity and biological activities. Each nonribosomal peptide synthetase is composed of an array of distinct modular sections, each of which is responsible for the incorporation of one defined monomer into the final peptide product. Biosynthesis of a nonribosomal peptide by NRPSs involves a series of repeating reactions that are catalyzed by the coordinated actions of modules and their core catalytic domains. Each enzyme module contains three catalytic domains: adenylation domain (A), peptidyl-carrier (PCP) domain and condensation domain (C). A final peptide product released from the enzyme through cyclization or hydrolysis that takes place by thioesterase domain (TE) which is located in the final NRPSs module (Figures 4A,B; Mankelow and Neilan, 2000; Finking and Marahiel, 2004). For recent example, Thiocoraline, an anticancer nonribosomal peptide (NRP) synthesis by marine bacteria Cromonospora marina contains peptidic backbone of two S-methylated Lcysteine residues. S-Methylation occurs very rarely in nature, and is observed extremely rarely in nonribosomal peptide scaffold. The four modules TioJ, TioO, TioR, and TioS of thiocoraline NRPSs are responsible for the thiocoraline-backbone biosynthesis. TioR and TioS would most probably constitute the NRPSs involved in the biosynthesis of the thiocoraline, according to the colinearity of the respective modules (Figure 5; Lombó et al., 2006; Al-Mestarihi et al., 2014). The potentials of marine microbes to produce NRP's with antimicrobial and anticancer activity are reported in this review. The data referring to these activities are depicted in Tables 2–4 and the structures are given in Supplementary Materials (Figures S1–S17).
Figure 4.
(A) Organization of modules and their domains in nonribosomal peptide synthetase enzyme. Each module contains their catalytic domains that catalyze activities, substrate activation (A-domain), covalent loading (CP-domain), and peptide bond formation (C-domain). The first modules always lacks a C domain and is used to initiate nonribosomal peptide synthesis, while those harboring a C-domain qualify for elongation and modules with thioesterase domains (TE) usually in the last domain, for termination of peptide product from enzyme through cyclization or hydrolysis (Prieto et al., 2012). (B) Mechanism of nonribosomal peptide (NRP) synthesis Adenylation domain (A) activates amino acid as aminoacyl-AMP and transfer to PCP domain which condenses coming amino acids by forming peptide bonds. Structural modifications mostly operate by epimerization domains which converts L-amino acid to D-amino acid and vice a versa. Peptide chain thus transfers to TE domain by transesterification reaction by PCP. Finally, TE domain catalyzed product release (NRPs) by either hydrolysis or macrocyclization (Condurso and Bruner, 2012).
Figure 5.
Structural organization of the thiocoraline NRPSs. L, AMP-ligase; P, peptidyl-carrier protein domain; C, condensation domain; A, adenylation domain; E, epimerization domain; M, N-methyltransferase domain; TE, thioesterase domain.
Table 2.
Antimicrobial NRPs from marine microbes.
| Sr. No. | NRPs | Chemical architecture | Source | Biological target | Biological active value (MIC/IC50/GI50/ID50/ED50) | References |
|---|---|---|---|---|---|---|
| BACTERIA | ||||||
| 1. | Bogorol A (1) | Linear peptide | Bacillus laterosporus | MRSA, VRE | 2 μg/mL, 10 μg/mL | Barsby et al., 2001 |
| 2. | Nocathiacins I–III (2–4) | Cyclic peptide | Nocardia sp. | multiple-drug resistant pathogens | 0.001–0.015, 0.0005-0.25, 0.002-0.06 μg/mL | Leet et al., 2003; Li et al., 2003 |
| 3. | YM- 266183 and YM- 266184 (5–6) | Cyclic peptide | Bacillus cereus | Staphylococci, Enterococci | 0.05–0.2 μg/mL, 0.013–0.025 μg/mL. 0.02–0.05 μg/mL, 0.006–0.01 μg/mL | Nagai et al., 2003; Suzumura et al., 2003 |
| 4. | cyclo-(glycyl-l-seryl-l-prolyl-l- glutamyl) (7) and cyclo-(glycyl-l-prolyl-l-glutamyl) (8) | Cyclic peptide | Ruegeria sp. | Bacillus subtilis | 25 and 50 μg/mL | Mitova et al., 2004 |
| 5. | Tauramamide (9) | Lipopeptide | B. laterosporus | Enterococcus sp. | 0.1 μg/mL | Desjardine et al., 2007 |
| 6 | Tetrapeptide cyclo-isoleucyl-prolyl-leucyl alanyl (10) and cyclo-phenylalanyl-prolyl-leucyl prolyl (11) | Cyclic tetra peptide | Pseudomonas sp. | Marine bacterial | – | Rungprom et al., 2008 |
| 7. | Unnarmicin A and C (12–13) | Depsipeptide | Photobacterium sp. | Pseudovibrio | 7–8 μg/mL | Oku et al., 2008b |
| 8. | Thiopeptide TP-1161 (14) | Cyclic peptide | Nocardiopsis sp. | Gram-positive bacteria | 0.25–4 μg/mL | Engelhardt et al., 2010 |
| 9. | Solonamide A–B (15–16) | Cyclo depsipeptide | Photobacterium halotolerans | S. aureus | – | Mansson et al., 2011 |
| 10. | Fijimycin A–C (17–19) | Depsipeptide | Streptomyces sp. | MRSA | 4–16 μg/mL | Sun et al., 2011 |
| 11. | Peptidolipins B–F (20–24) | Lipopeptide | Nocardia sp. | MRSA, MSSA | 64 μg/mL | Wyche et al., 2012 |
| 12. | Kocurin (25) | Cyclic peptide | Kocuria palustris | MRSA | 0.25 μg/mL | Martín et al., 2013 |
| 13. | Champacyclin (26) | Octapeptide | Streptomyces champavatii | Erwinia amylovora | 25 μM | Pesic et al., 2013 |
| 14. | Ngercheumicin F–I (27–30) | Cyclo depsipeptide | P. halotolerans | S. aureus | 5 μg/mL | Kjaerulff et al., 2013 |
| CYANOBACTERIA | ||||||
| 15. | Lobocyclamide B (31) | Cyclododecapeptide | Lyngbya confervoides | Fluconazole-resistant C. albicans | – | MacMillan and Molinski, 2002 |
| 16. | Brunsvicamide A–C (32–34) | Cyclic hexapeptide | Tychonema sp. | Mycobacterium tuberculosis (MptpB) | 7.3 μM | Müller et al., 2006 |
| FUNGI | ||||||
| 17. | Guangomides A–B (35–36) | Cyclic depsipeptide | Unidentified fungus | S. epidermis, E. durans | 100 μg/mL 100 μg/mL | Amagata et al., 2006 |
| 18. | 11-O-methylpseurotin A (37) | Linear peptide | Aspergillus fumigatus | Saccharomyces cerevisiae | – | Boot et al., 2007 |
| 19. | Emericellamides A–B (38–39) | Cyclic depsipeptides | Emericella sp. | MRSA | 3.8 and 6.0 μM | Oh et al., 2007 |
| 20. | Scopularides A–B (40–41) | Cyclododecapeptide | Scopulariopsis brevicaulis | Gram-positive bacteria | – | Yu et al., 2008 |
| 21. | Alternaramide (42) | Cyclic Penta depsipeptide | Alternaria sp. SF,5016 | B. subtilis and S. aureus | (ZOI 8 mm), (ZOI 13 mm) | Kim et al., 2009 |
| 22. | Trichoderins A, A', B2 (43–45) | Lipopeptide | Trichoderma sp. | Mycobacterium tuberculosis | 0.02–2.0 l g/mL | Pruksakorn et al., 2010 |
| 23. | Unguisin E (46) | Cyclic heptapeptide | Aspergillus sp. | Antibacterial | – | Liu and Shen, 2011 |
| 24. | Sclerotides A–B (47–48) | Cyclic hexapeptide | Aspergillus sclerotiorum | C. albicans, Pseudomonas aeruginosa | 7.0 and 3.5 μM. nil and, 35.3 μM | Zheng et al., 2009 |
| 25. | Sclerotiotides A–K (49–59) | Cyclic tripeptide | A. sclerotiorum PT06-1 | C. albicans | 7.5, 3.8, 30, 6.7μM | Zheng et al., 2010 |
Table 4.
NRPs with both antimicrobial and anticancer potential from marine microbes.
| Sr. No. | NRPs | Chemical architecture | Source | Biological target | Biological active value (MIC/IC50/GI50/ID50/ED50) | References |
|---|---|---|---|---|---|---|
| 1. | Lajollamycin (235) | Spiro-β-lactone-γ-lactam | Streptomyces nodosus | Drug resistant gram-positive bacteria/ | 2–20 μg/mL | Manam et al., 2005 |
| murine melanoma cell line B16-F10 | EC50 of 9.6 μM | |||||
| 2. | Ariakemicins A-B (236-237) | Linear peptide | Rapidithrix sp | S. aureus/ | 0.46 μg/mL | Oku et al., 2008a |
| human lung cancer cells (A549) and baby hamster kidney cells | IC50 values of 25 and 15 μg/mL | |||||
| 3. | Ieodoglucomide A-B (238-239) | Glycolipopeptide | Bacillus licheniformis | Antibacterial/lung cancer and stomach cancer cell lines | GI50 values of 25.18 and 17.78 μg/mL | Tareq et al., 2012 |
| 4. | Halolitoralin A (240) Halolitoralins B (241) C (242) | Cyclic hexapeptide Cyclic tetrapeptide | Halobacillus litoralis | Candida albicans and Tricophyton rubrum/human gastric tumor | 20, 30, 30 μg/mL and 25, 35, 40 μg/mL | Yang et al., 2002 |
| 5. | Mojavensin A (243) | Lipopeptide | Bacillus mojavensis | Valsa mali, cucumerinum, and Fusarium verticillioides/ | 2 mg /mL | Ma et al., 2012 |
| HL-60 | IC50 of 100 | |||||
| 6. | Marthiapeptide A (244) | Cyclic peptide | Marinactinospora thermotolerans | SF-268 (glioblastoma), MCF-7 (breast), NCI-H460 (lung), HepG2 (hepatocarcinoma)/ | 0.38–0.52 μM | Zhou et al., 2012 |
| gram-positive bacteria | ||||||
| 7. | Linear heptapeptide (245) | Heptapeptide | Paenibacillus profundus | SK-MEL-28 cell / | 3.07 μM/ | Kalinovskaya et al., 2013 |
| S. aureus, S. epidermis B. subtilis and Enterococcus faecium | ZOI = 24 mm, 20 mm,28 mm, 11 mm | |||||
Table 3.
Anticancer NRPs from marine microbes.
| Sr. No. | NRPs | Chemical architecture | Source | Biological target | Biological active value (MIC/IC50/GI50/ID50/ED50) | References |
|---|---|---|---|---|---|---|
| BACTERIA | ||||||
| 1. | Mixirins A–C (60–62) | Cyclopeptide | Bacillus sp. | HCT-116 cells (colon) | 0.68, 1.6, 1.3 μg/ml | Zhang et al., 2004 |
| 2. | Mechercharmycin A (63) Mechercharmycin B (64) | Cyclic peptide | Thermoactinomyces sp. | A549 cells (lung), Jurkat cells | 4.0 × 10−8 M 4.6 × 10−8 M | Kanoh et al., 2005 |
| 3. | Bromoalterochromide A (65) and A' (66) | Chromo peptide | Pseudoalteromonas maricaloris | Strongylocentrotus intermedius eggs | – | Speitling et al., 2007 |
| 4. | Lucentamycins A–D (67–70) | – | Nocardiopsis lucentensis | HCT-116 cells (colon) | 0.20 and 11 μM | Cho et al., 2007 |
| 5. | Piperazimycins A–C (71–73) | Cyclic hexadepsipeptide | Streptomyces sp. | HCT-116 cells (colon) | 76 ng/mL | Miller et al., 2007 |
| 6. | Urukthapelstatin A (74) | Cyclic thiopeptide | Mechercharimyces asporophorigenens | A549 cells (lung) | 12 nM | Matsuo et al., 2007 |
| 7. | Arenamides A–C (75–77) | Cyclo hexadepsipeptide | Salinispora arenicola | Tumor necrosis factor | 3.7 and 1.7 μM | Asolkar et al., 2008 |
| 8. | Bacillistatins 1-2 (78–79) | Cyclodepsipeptide | Bacillus silvestris | P388 (murine lymphocytic leukemia); BXPC-3 (pancreas); MCF-7 (breast); SF-268 (CNS); NCI- H460 (lung); KM20L2 (colon); DU- 145 (prostate) | 10−4−10−5 μg/mL | Pettit et al., 2009 |
| 9. | Turnagainolides A–B (80–81) | Cyclic peptide | Bacillus sp. | PI3K pathway | – | Li et al., 2011 |
| 10. | Padanamides A–B (82–83) | Linear tetrapeptide | Streptomyces sp. | Jurkat cells | 20 μg/mL | Williams et al., 2011 |
| 11. | Ohmyungsamycins A–B (84–85) | Cyclic peptide | Streptomyces sp. | Cytotoxic | 359–816 nM and 12.4–16.8 μM | Um et al., 2013 |
| 12. | Proximicin A–C (86–88) | – | Verrucosispora strain MG-37 | AGS (gastric), HepG2 (hepatocellular), MCF 7 (breast) | 0.6, 1.5, 0.25 μg/mL 0.8, 9.5, 0.7 μg/mL 7.2, 5.0, 9.0 μg/mL | Fiedler et al., 2008 |
| CYANOBACTERIA | ||||||
| 13. | Hoiamide A (89) | Cyclic depsipeptide | An assemblage of L. majuscula and Phormidium gracile | Voltage-gated sodium channel | 92.8 nM | Pereira et al., 2009 |
| 14. | Yanucamides A–B (90–91) | Cyclic depsipeptide | Lyngbya majuscule and Schizothrix sp. | Brine shrimp toxicity | 5 ppm | Sitachitta et al., 2000 |
| 15. | Lyngbyabellins A (92) | Cyclic depsipeptide | L. majuscula | KB cells LoVo cells | 0.03 and 0.50 μg/mL | Luesch et al., 2000 |
| 16. | Lyngbyabellin B (93) | Cyclic depsipeptide | L. majuscula | Brine shrimp (Artemia salina) | 3.0 ppm | Milligan et al., 2000 |
| 17. | Microcyclamide (94) | Cyclic hexapeptide | Microcystis aeruginosa | P388 murine leukemia cells | 24–30 μg/mL | Ishida et al., 2000 |
| 18. | Apratoxin A (95) | – | L. majuscula | KB cells and LoVo cancer cells | 0.52 nM 0.36 nM | Luesch et al., 2001b |
| 19. | Pitipeptolides A–B (96–97) | Cyclic depsipeptide | L. majuscula | LoVo cells | 2.25 and 1.95 μg/mL | Luesch et al., 2001a |
| 20. | Ulongamides A–F (98–103) | Cyclic depsipeptide | Lyngbya sp. | KB and LoVo cells | 1 μM 5 μM | Luesch et al., 2002 |
| 21. | Obyanamide (104) | Cyclic depsipeptide | L. confervoides | KB cells | 0.58 μg/mL | Williams et al., 2002a |
| 22. | Malevamide D (105) | Linear peptide | Symploca hydnoides | P-388, A-549, HT-29, and MEL-28 | 0.7 and 0.7 nM | Horgen et al., 2002 |
| 23. | Tasiamide (106) | Linear peptide | Symploca sp. | KB and LoVo cells | 0.48 and 3.47 μg/mL | Williams et al., 2002b |
| 24. | Tasiamide B (107) | Linear peptide | KB | KB cells | 0.8 μM | Williams et al., 2003a |
| 25. | Guineamides A–F (108–113) | Cyclic depsipeptide | L. majuscula | Mouse neuroblastoma cells | 15 and 16 μM | Tan et al., 2003b |
| 26. | Homodolastatin 16 (114) | Cyclic depsipeptide | L. majuscula | Oesophageal Cervical cells | 4.3 μg/mL 1 μg/mL | Davies-Coleman et al., 2003 |
| 27. | Lyngbyastatin 3 (115) | Cyclic peptide | L. majuscula | KB and LoVo cells | 32 and 400 nM | Williams et al., 2003a |
| 28. | Ulongapeptin (116) | Cyclic depsipeptide | Lyngbya sp. | KB cells | 0.63 μM | Williams et al., 2003d |
| 29. | Tasipeptins A–B (117–118) | Depsipeptide | Symploca sp. | KB cells | 0.93 and 0.82 μM | Williams et al., 2003a |
| 30. | Jamaicamides A–C (119–121) | Lipopeptide | L. majuscula | H-460 (lung), Neuro-2a. Sodium channel blocking | 15 μM 5 μM | Williams et al., 2003d |
| 31. | Wewakpeptins A–D (122–125) | Depsipeptide | Lyngbya semiplena | NCI-H460 neuro-2a | 0.4 μM | Williams et al., 2003b |
| 32. | Trungapeptin A–C (126–128) | Cyclic depsipeptide | L. majuscula | Ichthyotoxic Brine shrimp toxicity | 6.25 ppm 10 ppm | Bunyajetpong et al., 2006 |
| 33. | Aurilides B and C (129–130) | Cyclic peptide | L. majuscula | NCI-H460 (lung), neuro-2a | 0.01 and 0.13 μM | Han et al., 2006 |
| 34. | Belamide A (131) | Tetra peptide | Symploca sp. | MCF7 (breast) HCT-116 (colon) | 0.74 μM | Simmons et al., 2006 |
| 35. | Lyngbyastatins 5–7 (132–134) | – | Lyngbya sp. | Potent Elastase Inhibitors | 3–10 nM | Taori et al., 2007 |
| 36. | Mitsoamide (135) | Linear peptide | Geitlerinema sp. | NCI-H460 (lung) | 460 nM | Andrianasolo et al., 2007 |
| 37. | Apratoxin D (136) | Cyclic depsipeptide | L. majuscule and Lyngbya sordida | H-460 (lung) | 2.6 nM | Gutiérrez et al., 2008 |
| 38. | Apratoxin E (137) | – | Lyngbya bouillonii | HT29 (colon), HeLa (cervical), | 21–72 nM | Matthew et al., 2008 |
| 39. | Dragonamide C and D (138–139) | Linear lipopeptide | Lyngbya polychroa | U2OS (osteosarcoma), HT29 (colon), IMR-32 (neuroblastoma) | 56 and 59 μM 22 and 32 μM 49 and 51 μM | Gunasekera et al., 2008 |
| 40. | Coibamide A (140) | Cyclic depsipeptide | Leptolyngbya sp. | NCI 60 cancer cell | < 23 nM | Medina et al., 2008 |
| 41. | Symplostatin 4 (141) | Linear peptide | Symploca sp. | Antimitotic to H-460 (lung) neuro-2a | 40 nM 29 nM | Taori et al., 2009 |
| 42. | Itralamides A–B (142–143) | Depsipeptide | L. majuscula | HEK293 cells | 6 ± 1 μM | Jiménez et al., 2009 |
| 43. | Desmethoxymajusculamide C (144) | Cyclic and Liner peptide | L. majuscula | HCT-116 | 20 nM | Simmons et al., 2009 |
| 44. | Hantupeptin A (145) | Cyclic depsipeptide | L. majuscula | MOLT-4 (leukemia) MCF-7 (breast) | 32 μM 4.0μM | Tripathi et al., 2008 |
| 45. | Desacetyl-Microcolin B (146) | Linear peptide | Lyngbya cf. polychroa | HT-29 andIMR-32 cells | 14 nM 14 nM | Meickle et al., 2009 |
| 46. | Palmyramide A (147) | Cyclic depsipeptide | L. majuscula | Neuro-2a and H-460 | 17.2μM 39.7μM | Taniguchi et al., 2009 |
| 47. | Bisebromoamide (148) | Linear peptide | Lyngbya sp. | Protein kinase inhibitor (HeLa S3 cells) | 04 μg/mL | Teruya et al., 2009 |
| 48. | Lyngbyabellin J (149) Lyngbyapeptin D (150) | Linear peptide | L. bouillonii | Actin-disrupting | 0.041 μM | Matthew et al., 2010 |
| 49. | Grassypeptolides A–C (151–153) | Cyclic depsipeptide | L. confervoides | HT29 cells HeLa cells | 1.22μM, 4.97μM, 76.7nM 1.01μM, 2.93μM, 44.6 nM | Kwan et al., 2010 |
| 50. | Hantupeptin B and C (154–155) | Cyclodepsipeptide | L. majuscula | MOLT-4 cells MCF-7 cells | 0.2 μM 0.5 μM | Tripathi et al., 2010 |
| 51. | Lyngbyacyclamides A–B (156–157) | Cyclic peptide | Lyngbya sp | B16 mouse melanoma | 0.7 μM | Maru et al., 2010 |
| 52. | Grassypeptolides D–E (158–159) | Cyclic depsipeptide | Leptolyngbya sp. | HeLa and neuro-2a blastoma | 335 and 192 nM 599 and 407 nM | Thornburg et al., 2011 |
| 53. | Grassypeptolides F and G (160–161) | Cyclic depsipeptide | L. majuscula | Inhibit transcription (AP-1 cell) | 5.2 and 6.0 μM | Popplewell et al., 2011 |
| 54. | Veraguamides A–H and J-L (162–172) | – | Symploca hydnoides | H-460 (lung) | 141 nM | Mevers et al., 2011; Salvador et al., 2011 |
| 55. | Malyngamide 3 (173) and Cocosamide A–B (174–175) | Cyclic peptide | L. majuscula | HT-29 cells | 48 μM, 24 μM, 11μM | Gunasekera et al., 2011 |
| 56. | Cyclodepsipeptide (176–180) | Cyclodepsipeptide | L. majuscula | HT-29 and MCF7 | – | Montaser et al., 2010 |
| 57. | Lagunamide C (181) | Cyclodepsipeptide | L. majuscula | HT-29 and MCF7 | 2.1–24.4 nM | Tripathi et al., 2011 |
| 58. | Wewakamide A (182) Guineamide G (183) | Cyclic depsipeptide | L. semiplena and L. majuscula | Neuro-2a blastoma | 2.7 μM | Nan et al., 2011 |
| 59. | Porpoisamide A–B (184–185) | Cyclic depsipeptide | Lyngbya sp. | HCT-116 (colon) cells U2OS (osteosarcoma) cells | 25, 21μM 28, 22 μM | Meickle et al., 2011 |
| 60. | Lyngbyabellin K and L (186–187), 7-epi-lyngbyabellin L (188) and Lyngbyabellin M-N (189–190) | Lipopeptide | Moorea bouillonii | HCT116 (colon) cells | 40.9 ± 3.3 nM | Choi et al., 2012 |
| 61. | Viequeamides A–D (191–194) | Cyclic depsipeptide | Rivularia sp. | H460 (lung) cells | 60 ± 10 nM | Boudreau et al., 2012 |
| 62. | Symplocin A (195) | Linear peptide | Symploca sp. | Cathepsin E inhibitor | 300 pM | Molinski et al., 2012 |
| 63. | Bouillonamide (196) | Cyclic depsipeptide | M. bouillonii | Neuron 2a cells | 6.0 μM | Tan et al., 2013 |
| 64. | Malyngamide 4 (197) | Lipopeptide | Moorea producens | HTCLs | – | Shaala et al., 2013 |
| 65. | Kurahyne B (198) | – | Okeania sp. | HeLa and HL60 | 8.1 and 9.0 μM | Okamoto et al., 2015 |
| FUNGI | ||||||
| 66. | N-Methylsansalvamide (199) | Cyclic depsipeptide | Fusarium strain CNL-619 | Tumor cell | 8.3 μM | Cueto et al., 2000 |
| 67. | Dictyonamides A–B (200–201) | Linear dodecapeptide | Fungus K063 | Kinase 4 inhibitor | 16.5 μg/mL | Komatsu et al., 2001 |
| 68. | Scytalidamides A–B (202–203) | Cyclic heptapeptide | Scytalidium sp. | HCT-116 (colon) | 7 and 11.0 μM | Tan et al., 2003a |
| 69. | Trichodermamides A–B (204–205) | Dipeptide | Trichoderma virens | HCT-116 (colon) | 0.32 μg/mL | Garo et al., 2003 |
| 70. | Rostratins A–D (206–209) | Cyclic dipeptide | Exserohilum rostratum | HCT-116 (colon) | 8.5, 1.9, 0.76 and 16.5 μg/mL | Tan et al., 2004 |
| 71. | IB-01212 (210) | Cyclic depsipeptide | Clonostachys sp. ESNA,A009 | LN-caP (prostrate), SK-BR3 (breast), HT29 (colon) and HELA (cervix) | 10-8 M | Cruz et al., 2006 |
| 72. | Zygosporamide (211) | Cyclic Penta depsipeptide | Zygosporium masonii | SF-26 (CNS) RXF 393 (renal) | 6.5 nM ≤ 5.0 nM | Oh et al., 2006 |
| 73. | Trichoderide A (212) | – | Trichoderma reesei | A375-S2 melanoma | 18.5 mg/mL | Sun et al., 2006 |
| 74. | Spicellamide A–B (213–214) | Cyclo hexadepsipeptide | Spicellum roseum | Neuron 2a cells | 30 μg/mL and 6.2 μg/mL | (Kralj et al., 2007 |
| 75. | 1962A and 1962B (215–216) | Cyclic depsipeptide | Strain No. 1962 | MCF-7 (breast) | 100 μg/mL | Huang et al., 2007) |
| 76. | Microsporins A–B (217–218) | Cyclic tetrapeptide | Microsporum cf. gypseum | HCT-116, Potent inhibitors of HDAC | 0.6 μg/mL and 8.5 μg/mL | Gu et al., 2007 |
| 77. | Efrapeptin J (219) | Penta decapeptide | Tolypocladium sp. | HT1080 | – | Hayakawa et al., 2008 |
| 78. | Cotteslosin A–B (220–221) | Cyclo pentapeptide | Aspergillus versicolor | MM418c5 (melanoma), DU145 (prostate), T47D (breast) | 66 μg/mL 94 μg/mL 90 μg/mL | Fremlin et al., 2009 |
| 79. | Versicotide A–B (222–223) | Cyclo pentapeptide | A. versicolor | Anti-tumor | – | Zhou et al., 2011 |
| 80. | Fellutamide F (224) | Lipopeptide | A. versicolor | SK-MEL-2 (skin) XF498 (CNS) HCT15 (Colon) | 0.67 μM 0.14 μM 0.13 μM | Lee et al., 2011 |
| 81. | Cordyheptapeptides C–E (225–227) | Cyclo heptapeptide | Acremonium persicinum | SF-268, MCF-7, and NCI-H460 | 2.5–12.1 μM | Chen et al., 2012 |
| 82. | Pullularin E and F (228–229) | – | Bionectria ochroleuca | L5178Y (lymphoma) | 0.1 and 6.7 μg/mL | Ebrahim et al., 2012 |
| 83. | Clavatustides A–C (230–232) | Cyclodepsipeptide | Aspergillus clavatus | Hepatocellular Carcinoma Cycle Inhibitory | – | Jiang et al., 2013; Ye et al., 2014 |
| 84. | Asperterrestide A (233) | Cyclic tetrapeptide | Aspergillus terreus | U937 (carcinoma) and MOLT4 cell | 6.4 and 6.2 μM | He et al., 2013 |
| 85. | Similanamide (234) | Cyclohexapeptide | Aspergillus similanensis | MCF-7 (breast), NCI-H460 (lung) and A373 (melanoma) | 125 ± 0 117.50 ± 3.55 115 ± 7.07 | Prompanya et al., 2015 |
NRPs with antimicrobial potential
Antibiotic resistance in bacteria, parasites, viruses and fungi necessitates the continuous discovery of new drugs for the effective prevention and treatment of an ever-increasing range of infections caused by them (Organization, 2014). Natural products are the principal source for primary health care. Natural products are observed as a diverse group of molecules which have evolved to interact with a wide variety of protein targets for specific purposes. Also the same protein structure with little or no variation serves different purposes in different organisms. As a result, it is believed that the search for novel antimicrobial entity from natural sources will yield better results than from combinatorial chemistry and other synthetic procedures (Ngwoke et al., 2011). Here we described NRPs from marine microbial sources with antimicrobial potential.
Bacteria
A cationic antibiotic Bogorol A (1) (Figure S1), displaying potent activity against both methicillin-resistant Staphylococcus aureus (MRSA, MIC 2 μg/mL) and vancomycin-resistant enterococcal strains (VRE, MIC 10 μg/mL) of bacteria, has been isolated from cultures of a marine Bacillus laterosporus collected in Papua New Guinea (Barsby et al., 2001). A new broad spectrum thiazolyl peptide antibiotic, Nocathiacins I–III (2–4) (Figure S1), was isolated from the cultured broth of Nocardia sp. They share structural similarities to glycothiohexide-alpha (Li et al., 2003). All compounds exhibit potent in vitro activity against several multiple-drug resistant pathogens with MIC of 0.001–0.015, 0.0005–0.25, 0.002–0.06 μg/mL respectively. They demonstrate excellent in vivo efficacy in a systemic S. aureus infection mouse model at PD50 0.8, 0.6, 0.6 mg/kg/day respectively (Leet et al., 2003). The marine sponge Halichondria japonica was the source of Bacillus cereus which gave two cyclic thiopeptide antibiotics, YM-266183 (5) and YM-266184 (6) (Figure S1). They exhibited potent antibacterial activity against Staphylococci (MIC 0.05–0.2 μg/mL, 0.013–0.025 μg/mL) and Enterococci including multiple drug resistant strains (MIC 0.02–0.05 μg/mL, 0.006–0.01 μg/mL), whereas, they were inactive against gram-negative bacteria (Nagai et al., 2003). These structures contain thiazole and pyridine moieties and several unusual amino acids (Suzumura et al., 2003). A bacterial Ruegeria species isolated from a sponge Suberites domuncula (Gulf of Naples, Italy), gave two new cyclic peptides, cyclo-(glycyl-l-seryl-l-prolyl-l- glutamyl) (7) and cyclo-(glycyl-l-prolyl-l-glutamyl) (8) (Figure S1) with moderate antimicrobial activity against Bacillus subtilis at MIC of 25 and 50 μg/mL, respectively (Mitova et al., 2004). B. laterosporus PNG276 obtained from Papua New Guinea was the source of a new lipopeptide antibiotic, Tauramamide (9) (Figure S1). Anti-pathogenic activity against Enterococcus sp. were reported for tauramamide and its ethyl ester at MIC 0.1 μg/mL (Desjardine et al., 2007). Pseudomonas sp. separated out from the seaweed Diginea sp. (Ishigaki Is., Okinawa, Japan) were the source of cyclic tetrapeptides cyclo-[phenylalanyl-prolyl-leucyl-prolyl] (10) and cyclo-[isoleucyl-prolyl-leucyl-alanyl] (11) (Figure S1). The crude extract of this bacterial culture was found to inhibit the growth of other marine bacterial strains (Rungprom et al., 2008).
Unnarmicin A (12) and C (13) (Figure S2) are two depsipeptides isolated from a culture of a marine bacterium, Photobacterium sp. strain MBIC06485 having selective inhibitory effect on Pseudovibrio bacterial strains (Oku et al., 2008b). A strong antibacterial thiopeptide antibiotic TP-1161 (14) (Figure S2) with a rare aminoacetone moiety, have been isolated from Nocardiopsis sp. MICs value of TP-1161, ranging from 0.25 to 4 μg/ml for most gram-positive strain. The gene cluster for the biosynthesis of (14) was identified by PCR screening using degenerate primers (Engelhardt et al., 2010). Marine Photobacterium halotolerant yielded two novel cyclodepsipeptides Solonamide A (15) and B (16) (Figure S2) with inhibitory effect on virulence gene expression in S. aureus (Mansson et al., 2011). The fermentation broth of Streptomyces strain isolated from a marine sediment sample collected off Nasese, Fiji were the source of three new depsipeptides, Fijimycins A–C (17–19) (Figure S2). Fijimycins A–C were shown significant activity against three MRSA strains with MIC100 values between 4 and 16 μg/mL (Sun et al., 2011). Peptidolipins B–F (20–24) (Figure S2), antibacterial lipopeptides were obtained from an ascidian-derived Nocardia sp. Peptide (20) and (23) were moderately antibacterial against MRSA and methicillin-sensitive S. aureus (MSSA) (Wyche et al., 2012). A marine-derived bacterium Kocuria palustris was the source of a new thiazolyl peptide, Kocurin (25) (Figure S2). Kocurin strongly inhibits MRSA MB5393 with a MIC value of 0.25 μg/mL (Martín et al., 2013). An octapeptide, Champacyclin (26) (Figure S2) was isolated from three strains of Streptomyces champavatii (sediment, Gotland Deep and Kiel Bight, Baltic Sea and Urania Basin, Eastern Mediterranean) as an inhibitor of blight disease causing bacterium Erwinia amylovora (Pesic et al., 2013). Cyclodepsipeptides Ngercheumicin F–I (27–30) (Figure S2) isolated from P. halotolerans, inhibited quorum sensing in S. aureus (Kjaerulff et al., 2013).
Cyanobacteria
Lobocyclamide B (31) (Figure S3) a cyclododecapeptide containing five beta-hydroxy-alpha-amino acid residues, was discovered from Lyngbya confervoides which was active against fluconazole-resistant C. albicans. The absolute stereochemistry was determined by chiral chromatography of Marfey's reaction (MacMillan and Molinski, 2002). Brunsvicamides A–C (32–34) (Figure S3), three new cyclic hexapeptides have been isolated from cyanobacterium Tychonema sp. Brunsvicamide C contains an N-methylated N'-formylkynurenine moiety. Brunsvicamide B selectively inhibits the Mycobacterium tuberculosis protein tyrosine phosphatase B (MptpB, IC50 7.3 μM) (Müller et al., 2006).
Fungi
An extraction of a saltwater culture of an unidentifiable sponge-derived fungus leads to discovering two novel cyclic depsipeptides, Guangomides A (35) and B (36) (Figure S4). Both compounds had weak antibacterial activity against Staphylococcus epidermis (MIC = 100 μg/mL, each) and Enterococcus durans (MIC = 100 μg/mL, each) (Amagata et al., 2006). A marine-derived Aspergillus fumigatus yielded to 11-O-methylpseurotin A (37) (Figure S4) (PKS/NRPS), which selectively inhibited a Hof1 deletion strain of the yeast Saccharomyces cerevisiae (Boot et al., 2007). Marine-derived fungus Emericella sp., and marine actinomycete Salinispora arenicola were co-cultured to induce production of Emericellamides A (38) and B (39) (Figure S4) by fungi. Emericellamides A and B displayed modest antibacterial activities against MRSA with MIC values of 3.8 and 6.0 μM, respectively (Oh et al., 2007).
The fungus Scopulariopsis brevicaulis, isolated from marine sponge Tethya aurantium was the source of two novel cyclodepsipeptides, Scopularides A (40) and B (41) (Figure S4), which were weak inhibitors of gram-positive bacteria (Yu et al., 2008). Alternaramide (42) (Figure S4), a cyclic Penta depsipeptide, was produced by culture of Alternaria sp. which was isolated from sediment, Masan Bay, S. Korea. It's showed weak antimicrobial activity against B. subtilis (ZOI 8 mm) and S. aureus (ZOI 13 mm) (Kim et al., 2009). Trichoderins A (43), A1 (44), and B (45) (Figure S4) are three new amino lipopeptides reported from marine sponge-derived fungus of Trichoderma sp. All trichoderins have shown potent anti-mycobacterial activity against Mycobacterium smegmatis, Mycobacterium bovis BCG, and M. tuberculosis H37Rv under aerobic and dormancy-inducing hypoxic growth conditions with MIC values in the range of 0.02–2.0 l g/mL (Pruksakorn et al., 2010).
Aspergillus sp. AF119 was the source of γ-aminobutyric acid containing cyclic heptapeptide Unguisin E (46) (Figure S4) (Liu and Shen, 2011). Marine-derived halotolerant Aspergillus sclerotiorum PT06-1 gave to two novel cyclic hexapeptides Sclerotides A (47) and B (48) (Figure S4) in a nutrient-limited hypersaline medium. Both of these peptides were photo inter convertible, containing anthranilic acid, dehydroamino acid units and showed moderate antifungal activity against C. albicans. Compound (47) also inhibited P. aeruginosa growth (Zheng et al., 2009). A halotolerant A. sclerotiorum PT06-1 isolated from salt sediments from the Putian Sea Salt Field, Fujian, China was the source of 11 new aspochracin-type cyclic tripeptides, Sclerotiotides A–K (49–59) (Figure S4). Only sclerotiotides A, B, F and I showed selective antifungal activity against C. albicans with MIC values of 7.5, 3.8, 30, and 6.7 μM, respectively (Zheng et al., 2010).
NRPs with anticancer potential
Cancer is the second leading cause of death worldwide. Present therapies cause serious side effects. Therefore there is need to employ alternative concepts including natural products therapy, complementary or alternative medicine, surgery, radiation therapy used alone or in combination to the prevention of cancer (Reddy et al., 2003). Here we focus on the marine natural products specially NRPs that have been evaluated for cancer prevention.
Bacteria
Three new cytotoxic cyclopeptides, Mixirins A–C (60–62) (Figure S5) belonging to iturin class have been isolated from marine bacterium Bacillus sp. obtained from the mud near the Arctic pole. All compounds inhibited the growth of human colon tumor cells (HCT-116) with IC50 of 0.68, 1.6, 1.3 μg/ml. (Zhang et al., 2004). A Thermoactinomyces specie YM3-251 have been isolated from mud (Mecherchar, Republic of Palau), which was the source of a cyclic peptide Mechercharmycin A (63) as well as the linear congener Mechercharmycin B (64) (Figure S5). Mechercharmycin A exhibited relatively strong antitumor activity against A549 cells (human lung cancer) and Jurkat cells (human leukemia) with IC50 value of 4.0 × 10−8 M and 4.6 × 10−8 M respectively, whereas mechercharmycin B exhibited no activity (Kanoh et al., 2005). Bromoalterochromides A and A' (65–66) (Figure S5), an unprecedented chromo peptide was produced by a marine Pseudoalteromonas maricaloris strain KMM 636 which was isolated from sponge Fascaplysinopsis reticulata. Chemically both of these compounds are brominated yellow colored Thr-Val-Asn-Asn-X pentapeptide lactones, where the amino group of Thr is acylated with 9-(3-bromo-4-hydroxyphenyl)-nona-2, 4,6,8-tetraenoic acid, and X is aIle and Leu, respectively. They showed moderate cytotoxic effects on developing eggs of the sea urchin Strongylocentrotus intermedius (Speitling et al., 2007). New 3-methyl-4-ethylideneproline-containing cytotoxic peptides, Lucentamycins A–D (67–70) (Figures S5, S6) have been isolated from the broth of a marine-derived actinomycete Nocardiopsis lucentensis (strain CNR-712). Only lucentamycins A and B showed significant in vitro cytotoxicity against HCT-116 human colon carcinoma with IC50 values of 0.20 and 11 μM, respectively (Cho et al., 2007).
Three cyclic hexadepsipeptides Piperazimycins A–C (71–73) (Figure S6) have been isolated from the fermentation broth of a Streptomyces sp. (sediment, Guam). The structures of these cyclic hexadepsipeptides have shown presence of rare amino acids, including hydroxyacetic acid, α-methylserine, γ-hydroxypiperazic acid, γ-chloropiperazic acid 2-amino-8-methyl-4, 6-nonadienoic acid, and 2-amino-8-methyl-4,6-decadienoic acid and were all significantly cytotoxic against multiple tumor cell lines with an average GI50 76 ng/mL for each (Miller et al., 2007). The cultured mycelia of marine bacterium Mechercharimyces asporophorigenens (marine lake sediment, Urukthapel Island, Palau) was the source of Urukthapelstatin A (74) (Figure S6), a cyclic thiopeptide that displayed potent activity against a human cancer cell line panel. Urukthapelstatin A has also shown growth inhibition of human lung cancer A549 cells in dose-dependent manner with an IC value of 12 nM (Matsuo et al., 2007). The culture of Salinispora arenicola isolated from sea sediment (Great Astrolabe Reef, Fijiy) yielded three new cyclohexadepsipeptides, Arenamides A–C (75–77) (Figure S6). The absolute structures and configuration of these compounds were established by the spectroscopic technique. Arenamides A (75) and B (76) blocked tumor necrosis factor (TNF)-induced activation with IC50 values of 3.7 and 1.7 μM respectively. In addition, they also inhibited nitric oxide and prostaglandin E2 production and were moderately cytotoxic to HCT-116 cells (Asolkar et al., 2008). Bacillus silvestris that was isolated from a Pacific Ocean (southern Chile) crab yields two new cyclodepsipeptides, Bacillistatins 1-2 (78–79) (Figure S6) with strong anti-cancer (GI50 of 10−4-10−5 μg/mL) activity (Pettit et al., 2009). The epimeric cyclic peptides Turnagainolides A (80) and B (81) (Figure S6), isolated from marine Bacillus sp. (sediment, Turnagain Is., British Columbia, Canada), had indirect inhibitory effect on phosphatidylinositol-3-kinase (PI3K) pathway (Li et al., 2011). A Streptomyces sp. obtained from marine sediment produced two highly modified linear tetrapeptides, Padanamides A (82), and B (83) (Figure S6). They inhibit cysteine and methionine biosynthesis and are cytotoxic to Jurkat cells (IC50 of 20 μg/mL) respectively (Williams et al., 2011). Chemical genomics was performed to discover the mode of action of compounds, which suggested that padanamide A inhibits cysteine and methionine biosynthesis.
Streptomyces sp. isolated from volcanic island produced new cyclic peptides Ohmyungsamycin A (84) and B (85) (Figure S7). The presence of unusual amino acid units, including N-methyl-4-methoxytrytophan, β-hydroxyphenylalanine, and N, N-dimethylvaline in compound (84–85) have been determined by interpretation of the NMR, UV, and IR spectroscopic and MS data. Both exhibited inhibitory activities against diverse cancer cells with IC50 values ranging from 359 to 816 nM and 12.4 to 16.8 μM respectively. However, compound (84) was more active in this regard interestingly; these compounds exhibit relatively selective anti-proliferative activity against cancer cells compared to normal cells. This may be due to the consequence of genetic background or of the biologically various characteristics between cancer and normal cells. However, the exact molecular mechanism behind the selectivity should be further investigated (Um et al., 2013). Proximicins A–C (86–88) (Figure S7) are novel aminofuran antibiotics with anticancer activity, isolated from marine strains of verrucosispora sp. Compounds (86–88) showed inhibitory activity against gastric adenocarcinoma (AGS, IG50 = 0.6, 1.5, 0.25 μg/mL respectively), hepatocellular carcinoma (HepG2, IG50 = 0.8, 9.5, 0.7 μg/mL respectively) and breast carcinoma cells (MCF 7, IG50 = 7.2, 5.0, 9.0 μg/mL respectively). A cell-cycle analysis in AGS cells revealed that Proximicin C produced cell arrest in the G0/G1 phase after incubation for 24 h. After 40 h, there was an increase in the number of cells in the sub-G1 phase, that is, apoptotic cells (+2.9%). It was also found that proximicin C induce upregulation of p53 and of the cyclin kinase inhibitor p21 in AGS cells (Fiedler et al., 2008).
Cyanobacteria
An assemblage of Lyngbya majuscula and Phormidium gracile collected in Papua New Guinea produced a cyclic depsipeptide Hoiamide A (89) (Figure S8). The highly unusual structure of hoiamide A synthesized by mixed peptide–polyketide biosynthetic pathway showed moderate cytotoxicity to cancer cells and partial agonist of site 2 on the voltage-gated sodium channel as it produced a rapid and concentration-dependent elevation of neuronal [Na+] in neocortical neurons (IC50 = 92.8 nM) (Pereira et al., 2009). An assemblage of the marine cyanobacteria L. majuscula and Schizothrix species collected from Fiji was the source of cyclic depsipeptides Yanucamides A (90) and B (91) (Figure S8), which contain a 2, 2-dimethyl-3-hydroxyoct-7-ynoic acid moiety. Both compounds exhibited strong brine shrimp toxicity (LD50, 5 ppm) (Sitachitta et al., 2000). The cyclic depsipeptides named Lyngbyabellins A (92) (Figure S8), contain a 7,7-dichloro-2,2-dimethyl-3-hydroxyoctanoic acid moiety have been isolated from the cytotoxic fraction of L. majuscula collected from Guam and the Dry Tortugas National Park, Florida. Compound (92) have moderate cytotoxicity against human nasopharyngeal carcinoma cell line (KB cells) and human colon adenocarcinoma cell line (LoVo cells), with IC50 values of 0.03 and 0.50 μg/mL, respectively and also showed cellular microfilament network in A-10 cells at 0.01–5.0 μg/mL concentrations (Luesch et al., 2000). Another collection from Tortugas National Park, Florida was the source of cytotoxic and antifungal cyclic depsipeptide Lyngbyabellin B (93) (Figure S8). Lyngbyabellin B was toxic to brine shrimp (LD50 = 3.0 ppm) (Milligan et al., 2000). A marine cyanobacterium Microcystis aeruginosa contained the cyclic hexapeptide Microcyclamide (94) (Figure S8), which showed moderate cytotoxicity against P388 murine leukemia cells at 24–30 μg/mL (Ishida et al., 2000).
The cyanobacterium L. majuscule collected from Guam was the source of Apratoxin A (95) (Figure S8). This cyclodepsipeptide of mixed peptide-polyketide biogenesis exhibited in vitro cytotoxicity against human tumor cell lines at IC50 of 0.36–0.52 nM. Apratoxin A induces G1 phase cell arrest and apoptosis, which is at least particularly initiated through antagonism of FGF signaling via STAT3 (Luesch et al., 2001b). Another collection of L. majuscule from Guam gave two cyclic depsipeptides, Pitipeptolides A (96) and B (97) (Figure S8) with anti-mycobacterial and weak cytotoxicity against LoVo cells with IC50 values of 2.25 and 1.95 μg/mL, respectively. Pitipeptolides A and B also stimulated elastase activity. It is suggested that this activity is due to the presence of hydrophobic portions in the molecule (Luesch et al., 2001a). Marine cyanobacterium Lyngbya sp. collected from Palauan was the source of six new β-amino acid-containing cyclic depsipeptides, the Ulongamides A–F (98–103) (Figure S8). All peptides were found to be weakly cytotoxic against KB and LoVo cells with IC50 values of ca. 1 μM and ca. 5 μM respectively except compound Ulongamides F (Luesch et al., 2002). Examination of a L. confervoides collection from Saipan, Commonwealth of the Northern Mariana Islands, led to the isolation of a novel cytotoxic cyclic depsipeptide Obyanamide (104) (Figure S8). Obyanamide was cytotoxic against KB cells with an IC50 of 0.58 μg/mL. According to the results, the β-amino acid residue was found to play a critical role in the biological activities. Additionally, the ester bond along with the Ala (Thz) moiety was also essential for biological activities (Williams et al., 2002a). Malevamide D (105) (Figure S8), a highly cytotoxic peptide ester have been isolated from marine cyanobacterium Symploca hydnoides (Horgen et al., 2002). A culture Symploca sp. yielded Tasiamide (106) (Figure S8), an acyclic peptide. Tasiamide demonstrated cytotoxic activity against KB and LoVo cells with IC50 values of 0.48 and 3.47 μg/mL, respectively (Williams et al., 2002b). A new cytotoxic peptide Tasiamide B (107) (Figure S9) which contain the unusual amino acid-derived residue 4-amino-3-hydroxy-5-phenylpentanoic acid (Ahppa) have been isolated from cyanobacterium Symploca sp. This peptide displayed an IC50 value of 0.8 μM against KB cells (Williams et al., 2003b).
A Papua New Guinea collection of the marine cyanobacterium L. majuscule was the source of six cyclic depsipeptides, Guineamides A–F (108–113) (Figure S9). The presence of beta-amino or beta-hydroxy carboxylic acid residues in all peptides was determined using a combination of chemical manipulations as well as Marfey's method. Guineamides B and C showed moderate cytotoxicty to a mouse neuroblastoma cell line with IC50 values of 15 and 16 μM, respectively (Tan et al., 2003b). A new bioactive cyclic depsipeptide, Homodolastatin 16 (114) (Figure S9) have been isolated from L. majuscula, collected from Wasini Island off the southern Kenyan coast. Homodolastatin 16 showed moderate activity against oesophageal (IC50 = 4.3 μg/mL) and cervical cancer cell lines (IC50 = 1 μg/mL) (Davies-Coleman et al., 2003). An examination of an organic extract of a cyanobacterium L. majuscula, collected from Guam, led to the isolation of the cyclic peptide Lyngbyastatin 3 (115) (Figure S9). The presence of two unusual amino acid units, 3-amino-2-methylhexanoic acid (Amha) and 4-amino-2, 2-dimethyl-3-oxopentanoic acid units (Ibu) was determined by standard methods. Lyngbyastatin 3 displayed in vitro activity against KB and LoVo cell lines with IC50 values of 32 and 400 nM respectively (Williams et al., 2003a).
A collection of Lyngbya sp. from Palauan was the source of cytotoxic cyclic depsipeptide Ulongapeptin (116) (Figure S9) with an IC50 value of 0.63 μM against KB cells (Williams et al., 2003d). Two new depsipeptides Tasipeptins A (117) and B (118) (Figure S9) have been isolated from Palau collection of Symploca sp. The gross structure of all peptides (117–118) were determined by standard methods and was found to contain unusual amino acid-derived residue 4-amino-3-hydroxy-5-phenylpentanoic acid (Ahppa) and 3-amino-6-hydroxy-2-piperidone (Ahp) moiety respectively. Both were cytotoxic toward KB cells with IC50 values of 0.93 and 0.82 μM, respectively (Williams et al., 2003c). Lyngbya majuscula collected from Hector's Bay, Jamaica was found to contain three lipopeptides, Jamaicamides A–C (119–121) (Figure S9). Further biological investigation of the jamaicamides has revealed that they are generated by iterative hybrid PKS-NRPS assembly and exhibited cytotoxicity to both the H-460 human lung and Neuro-2a mouse neuroblastoma cell lines (IC50 = 15 μM for all), sodium channel blocking activity at 5 μM and ichthyotoxic activities (Edwards et al., 2004). Four new depsipeptides, Wewakpeptins A–D (122–125) (Figures S9, S10), were found cytotoxic to brine shrimp and to the NCI-H460 and neuro-2a cell lines (LC50 of approximately 0.4 μM). These were isolated from Lyngbya semiplena collected from Wewak Bay, Papua New Guinea (Han et al., 2005).
Trungapeptins A-C (126–128) (Figure S10), cyclodepsipeptides have been isolated from marine cyanobacterium L. majuscula. Trungapeptin A exhibited mild icthyotoxicity (6.25 ppm) and weak toxicity to brine shrimp (10 ppm) (Bunyajetpong et al., 2006). Cytotoxic cyclic peptides, Aurilides B (129) and C (130) (Figure S10) were produced by marine cyanobacterium L. majuscula collected from Papua New Guinea. Both aurilides B and C described to induce a dysfunction in mitochondria in NCI-H460 human lung tumor and the neuro-2a mouse neuroblastoma cell lines, with LC50 values between 0.01 and 0.13 μM (Han et al., 2006). A highly methylated tetrapeptide Belamide A (131) (Figure S10) was isolated from Symploca sp. (Salmedina Reef, Panama) which was shown antimitotic and cytotoxic to MCF7 breast cancer and HCT-116 cell lines (IC50 0.74 μM) by microtubule disruption with structural analogy to the important linear peptides dolastatins 10 and 15 (Simmons et al., 2006). Three new analogs of dolastatin 13, Lyngbyastatins 5-7 (132–134) (Figure S10), have been isolated from two different collections of marine cyanobacteria, Lyngbya sp., from South Florida with previously reported cyclodepsipeptide somamide B. Compounds (132–134) were found to selectively inhibit elastase over several other serine proteases, with IC50 values for porcine pancreatic elastase ranging from 3 to 10 nM (Taori et al., 2007). A new linear peptide Mitsoamide (135) (Figure S10) was produced by marine cyanobacterium Geitlerinema sp. collected from Mitso-Ankaraha Island. Mitsoamide has shown strong activity against NCI-H460 human lung tumor cells with LC50 of 460 nM (Andrianasolo et al., 2007).
A Papua New Guinea collection of the marine cyanobacteria L. majuscula and Lyngbya sordida was the source of potent cytotoxic cyclodepsipeptide Apratoxin D (136) (Figure S10). Compound (136) possesses 3, 7-dihydroxy-2, 5, 8, 10, 10-pentamethylundecanoic acid as the polyketide moiety and potent in vitro cytotoxicity against H-460 human lung cancer cells with an IC50 value of 2.6 nM (Gutiérrez et al., 2008). Another collection of marine cyanobacterium Lyngbya bouillonii from a Guamanian was found to contain Apratoxin E (137) (Figure S10), which was strongly cytotoxic to several cancer cell lines at IC50 21–72 nM (Matthew et al., 2008). Dragonamides C (138) and D (139) (Figures S10, S11) are linear lipopeptides isolated from the marine cyanobacterium brown Lyngbya polychroa. Both peptides are weak cytotoxic against several cancer cell lines with GI50 values of 56 and 59 μM against U2OS osteosarcoma cells, 22 and 32 μM against HT29 colon adenocarcinoma cells, and 49 and 51 μM against IMR-32 neuroblastoma cells, respectively (Gunasekera et al., 2008). Coibamide A (140) (Figure S11), a potent anti-proliferative highly methylated cyclic depsipeptide was isolated from a culture of Leptolyngbya sp. which was collected from the Coiba National Park, Panama. Compound (140) showed an unprecedented selectivity profile in the NCI 60 cancer cell line panel (LC50 < 23 nM). It causes S phase inhibition in cell cycle (Medina et al., 2008). Symploca sp. yielded dolastatin 10/15 hybrid linear peptide Symplostatin 4 (141) (Figure S11) which was shown to be antimitotic activity via microtubule depolymerization to H-460 lung cancer cells (IC50 = 40 nM) as well as neuro-2a neuroblastoma cells (IC50 = 29 nM) (Taori et al., 2009).
A culture of L. majuscule obtained from True Blue Bay, eastern Caribbean yielded two new depsipeptides, Itralamides A (142) and B (143) (Figure S11). Only itralamide B was found to cytotoxic to HEK293 cells IC50 6 ± 1 μM (Jiménez et al., 2009). An active peptide metabolite Desmethoxymajusculamide C (144) (Figure S11) (DMMC) have been isolated from Fijian collection of L. majuscule. Both cyclic and liner version of DMMC were found potent and showed selective anti-solid tumor activity at IC50 = 20 nM against HCT-116 through disruption of cellular microfilament networks (Simmons et al., 2009). A Singapore collection of L. majuscule has been shown to produce cyclodepsipeptide, Hantupeptin A (145) (Figure S11) with cytotoxicity to MOLT-4 leukemia cells (IC50 32 μM) and MCF-7 breast cancer cells (IC50 4.0 μM) (Tripathi et al., 2008). Chemical investigation of Lyngbya cf. polychroa resulted in isolation of a linear peptide desacetylmicrocolin B (146) (Figure S11), was a growth inhibitor of HT-29 (IC50 14 nM) and IMR-32 cells (IC50 14 nM) (Meickle et al., 2009).
Palmyramide A (147) (Figure S11), a cyclic depsipeptide found to block sodium channel in neuro-2a cells (IC50 17.2 μM) and modest cytotoxicity to H-460 cells (IC50 39.7 μM) and was isolated from from a Palmyra Atoll Collection of the marine cyanobacterium L. majuscule (Taniguchi et al., 2009). Lyngbya sp. was the source of a potent cytotoxic peptide Bisebromoamide (148) (Figure S11) which potentially inhibit protein kinase and is cytotoxic to HeLa S3 cells with an IC50 value of 0.04 μg/mL (Teruya et al., 2009). An examination of an organic extract of the cyanobacterium L. bouillonii, collected from Guam, led to the isolation of cytoskeletal actin-disrupting and cytotoxic (IC50 = 0.041 μM) Lyngbyabellin J (149) and a linear modified peptide, lyngbyapeptin D (150) (Figure S11) (Matthew et al., 2010). Marine cyanobacterium L. confervoides gave bis-thiazoline containing cyclic depsipeptides, Grassypeptolides A–C (151-153) (Figure S12). All grassypeptolides cause G1 phase cell cycle arrest in HT29 (IC50 = 1.22, 4.97 μM, 76.7 nM) and HeLa cell lines (IC50 = 1.01, 2.93 μM, 44.6 nM) (Kwan et al., 2010). Hantupeptins B (154) and C (155) (Figure S12), two cytotoxic cyclodepsipeptides were obtained from a marine cyanobacterium L. majuscule collected from Pulau Hantu Besar, Singapore. Compound (154) gave an IC50 of 0.2 μM against MOLT-4 and 0.5 μM against MCF-7 cancer cell lines however compound (155) showed moderate cytotoxicity against the MOLT-4 and MCF-7 cancer cell lines with IC50 values of 3.0 μM and 1.0 μM, respectively (Tripathi et al., 2010). Marine cyanobacteria Lyngbya sp. was the source of novel cyclic peptides Lyngbyacyclamides A (156) and B (157) (Figure S12) which moderately inhibited the growth of B16 mouse melanoma cells (IC50 of 0.7 μM) (Maru et al., 2010).
The marine cyanobacterium Leptolyngbya sp. collected from the SS Thistlegorm shipwreck in the Red Sea offered two cyclic depsipeptides, Grassypeptolides D (158) and E (159) (Figure S12). Both of these peptides were cytotoxic to HeLa (IC50 = 335 and 192 nM, respectively) and mouse neuro-2a blastoma cells (IC50 = 599 and 407 nM, respectively) (Thornburg et al., 2011). Bis-thiazoline-containing cyclic depsipeptides Grassypeptolides F (160) and G (161) (Figure S12) which contains rare β-amino acid, extensive N-methylation and a large number of d-amino acids was isolated from an extract of Palauan cyanobacterium L. majuscule. Both (160) and (161) were found to have moderate inhibitory activity against the transcription factor AP-1 (IC50 = 5.2 and 6.0 μM, respectively) (Popplewell et al., 2011). An examination of an organic extract of the cyanobacterium Symploca cf. hydnoides sampled from Cetti Bay, Guam, led to the isolation of the eleven new peptides Veraguamides A-G (162–168), Veraguamide H (169) and J–L (170–172) (Figure S12). Veraguamide A showed potent cytotoxicity to H-460 human lung cancer cell line at LD50 = 141 nM whilst the others were weak inhibitors. Their structures were elucidated by combining various techniques in spectroscopy, chromatography, and synthetic chemistry (Mevers et al., 2011; Salvador et al., 2011).
Three new cyclic peptides Malyngamide 3 (173) and Cocosamides A (174) and B (175) (Figure S13) have been isolated from the lipophilic extract of marine cyanobacteria L. majuscula collected from Cocos Lagoon, Guam and were found to modestly cytotoxic to HT-29 cells with IC50 value of 48, 24, and 11 μM respectively (Gunasekera et al., 2011). L. majuscula (Piti Bomb Holes, Guam) was the source of proline rich unusual cyclic depsipeptide Pitiprolamide (176) (Figure S13). Further investigation yielded four more peptides Pitipeptolides C–F (177–180) (Figure S13). All peptides were found moderately cytotoxic against two HTCLs, however, pitipeptolides C–F were more active against M. tuberculosis and B. cereus as compared to compound (176) (Montaser et al., 2010). The marine cyanobacterium L. majuscule collected from western lagoon of Pulau Hantu Besar, Singapore was the source of cyclodepsipeptide Lagunamide C (181) (Figure S13). Lagunamide C exhibited potent cytotoxic activity against HTCLs with IC50 values ranging from 2.1 to 24.4 nM, antimalarial activity against Plasmodium falciparum (IC50 0.29 μM) and weak anti-swarming activity against P. aeruginosa (Tripathi et al., 2011).
A collection of marine cyanobacterium L. semiplena and L. majuscule from Papua New Guinea led to isolation of the cyclic depsipeptides Wewakamide A (182) and Guineamide G (183) (Figure S13) were respectively. Both of these peptides displayed potent toxicity against brine shrimp and only guineamide G showed cytotoxicity to a mouse neuroblastoma cell line with LC50 values of 2.7 μM (Nan et al., 2011). A Lyngbya sp. collected in Florida Keys was found to contain epimeric cyclic depsipeptides Porpoisamide A (184) and B (185) (Figure S13) which was weakly cytotoxic to HCT-116 (IC50 = 25, 21 μM respectively) and osteosarcoma U2OS cells (IC50 = 28, 22 μM respectively) (Meickle et al., 2011). Moorea bouillonii (Strawn Is., Palmyra Atoll, Central Pacific Ocean) gave five lipopeptides Lyngbyabellin K (186) and L (187), 7-epi-lyngbyabellin L (188) and Lyngbyabellin M (189) and N (190) (Figure S13). Of note, cyclic metabolites (189) and (190) possess rare monochlorination on the 3-acyloxy-2-methyloctanoate residue, whereas unusual N, N-dimethylvaline containing lyngbyabellin N was strongly cytotoxic to HCT116 colon cancer cell line (IC50 = 40.9 ± 3.3 nM) (Choi et al., 2012). Viequeamides are novel 2, 2-dimethyl-3-hydroxy-7-octynoic acid (Dhoya)-containing cyclic depsipeptides isolated from a shallow subtidal collection of a “button” cyanobacterium Rivularia sp. (Vieques, Puerto Rico). The absolute structures and configurations of major components Viequeamide A-D (191–194) (Figures S13, S14) of the mixture were established by spectroscopic technique. However, viequeamides B–F were not separated out and only viequeamide A showed high cytotoxicity against H460 human lung cancer cells at IC50 60 ± 10 nM (Boudreau et al., 2012). A new N, N-dimethyl-terminated linear peptide Symplocin A (195) (Figure S14) was produced by Bahamian collection of cyanobacterium Symploca sp. The absolute configuration of symplocin A was done by chiral-phase HPLC of the corresponding 2-naphthacyl esters. Symplocin A showed potent inhibitory effect on protease enzyme cathepsin E with IC50 300 pM (Molinski et al., 2012). A collection of tropical marine cyanobacterium, M. bouillonii, from New Britain, Papua New Guinea resulted in isolation of a novel cytotoxic cyclic depsipeptide, Bouillonamide (196) (Figure S14). Compound (196) which contains two unique polyketide-derived moieties, a 2-methyl-6-methylamino-hex-5-enoic acid residue and a unit of 3-methyl-5-hydroxy-heptanoic acid have shown mild toxicity against neuron 2a mouse neuroblastoma cells with IC50 6.0 μM (Tan et al., 2013). A new lipopeptides, Malyngamide 4 (197) (Figure S14) as a moderate inhibitor of several HTCLs have been isolated from marine cyanobacterium Moorea producens collected from the Red Sea, Saudi Arabia (Shaala et al., 2013). The marine cyanobacterium Okeania sp. collected from the coast near Jahana, Okinawa, was the source of Kurahyne B (198) (Figure S14). It showed growth inhibition against HeLa and HL60 cells, with IC50 values of 8.1 and 9.0 μM, respectively (Okamoto et al., 2015).
Fungi
A culture of marine fungi Fusarium CNL-619 was the source of a new cyclic depsipeptide N-Methylsansalvamide (199) (Figure S15), which showed weak in vitro cytotoxicity against NCI human tumor cell lines (GI50 8.3 μM) (Cueto et al., 2000). An unidentified fungus isolated from the red alga, Ceradictyon spongiosum (Okinawa) have been shown to produce two linear dodecapeptides, Dictyonamides A (200) and B (201) (Figure S15). Only the compound (200) showed inhibitory effect on cyclin-dependent kinase 4 with IC50 value of 16.5 μg/mL (Komatsu et al., 2001). A culture of marine fungus, Scytalidium sp., collected from Bahamas was the source of two new cyclic heptapeptides Scytalidamides A (202) and B (203) (Figure S15) and both compounds displayed moderate cytotoxicity to the HCT-116 cell line in vitro with IC50 values of 2.7 and 11.0 μM, respectively (Tan et al., 2003a). A strain of Trichoderma virens was isolated from ascidian Didemnum molle and from the surface of a green alga of genus Halimeda from Papua New Guinea, which was the source of two modified dipeptides Trichodermamides A (204) and B (205) (Figure S15). Trichodermamide B has showed significant in vitro cytotoxicity against HCT-116 cells (colon carcinoma) with an IC50 of 0.32 μg/mL (Garo et al., 2003). A fungal strain Exserohilum rostratum associated with a marine cyanobacterial mat produced four moderately cytotoxic cyclic dipeptides Rostratins A–D (206–209) (Figure S15). The structures and absolute configurations of peptides were determined by two-dimensional NMR techniques and Mosher method respectively. Compounds (206–209) exhibit activity against colon carcinoma (HCT-116) with IC50 values of 8.5, 1.9, 0.76, and 16.5 μg/mL, respectively (Tan et al., 2004).
A new cytotoxic cyclodepsipeptide, IB-01212 (210) (Figure S15) was produced by filamentous fungus Clonostachys sp., ESNA-A009 isolated from an unidentified Japanese sponge. IB-01212 was potent cytotoxic to several human tumor cell lines which includes LN-caP (prostrate), SK-BR3 (breast), HT29 (colon), and HELA (cervix) cell lines with GI50 (growth inhibition) in order of 10−8 M (Cruz et al., 2006). A culture of Zygosporium masonii isolated from a marine cyanobacterium afforded a new cyclic Penta depsipeptide, Zygosporamide (211) (Figure S15), which had significant cytotoxicity in the NCI's 60 cell line panel, CNS cancer cell line SF-268 (GI50 = 6.5 nM) and the renal cancer cell line RXF 393 (GI50 ≤ 5.0 nM) (Oh et al., 2006). Trichoderma reesei isolated from China, Lianyungang collection of sea mud produced moderately cytotoxic Trichoderide A (212) (Figure S15) (Sun et al., 2006). Two new cyclohexadepsipeptides, Spicellamide A (213) and Spicellamide B (214) (Figure S15) obtained from fermentation of Spicellum roseum (Ectyplasia perox, Dominica), exhibited cytotoxicity to neuroblastoma cells with IC50 value of 30 and 6.2 μg/mL respectively (Kralj et al., 2007). Two new cyclic depsipeptides 1962A, cyclo-(d-Leu-Gly-l-Tyr-l-Val-Gly-S-O-Leu) (215), and 1962B, cyclo-(d-Leu-Gly-l-Phe-l-Val-Gly-S-O-Leu) (216) (Figure S15) have been isolated from the fermentation broth of the mangrove endophytic fungus isolated from the leaf of Kandelia candel. Compound (215) only showed activity against human breast cancer MCF-7 cells with an IC50 value of 100 μg/mL (Huang et al., 2007).
Two new cyclic tetrapeptides Microsporins A (217) and B (218) (Figure S15) with potent inhibitors of histone deacetylase (HDAC), cytotoxic to HCT-116 cells (IC50 0.6 and 8.5 μg/mL) was isolated from the marine-derived fungus Microsporum gypseum (Gu et al., 2007). A Penta decapeptide, Efrapeptin J (219) (Figure S16), a down-regulator of the molecular chaperone GRP78 have been isolated from Tolypocladium sp. (sea mud, Aomori Prefecture, Japan) (Hayakawa et al., 2008). An Australian marine isolate of Aspergillus versicolor (MST-MF495) offered two cyclo pentapeptides, Cotteslosins A (220) and B (221) (Figure S16) (Fremlin et al., 2009). Two new cyclic pentapeptides, Versicotides A (222) and B (223) (Figure S16) came from marine fungus strain ZLN-60, identified as A. versicolor (Zhou et al., 2011). A Cytotoxic lipopeptide Fellutamide F (224) (Figure S16) have been isolated from the sponge-derived fungus A. versicolor with cytotoxicity to several human tumor cells, especially SK-MEL-2 (skin, IC50 0.67 μM), XF498 (CNS, IC50 0.14 μM) and HCT15 (Colon, IC50 0.13 μM) (Lee et al., 2011). Fermentation extract of the marine-derived fungus Acremonium persicinum SCSIO 115 resulted in the discovery of three new cyclo heptapeptides, Cordyheptapeptides C–E (225–227) (Figure S16) with cytotoxicity against SF-268, MCF-7, and NCI-H460 tumor cell line with IC50 values ranging from 2.5 to 12.1 μM (Chen et al., 2012). Chemical investigation of endophytic fungus Bionectria ochroleuca isolated from the inner leaf tissues of the plant Sonneratia caseolaris (Sonneratiaceae) from Hainan Island (China), lead to discover two new peptides, Pullularins E and F (228-229) (Figure S16). Both compounds exhibited moderate cytotoxic activity against the mouse lymphoma cells (L5178Y) with EC50 values ranging between 0.1 and 6.7 μg/mL (Ebrahim et al., 2012). An unusual anthranilic acid dimer and a d-phenyllactic acid residue containing cyclodepsipeptides Clavatustides A–C (230–232) (Figure S16) were discovered from cultured mycelia and broth of Aspergillus clavatus C2WU isolated from Xenograpsus testudinatus and suppressed proliferation of HTCLs (Jiang et al., 2013; Ye et al., 2014). Aspergillus terreus SCSGAF0162 gave a new cytotoxic (HTCLs) and antiviral (H1N1 and H3N2) cyclic tetrapeptide, Asperterrestide A (233) (Figure S16). Which was cytotoxic toward human carcinoma U937 and MOLT4 cell lines with IC50 values of 6.4 and 6.2 μM, respectively, and also showed inhibitory effects on the influenza virus strains A/WSN/33 (H1N1) and A/Hong Kong/8/68 (H3N2) with IC50 values of 15 and 8.1 μM, respectively (He et al., 2013). A new cyclohexapeptide, Similanamide (234) (Figure S16) was isolated from sponge-associated fungus Aspergillus similanensis KUFA 0013 with weak anticancer activity (Prompanya et al., 2015).
Marine microorganisms have been recognized as one of the most promising groups of organisms from which novel pharmacologically active molecules, with potential benefits against cancer, can be isolated. Recently, several compounds have been emerged as templates for the development of novel anticancer drugs. However the mechanisms implicated in the cytotoxicity of these compounds in tumor cell lines are still largely overlooked but several studies point to an implication in apoptosis. For instance, several compounds were found to inhibit cell growth in a large variety of cancer cell lines, the pathways by which cancer cells are inhibited are still poorly elucidated. In some cases, compounds were found to induce cell death by activation of the apoptotic process; nevertheless the mechanisms underlying the apoptosis still need more investigations. Some compounds were found to create an imbalance in cellular redox potential, with mitochondria representing a central role in the process. However, more studies are needed in order to clarify it. Cell cycle is another disturbed process, mainly due to disruption of the microtubules and actin filaments; however there are only a few studies connecting marine NRPs with alterations in cell cycle and more studies are needed in order to clarify the involvement of these compounds in the process. Even membrane sodium channels can establish interactions with the compounds, revealing its potentially important role in the observed effects. In summary, more investigations are needed in order to clarify the specific targets and the mechanisms that are behind cancer cell cytotoxicity, namely the involvement of the apoptotic process by the implication of functional genomics.
NRPs with both antimicrobial and anticancer potential
Lajollamycin, (235) (Figure S17) a nitro-tetraene spiro-β-lactone-γ-lactam antibiotic have been isolated from marine actinomycete Streptomyces nodosus. In vitro lajollamycin inhibited the growth of the murine melanoma cell line B16-F10 with an EC50 of 9.6 μM and also displayed antimicrobial activity against both drug resistant and sensitive gram-positive bacteria with MIC 2–20 μg/mL (Manam et al., 2005). Two unusual linear hybrid polyketide-nonribosomal peptide antibiotics, Ariakemicins A-B (236–237) (Figure S17) have been isolated from the fermentation broth of the marine gliding bacterium Rapidithrix sp., (Ariake Inland Sea, Japan). These antibiotics contain threonine, two omega-amino-(omega-3)-methyl carboxylic acids with diene or triene units, and delta-isovanilloylbutyric acid and selectively inhibited Gram-positive bacteria among which S. aureus was the most affected (MIC 0.46 μg/mL) and were slightly cytotoxic to human lung cancer cells (A549) and baby hamster kidney cells with IC50 values of 25 and 15 μg/mL respectively (Oku et al., 2008a). Glycolipopeptides Ieodoglucomide A (238) and B (239) (Figure S17) have been isolated from marine-derived bacterium Bacillus licheniformis (sediment, Ieodo Reef, S. Korea). Compounds (238) and (239) displayed moderately in vitro antimicrobial activity. However, ieodoglucomide B also displayed cytotoxic activity against lung cancer and stomach cancer cell lines with GI50 values of 25.18 and 17.78 μg/mL, respectively (Tareq et al., 2012).
Halolitoralin A (a cyclic hexapeptide) (240) (Figure S17), Halolitoralin B and C, two cyclic tetrapeptides (241–242) (Figure S17) were isolated from the marine sediment-derived Halobacillus litoralis YS3106. All three cyclopeptides show surprisingly simple architectures with highly repeated residue units. Compounds (241–242) have shown antifungal activity against two human fungi Candida albicans and Tricophyton rubrum with MIC of 20, 30, 30 μg/mL and 25, 35, 40 μg/mL respectively. In addition, these three cyclopeptides showed moderate anti-human gastric tumor activities in vitro (with a cell line of BGC) (Yang et al., 2002). Bioactivity-guided fractionation from the fermentation broth of Bacillus mojavensis B0621A (Pearl oyster Pinctada martensii, Weizhou Is., South China Sea) was the source of antifungal iturinic lipopeptide Mojavensin A (243) (Figure S17). The Marfey's analysis of mojavensin A provides the novel peptide backbone of L-Asn1, D-Tyr2, D-Asn3, L-Gln4, L-Pro5, D-Asn6, L-Asn7 and an anteiso-type of the saturated β-fatty acid side chain. Compound 243 also inhibited the growth of HL-60 with IC50 of 100 (Ma et al., 2012). A new sequential tristhiazole-thiazoline-containing cyclic peptide, Marthiapeptide A (244) (Figure S17), have been isolated from a culture of the deep South China Sea-derived strain Marinactinospora thermotolerans SCSIO 00652. Marthiapeptide A exhibited inhibition against a panel of gram-positive bacteria, with MIC values ranging from 2.0 to 8.0 μg/mL, and displayed strong cytotoxic activity against a panel of human cancer cell lines with IC50 values ranging from 0.38 to 0.52 μM (Zhou et al., 2012). A new linear glyceryl acid derived heptapeptide (Glyceryl-D-leucyl-D-alanyl-D-leucyl-D-leucyl-L-valyl-D-leucyl-D-alanine, (245) (Figure S17), were produced by the culture of marine deep sediment strain Paenibacillus profundus Sl 79. The compound (245) was cytotoxic to SK-MEL-28 cell line (IC50 = 3.07 μM after 72 h) and also inhibited the growth of S. aureus (ZOI 24 mm), S. epidermis (ZOI 20 mm), B. subtilis (ZOI 28 mm) and Enterococcus faecium (ZOI 11 mm) (Kalinovskaya et al., 2013).
Role of genomics, proteomics and bioinformatics in discovery and development of nonribosomal peptides drugs
The non-ribosomal peptides (NRPs) are an essential source of chemical diversity for drug discovery and development. At present, there are more than 1,164 different non-ribosomal peptides known in public database (NCBI) which consists of over 500 unique monomers, including both proteinogenic and non-proteinogenic L- and D-amino acids as well as carboxylic acids and amines (Caboche et al., 2010). Due to great structural diversity (linear, cyclic and branched or other complex primary structures) these complex secondary metabolites had impact on all therapeutic area, as making them suitable to be used as clinical agents. However, such potential NRPs often need to be modified to improve their clinical properties and/or bypass resistance mechanisms (Bush, 2012). For instance, FDA approved Oritavancin has been developed by using semi-synthesis strategy from Vancomycin for treatment of drug resistant skin infections (Markham, 2014). Indeed, modification in the nucleotide sequence of a natural NRPS gene or combining modules of different NRPSs may potentially lead them to be more effective with unique pharmacological activity. However, this requires in-depth understanding of both the assembly line and the resulting products. Over the last few decades several bioengineering approaches have been developed to increase the yields of NRPs and generating modified peptides with altered bioactivity or improved physicochemical properties (Winn et al., 2016). Earlier, biosynthetic generation of novel NRPs analogs focused on precursor directed biosynthesis (PDB) or mutasynthesis. In PDB, a wild-type organism is provided with modified or synthetic amino acids with the prospect that the substrate specificity of the relevant NRPS shall be flexible enough to allow addition of the modified precursors into the final peptide. However, mutasynthesis is the exact opposite. The modified substrates are fed to an engineered organism which lacks the enzyme(s) required for the biosynthesis of a specific natural precursor, so that a modified substrate or precursor analog may be effectively incorporated (Weist et al., 2004). These methods are important because they generate natural product analogs rapidly.
In earlier reviews many examples of precursor directed biosynthesis of NRPs are available (Thiericke and Rohr, 1993). Other methods being adopted for the production of new nonribosomal peptides is engineering of precursor supply in vivo or introducing tailoring enzymes from other pathways with new glycosylation, halogenation and sulfation enzymes being applied outside of their native clusters to create structural diversity. Although it's similar to precursor directed biosynthesis, it focuses mainly on endogenous biosynthesis rather than exogenous feeding. The introduction of halogen unit into NRP scaffolds has been a common target. For example, when the enzyme PrnA (a favin-dependent tryptophan-7-halogenase) from Pseudomonas fuorescens Pf-5 was expressed alongside the NRPS genes for the uridyl peptide antibiotic pacidamycin, produced by Streptomyces coeruleorubidus, a new halogenated analog was generated (Roy et al., 2010). Using such a technique enduracidin analogs have been produced by altering halogenase in wild-type Streptomyces fungicidicus (Yin et al., 2010). An alternative but complicated strategy has also been developed to generate novel NRPs. It exchanges NRPS subunit, module, and domain of the core peptide itself. Initially, this method was applied by Cubist Pharmaceuticals for the development and marketing of nonribosomal peptide antibiotic daptomycin, first natural product antibiotic that gained approval for clinical use in over 30 years (Baltz et al., 2006). Unfortunately, Cubist Pharmaceuticals failed to identify any daptomycin variants with better antibacterial property than parent daptomycin. Another route that has also been explored which involves modifying the length of the peptide chain by deletion or insertion of one or more modules (Mootz et al., 2002; Butz et al., 2008). A recent study indicates that the introduction of individual or combined point mutations in the binding pocket of an NRPS adenylation domain generates new diversity of NRPs (Han et al., 2012).
A latest technique called heterologous expression offer considerable promise especially for natural hosts which are slow growing, genetically difficult to handle, unculturable, or even unknown. The transfer of biosynthetic genes from the original microbial organisms to more amenable heterologous host bacteria is more amenable to large-scale fermentation production would overcome the limitation of procurement of the drug from the ocean (which is currently limited to expensive aquaculture or field harvesting) and ensure supply (Ongley et al., 2013). The gene cluster responsible for polyketide epothilone (a potential anticancer agent) biosynthesis in the myxobacterium Sorangium cellulosum was cloned and completely sequenced by Tang et al. (2000). Concomitant expression of these genes in the actinomycete Streptomyces coelicolor produced epothilones A and B (Tang et al., 2000). After this heterologous expression system portends a plentiful supply of this medically relevant agent. Similarly A novel gene (amyZ) encoding a cold-active and salt-tolerant α-amylase (AmyZ) was cloned from marine bacterium Zunongwangia profunda (MCCC 1A01486) and the protein was expressed in Escherichia coli (Qin et al., 2014). The Ptchi19 gene of the marine Pseudoalteromonas tunicata CCUG 44952T was cloned and expressed in E.coli (García-Fraga et al., 2015). A new κ-carrageenase gene from marine bacterium Zobellia sp. ZM-2 was cloned and expressed in E.coli (Liu et al., 2013). Heterologous expression of the barbamide biosynthetic gene cluster from the marine cyanobacterium Moorea producens in the terrestrial also led to the production of a new barbamide congener 4-O-demethylbarbamide (Kim et al., 2012). The biosynthetic pathway for bacitracin was successfully transferred from Bacillus licheniformis to the related species B. subtilis (Eppelmann et al., 2001). The polyketide biosynthesis pathway for the marine-derived telomerase inhibitor griseorhodin A was productively transferred to Streptomyces lividans from an environmental Streptomyces isolate (Li and Piel, 2002). Ugai et al. (2016) got success in heterologous expression of the cryptic gene cluster found in A. solani to obtain a marine-derived antifungal agent didymellamide B from the A. oryzae transformant introducing PKS–NRPS, trans-ER, and P450 genes asolSCA (Ugai et al., 2016). Likewise many other successful examples are available in literature (Fortman and Sherman, 2005; Luo et al., 2016; Winn et al., 2016).
All studies presented above for production of novel NRPs and engineering NRPS assembly lines in the native host are laborious having low throughput and low yield. Recent advances in genome sequencing, gene synthesis, metabolomics and bioinformatics revolutionized the process of NRPS engineering. In-silico based bioprospecting of available microbial genome sequences gives us a quick look at the hidden biosynthetic capacity of natural products in the microbial species. Several active as well as silent enzymes have been identified in fungal and bacterial genomes which are involved in the biosynthesis of NRPs. The corresponding secondary metabolites of these enzymes have not been identified to date (Brakhage, 2013; Doroghazi and Metcalf, 2013). Various powerful computational algorithms and tools have been developed to analyze BGC and to determine whether they are likely to encode unique compounds (Medema and Fischbach, 2015). Comprehensive ranges of software tools are available for identification of BGC in genome sequences. These tools are generally divided into two categories: high-confidence/low-novelty and low-confidence/high-novelty. High-confidence/low-novelty includes tools such as CLUSEAN13, ClustScan14, np.searcher15, SMURF16 and antiSMASH (Medema et al., 2011). These tools analyze Hidden Markov Models (HMMs) with manually curated cutoffs to identify signature genes or domains that are highly specific for known classes of biosynthetic pathways. Such strategies give a quick and reliable interpretation of NRPs gene cluster of a single strain from its genome sequence. Low-confidence/high-novelty mainly focuses on the identification of new BGC types by applying three approaches; namely pattern-based mining, phylogenetic mining and comparative genomic mining. These further include Cluster Finder, EvoMining, and Algorithim, respectively. Tools for identification of BGCs with respect to metagenomes include PCR-based sequence-tag and the shotgun assembly approach. The sequence-tag approach identifies clones from selected harbor pathways in metagenomic libraries by amplifying known biosynthetic domains using PCR. This is particularly useful for identifying variants of known pathway types. This has been also used to identify gene clusters encoding close relatives of molecule such as rapamycin, teicoplanin, and thiocoraline (Owen et al., 2013). However, the tag-based approach can be used to find entirely new molecules that are produced by known BGC classes, especially when coupled with phylogenomic tools such as NaPDoS (Ziemert et al., 2012). These tools find application in identifying domains that represent new areas of the extant biosynthetic diversity. A range of systems have been developed to predict the substrate specificities of NRPS adenylation domains (Röttig et al., 2011; Prieto et al., 2012). Tools such as NP.searcher and antiSMASH individual monomer predictions are then combined to give a rough idea of the core scaffold of a nonribosomal peptide. Simultaneously, advancement in mass spectrometry gives efficient dereplication for analysis of small-molecule products of biosynthetic pathways (Nielsen and Larsen, 2015). NRPQuest algorithm uses molecular networking approach to identify potential gene clusters for observed tandem mass spectra of NRPs (Mohimani et al., 2014). The search database for NRPquest generates all possible orders of NRPS assembly lines within each detected NRP BGC hence, predicting the amino acids encoded by each of its module using NRPSPredictor2 (Röttig et al., 2011). A chemoinformatic based library and informatic search strategy for natural products (iSNAP) has also been doveloped for true nontargeted dereplication across a spectrum of nonribosomal peptides and within natural product extracts (Ibrahim et al., 2012). It is clear that the tools and techniques discussed above have accelerated the discovery and development of novel NRPs with desirable biological activities.
Conclusion and future prospects of marine derived nonribosomal peptides
Marine chemicals often possess quite novel structures which in turn lead to pronounced biological activity and novel pharmacology. The study of such chemicals, therefore, is a very promising endeavor. There are three parallel branches in marine natural products chemistry: marine biomedicinals, marine chemical ecology and marine toxins. Integration of these three fields of study gives marine natural products chemistry its exclusive character and vigor. The search among marine chemicals for medically useful agents involves two steps, discovering the type of biological activity and studying the pharmacological mechanism of the activity. It is now clear that efforts to date in marine natural product chemistry have largely focused on easily collected microorganisms and their major metabolites, and while there has been a recent shift to, as detailed above, minor metabolites present in very small quantities are a challenge for analytical and biological evaluations.
As has been demonstrated in this review, the potential for nonribosomal peptides from marine as sources and/or leads to drugs that have pharmacological effects (i.e., cancer and anti-infective) is only now being realized. Combining enzyme technology and solid phase peptide synthesis, it is possible to generate a vast variety of unique peptides composed of non-proteinogenic amino acids with unique pharmacological and biotherapeutic potential. It is possible that in coming years at least one or more marine derived novel nonribosomal peptide will enter into commerce as a drug. In concluding, the huge ranges of nonribosomal peptides that have so far been identified from marine resources frequently have no comparable equivalent in terrestrial organisms. The work by (predominately) young investigators on the many aspects of nonribosomal peptides (like biosynthesis) in the commensal and/or symbiotic microbes associated with these invertebrates, or in the microbes isolated from shallow and deep sediments will increase the numbers of nonribosomal peptides from marine for further work. The marine system has hardly been scratched as yet!
Author contributions
SA collected the available bibliographic information and wrote the manuscript. AA and CB conceived the study. SD and DA reviewed the collected information critically.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to TERI Deakin Nano biotechnology Centre, Biotechnology and Bioresources Division, The Energy and Resources Institute, India for continuous support.
Footnotes
Funding. Deakin University provided a postgraduate scholarship to SA.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2017.00828/full#supplementary-material
References
- Agrawal S., Adholeya A., Deshmukh S. K. (2016). The pharmacological potential of non-ribosomal peptides from marine sponge and tunicates. Front. Pharmacol. 7:333. 10.3389/fphar.2016.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mestarihi A. H., Villamizar G. N., Fernández J., Zolova O. E., Lombó F., Garneau-Tsodikova S. (2014). Adenylation and S-methylation of cysteine by the bifunctional enzyme TioN in thiocoraline biosynthesis. J. Am. Chem. Soc. 136, 17350–17354. 10.1021/ja510489j [DOI] [PubMed] [Google Scholar]
- Amagata T., Morinaka B. I., Amagata A., Tenney K., Valeriote F. A., Lobkovsky E., et al. (2006). A chemical study of cyclic depsipeptides produced by a sponge-derived fungus. J. Nat. Prod. 69, 1560–1565. 10.1021/np060178k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianasolo E. H., Goeger D., Gerwick W. H. (2007). Mitsoamide: a cytotoxic linear lipopeptide from the Madagascar marine cyanobacterium Geitlerinema sp. Pure Appl. Chem. 79, 593–602. 10.1351/pac200779040593 [DOI] [Google Scholar]
- Asolkar R. N., Freel K. C., Jensen P. R., Fenical W., Kondratyuk T. P., Park E.-J., et al. (2008). Arenamides A– C, Cytotoxic NFκB Inhibitors from the Marine Actinomycete Salinispora arenicola? J. Nat. Prod. 72, 396–402. 10.1021/np800617a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H., Brian P., Miao V., Wrigley S. K. (2006). Combinatorial biosynthesis of lipopeptide antibiotics in Streptomyces roseosporus. J. Indus. Microbiol. Biotechnol. 33, 66–74. 10.1007/s10295-005-0030-y [DOI] [PubMed] [Google Scholar]
- Barsby T., Kelly M. T., Gagné S. M., Andersen R. J. (2001). Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Organ. Lett. 3, 437–440. 10.1021/ol006942q [DOI] [PubMed] [Google Scholar]
- Blunt J. W., Copp B. R., Keyzers R. A., Munro M., Prinsep M. R. (2015). Marine natural products. Nat. Prod. Rep. 32, 116–211. 10.1039/C4NP00144C [DOI] [PubMed] [Google Scholar]
- Boot C. M., Gassner N. C., Compton J. E., Tenney K., Tamble C. M., Lokey R. S., et al. (2007). Pinpointing pseurotins from a marine-derived Aspergillus as tools for chemical genetics using a synthetic lethality yeast screen. J. Nat. Prod. 70, 1672–1675. 10.1021/np070307c [DOI] [PubMed] [Google Scholar]
- Boudreau P. D., Byrum T., Liu W.-T., Dorrestein P. C., Gerwick W. H. (2012). Viequeamide A, a cytotoxic member of the kulolide superfamily of cyclic depsipeptides from a marine button cyanobacterium. J. Nat. Prod. 75, 1560–1570. 10.1021/np300321b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. 10.1038/nrmicro2916 [DOI] [PubMed] [Google Scholar]
- Bunyajetpong S., Yoshida W. Y., Sitachitta N., Kaya K. (2006). Trungapeptins AC, cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 69, 1539–1542. 10.1021/np050485a [DOI] [PubMed] [Google Scholar]
- Bush K. (2012). Improving known classes of antibiotics: an optimistic approach for the future. Curr. Opin. Pharmacol. 12, 527–534. 10.1016/j.coph.2012.06.003 [DOI] [PubMed] [Google Scholar]
- Butz D., Schmiederer T., Hadatsch B., Wohlleben W., Weber T., Süssmuth R. D. (2008). Module extension of a non-ribosomal peptide synthetase of the glycopeptide antibiotic balhimycin produced by Amycolatopsis balhimycina. Chembiochem 9, 1195–1200. 10.1002/cbic.200800068 [DOI] [PubMed] [Google Scholar]
- Caboche S., Leclère V., Pupin M., Kucherov G., Jacques P. (2010). Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. J. Bacteriol. 192, 5143–5150. 10.1128/JB.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche S., Pupin M., Leclere V., Fontaine A., Jacques P., Kucherov G. (2008). NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 36, D326–D331. 10.1093/nar/gkm792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche S., Pupin M., Leclère V., Jacques P., Kucherov G. (2009). Structural pattern matching of nonribosomal peptides. BMC Struct. Biol. 9:1. 10.1186/1472-6807-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Song Y., Chen Y., Huang H., Zhang W., Ju J. (2012). Cyclic Heptapeptides, Cordyheptapeptides C–E, from the Marine-Derived Fungus Acremonium persicinum SCSIO 115 and Their Cytotoxic Activities. J. Nat. Prod. 75, 1215–1219. 10.1021/np300152d [DOI] [PubMed] [Google Scholar]
- Cho J. Y., Williams P. G., Kwon H. C., Jensen P. R., Fenical W. (2007). Lucentamycins AD, cytotoxic peptides from the marine-derived actinomycete Nocardiopsis lucentensis. J. Nat. Prod. 70, 1321–1328. 10.1021/np070101b [DOI] [PubMed] [Google Scholar]
- Choi H., Mevers E., Byrum T., Valeriote F. A., Gerwick W. H. (2012). Lyngbyabellins K–N from two Palmyra atoll collections of the marine cyanobacterium Moorea bouillonii. European J. Org. Chem., 2012, 5141–5150. 10.1002/ejoc.201200691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condurso H. L., Bruner S. D. (2012). Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat. Prod. Rep. 29, 1099–1110. 10.1039/c2np20023f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L. J., Insua M. M., Baz J. P., Trujillo M., Rodriguez-Mias R. A., Oliveira E., et al. (2006). IB-01212, a new cytotoxic cyclodepsipeptide isolated from the marine fungus Clonostachys sp. ESNA-A009. J. Organ. Chem. 71, 3335–3338. 10.1021/jo051600p [DOI] [PubMed] [Google Scholar]
- Cueto M., Jensen P. R., Fenical W. (2000). N-Methylsansalvamide, a cytotoxic cyclic depsipeptide from a marine fungus of the genus Fusarium. Phytochemistry 55, 223–226. 10.1016/S0031-9422(00)00280-6 [DOI] [PubMed] [Google Scholar]
- Davies-Coleman M. T., Dzeha T. M., Gray C. A., Hess S., Pannell L. K., Hendricks D. T., et al. (2003). Isolation of Homodolastatin 16, a New Cyclic Depsipeptide from a Kenyan Collection of Lyngbya m ajuscula. J. Nat. Prod. 66, 712–715. 10.1021/np030014t [DOI] [PubMed] [Google Scholar]
- Desjardine K., Pereira A., Wright H., Matainaho T., Kelly M., Andersen R. J. (2007). Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: structure elucidation and synthesis. J. Nat. Prod. 70, 1850–1853. 10.1021/np070209r [DOI] [PubMed] [Google Scholar]
- Doroghazi J. R., Metcalf W. W. (2013). Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14:611. 10.1186/1471-2164-14-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim W., Kjer J., El Amrani M., Wray V., Lin W., Ebel R., et al. (2012). Pullularins E and F, two new peptides from the endophytic fungus Bionectria ochroleuca isolated from the mangrove plant Sonneratia caseolaris. Mar. Drugs 10, 1081–1091. 10.3390/md10051081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. J., Marquez B. L., Nogle L. M., Mcphail K., Goeger D. E., Roberts M. A., et al. (2004). Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem. Biol. 11, 817–833. 10.1016/j.chembiol.2004.03.030 [DOI] [PubMed] [Google Scholar]
- Engelhardt K., Degnes K. F., Kemmler M., Bredholt H., Fjaervik E., Klinkenberg G., et al. (2010). Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 76, 4969–4976. 10.1128/AEM.00741-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppelmann K., Doekel S., Marahiel M. A. (2001). Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 276, 34824–34831. 10.1074/jbc.M104456200 [DOI] [PubMed] [Google Scholar]
- Felnagle E. A., Jackson E. E., Chan Y. A., Podevels A. M., Berti A. D., Mcmahon M. D., et al. (2008). Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5, 191–211. 10.1021/mp700137g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenical W. (2006). Marine pharmaceuticals: past, present, and future. Oceanography 2, 110–119. 10.5670/oceanog.2006.74 [DOI] [Google Scholar]
- Fiedler H.-P., Bruntner C., Riedlinger J., Bull A. T., Knutsen G., Goodfellow M., et al. (2008). Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J. Antibiot. 61, 158–163. 10.1038/ja.2008.125 [DOI] [PubMed] [Google Scholar]
- Finking R., Marahiel M. A. (2004). Biosynthesis of nonribosomal peptides1. Annu. Rev. Microbiol. 58, 453–488. 10.1146/annurev.micro.58.030603.123615 [DOI] [PubMed] [Google Scholar]
- Fortman J., Sherman D. H. (2005). Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. ChemBioChem 6, 960–978. 10.1002/cbic.200400428 [DOI] [PubMed] [Google Scholar]
- Fremlin L. J., Piggott A. M., Lacey E., Capon R. J. (2009). Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus versicolor. J. Nat. Prod. 72, 666–670. 10.1021/np800777f [DOI] [PubMed] [Google Scholar]
- García-Fraga B., Da Silva A. F., López-Seijas J., Sieiro C. (2015). A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 93, 84–93. 10.1016/j.bej.2014.09.014 [DOI] [Google Scholar]
- Garo E., Starks C. M., Jensen P. R., Fenical W., Lobkovsky E., Clardy J. (2003). Trichodermamides A and B, cytotoxic modified dipeptides from the marine-derived fungus Trichoderma virens. J. Nat. Prod. 66, 423–426. 10.1021/np0204390 [DOI] [PubMed] [Google Scholar]
- Grünewald J., Marahiel M. A. (2006). Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70, 121–146. 10.1128/MMBR.70.1.121-146.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Cueto M., Jensen P. R., Fenical W., Silverman R. B. (2007). Microsporins A and B: new histone deacetylase inhibitors from the marine-derived fungus Microsporum cf. gypseum and the solid-phase synthesis of microsporin A. Tetrahedron 63, 6535–6541. 10.1016/j.tet.2007.04.025 [DOI] [Google Scholar]
- Gunasekera S. P., Owle C. S., Montaser R., Luesch H., Paul V. J. (2011). Malyngamide 3 and cocosamides A and B from the marine cyanobacterium Lyngbya majuscula from Cocos Lagoon, Guam. J. Nat. Prod. 74, 871–876. 10.1021/np1008015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera S. P., Ross C., Paul V. J., Matthew S., Luesch H. (2008). Dragonamides C and D, linear lipopeptides from the marine cyanobacterium brown Lyngbya polychroa. J. Nat. Prod. 71, 887–890. 10.1021/np0706769 [DOI] [PubMed] [Google Scholar]
- Gutiérrez M., Suyama T. L., Engene N., Wingerd J. S., Matainaho T., Gerwick W. H. (2008). Apratoxin D, a potent cytotoxic cyclodepsipeptide from Papua New Guinea collections of the marine cyanobacteria Lyngbya majuscula and Lyngbya sordida. J. Nat. Prod. 71, 1099–1103. 10.1021/np800121a [DOI] [PubMed] [Google Scholar]
- Han B., Goeger D., Maier C. S., Gerwick W. H. (2005). The Wewakpeptins, Cyclic Depsipeptides from a Papua New Guinea Collection of the Marine Cyanobacterium Lyngbya s emiplena. J. Org. Chem. 70, 3133–3139. 10.1021/jo0478858 [DOI] [PubMed] [Google Scholar]
- Han B., Gross H., Goeger D. E., Mooberry S. L., Gerwick W. H. (2006). Aurilides B and C, Cancer Cell Toxins from a Papua New Guinea Collection of the Marine Cyanobacterium Lyngbya m ajuscula. J. Nat. Prod. 69, 572–575. 10.1021/np0503911 [DOI] [PubMed] [Google Scholar]
- Han J. W., Kim E. Y., Lee J. M., Kim Y. S., Bang E., Kim B. S. (2012). Site-directed modification of the adenylation domain of the fusaricidin nonribosomal peptide synthetase for enhanced production of fusaricidin analogs. Biotechnol. Lett. 34, 1327–1334. 10.1007/s10529-012-0913-8 [DOI] [PubMed] [Google Scholar]
- Hayakawa Y., Hattori Y., Kawasaki T., Kanoh K., Adachi K., Shizuri Y., et al. (2008). Efrapeptin J, a new down-regulator of the molecular chaperone GRP78 from a marine Tolypocladium sp. J. Antibiot. 61, 365–371. 10.1038/ja.2008.51 [DOI] [PubMed] [Google Scholar]
- He F., Bao J., Zhang X.-Y., Tu Z.-C., Shi Y.-M., Qi S.-H. (2013). Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 76, 1182–1186. 10.1021/np300897v [DOI] [PubMed] [Google Scholar]
- Hedgepeth J. (1957). Sandy Beaches. Penrose: Geological Society of America. [Google Scholar]
- Horgen F. D., Kazmierski E. B., Westenburg H. E., Yoshida W. Y., Scheuer P. J. (2002). Malevamide D: isolation and structure determination of an isodolastatin H Analogue from the Marine Cyanobacterium Symploca h ydnoides. J. Nat. Prod. 65, 487–491. 10.1021/np010560r [DOI] [PubMed] [Google Scholar]
- Hoyer K. M., Mahlert C., Marahiel M. A. (2007). The iterative gramicidin s thioesterase catalyzes peptide ligation and cyclization. Chem. Biol. 14, 13–22. 10.1016/j.chembiol.2006.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranueli D., Žučko J., Diminić J., Starčević A. (2010). From DNA sequences to chemical structures–Methods for mining microbial genomic and metagenomic datasets for new natural products. Food Technol. Biotechnol. 48, 234–242. [Google Scholar]
- Huang H., She Z., Lin Y., Vrijmoed L., Lin W. (2007). Cyclic peptides from an endophytic fungus obtained from a mangrove leaf (Kandelia candel). J. Nat. Prod. 70, 1696–1699. 10.1021/np0605891 [DOI] [PubMed] [Google Scholar]
- Hur G. H., Vickery C. R., Burkart M. D. (2012). Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 29, 1074–1098. 10.1039/c2np20025b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A., Yang L., Johnston C., Liu X., Ma B., Magarvey N. A. (2012). Dereplicating nonribosomal peptides using an informatic search algorithm for natural products (iSNAP) discovery. Proc. Natl. Acad. Sci. U.S.A. 109, 19196–19201. 10.1073/pnas.1206376109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K., Nakagawa H., Murakami M. (2000). Microcyclamide, a Cytotoxic Cyclic Hexapeptide from the Cyanobacterium Microcystis a eruginosa. J. Nat. Prod. 63, 1315–1317. 10.1021/np000159p [DOI] [PubMed] [Google Scholar]
- Jiang W., Ye P., Chen C.-T. A., Wang K., Liu P., He S., et al. (2013). Two novel hepatocellular carcinoma cycle inhibitory cyclodepsipeptides from a hydrothermal vent crab-associated fungus Aspergillus clavatus C2WU. Mar. Drugs 11, 4761–4772. 10.3390/md11124761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Vansach T., Yoshida W. Y., Sakamoto B., PöRzgen P., Horgen F. D. (2009). Halogenated fatty acid amides and cyclic depsipeptides from an eastern caribbean collection of the cyanobacterium Lyngbya majuscula†. J. Nat. Prod. 72, 1573–1578. 10.1021/np900173d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J. T., Sturdíkova M., Studik E. (2009). Natural products of marine origin and their perspectives in the discovery of new anticancer drugs. Acta Chim. Slov. 2, 63–74. [Google Scholar]
- Jimeno J., Faircloth G., Sousa-Faro J., Scheuer P., Rinehart K. (2004). New marine derived anticancer therapeutics–a journey from the sea to clinical trials. Mar. Drugs 2, 14–29. 10.3390/md201014 [DOI] [Google Scholar]
- Kalinovskaya N. I., Romanenko L. A., Kalinovsky A. I., Dmitrenok P. S., Dyshlovoy S. A. (2013). A new antimicrobial and anticancer peptide producing by the marine deep sediment strain “Paenibacillus profundus” sp. nov. Sl 79. Nat. Prod. Commun. 8, 381–384. [PubMed] [Google Scholar]
- Kanoh K., Matsuo Y., Adachi K., Imagawa H., Nishizawa M., Shizuri Y. (2005). Mechercharmycins A and B, cytotoxic substances from marine-derived Thermoactinomyces sp. YM3-251. J. Antib. 58, 289–292. 10.1038/ja.2005.36 [DOI] [PubMed] [Google Scholar]
- Kim E. J., Lee J. H., Choi H., Pereira A. R., Ban Y. H., Yoo Y. J., et al. (2012). Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Org. Lett. 14, 5824–5827. 10.1021/ol302575h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-Y., Sohn J. H., Ahn J. S., Oh H. (2009). Alternaramide, a cyclic depsipeptide from the marine-derived fungus Alternaria sp. SF-5016. J. Natl. Products 72, 2065–2068. 10.1021/np900464p [DOI] [PubMed] [Google Scholar]
- Kjaerulff L., Nielsen A., Mansson M., Gram L., Larsen T. O., Ingmer H., et al. (2013). Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar. Drugs, 11, 5051–5062. 10.3390/md11125051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K., Shigemori H., Kobayashi J. I. (2001). Dictyonamides A and B, new peptides from marine-derived fungus. J. Org. Chem. 66, 6189–6192. 10.1021/jo0156767 [DOI] [PubMed] [Google Scholar]
- Kralj A., Kehraus S., Krick A., Van Echten-Deckert G., König G. M. (2007). Two New Depsipeptides from the Marine Fungus Spicellum roseum. Planta Med. 73, 366–371. 10.1055/s-2007-967131 [DOI] [PubMed] [Google Scholar]
- Kubanek J., Jensen P. R., Keifer P. A., Sullards M. C., Collins D. O., Fenical W. (2003). Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. U.S.A. 100, 6916–6921. 10.1073/pnas.1131855100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan J. C., Ratnayake R., Abboud K. A., Paul V. J., Luesch H. (2010). Grassypeptolides A–C, cytotoxic bis-thiazoline containing marine cyclodepsipeptides. J. Org. Chem. 75, 8012–8023. 10.1021/jo1013564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-M., Li J., Zhang P., Hong J.-K., Lee C.-O., Jung J.-H. (2011). A cytotoxic fellutamide analogue from the sponge-derived fungus Aspergillus versicolor. Bullet. Korean Chem. Soc. 32, 3817–3820. 10.5012/bkcs.2011.32.10.3817 [DOI] [Google Scholar]
- Leet J. E., Li W., Ax H. A., Matson J. A., Huang S., Huang R., et al. (2003). Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. II. Isolation, characterization, and structure determination. J. Antib. 56, 232–242. 10.7164/antibiotics.56.232 [DOI] [PubMed] [Google Scholar]
- Li A., Piel J. (2002). A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 9, 1017–1026. 10.1016/S1074-5521(02)00223-5 [DOI] [PubMed] [Google Scholar]
- Li D., Carr G., Zhang Y., Williams D. E., Amlani A., Bottriell H., et al. (2011). Turnagainolides A and B, cyclic depsipeptides produced in culture by a Bacillus sp.: isolation, structure elucidation, and synthesis. J. Nat. Prod. 74, 1093–1099. 10.1021/np200033y [DOI] [PubMed] [Google Scholar]
- Li W., Leet J. E., Ax H. A., Gustavson D. R., Brown D. M., Turner L., et al. (2003). Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. I. Taxonomy, fermentation and biological activities. J. Antib. 56, 226–231. 10.7164/antibiotics.56.226 [DOI] [PubMed] [Google Scholar]
- Liu S., Shen Y. (2011). A new cyclic peptide from the marine fungal strain Aspergillus sp. AF119. Chem. Natl. Compounds 47, 786–788. 10.1007/s10600-011-0059-2 [DOI] [Google Scholar]
- Liu Z., Li G., Mo Z., Mou H. (2013). Molecular cloning, characterization, and heterologous expression of a new κ-carrageenase gene from marine bacterium Zobellia sp. ZM-2. Appl. Microbiol. Biotechnol. 97, 10057–10067. 10.1007/s00253-013-5215-0 [DOI] [PubMed] [Google Scholar]
- Lombó F., Velasco A., Castro A., de la Calle F., Braña A. F., Sánchez-Puelles J. M., et al. (2006). Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. Chembiochem 7, 366–376. 10.1002/cbic.200500325 [DOI] [PubMed] [Google Scholar]
- Luesch H., Pangilinan R., Yoshida W. Y., Moore R. E., Paul V. J. (2001a). Pitipeptolides A and B, new cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 64, 304–307. 10.1021/np000456u [DOI] [PubMed] [Google Scholar]
- Luesch H., Williams P. G., Yoshida W. Y., Moore R. E., Paul V. J. (2002). Ulongamides AF, New β-Amino Acid-Containing Cyclodepsipeptides from Palauan Collections of the Marine Cyanobacterium Lyngbya sp. J. Nat. Prod. 65, 996–1000. 10.1021/np0200461 [DOI] [PubMed] [Google Scholar]
- Luesch H., Yoshida W. Y., Moore R. E., Paul V. J., Corbett T. H. (2001b). Total Structure Determination of Apratoxin A, a Potent Novel Cytotoxin from the Marine Cyanobacterium Lyngbya m ajuscula. J. Am. Chem. Soc. 123, 5418–5423. 10.1021/ja010453j [DOI] [PubMed] [Google Scholar]
- Luesch H., Yoshida W. Y., Moore R. E., Paul V. J., Mooberry S. L. (2000). Isolation, Structure Determination, and Biological Activity of Lyngbyabellin A from the Marine Cyanobacterium Lyngbya m ajuscula. J. Nat. Prod. 63, 611–615. 10.1021/np990543q [DOI] [PubMed] [Google Scholar]
- Luo Y., Enghiad B., Zhao H. (2016). New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat. Prod. Rep. 33, 174–182. 10.1039/C5NP00085H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Wang N., Hu J., Wang S. (2012). Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J. Antibiot. 65, 317–322. 10.1038/ja.2012.19 [DOI] [PubMed] [Google Scholar]
- Mach B., Reich E., Tatum E. (1963). Separation of the biosynthesis of the antibiotic polypeptide tyrocidine from protein biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 50:175. 10.1073/pnas.50.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan J. B., Molinski T. F. (2002). Lobocyclamide B from Lyngbya confervoides. Configuration and Asymmetric Synthesis of β-Hydroxy-α-amino Acids by (-)-Sparteine-Mediated Aldol Addition. Organ. Lett. 4, 1883–1886. 10.1021/ol025876k [DOI] [PubMed] [Google Scholar]
- Manam R. R., Teisan S., White D. J., Nicholson B., Grodberg J., Neuteboom S. T., et al. (2005). Lajollamycin, a Nitro-tetraene Spiro-β-lactone-γ-lactam Antibiotic from the Marine Actinomycete Streptomyces n odosus. J. Nat. Prod. 68, 240–243. 10.1021/np049725x [DOI] [PubMed] [Google Scholar]
- Mankelow D. P., Neilan B. A. (2000). Non-ribosomal peptide antibiotics. Exp. Opin. Ther. Pat. 10, 1583–1591. 10.1517/13543776.10.10.1583 [DOI] [Google Scholar]
- Mansson M., Nielsen A., Kjærulff L., Gotfredsen C. H., Wietz M., Ingmer H., et al. (2011). Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine Photobacterium. Mar. Drugs 9, 2537–2552. 10.3390/md9122537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham A. (2014). Oritavancin: first global approval. Drugs 74, 1823–1828. 10.1007/s40265-014-0295-4 [DOI] [PubMed] [Google Scholar]
- Martín J., Da S Sousa T., Crespo G., Palomo S., González I., Tormo J. R., et al. (2013). Kocurin, the true structure of PM181104, an anti-methicillin-resistant Staphylococcus aureus (MRSA) thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar. Drugs, 11, 387–398. 10.3390/md11020387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru N., Ohno O., Uemura D. (2010). Lyngbyacyclamides A and B, novel cytotoxic peptides from marine cyanobacteria Lyngbya sp. Tetrahedron Lett. 51, 6384–6387. 10.1016/j.tetlet.2010.06.105 [DOI] [Google Scholar]
- Matsunaga S., Fusetani N. (2003). Nonribosomal peptides from marine sponges. Curr. Org. Chem. 7, 945–966. 10.2174/1385272033486648 [DOI] [Google Scholar]
- Matsuo Y., Kanoh K., Yamori T., Kasai H., Katsuta A., Adachi K., et al. (2007). Urukthapelstatin A, a novel cytotoxic substance from marine-derived Mechercharimyces asporophorigenens YM11-542. J. Antibiot. 60, 251–255. 10.1038/ja.2007.30 [DOI] [PubMed] [Google Scholar]
- Matthew S., Salvador L. A., Schupp P. J., Paul V. J., Luesch H. (2010). Cytotoxic halogenated macrolides and modified peptides from the apratoxin-producing marine cyanobacterium Lyngbya bouillonii from Guam. J. Nat. Prod. 73, 1544–1552. 10.1021/np1004032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew S., Schupp P. J., Luesch H. (2008). Apratoxin E, a cytotoxic peptolide from a Guamanian collection of the marine cyanobacterium Lyngbya bouillonii. J. Nat. Prod. 71, 1113–1116. 10.1021/np700717s [DOI] [PubMed] [Google Scholar]
- Medema M. H., Fischbach M. A. (2015). Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648. 10.1038/nchembio.1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H., Blin K., Cimermancic P., De Jager V., Zakrzewski P., Fischbach M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. 10.1093/nar/gkr466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R. A., Goeger D. E., Hills P., Mooberry S. L., Huang N., Romero L. I., et al. (2008). Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 130, 6324–6325. 10.1021/ja801383f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meickle T., Gunasekera S. P., Liu Y., Luesch H., Paul V. J. (2011). Porpoisamides A and B, two novel epimeric cyclic depsipeptides from a Florida Keys collection of Lyngbya sp. Bioorg. Med. Chem. 19, 6576–6580. 10.1016/j.bmc.2011.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meickle T., Matthew S., Ross C., Luesch H., Paul V. (2009). Bioassay-guided isolation and identification of desacetyl-microcolin B from Lyngbya cf. polychroa. Planta Med. 75:1427 10.1055/s-0029-1185675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevers E., Liu W.-T., Engene N., Mohimani H., Byrum T., Pevzner P. A., et al. (2011). Cytotoxic veraguamides, alkynyl bromide-containing cyclic depsipeptides from the marine cyanobacterium cf. Oscillatoria margaritifera. J. Natl. Products 74, 928–936. 10.1021/np200077f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. D., Kauffman C. A., Jensen P. R., Fenical W. (2007). Piperazimycins: cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J. Org. Chem. 72, 323–330. 10.1021/jo061064g [DOI] [PubMed] [Google Scholar]
- Milligan K. E., Marquez B. L., Williamson R. T., Gerwick W. H. (2000). Lyngbyabellin B, a Toxic and Antifungal Secondary Metabolite from the Marine Cyanobacterium Lyngbya m ajuscula. J. Nat. Prod. 63, 1440–1443. 10.1021/np000133y [DOI] [PubMed] [Google Scholar]
- Mitova M., Popov S., De Rosa S. (2004). Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod. 67, 1178–1181. 10.1021/np049900+ [DOI] [PubMed] [Google Scholar]
- Mohimani H., Liu W.-T., Kersten R. D., Moore B. S., Dorrestein P. C., Pevzner P. A. (2014). NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909. 10.1021/np500370c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinski T. F., Reynolds K. A., Morinaka B. I. (2012). Symplocin A, a linear peptide from the bahamian cyanobacterium Symploca sp. configurational analysis of N, N-Dimethylamino acids by chiral-phase HPLC of naphthacyl esters. J. Natl. Products 75, 425–431. 10.1021/np200861n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaser R., Abboud K. A., Paul V. J., Luesch H. (2010). Pitiprolamide, a proline-rich dolastatin 16 analogue from the marine cyanobacterium Lyngbya majuscula from Guam. J. Nat. Prod. 74, 109–112. 10.1021/np1006839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz H. D., Kessler N., Linne U., Eppelmann K., Schwarzer D., Marahiel M. A. (2002). Decreasing the ring size of a cyclic nonribosomal peptide antibiotic by in-frame module deletion in the biosynthetic genes. J. Am. Chem. Soc. 124, 10980–10981. 10.1021/ja027276m [DOI] [PubMed] [Google Scholar]
- Mootz H. D., Schwarzer D., Marahiel M. A. (2000). Construction of hybrid peptide synthetases by module and domain fusions. Proc. Natl. Acad. Sci. U.S.A. 97, 5848–5853. 10.1073/pnas.100075897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Krick A., Kehraus S., Mehner C., Hart M., Küpper F. C., et al. (2006). Brunsvicamides AC: sponge-related cyanobacterial peptides with Mycobacterium tuberculosis protein tyrosine phosphatase inhibitory activity. J. Med. Chem. 49, 4871–4878. 10.1021/jm060327w [DOI] [PubMed] [Google Scholar]
- Nagai K., Kamigiri K., Arao N., Suzumura K.-I., Kawano Y., Yamaoka M., et al. (2003). YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J. Antibiot. 56, 123–128. 10.7164/antibiotics.56.123 [DOI] [PubMed] [Google Scholar]
- Nan H. B., Mcphail K. L., Maier C. S. (2011). Wewakamide A and guineamide G, cyclic depsipeptides from the marine cyanobacteria lyngbya semiplena and lyngbya majuscula. J. Microbiol. Biotechnol. 21, 930–936. 10.4014/jmb.1105.05011 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2004). Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 67, 1216–1238. 10.1021/np040031y [DOI] [PubMed] [Google Scholar]
- Ngwoke K. G., Odimegwu D. C., Esimone C. (2011). Antimicrobial Natural Products. Science agaianst Microbial Pathogens: Communicating Current Research and Technology Advances. Badajoz: FORMATEX. [Google Scholar]
- Nielsen K. F., Larsen T. O. (2015). The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front. Microbiol. 6:71. 10.3389/fmicb.2015.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolouli K., Mossialos D. (2012). Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol. Lett. 34, 1393–1403. 10.1007/s10529-012-0919-2 [DOI] [PubMed] [Google Scholar]
- Oh D.-C., Jensen P. R., Fenical W. (2006). Zygosporamide, a cytotoxic cyclic depsipeptide from the marine-derived fungus Zygosporium masonii. Tetrahedr. Lett. 47, 8625–8628. 10.1016/j.tetlet.2006.08.113 [DOI] [Google Scholar]
- Oh D.-C., Kauffman C. A., Jensen P. R., Fenical W. (2007). Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Natl. Products 70, 515–520. 10.1021/np060381f [DOI] [PubMed] [Google Scholar]
- Okamoto S., Iwasaki A., Ohno O., Suenaga K. (2015). Isolation and structure of kurahyne B and total synthesis of the kurahynes. J. Nat. Prod. 78, 2719–2725. 10.1021/acs.jnatprod.5b00662 [DOI] [PubMed] [Google Scholar]
- Oku N., Adachi K., Matsuda S., Kasai H., Takatsuki A., Shizuri Y. (2008a). Ariakemicins A and B, novel polyketide-peptide antibiotics from a marine gliding bacterium of the genus Rapidithrix. Org. Lett. 10, 2481–2484. 10.1021/ol8007292 [DOI] [PubMed] [Google Scholar]
- Oku N., Kawabata K., Adachi K., Katsuta A., Shizuri Y. (2008b). Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J. Antibiot. 61, 11–17. 10.1038/ja.2008.103 [DOI] [PubMed] [Google Scholar]
- Ongley S. E., Bian X., Neilan B. A., Müller R. (2013). Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 30, 1121–1138. 10.1039/c3np70034h [DOI] [PubMed] [Google Scholar]
- Organization W. H. (2014). Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization. [Google Scholar]
- Owen J. G., Reddy B. V. B., Ternei M. A., Charlop-Powers Z., Calle P. Y., Kim J. H., et al. (2013). Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. U.S.A. 110, 11797–11802. 10.1073/pnas.1222159110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A., Cao Z., Murray T. F., Gerwick W. H. (2009). Hoiamide a, a sodium channel activator of unusual architecture from a consortium of two papua new Guinea cyanobacteria. Chem. Biol. 16, 893–906. 10.1016/j.chembiol.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesic A., Baumann H. I., Kleinschmidt K., Ensle P., Wiese J., Süssmuth R. D., et al. (2013). Champacyclin, a New Cyclic Octapeptide from Streptomyces Strain C42 Isolated from the Baltic Sea. Mar. Drugs 11, 4834–4857. 10.3390/md11124834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit K., Biard J.-F. (2013). Marine natural products and related compounds as anticancer agents: an overview of their clinical status. Anticancer Agents Med. Chem. 13, 603–631. 10.2174/1871520611313040010 [DOI] [PubMed] [Google Scholar]
- Pettit G. R., Knight J. C., Herald D. L., Pettit R. K., Hogan F., Mukku V. J., et al. (2009). Antineoplastic Agents. 570. Isolation and Structure Elucidation of Bacillistatins 1 and 2 from a Marine Bacillus silvestris†,‡. J. Natl. Products 72, 366–371. 10.1021/np800603u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig S., Stubbs M. T. (2012). Flexing and stretching in nonribosomal Peptide synthetases. Chem. Biol. 19, 167–169. 10.1016/j.chembiol.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Popplewell W. L., Ratnayake R., Wilson J. A., Beutler J. A., Colburn N. H., Henrich C. J., et al. (2011). Grassypeptolides F and G, cyanobacterial peptides from Lyngbya majuscula. J. Nat. Prod. 74, 1686–1691. 10.1021/np2005083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto C., García-Estrada C., Lorenzana D., Martín J. F. (2012). NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28, 426–427. 10.1093/bioinformatics/btr659 [DOI] [PubMed] [Google Scholar]
- Prompanya C., Fernandes C., Cravo S., Pinto M. M., Dethoup T., Silva A., et al. (2015). A new cyclic hexapeptide and a new isocoumarin derivative from the marine sponge-associated fungus Aspergillus similanensis KUFA 0013. Mar. Drugs 13, 1432–1450. 10.3390/md13031432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruksakorn P., Arai M., Kotoku N., Vilcheze C., Baughn A. D., Moodley P., et al. (2010). Trichoderins, novel aminolipopeptides from a marine sponge-derived Trichoderma sp., are active against dormant mycobacteria. Bioorg. Med. Chem. Lett. 20, 3658–3663. 10.1016/j.bmcl.2010.04.100 [DOI] [PubMed] [Google Scholar]
- Qin Y., Huang Z., Liu Z. (2014). A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: molecular cloning, heterologous expression and biochemical characterization. Extremophiles 18, 271–281. 10.1007/s00792-013-0614-9 [DOI] [PubMed] [Google Scholar]
- Reddy L., Odhav B., Bhoola K. (2003). Natural products for cancer prevention: a global perspective. Pharmacol. Ther. 99, 1–13. 10.1016/S0163-7258(03)00042-1 [DOI] [PubMed] [Google Scholar]
- Röttig M., Medema M. H., Blin K., Weber T., Rausch C., Kohlbacher O. (2011). NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367. 10.1093/nar/gkr323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. D., GrüSchow S., Cairns N., Goss R. J. (2010). Gene expression enabling synthetic diversification of natural products: chemogenetic generation of pacidamycin analogs. J. Am. Chem. Soc. 132, 12243–12245. 10.1021/ja1060406 [DOI] [PubMed] [Google Scholar]
- Rungprom W., Siwu E. R., Lambert L. K., Dechsakulwatana C., Barden M. C., Kokpol U., et al. (2008). Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 64, 3147–3152. 10.1016/j.tet.2008.01.089 [DOI] [Google Scholar]
- Salvador L. A., Biggs J. S., Paul V. J., Luesch H. (2011). Veraguamides A– G, cyclic hexadepsipeptides from a dolastatin 16-producing cyanobacterium Symploca cf. hydnoides from Guam. J. Natl. Products 74, 917–927. 10.1021/np200076t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaala L. A., Youssef D. T., Mcphail K. L., Elbandy M. (2013). Malyngamide 4, a new lipopeptide from the Red Sea marine cyanobacterium Moorea producens (formerly Lyngbya majuscula). Phytochem. Lett. 6, 183–188. 10.1016/j.phytol.2013.01.002 [DOI] [Google Scholar]
- Sieber S. A., Marahiel M. A. (2003). Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J. Bacteriol. 185, 7036–7043. 10.1128/JB.185.24.7036-7043.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons T. L., Mcphail K. L., Ortega-Barría E., Mooberry S. L., Gerwick W. H. (2006). Belamide A, a new antimitotic tetrapeptide from a Panamanian marine cyanobacterium. Tetrahedron Lett. 47, 3387–3390. 10.1016/j.tetlet.2006.03.082 [DOI] [Google Scholar]
- Simmons T. L., Nogle L. M., Media J., Valeriote F. A., Mooberry S. L., Gerwick W. H. (2009). Desmethoxymajusculamide C, a cyanobacterial depsipeptide with potent cytotoxicity in both cyclic and ring-opened forms. J. Nat. Prod. 72, 1011–1016. 10.1021/np9001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitachitta N., Williamson R. T., Gerwick W. H. (2000). Yanucamides A and B, Two New Depsipeptides from an Assemblage of the Marine Cyanobacteria Lyngbya m ajuscula and Schizothrix Species. J. Nat. Prod. 63, 197–200. 10.1021/np990466z [DOI] [PubMed] [Google Scholar]
- Speitling M., Smetanina O. F., Kuznetsova T. A., Laatsch H. (2007). Bromoalterochromides A and A′, Unprecedented Chromopeptides from a Marine Pseudoalteromonas maricaloris Strain KMM 636T†. J. Antibiot. 60, 36–42. 10.1038/ja.2007.5 [DOI] [PubMed] [Google Scholar]
- Strieker M., Tanovic A., Marahiel M. A. (2010). Nonribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 20, 234–240. 10.1016/j.sbi.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Sun P., Maloney K. N., Nam S.-J., Haste N. M., Raju R., Aalbersberg W., et al. (2011). Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 19, 6557–6562. 10.1016/j.bmc.2011.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Tian L., Huang Y.-F., Sha Y., Pei Y.-H. (2006). A new cyclotetrapeptide from marine fungus Trichoderma reesei. Die Pharm. Int. J. Pharm. Sci. 61, 809–810. [PubMed] [Google Scholar]
- Suzumura K.-I., Yokoi T., Funatsu M., Nagai K., Tanaka K., Zhang H., et al. (2003). YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge II. Structure elucidation. J. Antibiot. 56, 129–134. 10.7164/antibiotics.56.129 [DOI] [PubMed] [Google Scholar]
- Tan L. T., Cheng X. C., Jensen P. R., Fenical W. (2003a). Scytalidamides A and B, new cytotoxic cyclic heptapeptides from a marine fungus of the genus Scytalidium. J. Org. Chem. 68, 8767–8773. 10.1021/jo030191z [DOI] [PubMed] [Google Scholar]
- Tan L. T., Okino T., Gerwick W. H. (2013). Bouillonamide: a mixed polyketide–peptide cytotoxin from the Marine Cyanobacterium Moorea bouillonii. Mar. Drugs 11, 3015–3024. 10.3390/md11083015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. T., Sitachitta N., Gerwick W. H. (2003b). The Guineamides, Novel Cyclic Depsipeptides from a Papua New Guinea Collection of the Marine Cyanobacterium Lyngbya m ajuscula. J. Nat. Prod. 66, 764–771. 10.1021/np020492o [DOI] [PubMed] [Google Scholar]
- Tan R. X., Jensen P. R., Williams P. G., Fenical W. (2004). Isolation and Structure Assignments of Rostratins AD, Cytotoxic Disulfides Produced by the Marine-Derived Fungus Exserohilum r ostratum. J. Nat. Prod. 67, 1374–1382. 10.1021/np049920b [DOI] [PubMed] [Google Scholar]
- Tang L., Shah S., Chung L., Carney J., Katz L., Khosla C., et al. (2000). Cloning and heterologous expression of the epothilone gene cluster. Science 287, 640–642. 10.1126/science.287.5453.640 [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Nunnery J. K., Engene N., Esquenazi E., Byrum T., Dorrestein P. C., et al. (2009). Palmyramide A, a Cyclic Depsipeptide from a Palmyra Atoll Collection of the Marine Cyanobacterium Lyngbya majuscula†. J. Nat. Prod. 73, 393–398. 10.1021/np900428h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taori K., Liu Y., Paul V. J., Luesch H. (2009). Combinatorial strategies by marine cyanobacteria: symplostatin 4, an antimitotic natural dolastatin 10/15 hybrid that synergizes with the coproduced HDAC inhibitor largazole. Chembiochem 10, 1634–1639. 10.1002/cbic.200900192 [DOI] [PubMed] [Google Scholar]
- Taori K., Matthew S., Rocca J. R., Paul V. J., Luesch H. (2007). Lyngbyastatins 5–7, Potent Elastase Inhibitors from Floridian Marine Cyanobacteria, Lyngbya spp. J. Nat. Prod. 70, 1593–1600. 10.1021/np0702436 [DOI] [PubMed] [Google Scholar]
- Tareq F. S., Kim J. H., Lee M. A., Lee H.-S., Lee Y.-J., Lee J. S., et al. (2012). Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 14, 1464–1467. 10.1021/ol300202z [DOI] [PubMed] [Google Scholar]
- Teruya T., Sasaki H., Fukazawa H., Suenaga K. (2009). Bisebromoamide, a potent cytotoxic peptide from the marine cyanobacterium Lyngbya sp.: isolation, stereostructure, and biological activity. Org. Lett. 11, 5062–5065. 10.1021/ol9020546 [DOI] [PubMed] [Google Scholar]
- Thiericke R., Rohr J. (1993). Biological variation of microbial metabolites by precursor-directed biosynthesis. Nat. Prod. Rep. 10, 265–289. 10.1039/np9931000265 [DOI] [PubMed] [Google Scholar]
- Thornburg C. C., Thimmaiah M., Shaala L. A., Hau A. M., Malmo J. M., Ishmael J. E., et al. (2011). Cyclic depsipeptides, grassypeptolides D and E and Ibu-epidemethoxylyngbyastatin 3, from a Red Sea Leptolyngbya cyanobacterium. J. Nat. Prod. 74, 1677–1685. 10.1021/np200270d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Puddick J., Prinsep M. R., Lee P. P. F., Tan L. T. (2008). Hantupeptin A, a cytotoxic cyclic depsipeptide from a Singapore collection of Lyngbya majuscula. J. Nat. Prod. 72, 29–32. 10.1021/np800448t [DOI] [PubMed] [Google Scholar]
- Tripathi A., Puddick J., Prinsep M. R., Lee P. P. F., Tan L. T. (2010). Hantupeptins B and C, cytotoxic cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 71, 307–311. 10.1016/j.phytochem.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Tripathi A., Puddick J., Prinsep M. R., Rottmann M., Chan K. P., Chen D. Y.-K., et al. (2011). Lagunamide C, a cytotoxic cyclodepsipeptide from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 72, 2369–2375. 10.1016/j.phytochem.2011.08.019 [DOI] [PubMed] [Google Scholar]
- Ugai T., Minami A., Gomi K., Oikawa H. (2016). Genome mining approach for harnessing the cryptic gene cluster in Alternaria solani: production of PKS–NRPS hybrid metabolite, didymellamide B. Tetrahedr. Lett. 57, 2793–2796. 10.1016/j.tetlet.2016.05.043 [DOI] [Google Scholar]
- Um S., Choi T. J., Kim H., Kim B. Y., Kim S.-H., Lee S. K., et al. (2013). Ohmyungsamycins A and B: Cytotoxic and Antimicrobial Cyclic Peptides Produced by Streptomyces sp. from a Volcanic Island. J. Organic Chem. 78, 12321–12329. 10.1021/jo401974g [DOI] [PubMed] [Google Scholar]
- Vignesh S., Raja A., James R. A. (2011). Marine drugs: implication and future studies. Int. J. Pharmacol. 7, 22–30. 10.3923/ijp.2011.22.30 [DOI] [Google Scholar]
- Vinothkumar S., Parameswaran P. (2013). Recent advances in marine drug research. Biotechnol. Adv. 31, 1826–1845. 10.1016/j.biotechadv.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Weist S., Kittel C., Bischoff D., Bister B., Pfeifer V., Nicholson G. J., et al. (2004). Mutasynthesis of glycopeptide antibiotics: variations of vancomycin's AB-ring amino acid 3, 5-dihydroxyphenylglycine. J. Am. Chem. Soc. 126, 5942–5943. 10.1021/ja0499389 [DOI] [PubMed] [Google Scholar]
- Williams D. E., Dalisay D. S., Patrick B. O., Matainaho T., Andrusiak K., Deshpande R., et al. (2011). Padanamides A and B, highly modified linear tetrapeptides produced in culture by a Streptomyces sp. isolated from a marine sediment. Organ. Lett. 13, 3936–3939. 10.1021/ol2014494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. G., Moore R. E., Paul V. J. (2003a). Isolation and structure determination of lyngbyastatin 3, a lyngbyastatin 1 homologue from the marine cyanobacterium lyngbya m ajuscula. Determination of the configuration of the 4-Amino-2, 2-dimethyl-3-oxopentanoic acid unit in majusculamide C, dolastatin 12, lyngbyastatin 1, and lyngbyastatin 3 from cyanobacteria. J. Natl. Prod. 66, 1356–1363. 10.1021/np0302145 [DOI] [PubMed] [Google Scholar]
- Williams P. G., Yoshida W. Y., Moore R. E., Paul V. J. (2002a). Isolation and structure determination of obyanamide, a novel cytotoxic cyclic depsipeptide from the marine cyanobacterium Lyngbya confervoides. J. Nat. Prod. 65, 29–31. 10.1021/np0102253 [DOI] [PubMed] [Google Scholar]
- Williams P. G., Yoshida W. Y., Moore R. E., Paul V. J. (2002b). Tasiamide, a cytotoxic peptide from the marine cyanobacterium Symploca sp. J. Nat. Prod. 65, 1336–1339. 10.1021/np020184q [DOI] [PubMed] [Google Scholar]
- Williams P. G., Yoshida W. Y., Moore R. E., Paul V. J. (2003b). The isolation and structure elucidation of tasiamide B, a 4-amino-3-hydroxy-5-phenylpentanoic acid containing peptide from the marine cyanobacterium Symploca sp. J. Nat. Prod. 66, 1006–1009. 10.1021/np030114z [DOI] [PubMed] [Google Scholar]
- Williams P. G., Yoshida W. Y., Moore R. E., Paul V. J. (2003c). Tasipeptins A and B: new cytotoxic depsipeptides from the marine cyanobacterium Symploca sp. J. Nat. Prod. 66, 620–624. 10.1021/np020582t [DOI] [PubMed] [Google Scholar]
- Williams P. G., Yoshida W. Y., Quon M. K., Moore R. E., Paul V. J. (2003d). Ulongapeptin, a cytotoxic cyclic depsipeptide from a Palauan marine cyanobacterium Lyngbya sp. J. Nat. Prod. 66, 651–654. 10.1021/np030050s [DOI] [PubMed] [Google Scholar]
- Winn M., Fyans J., Zhuo Y., Micklefield J. (2016). Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 33, 317–347. 10.1039/C5NP00099H [DOI] [PubMed] [Google Scholar]
- Wu X., Bu X., Wong K. M., Yan W., Guo Z. (2003). Biomimetic synthesis of gramicidin S and analogues by enzymatic cyclization of linear precursors on solid support. Org. Lett. 5, 1749–1752. 10.1021/ol034437y [DOI] [PubMed] [Google Scholar]
- Wyche T. P., Hou Y., Vazquez-Rivera E., Braun D., Bugni T. S. (2012). Peptidolipins B–F, Antibacterial Lipopeptides from an Ascidian-Derived Nocardia sp. J. Nat. Prod. 75, 735–740. 10.1021/np300016r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Tan R.-X., Wang Q., Huang W.-Y., Yin Y.-X. (2002). Antifungal cyclopeptides from Halobacillus litoralis YS3106 of marine origin. Tetrahedron Lett. 43, 6545–6548. 10.1016/S0040-4039(02)01458-2 [DOI] [Google Scholar]
- Ye P., Shen L., Jiang W., Ye Y., Chen C.-T. A., Wu X., et al. (2014). Zn-Driven Discovery of a Hydrothermal Vent Fungal Metabolite Clavatustide C, and an experimental study of the anti-cancer mechanism of clavatustide B. Mar. Drugs 12, 3203–3217. 10.3390/md12063203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Chen Y., Zhang L., Wang Y., Zabriskie T. M. (2010). Enduracidin analogues with altered halogenation patterns produced by genetically engineered strains of Streptomyces fungicidicus. J. Nat. Prod. 73, 583–589. 10.1021/np900710q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Lang G., Kajahn I., Schmaljohann R., Imhoff J. F. (2008). Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J. Nat. Prod. 71, 1052–1054. 10.1021/np070580e [DOI] [PubMed] [Google Scholar]
- Zhang H. L., Hua H. M., Pei Y. H., Yao X. S. (2004). Three new cytotoxic cyclic acylpeptides from marine Bacillus sp. Chem. Pharm. Bullet. 52, 1029–1030. 10.1248/cpb.52.1029 [DOI] [PubMed] [Google Scholar]
- Zheng J., Xu Z., Wang Y., Hong K., Liu P., Zhu W. (2010). Cyclic tripeptides from the halotolerant fungus Aspergillus sclerotiorum PT06-1. J. Nat. Prod. 73, 1133–1137. 10.1021/np100198h [DOI] [PubMed] [Google Scholar]
- Zheng J., Zhu H., Hong K., Wang Y., Liu P., Wang X., et al. (2009). Novel cyclic hexapeptides from marine-derived fungus, Aspergillus sclerotiorum PT06-1. Org. Lett. 11, 5262–5265. 10.1021/ol902197z [DOI] [PubMed] [Google Scholar]
- Zhou L. N., Gao H. Q., Cai S. X., Zhu T. J., Gu Q. Q., Li D. H. (2011). Two New Cyclic Pentapeptides from the Marine-Derived Fungus Aspergillus versicolor. Helv. Chim. Acta 94, 1065–1070. 10.1002/hlca.201000408 [DOI] [Google Scholar]
- Zhou X., Huang H., Chen Y., Tan J., Song Y., Zou J., et al. (2012). Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 75, 2251–2255. 10.1021/np300554f [DOI] [PubMed] [Google Scholar]
- Ziemert N., Podell S., Penn K., Badger J. H., Allen E., Jensen P. R. (2012). The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7:e34064. 10.1371/journal.pone.0034064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.