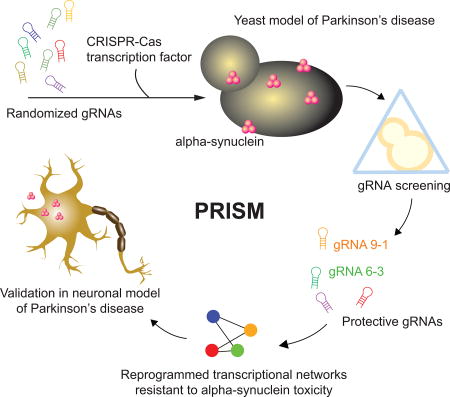
SUMMARY
The genome-wide perturbation of transcriptional networks with CRISPR-Cas technology has primarily involved systematic and targeted gene modulation. Here, we developed PRISM (Perturbing Regulatory Interactions by Synthetic Modulators), a screening platform that uses randomized CRISPR-Cas transcription factors (crisprTFs) to globally perturb transcriptional networks. By applying PRISM to a yeast model of Parkinson’s disease (PD), we identified guide RNAs (gRNAs) that modulate transcriptional networks and protect cells from alpha-synuclein (αSyn) toxicity. One gRNA identified in this screen outperformed the most protective suppressors of αSyn toxicity reported previously, highlighting PRISM’s ability to identify modulators of important phenotypes. Gene expression profiling revealed genes differentially modulated by this strong protective gRNA that rescued yeast from αSyn toxicity when overexpressed. Human homologs of top-ranked hits protected against αSyn-induced cell death in a human neuronal PD model. Thus, high-throughput and unbiased perturbation of transcriptional networks via randomized crisprTFs can reveal complex biological phenotypes and effective disease modulators.
Graphical abstract
INTRODUCTION
The systematic perturbation of transcriptional networks enables the elucidation of gene functions and regulatory networks that underlie biological processes. Current methods of modulating transcriptional networks mainly rely on targeted single-gene overexpression, knockout, and knockdown (Boutros and Ahringer, 2008; Carpenter and Sabatini, 2004; Costanzo et al., 2016; Forsburg, 2001). With the advent of artificial transcription factors, such as zinc-finger-, Transcriptional Activator-Like Effector (TALE)-, and CRISPR-Cas9-based transcription factors (crisprTFs), customized transcriptional perturbations are possible (Blancafort et al., 2005; Carroll, 2014; Park et al., 2003; Zhang et al., 2011). For example, crisprTF-based platforms enable bi-directional gene activation and repression in eukaryotic systems (Chavez et al., 2015; Farzadfard et al., 2013; Gilbert et al., 2013; Mali et al., 2013; Nishimasu et al., 2014; Zalatan et al., 2015) and have been used for genome-wide targeted screens owing to the ease of designing and synthesizing guide RNAs (gRNAs) (Gilbert et al., 2014; Horlbeck et al., 2016; Konermann et al., 2015). In addition, strategies for higher-order perturbations using barcoded combinatorial genetic screens in human cells have been adapted to be compatible with CRISPR-Cas9 screens (Wong et al., 2015; Wong et al., 2016). Existing CRISPR-Cas9-based screening strategies rely on gRNAs designed to target individual genes while minimizing off-target effects (Cencic et al., 2014; Frock et al., 2015; Gilbert et al., 2014; O'Geen et al., 2015; Parnas et al., 2015; Shalem et al., 2014; Wang et al., 2014; Wu et al., 2014). Although these technologies provide powerful strategies for perturbing individual genes, they may not be suitable for global or combinatorial perturbation of transcriptional networks. Many complex diseases, as well as treatments required to counteract those conditions, may involve simultaneous or dynamic changes in the expression levels of many genes, which are not accessible by screens that target genes one at a time (Khurana et al., 2017; Yeger-Lotem et al., 2009).
To address this limitation, we explored the use of randomized gRNAs and crisprTFs in an approach called PRISM (Perturbing Regulatory Interactions by Synthetic Modulators) in order to effect global transcriptional perturbations conferring enhanced cellular resistance to alpha-synuclein (αSyn). The aggregation of misfolded αSyn in intraneuronal Lewy bodies is one of the pathological hallmarks of Parkinson’s disease (PD) (Goedert et al., 2013; Spillantini et al., 1997). The overexpression of αSyn in various eukaryotic model organisms has been used to elucidate the complex cellular processes associated with PD (Lashuel et al., 2013; Maries et al., 2003; Wong and Krainc, 2017). Because of its conserved molecular mechanisms and the availability of genetic tools, Saccharomyces cerevisiae has been extensively used as a model to systematically study and identify genes involved in neurodegenerative diseases such as PD and Alzheimer's disease (Khurana and Lindquist, 2010; Tardiff et al., 2014).
Here, we demonstrate that one of the strongest protective gRNAs identified in our PRISM screens outperformed any individual overexpressed gene that we tested in suppressing αSyn toxicity, including the strongest protective genes found in previous genome-wide screens (Cooper et al., 2006; Gitler et al., 2009; Tenreiro et al., 2016; Yeger-Lotem et al., 2009). These results highlight that randomized gRNA/crisprTF perturbations can achieve powerful phenotypic modulation compared with other targeted gene perturbation methods.
RANDOMIZED gRNA SCREENING DESIGN
We cloned a dCas9-VP64 expression cassette under the control of a doxycycline (Dox)-inducible (Tet-ON) promoter. To build the yeast screen strain, this construct was integrated into the genome of an αSyn-expressing S. cerevisiae strain (hereafter referred to as the yeast parental strain) in which two copies of the human wild-type αSyn (SNCA) gene fused to yellow fluorescent protein (YFP) are overexpressed under the control of a galactose (Gal)-inducible promoter (Cooper et al., 2006) (Figure 1A). Both the yeast parental strain and the screen strains showed significant cellular growth defects in the presence of Gal due to overexpression of αSyn. The expression of dCas9-VP64 with no gRNA in the screen strain did not interfere with normal cellular growth or αSyn-associated toxicity (Figure S1A).
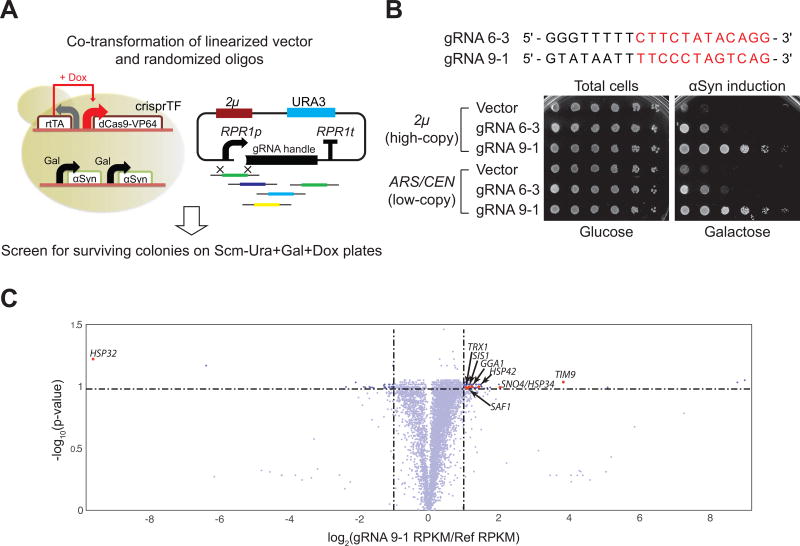
Figure 1. Randomized gRNA/crisprTF screens identify genetic modifiers of αSyn toxicity in S. cerevisiae.
(A) Schematic illustration of the engineered yeast screen strain expressing αSyn and crisprTF (left) and the strategy used for building the randomized gRNA library (right). See Methods section and Figures S1–S2 for details. (B) Sequences of the two identified gRNAs (designated as gRNA 6-3 and 9-1) that could suppress αSyn-mediated toxicity. 5-fold serial dilutions of saturated cultures were spotted on Scm (Synthetic complete media)−Uracil (Ura)+Glucose+Doxycycline (Dox) plates to quantify the total number of viable cells and Scm−Ura+Galactose (Gal)+Dox plates to score cell viability upon αSyn induction. gRNA 9-1 is a strong suppressor of αSyn toxicity while gRNA 6-3 is a moderate suppressor. Both gRNAs performed better than the negative control (empty vector), and suppression levels were independent of gRNA plasmid copy number. See also Figure S1. (C) The transcriptome analysis of the screen strain harboring gRNA 9-1 in comparison with the reference strain (screen strain with no gRNA) represented as a volcano plot (x-axis: fold change versus y-axis: statistical significance). A list of differentially expressed genes is provided in Table S2.
A randomized gRNA-expressing plasmid library was built by co-transforming the screen strain with linearized high-copy 2μ plasmids, flanked by the RPR1 promoter (RPR1p) and gRNA handle at the ends, and a randomized oligo library encoding 20-mer randomized nucleotides flanked by homology arms to the ends of the vector (Figure 1A). We observed approximately 100 million colony forming units (CFUs) per library transformation, which is comparable with the theoretical diversity of the seed sequence, the protospacer adjacent motif (PAM)-proximal 12 nucleotides (412 = ~1.67 ×107). Library representation and sequence distributions were determined by deep sequencing (Figures S2A–S2G). After cells were transformed with the library, they were recovered in liquid culture with Dox (1 µg/mL) for 12 hours to amplify the library and induce crisprTF expression. The cultures were then plated on synthetic complete media (Scm)−Uracil (Ura)+Gal+Dox plates, and gRNAs from surviving colonies were characterized by colony PCR followed by Sanger sequencing.
RESULTS
gRNA Suppressors of αSyn Toxicity were Identified by a Randomized Screen in Saccharomyces cerevisiae
To validate the activity of the identified gRNAs, they were re-cloned in both high-copy 2μ and low-copy ARS/CEN plasmids, and transformed back into both parental and screen strains. We confirmed that two gRNAs (designated as gRNA 6-3 and gRNA 9-1), expressed from either high-copy and low-copy plasmids, could rescue the screen strain from αSyn toxicity (Figure 1B). gRNA 6-3 moderately suppressed αSyn toxicity whereas gRNA 9-1 strongly suppressed it; gRNA 9-1 was thus chosen for further characterization. Although no perfect match between the identified gRNAs and the yeast genome was found, relaxing the search criteria (to find up to two mismatches inside the seed region) revealed the presence of a few dozen sites that could potentially serve as off-target binding sites of these gRNAs, including one in the GAL4 gene (Table S1). As additional controls, we confirmed that gRNA 9-1-mediated suppression of αSyn toxicity relied on the presence of dCas9-VP64 (Figure S1B) and that GAL4 and αSyn expression levels were not directly affected by gRNA 9-1/crisprTF (Figures S1E–S1F). Furthermore, we re-encoded the putative gRNA 9-1 off-target binding site in GAL4 so that there were five matches in the seed sequence and found that gRNA 9-1 preserved its ability to rescue the screen yeast strain from αSyn toxicity (Figures S1G–S1I).
Gene Expression Profiling of αSyn-resistant Cells by gRNA 9-1/crisprTF Revealed Suppressors of αSyn Toxicity
We compared the transcriptome of screen cells expressing gRNA 9-1 and dCas9-VP64 to that of cells expressing dCas9-VP64 but no gRNA by using RNA sequencing to map transcriptional perturbations enacted by the αSyn-protective crisprTF (Figure 1C and Figures S2H–S2I). We identified 114 differentially expressed genes with at least two-fold changes in mRNA expression levels compared with the non-gRNA control (false discovery rate (FDR)-adjusted p-value ≤ 0.1) (Table 1 and Table S2). Most of these genes (93%) have not been previously identified in single gene knockout or overexpression screens as suppressors of αSyn toxicity (Cooper et al., 2006; Gitler et al., 2009; Yeger-Lotem et al., 2009). Intriguingly, they were enriched in Gene Ontology (GO) categories including protein quality control, ER/Golgi trafficking, lipid metabolism, mitochondrial function, and stress responses (Table S3). Almost all of the newly identified genes exhibited only modest changes in gene expression (109 out of 114 genes had fold-changes <5).
Table 1.
A summary of top-ranked genes that are differentially regulated by gRNA 9-1 and that suppressed αSyn toxicity in yeast when overexpressed. A complete list of genes differentially modulated by gRNA 9-1 is provided in Table S2.
| Yeast Gene | Human Homologs | Log2(fold change) |
Survival Score |
Fluorescent Foci Score |
Biological Function |
|---|---|---|---|---|---|
| SNO4/HSP34 | DJ-1/PARK7 | 2.035 | 6 | 7 | Chaperone and cysteine protease |
|
| |||||
| HSP32 | DJ-1/PARK7 | −9.593 | 6 | 7 | Chaperone and cysteine protease |
|
| |||||
| HSP42 | HSPB1, HSPB3, HSPB6, HSPB7, HSPB8, HSPB9 | 1.434 | 5 | 6 | Chaperone |
|
| |||||
| SIS1 | DNAJB1-B9 | 1.154 | 6 | 3 | Chaperone |
|
| |||||
| GGA1 | GGA1, GGA2, GGA3 | 1.241 | 6 | 6 | ER to Golgi vesicular trafficking |
|
| |||||
| SRN2 | 1.031 | 6 | 4 | Ubiquitin-dependent protein sorting | |
|
| |||||
| SAF1 | ALS2, RCC1 | 1.180 | 6 | 4 | Proteasome-dependent degradation |
|
| |||||
| TRX1 | TXN, TXNDC2, TXNDC8 | 1.072 | 6 | 5 | Thioredoxin |
|
| |||||
| TIM9 | TIMM9 | 3.846 | 6 | 5 | Mitochondrial intermembrane protein |
|
| |||||
| OXR1 | OXR1, NCOA7, TLDC2 | 1.003 | 5 | 3 | Oxidative damage resistance |
|
| |||||
| STF2 | SERBP1, HABP4 | 2.004 | 6 | 3 | mRNA stabilization |
|
| |||||
| gRNA 9-1 | 6 | 10 | |||
| UBP3 | 6 | 7 | |||
| Vector | 1 | 1 | |||
We systematically tested the effects of our differentially expressed genes on αSyn toxicity in the screen strain by overexpressing 95 of them that were found in the Yeast ORF Collection (Open Biosystems). Overexpression of 57 out of 95 (60%) genes (13 down-regulated and 44 up-regulated by gRNA 9-1/crisprTF) significantly suppressed αSyn toxicity (Figure S3A, summarized in Table S2; representative candidates are shown in Figure 2A), whereas only 5 out of 34 (14.7%) genes randomly chosen from the Yeast ORF Collection suppressed αSyn toxicity (Figure S3B and Table S4). Thus, our randomized gRNA/crisprTF screening approach enriched the search for αSyn-toxicity suppressors.
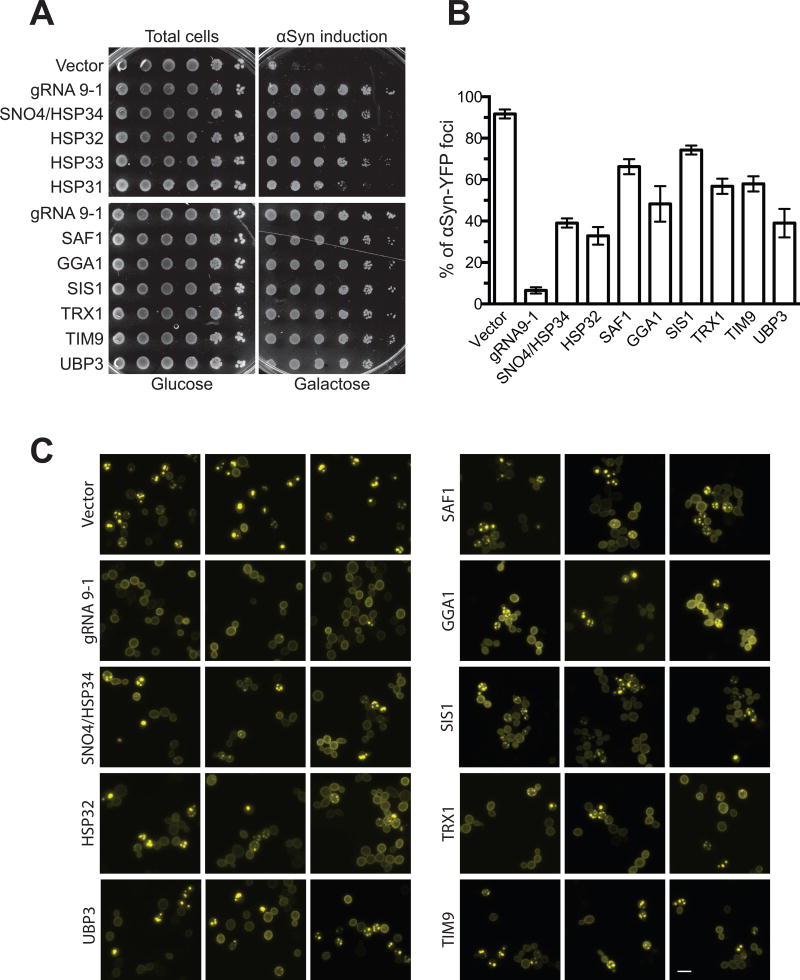
Figure 2. Overexpressing genes identified from the gRNA 9-1/crisprTF screen rescues αSyn-associated cellular defects in yeast.
(A) Survival of the screen strain harboring gRNA 9-1 (‘gRNA 9-1’) compared to cells expressing the empty vector (‘Vector’) and those overexpressing HSP31–34 (heat shock proteins) (top) as well as top-ranked αSyn suppressors identified in the screen (bottom). UBP3, a known strong αSyn suppressor, was used as a positive control. (B) Quantification of αSyn-YFP foci in the screen strain harboring either no gRNA or gRNA 9-1, or cells harboring plasmids that overexpress the indicated genes. Cytoplasmic YFP foci represent αSyn aggregates produced as a result of defects in vesicular trafficking. Cells expressing crisprTF and gRNA 9-1 robustly inhibited αSyn aggregates, evidenced by the absence of cytoplasmic YFP foci in these samples. Cells overexpressing UBP3 were used as a positive control in this assay. Data were presented as mean ± SEM of three biological replicates. (C) Representative images of αSyn-expressing cells mentioned in b. Bar = 10 µm. See also Figures S3–S4.
Furthermore, there was no significant correlation between observed αSyn expression levels and toxicity (Figures S1E–S1F). UBP3 (ubiquitin-specific protease) was used as a positive control. UBP3, previously shown to be a strong suppressor of αSyn toxicity, is known to participate in the degradation of misfolded proteins in the vesicular trafficking processes (Chung et al., 2013; Cooper et al., 2006; Tardiff et al., 2013). We found that 29 genes whose expression was modulated by gRNA 9-1 protected against αSyn-toxicity similarly to or better than UBP3. Notably, gRNA 9-1 alone outperformed the overexpression of any single gene in abrogating αSyn-associated phenotypes based on cell viability assay results, shown in Figure 2A and microscopy (see below and Figures S4A–S4B), suggesting that gRNA 9-1 plays a master role in mitigating αSyn stress.
Alterations in membrane trafficking and localization of αSyn from the plasma membrane into cytoplasmic foci are well-established hallmarks of PD (Outeiro and Lindquist, 2003). Owing to highly conserved mechanisms involved in membrane trafficking, yeast cells have been used to study αSyn-coupled vesicular trafficking defects, which has led to mechanistic insights into modifiers of αSyn toxicity, such as UBP3 and the Rab family GTPase YPT1 and their human homologs (Chung et al., 2013; Cooper et al., 2006; Tardiff et al., 2013). We quantitatively measured the effect of gRNA 9-1 on the localization of αSyn-YFP by microscopy. In this assay, aggregated αSyn-YFP is detected as cytoplasmic foci, which are distinguishable from the membrane-localized, non-aggregated form of the protein. As shown in Figures 2B and 2C, upon 6 hours of αSyn induction, 92% of yeast cells with dCas9-VP64 but no gRNA (negative control) contained aggregated αSyn-YFP foci. Overexpression of dCas9-VP64 along with gRNA 9-1 resulted in localization of αSyn-YFP to the plasma membrane such that aggregated αSyn-YFP foci were observed in only ~7% of cells. This was significantly lower than the percentage of cells overexpressing UBP3 (~39% cells with αSyn-YFP foci), which we used as a positive control.
Human DJ-1/PARK7, ALS2, GGA1, and DNAJB1 Homologs were Identified as Robust Protectors against αSyn Toxicity
One of the interesting functional categories identified in our screen involves the heat shock chaperones. Specifically, HSP31–34 heat shock proteins are homologs of the human DJ-1/PARK7 gene, in which autosomal recessive mutations are associated with early onset of familial PD (Bonifati et al., 2003). DJ-1 is thought to protect neurons from mitochondrial oxidative stress by acting as a redox-dependent chaperone to inhibit αSyn aggregates (Bonifati et al., 2003; Canet-Aviles et al., 2004). The roles of HSP31–34 in protecting yeast cells from αSyn toxicity have been previously investigated (Zondler et al., 2014); however, these genes have not been identified in previous genome-wide screens for modifiers of αSyn toxicity. We identified SNO4/HSP34 and HSP32 as two of the differentially expressed genes in our screen. As shown in Figure 2, either SNO4/HSP34 or HSP32, when overexpressed, significantly rescued αSyn-induced growth defects and membrane-trafficking abnormalities. Interestingly, SNO4/HSP34 was moderately up-regulated by gRNA 9-1, whereas HSP32 was extremely down-regulated (Figure 1C and Table 1), which could reflect evolutionarily conserved functions of these paralog proteins, despite their being under the control of different gene regulatory programs. Furthermore, overexpression either of the other two yeast DJ-1 homologs (HSP31 and HSP33) also significantly suppressed αSyn toxicity (Figure 2A), even though they were not significantly modulated by gRNA 9-1. This further supports the involvement of this class of paralog heat-shock proteins in suppressing αSyn toxicity. Consistently, HSP31 (which among HSP31–34 shows the least homology with DJ-1) was recently shown to be a chaperone involved in mitigating various protein misfolding stresses, including that of αSyn (Tsai et al., 2015).
Among other top αSyn-toxicity suppressors (Table 1 and Figure 2), yeast SAF1 encodes an F-Box protein that selectively targets unprocessed vacuolar/lysosomal proteins for proteasome-dependent degradation (Escusa et al., 2007; Mark et al., 2014). The homolog of this protein in mice and humans, ALS2/alsin, functions as a guanine nucleotide exchange factor (GEF) that activates the small GTPase Rab5, an evolutionarily conserved protein involved in membrane trafficking in endocytic pathways (Hadano et al., 2007). Mutations in human ALS2 have been shown to cause autosomal recessive motor neuron diseases (Chandran et al., 2007). In addition, we found that GGA1 and its paralog GGA2 could both alleviate αSyn toxicity (Figures 2 and S3), which was interesting because neither of them had been reported previously to have this activity. The yeast GGA1 protein has been implicated in binding ubiquitin, thus facilitating the sorting of cargo proteins from the trans-Golgi network to endosomal compartments (Takatsu et al., 2002; Zhdankina et al., 2001). Human GGA1 overexpression attenuates amyloidogenic processing of amyloid precursor proteins (APP) in Alzheimer’s disease and a rare inherited lipid-storage disease, Niemann-Pick type C (NPC) (Kosicek et al., 2014; von Einem et al., 2015). Finally, SIS1, the yeast Hsp40 homolog of human DNAJ/HSP40 family proteins, was identified as αSyn suppressor via PRISM. DNAJ family proteins play roles in priming the specificity of HSP70 chaperoning complexes. It has been shown that mammalian DNAJ and HSP70 are up-regulated in response to αSyn overexpression (Vos et al., 2008). In addition, the DNAJB subfamily has been shown to suppress polyglutamine (polyQ) aggregates (Gillis et al., 2013). These results demonstrate that randomized transcriptional perturbations with PRISM enable the discovery of modulators of disease-relevant phenotypes.
Verification of Human Homologs of the Identified Hits in a Neuronal PD Model
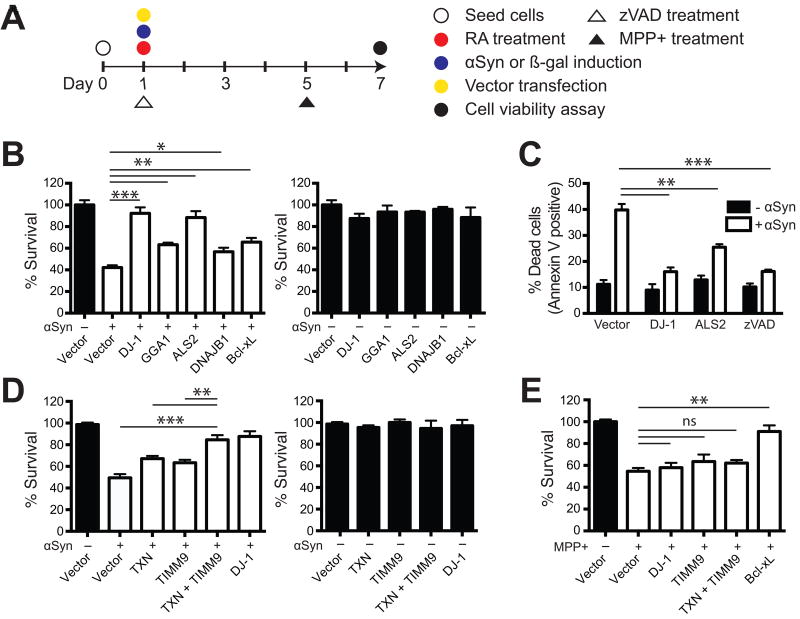
To investigate the neuroprotective effects of the human homologs of the protective yeast genes described above, we overexpressed DJ-1, ALS2, GGA1, and DNAJB1 in an αSyn-overexpressing human neuroblastoma cell line (SH-SY5Y), an established neuronal model of PD (Vekrellis et al., 2009). SH-SY5Y cells were differentiated into cells with dopaminergic neuron-like phenotypes upon retinoic acid (RA) treatment. When β-galactosidase (β-gal) was expressed in these cells, no toxicity was observed. In contrast, αSyn-expressing cells gradually exhibited neurite retraction and only 40–50% viability at 6 days of differentiation (Figures S5A–S5B). Expressing DJ-1 or ALS2 alone did not alter cell survival in the absence of αSyn, but strongly suppressed αSyn-inducible cell death (Figure 3B). αSyn-expressing cells that were transfected with GGA1 or DNAJB1 exhibited about 60% viability, which was similar to the effect of expressing the known anti-apoptotic gene Bcl-xL (positive control). Consistent with these results, overexpression of DJ-1 or ALS2 resulted in reductions in the dead cell populations, as did treatment with the apoptotic inhibitor zVAD (Figure 3C).
Figure 3. Human homologs of yeast αSyn-toxicity suppressors in a human neuronal PD model.
(A) A schematic representation of the experimental procedure used to test the human homologs of yeast αSyn-toxicity suppressors in differentiated neuronal cell lines. Different constructs expressing individual genes were transfected into the SH-SY5Y neuroblastoma cell line via transient transfection to examine their ability to protect against αSyn toxicity. αSyn expression was induced by removal of Dox from the media and retinoic acid (RA) treatment was used for neuronal differentiation over the course of a six-day period. The anti-cell-death drug zVAD and the toxin MPP+ were applied in control experiments. See also Figure S5. (B) Cell viability of differentiated cell lines overexpressing αSyn and the indicated constructs (white bars) were determined by the CellTiter-Glo luminescent assay. The expression of individual genes did not significantly affect cell survival of differentiated cells in the absence of αSyn induction (black bars). Constructs expressing human DJ-1 (homolog of yeast SNO4/HSP34 and HSP32), GGA1 (GGA1), ALS2 (SAF1), or DNAJB1 (SIS1) were tested. Bcl-xL, which protects against apoptotic neuronal death, was used a positive control (Dietz et al., 2008). (C) The percentage of dead cells upon αSyn induction was quantitated by FITC-Annexin V staining followed by flow cytometry. Effects of overexpressing DJ-1 or ALS2 via plasmid transfection were compared with effects of zVAD. (D) Constructs expressing human TXN (homolog of yeast TRX1) or TIMM9 (homolog of yeast TIM9) were transfected individually or co-transfected together to test for synergistic effects on protection from αSyn toxicity. TXN + TIMM9 synergistically rescued these cells from αSyn toxicity when co-transfected together. (E) Overexpression of DJ-1, TIMM9, or TXN + TIMM9 did not protect against MPP+ toxicity, in contrast with Bcl-xL overexpression. Transfected and differentiated cells were treated with 6 mM MPP+ and then tested for cell viability 48 hours afterwards. All data were presented as mean ± SEM of triplicate sets. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
Human TNX and TIMM9 Synergistically Protect Cells against αSyn Toxicity
Increased oxidative stresses and defective mitochondrial function are pathological mechanisms involved in sporadic PD (Henchcliffe and Beal, 2008). We identified that yeast thioredoxin TRX1, an oxidoreductase involved in maintaining the cellular redox potential, and TIM9, a mitochondrial chaperone involved in the transport of hydrophobic proteins across mitochondrial intermembrane space (Neupert and Herrmann, 2007), participate in the suppression of αSyn toxicity in yeast cells (Figure 2 and Figures S4C–S4E). Neuronal cells transfected with the human homologs of either of these genes, TXN or TIMM9, respectively, exhibited about ~60% survival upon αSyn induction compared with <50% with the vector control expressing no transgene. Intriguingly, co-expression of the two human genes TXN and TIMM9 led to enhanced survival in the presence of αSyn toxicity (~88 % survival) (Figure 3D). Furthermore, the neuroprotective effects of expressing DJ-1, TXN, and TIMM9 were specific to αSyn-associated toxicity, as these genes did not protect against 1-methyl-4-phenyl pyridinium (MPP+)-induced neurodegeneration (Dietz et al., 2008) (Figure 3E and Figures S5C–S5D).
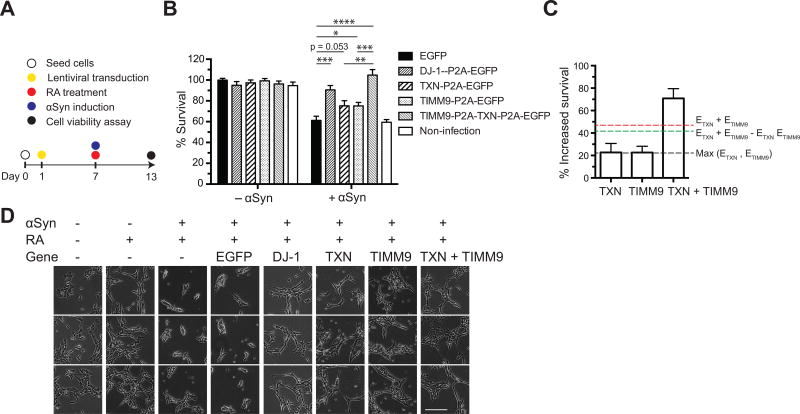
To further investigate these genes as potential therapeutic targets for neuroprotection in PD, we engineered lentiviral vectors expressing DJ-1, TXN, or TIMM9, or co-expressing TXN and TIMM9. We then used these vectors to stably infect cells prior to inducing αSyn stress. Consistent with our transient transfection experiments, DJ-1 reliably protected differentiated SH-SY5Y cells from αSyn-induced cell death and neuronal abnormalities, as did co-expression of TXN and TIMM9 (Figure 4). These results also suggest that activation of these endogenous genes could be explored as a potential therapeutic direction for neuroprotection in PD.
Figure 4. Lentiviral expression of human DJ-1, TXN, and TIMM9 protects against αSyn-associated toxicity in a neuronal PD model.
(A) The human homologs of yeast αSyn-toxicity suppressors were stably expressed via lentiviral vectors six days before RA treatment and αSyn induction, as indicated in the experimental procedure diagram. (B) Overexpression of DJ-1 or TXN + TIMM9 significantly increased neuronal viability in the presence of αSyn. The 2A peptide sequence (P2A) was used to achieve the simultaneous expression of multiple genes from a single promoter. (C) TXN and TIMM9 work synergistically protect neuronal cells from αSyn toxicity based on Highest Single Agent [Max(ETXN, ETIMM9)] (Borisy et al., 2003), Linear Interaction Effect (ETXN + ETIMM9) (Slinker, 1998), and Bliss Independence (ETXN + ETIMM9 − ETXN ETIMM9) (Greco et al., 1995) models (dashed lines). The effect of TXN + TIMM9 was greater than the threshold values obtained from these models. (D) Representative images of neuronal morphology and cell density of cells transfected with lentiviral vectors overexpressing the indicated human genes. Bar = 400 µm. All data were presented as mean ± SEM, n = 6. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DISCUSSION
In this study, we introduce the PRISM screening platform to probe mechanisms underlying cellular responses to αSyn stress. This platform takes advantage of the promiscuity of crisprTF activity with randomized gRNAs for global transcriptional network perturbations. In contrast to typical targeted CRISPR screens, in which gRNAs are designed to modulate individual genes, PRISM does not require any assumptions regarding potential targets and enables unbiased and high-level perturbations of cellular networks. Thus, randomized gRNA screening with PRISM is complementary to targeted screening strategies that have been used with CRISPR-Cas nucleases, crisprTFs, and RNA interference (Blancafort et al., 2005; Demir and Boutros, 2012; Gilbert et al., 2014; Konermann et al., 2015; Moffat et al., 2006; Park et al., 2003; Parnas et al., 2015; Paulsen et al., 2009; Root et al., 2006; Santos and Stephanopoulos, 2008; Shalem et al., 2014; Wang et al., 2014; Whitehurst et al., 2007; Wong et al., 2015; Wong et al., 2016). Randomized gRNA screening with crisprTFs involves a simple library construction procedure and enables global perturbations of transcriptional networks that might not be accessible by traditional single- or multiple-gene perturbations. Such high-order perturbations may be especially important when studying sophisticated phenotypes involving multi-layered regulatory networks, such as those associated with complex human diseases or stress tolerance.
As a proof of concept, we applied this system to a yeast model of PD and identified a diverse set of differentially expressed genes that could individually and collectively rescue αSyn-associated phenotypes. Transcriptomic analysis of the top αSyn-toxicity-protecting gRNA candidate (gRNA 9-1) revealed modest changes in the expression of multiple genes involved in various pathways associated with PD. Intriguingly, genes perturbed by gRNA 9-1 were enriched for αSyn toxicity suppressors versus random selection, most of which have not been reported in previous unbiased genome-wide screens. We verified that over half of these newly identified genes, when overexpressed, rescued yeast cells from αSyn-mediated toxicity. Moreover, gRNA 9-1/crisprTF ameliorated αSyn-associated phenotypes more than overexpression of any individual gene, highlighting the power of global transcription factor screening and suggesting that combinatorial or global effects can enhance desired phenotypes.
To verify the physiological relevance of our hits, we overexpressed the human homologs of validated yeast hits in a human neuronal model of αSyn toxicity. Interestingly, human genes TXN and TIMM9, homologous to yeast genes TRX1 and TIM9, respectively, worked synergistically to suppress αSyn toxicity in human cells (Figure 4). Thioredoxin is known to facilitate the mitochondrial import of TIM9 in yeast (Durigon et al., 2012) and to act as a neuroprotective agent against oxidative stress in neuronal cells (Lin and Beal, 2006). The observed synergistic effect of TXN and TIMM9, as well as the protective effect of redox-dependent chaperones, in suppressing αSyn toxicity further points to the potential importance of mitochondrial maintenance and oxidative stress in PD. Future efforts will be needed to determine whether combinatorial modulation of mitochondrial function pathways and cellular redox may help treat αSyn-associated dysfunction in animal models and clinical settings. In addition, future work should investigate the underlying mechanisms of neuroprotection from the hits identified in this study. These insights could help to develop neuroprotective strategies or engineer αSyn-resistant neuronal cells that could help prevent progressive neurodegeneration in PD patients diagnosed early.
LIMITATIONS
In this screening effort, we used a first-generation crisprTF that results in modest levels of gene activation or repression (Farzadfard et al., 2013; Gilbert et al., 2014). Stronger activation and repression could be achieved by recently improved variants of crisprTFs (Chavez et al., 2015; Konermann et al., 2015; Tanenbaum et al., 2014) while other types of perturbations could be introduced via dCas9 fused to epigenetic regulatory domains (Hilton et al., 2015; Kearns et al., 2015; Thakore et al., 2015).
We performed Sanger sequencing on the surviving colonies to identify gRNA 9-1 and gRNA 6-3, which were then tested in validation experiments. In several cases, we identified multiple different gRNAs within a single yeast cell. Performing high-throughput sequencing on amplicons obtained from surviving yeast colonies could reveal additional gRNAs that could rescue cells from αSyn toxicity when combinatorially expressed. However, for the purpose of this study, we chose to focus only on individual gRNAs that had suppressive effects against αSyn toxicity on their own. In future work, yeast single-copy centromeric (ARS/CEN) plasmids could be used for gRNA expression to ensure that each transformant receives only one gRNA variant.
Even though many genes were up- or down-regulated by gRNA 9-1, we validated them only through overexpression using the readily available Yeast ORF Collection. Thus, our current data suggests that genes modulated by gRNA hits from PRISM screens are enriched for effects on the desired phenotype, but does not indicate the impact of directionality of gene modulation on the phenotype. In future work, genes identified via PRISM screens can be tested via knockdown as well as overexpression in order to determine whether directionality makes a difference. Furthermore, we chose to overexpress genes from the GAL1 promoter in the Yeast ORF Collection rather than using CRISPR activation because efficient targeted CRISPR activation still requires tuning and optimization for each gene of interest. However, overexpression of ORFs by the strong GAL1 promoter can result in expression levels much higher than those achievable by our first-generation crisprTFs. In future work, assessing how different expression levels of identified genes modulate the phenotypes identified through PRISM screening will also be of interest. Finally, we envision that genetic interactions between genes identified by PRISM screening can be further mapped through combinatorial CRISPR technologies (Han et al., 2017; Shen et al., 2017; Wong et al., 2016) and both gene activation and inhibition technologies. Thus, high-throughput randomized crisprTF screens should provide access to a broader range of biological phenotypes across a wide range of organisms in the future.
We predicted 51 potential binding sites for gRNA 9-1 in the yeast genome (Table S1). Although direct crisprTF binding should be identifiable by Chromatin Immunoprecipitation Sequencing (ChIP-Seq) experiments, we were unable to achieve this despite trying multiple different approaches (Kuscu et al., 2014; O'Geen et al., 2015; Wu et al., 2014). We hypothesize that this may be due to weak binding of dCas9 in the absence of perfect-match binding sites for gRNA 9-1 in the genome. Furthermore, it remains challenging to infer transcription regulatory networks solely based on predicted crisprTF binding sites and changes in RNA levels without mapping the transient and indirect cascades involved in transcriptional perturbations (MacQuarrie et al., 2011). Therefore, drawing direct connections between crisprTFs and regulated genes in PRISM screens remains a challenge that needs to be addressed in future work.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Timothy K. Lu (timlu@mit.edu).
Experimental Model and Subject Details
Yeast Strains and Growth Condition
Strains used in this study are all derivatives of W303 (MATa ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3). The ITox2C yeast strain (Cooper et al., 2006) harboring two copies of wild-type αSyn (SNCA)-YFP under control of the Gal-inducible GAL1 promoter (hereafter referred to as the parental strain, a generous gift from Dr. Susan Lindquist, Whitehead Institute, USA) was used for the construction of the crisprTF-expressing screen strain. The Dox-inducible (Tet-ON) promoter was constructed by cloning the pTRE promoter and reverse tetracycline-controlled transactivator (rtTA, from Addgene plasmid #31797) upstream of a minimal CYC1 promoter in the pRS405 backbone. The dCas9-VP64 expression cassette (Addgene plasmid #49013) was then cloned into this vector using Gibson assembly. A sense mutation was introduced within the LEU2 ORF by using the QuikChange system (Stratagene) in order to generate a unique PstI site in the vector. The pRS405-pTetON-dCas9-VP64-PstI plasmid was linearized by PstI and transformed into ITox2C parental strain to build the screen strain. Leucine-positive integrants were verified by genomic PCRs as well as testing for the presence of αSyn-mediated defects by the survival assay and microscopy after Gal induction.
To build the GAL4* strain, a sequence containing full endogenous GAL4 promoter (−257 to 214) was first PCR amplified by oligos (forward: 5’- CCCAGTATTTTTTTTATTCTACAAACC -3’; reversed: 5’- AAATCAGTAGAAATAGCTGTTCCAGTCTTTCTAGCCTTGATTCCACTTCTGTCAGgTGaGCtCggGTtaaCGGAGACCTTTTGGTTTTGG -3’). This fragment was then assembled (by Gibson assembly) with a kanMX6 expression cassette amplified from pFA6a-kanMX (Addgene plasmid #39296) using oligos (forward: 5’- GGGGCGATTGGTTTGGGTGCGTGAGCGGCAAGAAGTTTCAAAACGTCCGCGTCCTTTGAGACAGCATTCGGAATTCGAGCTCGTTTAAAC -3’; reversed: 5’- GAAGGTTTGTAGAATAAAAAAAATACTGGGCGGATCCCCGGGTTAATTAA -3’). The assembled kanMX-GAL4* cassette was then purified and transformed into yeast cells and transformants were selected in presence of 200 mg/L G418 (Thermo Fisher Scientific). Integrants were confirmed by yeast colony PCR and Sanger sequencing.
Yeast cells were cultured in either YPD (1% yeast extract, 2% Bacto-peptone and 2% glucose) or Synthetic complete medium (Scm) supplemented with 2% glucose, raffinose, or galactose. Doxycycline (Sigma) was added directly to culture media or plates immediately before pouring (final concentration of 1 µg/mL).
Neuroblastoma Cell Culture and Gene Expression
Parental and engineered SH-SY5Y cell lines (Vekrellis et al., 2009) (kindly provided by Dr. Leonidas Stefanis, Biomedical Research Foundation Academy Of Athens, Greece) were grown in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) base medium plus 1% GlutaMAX™ supplemented with 15% heat-inactivated FBS (Fetal Bovine Serum) and 1× antibiotic-antimycotic (Invitrogen) at 37 °C with 5% CO2. Cells were seeded at an initial density of 104 cells/cm2 in culture dishes coated with 0.05 mg/mL collagen (Invitrogen). Cells were maintained with 2 µg/mL Dox as previously described (Vekrellis et al., 2009), in order to repress expression of αSyn and β-galactosidase (β-gal), which are driven by the Tet-OFF promoter (Gouarne et al., 2015; Vekrellis et al., 2009). The expression of αSyn and β-gal was induced by removing Dox from the media. Cells were differentiated by treating the cells with 10 µM all-trans Retinal (RA; Sigma) for 6 days. For transient expression of human genes, cells were transfected by adding 1 µg plasmid DNA/ 4 µL FuGENE® HD Transfection Reagent (Promega).
Lentivirus production and transduction were performed as previously described (Lois et al., 2002). Viral supernatants from HEK-293T fibroblasts were collected at 48-hr after transfection, and filtered through a 0.45 µm polyethersulfone membrane. For transduction with individual vector constructs, 2 ml filtered viral supernatant was used to infect 2 × 106 cells in the presence of 8 µg/mL polybrene (Sigma) overnight. Cells were washed with fresh culture medium 1 day after infection, and cultured for following 6 days before RA treatment and αSyn induction.
Method Details
Randomized gRNA Library construction and Screening
To build the randomized gRNA library, random oligos containing 20 bp random nucleotide flanked by homology arms to the vector were co-transformed into yeast with a linearized 2μ vector flanked by RPR1 promoter and gRNA handle at the ends into the screen yeast strain. Once inside the cells, a gRNA-expressing library was reconstituted by the yeast homologous recombination machinery. The randomized oligo library was synthesized by the IDT hand-mixed protocol for randomized oligos using the following template: 5'-GCTGGGAACGAAACTCTGGGAGCTGCGATTGGCAG(N1:32181832)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)(N1)GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3', where N1 indicates the hand-mixed nucleotide with the following ratio: A:C:G:T = 32:18:18:32. The GC content of the randomized portion of the oligo pool was set to 64% to match with the average GC content of yeast promoters (http://rulai.cshl.edu/SCPD/). The libraries were screened in the presence of both galactose and Dox, and the gRNA content of surviving colonies were characterized by colony PCR followed by Sanger sequencing. Individual gRNAs were verified by cloning each gRNA sequence into the empty gRNA vector and transforming these vectors back into the screen strain to validate gRNA activity in a clean background.
Yeast Growth and Viability Assays
The yeast screen strain was transformed with gRNAs or individual genes obtained from yeast ORF library (Open Biosystems). Single transformant colonies were grown overnight in Scm-Uracil (Ura)+raffinose media in the presence of Dox (1 µg/mL) to induce crisprTF expression. Saturated cultures were diluted to OD600 = 0.1 in Scm-Ura+Glucose+Dox and Scm-Ura+Galactose+Dox media and grown at 30 °C in a Synergy H1 Microplate Reader (BioTek). OD600 and fluorescence (excitation and emission spectrum at 508 and 534 nm, respectively) were monitored over the course of the experiments. For measuring cell viability by spotting assays (Chen et al., 2013), cultures were serially diluted (5-fold dilutions) and spotted on Scm-Ura+Glucose+Dox plates for visualizing total viable cells and on Scm-Ura+Galactose+Dox plates for measuring survival. Plates were incubated at 30 °C for 2 days.
RNA Preparation and Sequencing
The screen strain was transformed with either a vector expressing gRNA 9-1 or the empty gRNA vector. Two single-colony transformants from each sample were grown overnight in Scm-Ura+Glucose+Dox. These cultures were diluted into the same fresh media to OD600 = 0.1 and were incubated at 30 °C, 300 RPM. Samples were collected in mid-logarithmic phase (OD600 = 0.8) and flash-frozen in liquid nitrogen. Samples were kept in −80 °C until further processing. Total RNA samples were prepared using the MasterPure Yeast RNA Purification kit (Epicentre) following the manufacturer’s protocol. mRNA libraries were prepared using the Illumina TruSeq library preparation kit, barcoded, multiplexed and sequenced by Illumina HiSeq. The reads were processed by the MIT BioMicroCenter facility pipeline and mapped to the S. cerevisiae reference genome (sacCer3). RPKM values were calculated using ArrayStar and differentially expressed genes were identified by t-test (p-value ≤ 0.1, FDR correction(Benjamini, 1995)). Genes that exhibited at least twofold changes in expression in cells containing the gRNA 9-1 compared with the reference (empty gRNA vector) were considered as differentially expressed. Functional classification of the identified genes was performed using the FunSpec webserver (Robinson et al., 2002).
Western Blotting and Fluorescence Imaging
Yeast protein extracts were prepared for Western blotting by trichloroacetic acid extraction (Chen and Weinreich, 2010). Blots were probed in phosphate-buffered saline containing 0.1% Tween containing 1% (w/v) dried milk. Overexpression constructs containing a 6×His tag were detected using anti-His monoclonal antibody (1:2000; R93025, Life Technologies) followed by anti-mouse-HRP secondary antibody. αSyn (SNCA) was detected with mouse monoclonal anti-αSyn antibodies (1:1000; Syn-1/Clone 42, BD Biosciences).
αSyn-YFP expressing cells were directly visualized under an inverted fluorescence microscope (Zeiss) after 6 days of αSyn induction. The phenotypes were quantified by counting αSyn foci in at least 100 individual cells in multiple randomly chosen fields of view for three independent sets of experiments.
Neuroblastoma Cell Viability and Death Assays
Viable SH-SY5Y cells were quantified by using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Images were captured using the EVOS™ FL Cell Imaging System directly from culture plates under 10× magnification. Cell death was measured by the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) followed by LSR Fortessa II flow cytometry analysis. At least 10,000 cells were recorded per sample in each data set. In the cell death assay (Figure 3C), caspase inhibitor zVAD (Z-VAD-FMK; BD Biosciences) was added into the media upon αSyn induction (100 µM final concentration). For the cell survival assay (Figure 3E), MPP+ iodide (1-methyl-4-phenylpyridinium iodide; Sigma) was added into media of transfected cells 48 hours before processing for cell viability assay.
Quantification and Statistical Analysis
Potential Target Site Analysis
Potential target sites for gRNAs 6-3 and 9-1 in the S. cerevisiae genome were identified using CasOT CRISPR off-target search tool (Xiao et al., 2014). All potential target sites with up to two mismatches inside the seed region are presented in Table S1.
Scoring Strategy for αSyn-toxicity Suppression in Yeast Survival Assays
A defined scoring system, which quantified the numbers of total and full spots in the spotting assays with serial dilutions, was used to score yeast survival upon αSyn induction: cells expressing the empty vector (which showed the least survival upon αSyn induction; sick colonies in the first spot) were scored as 1, and samples overexpressing gRNA 9-1 and UBP3 (the positive control for αSyn-toxicity suppression) were scored as 6 (five full spots and healthy colonies in the sixth spot). Other samples were scored by visual inspection and comparing the spotting assay survival results with the two abovementioned reference points.
| Score | Number of total spots | Number of full spots | Score reference |
|---|---|---|---|
| 6 | 6 | 5 | UBP3 and gRNA 9-1 |
| 5 | 5 | 4 | |
| 4 | 4 | 3 | |
| 3 | 3 | 2 | |
| 2 | 2 | 1 | |
| 1 | 1 | 0 | Vector |
| 0 | 0 | 0 |
Scoring Strategy for αSyn Aggregate Suppression in Fluorescence Microscopy Assays
A defined scoring system, which distinguished the percentage of cell-containing αSyn-YFP foci, was used to score αSyn-aggregate suppression: cells expressing the empty vector were scored as 1 (91.7%), and the samples overexpressing gRNA 9-1 were scored as 10 (6.5%).
| Score | % cells with αSyn aggregates | Score reference |
|---|---|---|
| 10 | 0 – 10% | gRNA 9-1 |
| 9 | 10 – 20% | |
| 8 | 20 – 30% | |
| 7 | 30 – 40% | |
| 6 | 40 – 50% | |
| 5 | 50 – 60% | |
| 4 | 60 – 70% | |
| 3 | 70 – 80% | |
| 2 | 80 – 90% | |
| 1 | 90 – 100% | Vector |
Synergy Quantification
The increased survival against αSyn toxicity by overexpression of TXN, TIMM9, and TXN + TIMM9 was normalized to the vector control (Figure 3D) or the EGFP control (Figure 4B). We considered co-expression of TXN + TIMM9 to be interacting synergistically if the observed combination effect was greater than the expected effect given by Highest Single Agent (Borisy et al., 2003), Linear Interaction Effect (Slinker, 1998), and Bliss Independence (Greco et al., 1995) models. Synergy was calculated based on data presented in Figure 4B and tested by three models respectively, as illustrated in Figure 4C.
Data and Software Availability
The accession number for the RNA-Seq data reported in this paper is GEO: GSE87547. The sequences of recombinant DNA reported in this study have been deposited at Mendeley database (http://dx.doi.org/10.17632/wfskh3hjj5.1).
Additional Resources
Detailed Protocols
The protocols describe the procedures to construct and screen randomized gRNA library.
Supplementary Material
Acknowledgments
The ITox2C yeast strain was a generous gift from Susan Lindquist (Whitehead Institute, Cambridge, MA). We thank Leonidas Stefanis (Biomedical Research Foundation Academy of Athens, Athens, Greece) for SH-SY5Y cell lines. This work was supported by The Ellison Medical Foundation and the National Institutes of Health (1P50GM098792).
Footnotes
AUTHOR CONTRIBUTIONS
YCC, FF and TKL designed experiments, analyzed data, discussed results and wrote the manuscript. YCC and FF performed yeast experiments. FF and NG designed RNA-Seq experiments and analyzed data. YCC performed mammalian cell experiments. WC and JC helped with mammalian cell experiments.
References
- Benjamini Y, Yosef Hochberg. Controlling the false discovery rate: a practical and powerful approach to multiple testing. ournal of the royal statistical society. Series B (Methodological) 1995:289–300. [Google Scholar]
- Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, Brachat A, Brakenhoff RH, Quigley JP, Erdmann D, et al. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc Natl Acad Sci U S A. 2005;102:11716–11721. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nature reviews. Genetics. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Sabatini DM. Systematic genome-wide screens of gene function. Nature reviews. Genetics. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with targetable nucleases. Annual review of biochemistry. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Cencic R, Miura H, Malina A, Robert F, Ethier S, Schmeing TM, Dostie J, Pelletier J. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran J, Ding J, Cai H. Alsin and the molecular pathways of amyotrophic lateral sclerosis. Molecular neurobiology. 2007;36:224–231. doi: 10.1007/s12035-007-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, E PRI, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Kenworthy J, Gabrielse C, Hanni C, Zegerman P, Weinreich M. DNA replication checkpoint signaling depends on a Rad53-Dbf4 N-terminal interaction in Saccharomyces cerevisiae. Genetics. 2013;194:389–401. doi: 10.1534/genetics.113.149740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Weinreich M. Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction. J Biol Chem. 2010;285:41244–41254. doi: 10.1074/jbc.M110.155242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir K, Boutros M. Cell perturbation screens for target identification by RNAi. Methods Mol Biol. 2012;910:1–13. doi: 10.1007/978-1-61779-965-5_1. [DOI] [PubMed] [Google Scholar]
- Dietz GP, Stockhausen KV, Dietz B, Falkenburger BH, Valbuena P, Opazo F, Lingor P, Meuer K, Weishaupt JH, Schulz JB, et al. Membrane-permeable Bcl-xL prevents MPTP-induced dopaminergic neuronal loss in the substantia nigra. Journal of neurochemistry. 2008;104:757–765. doi: 10.1111/j.1471-4159.2007.05028.x. [DOI] [PubMed] [Google Scholar]
- Durigon R, Wang Q, Ceh Pavia E, Grant CM, Lu H. Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO reports. 2012;13:916–922. doi: 10.1038/embor.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa S, Laporte D, Massoni A, Boucherie H, Dautant A, Daignan-Fornier B. Skp1-Cullin-F-box-dependent degradation of Aah1p requires its interaction with the F-box protein Saf1p. J Biol Chem. 2007;282:20097–20103. doi: 10.1074/jbc.M702425200. [DOI] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS Synth Biol. 2013 doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. The art and design of genetic screens: yeast. Nature reviews. Genetics. 2001;2:659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van den Nieuwendijk R, van Veen H, Overkleeft H, Goedhart J, Kampinga HH, et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013;288:17225–17237. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, Caldwell KA, Caldwell GA, Cooper AA, Rochet JC, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nature genetics. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nature reviews. Neurology. 2013;9:13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- Gouarne C, Tracz J, Paoli MG, Deluca V, Seimandi M, Tardif G, Xilouri M, Stefanis L, Bordet T, Pruss RM. Protective role of olesoxime against wild-type alpha-synuclein-induced toxicity in human neuronally differentiated SHSY-5Y cells. British journal of pharmacology. 2015;172:235–245. doi: 10.1111/bph.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacological reviews. 1995;47:331–385. [PubMed] [Google Scholar]
- Hadano S, Kunita R, Otomo A, Suzuki-Utsunomiya K, Ikeda JE. Molecular and cellular function of ALS2/alsin: implication of membrane dynamics in neuronal development and degeneration. Neurochemistry international. 2007;51:74–84. doi: 10.1016/j.neuint.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchcliffe C, Beal MF. Mitochondrial biology and oxidative stress in Parkinson disease pathogenesis. Nature clinical practice. Neurology. 2008;4:600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5 doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker's yeast? Nature reviews. Neuroscience. 2010;11:436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- Khurana V, Peng J, Chung CY, Auluck PK, Fanning S, Tardiff DF, Bartels T, Koeva M, Eichhorn SW, Benyamini H, et al. Genome-Scale Networks Link Neurodegenerative Disease Genes to alpha-Synuclein through Specific Molecular Pathways. Cell systems. 2017;4:157–170. e114. doi: 10.1016/j.cels.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicek M, Wunderlich P, Walter J, Hecimovic S. GGA1 overexpression attenuates amyloidogenic processing of the amyloid precursor protein in Niemann-Pick type C cells. Biochemical and biophysical research communications. 2014;450:160–165. doi: 10.1016/j.bbrc.2014.05.083. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Overk CR, Oueslati A, Masliah E. The many faces of alpha-synuclein: from structure and toxicity to therapeutic target. Nature reviews. Neuroscience. 2013;14:38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends in genetics : TIG. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maries E, Dass B, Collier TJ, Kordower JH, Steece-Collier K. The role of alpha-synuclein in Parkinson's disease: insights from animal models. Nature reviews. Neuroscience. 2003;4:727–738. doi: 10.1038/nrn1199. [DOI] [PubMed] [Google Scholar]
- Mark KG, Simonetta M, Maiolica A, Seller CA, Toczyski DP. Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell. 2014;53:148–161. doi: 10.1016/j.molcel.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annual review of biochemistry. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic acids research. 2015;43:3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Santos CN, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017 doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinker BK. The statistics of synergism. Journal of molecular and cellular cardiology. 1998;30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Takatsu H, Yoshino K, Toda K, Nakayama K. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. The Biochemical journal. 2002;365:369–378. doi: 10.1042/BJ20020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, et al. Yeast reveal a "druggable" Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Khurana V, Chung CY, Lindquist S. From yeast to patient neurons and back again: powerful new discovery platform. Movement disorders : official journal of the Movement Disorder Society. 2014;29:1231–1240. doi: 10.1002/mds.25989. [DOI] [PubMed] [Google Scholar]
- Tenreiro S, Rosado-Ramos R, Gerhardt E, Favretto F, Magalhaes F, Popova B, Becker S, Zweckstetter M, Braus GH, Outeiro TF. Yeast reveals similar molecular mechanisms underlying alpha- and beta-synuclein toxicity. Hum Mol Genet. 2016;25:275–290. doi: 10.1093/hmg/ddv470. [DOI] [PubMed] [Google Scholar]
- Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Aslam K, Drendel HM, Asiago JM, Goode KM, Paul LN, Rochet JC, Hazbun TR. Hsp31 Is a Stress Response Chaperone That Intervenes in the Protein Misfolding Process. J Biol Chem. 2015;290:24816–24834. doi: 10.1074/jbc.M115.678367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekrellis K, Xilouri M, Emmanouilidou E, Stefanis L. Inducible over-expression of wild type alpha-synuclein in human neuronal cells leads to caspase-dependent non-apoptotic death. Journal of neurochemistry. 2009;109:1348–1362. doi: 10.1111/j.1471-4159.2009.06054.x. [DOI] [PubMed] [Google Scholar]
- von Einem B, Wahler A, Schips T, Serrano-Pozo A, Proepper C, Boeckers TM, Rueck A, Wirth T, Hyman BT, Danzer KM, et al. The Golgi-Localized gamma-Ear-Containing ARF-Binding (GGA) Proteins Alter Amyloid-beta Precursor Protein (APP) Processing through Interaction of Their GAE Domain with the Beta-Site APP Cleaving Enzyme 1 (BACE1) PLoS One. 2015;10:e0129047. doi: 10.1371/journal.pone.0129047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- Wong AS, Choi GC, Cheng AA, Purcell O, Lu TK. Massively parallel high-order combinatorial genetics in human cells. Nat Biotechnol. 2015;33:952–961. doi: 10.1038/nbt.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AS, Choi GC, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Krainc D. alpha-synuclein toxicity in neurodegeneration: mechanism and therapeutic strategies. Nature medicine. 2017;23:1–13. doi: 10.1038/nm.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nature genetics. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O, Strand NL, Redmond JM, Boman AL. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast (Chichester, England) 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zondler L, Miller-Fleming L, Repici M, Goncalves S, Tenreiro S, Rosado-Ramos R, Betzer C, Straatman KR, Jensen PH, Giorgini F, et al. DJ-1 interactions with alpha-synuclein attenuate aggregation and cellular toxicity in models of Parkinson's disease. Cell Death Dis. 2014;5:e1350. doi: 10.1038/cddis.2014.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.